Abstract
Osteoarticular tuberculosis (OAT) may cause severe complications and disability. Due to its indolent nature, OAT is difficult to diagnose in the early stages. Diagnosis by conventional culture is time-consuming and insensitive, and polymerase chain reaction-based molecular diagnostic methods are incapable of excluding co-infections. Metagenomic next-generation sequencing (mNGS) may identify a broad spectrum of microorganisms, including Mycobacterium, bacteria and fungi, in clinical specimens. Therefore, the diagnosis of OAT may be rapidly performed using mNGS. The present study reports on a case of OAT. The patient presented with right knee swelling and pain for 1 year; his C-reactive protein levels and erythrocyte sedimentation rate were markedly elevated. Although multiple pre-operative cultures were negative, mNGS was finally used to successfully detect the underlying pathogen. The result was confirmed by other molecular biology methods and Mycobacterium culture. Anti-tuberculosis therapy was administered accordingly and the patient finally recovered. In conclusion mNGS, with the ability to detect Mycobacterium and other microorganisms in a single assay, is an emerging approach for rapidly and accurately diagnosing OAT. This method may provide significant support to guide physicians in selecting the appropriate pharmacotherapy and surgical treatments.
Keywords: osteoarticular tuberculosis, mycobacterium tuberculosis, metagenomic next-generation sequencing
Introduction
Osteoarticular tuberculosis (OAT) may cause significant morbidity and disability if not treated properly. OAT is frequently difficult to diagnose in the early stage due to the absence of typical systematic symptoms, physical signs, blood parameters and radiographic indications (1). Conventional Mycobacterium tuberculosis (MTB) culture is a gold standard for diagnosis, but it has a relatively low sensitivity and is time-consuming (2). Polymerase chain reaction (PCR)-based genetic analyses are effective for rapidly diagnosing OAT. However, these methods specifically detect MTB sequences and are incapable of excluding mixed bacterial and fungal infections that commonly occur in OAT patients as a result of low immunity and invasive procedures (3).
Metagenomic next-generation sequencing (mNGS) is a novel technique for the clinical diagnosis of infectious diseases. mNGS may accurately detect MTB in sputum specimens from patients with suspected pulmonary tuberculosis (4). Its ability to identify multiple pathogens using a single detection method demonstrates its great potential for uncovering mixed infection. To date, no study has described the diagnosis of OAT using the mNGS method.
The present study reports on a case of non-specific progressive swelling and pain in the knee, which was diagnosed as OAT using mNGS. After surgical treatment and chemotherapy, the patient demonstrated functional recovery.
Case report
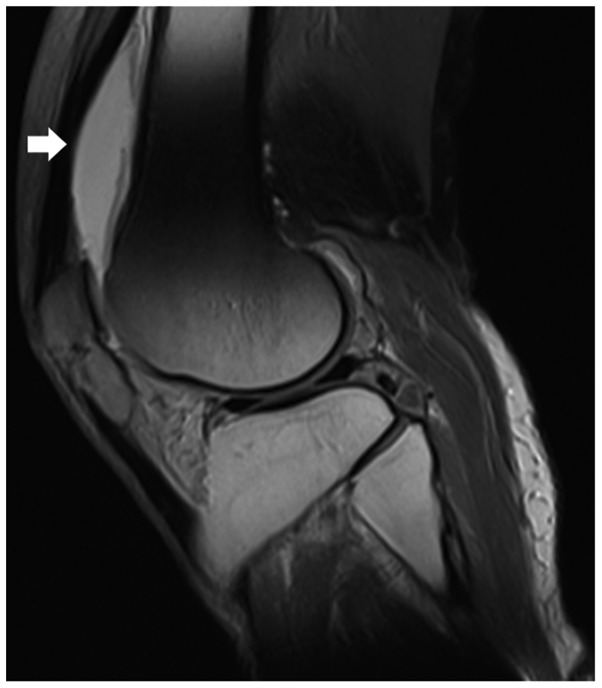
A 64-year-old male presented at the outpatient department of the First Affiliated Hospital of Fujian Medical University (Fuzhou, China) in October 2017 with a history of repeated progressive swelling and pain in the right knee for 1 year. The pain was usually exacerbated after movement. The patient exhibited other symptoms, including fever or walking instability. Magnetic resonance imaging scans performed at a local hospital indicated effusion of the suprapatellar bursa and based on this, early osteoarthritis was suspected (Fig. 1). Multiple knee aspirations and injections, as well as anti-microbial therapy, which were performed at a local clinic (specific details unknown), were ineffective.
Figure 1.
Magnetic resonance imaging scans revealing effusion of the suprapatellar bursa (white arrow) in T2-weighted images.
The patient's medical history included a right patella fracture due to a falling accident two years previously. Good union of the bone was achieved and function of the joint as regained by the application of a knee brace for 3 months. The patient denied multiple sexual partners or any recent travel to endemic areas.
On physical examination, right knee swelling was noted, with no redness of the local skin or elevated skin temperature. The patella floating test was positive. The anterior and posterior drawer tests and the McMurray test were negative. The range of motion of the knee was restricted to 0–90 degrees. The right quadriceps muscle exhibited mild atrophy, while lower extremity limb muscle strength and sensations were normal.
Routine blood and coagulation test results, as well as liver and kidney function, were normal. Tests for rheumatoid factor, anti-keratin antibody and anti-cyclic citrullinated peptide were all negative. The patient's C-reactive protein (CRP) level was elevated (62.10 mg/l) and the erythrocyte sedimentation rate (ESR) was 31 mm/h. A T-SPOT.TB assay (Oxford Immunotec) was positive with the following results: 86/250,000 peripheral blood mononuclear cells (PBMCs) expressing ESAT-6 (antigen A) and 89/250,000 PBMCs expressing CFP-10 (antigen B).
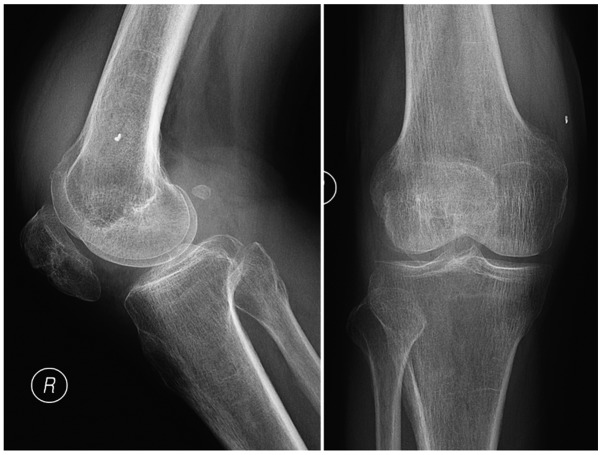
A CT scan of the lung was normal. Anterior and lateral plain images of the right knee indicated that the tibiofemoral joint space of the right knee was intact and contained a small number of osteophytes (Fig. 2). Aspiration of the right knee yielded 10 ml red, cloudy synovial fluid. No uric acid crystals were visible under a polarizing microscope. The white blood cell (WBC) count of the synovial fluid was 5895.00 106/l, with 55.4% polymorphonuclear cells. Acid-fast staining, as well as bacterial and fungal cultures, were negative, and a rapid MTB culture using the BACTEC-MGIT960 system (BD Biosciences) was negative within 2 weeks.
Figure 2.
Anterior and lateral plain images of the right knee revealing that the tibiofemoral joint space was intact and contained a small number of osteophytes.
At this time-point, the patient's serum CRP levels, ESR, synovial WBC count and polymorphonuclear cell percentage were all markedly elevated, and no extra-articular symptoms were observed. Immune disease indicators were negative; therefore, it was highly suspected that the patient had a right knee joint infection. While the cultures were negative, no low-virulence, difficult-to-grow bacteria or atypical pathogens (mycoplasma, MTB or nontuberculous mycobacteria) were excluded.
Therefore, the 16s ribosomal (r)RNA PCR method was used to detect pathogenic microorganisms (5). Ceramic beads were added to 200 µl synovial fluid to lyse the cell walls. Total DNA was extracted using the TIANAMP Micro DNA kit (cat. no. DP316; Tiangen Biotech) following the manufacturer's protocol. Primers targeting the V3-V4 region of 16S rRNA (forward, 5′-CGGCCCAGACTCCTACGGGAGGCAGCA-3′ and reverse, 5′-GCGTGGACTACCAGGGTATCTAATCC-3′) were used. Real-time PCR was performed on an ABI 7500 instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.) using the Takara SYBR Premix Ex Taq kit (Takara Bio, Inc.). No positive results were detected.
The patient was admitted for open surgery to perform aggressive debridement (6), during which adequate tissue samples for microbiological identification were obtained. The medial parapatellar approach was utilized. Except for a small cartilage defect on the patella due to a previous fracture, the surface of the tibiofemoral joint was smooth. The cruciate ligaments were intact. Extensive swelling and hyperplastic synovium were observed. The affected synovium was resected extensively. The synovial fluid and resected synovial tissue were transported to the laboratory within 30 min for further pathological and microbiological examination.
Tissue samples were fixed in 10% neutral formalin for 8–48 h, dehydrated by 75, 95 and 100% ethanol, embedded in paraffin and sliced into 4-µm-thick sections. The samples were stained with hematoxylin for 5–10 min and eosin for 1–3 min at the room temperature. The sections were observed using an optical microscope at a magnification of ×300. Histopathological examination with hematoxylin and eosin revealed an inflammatory reaction, including massive granuloma with central necrosis, lymphocytic infiltration, and a small amount of neutrophil infiltration (Fig. 3). Bacterial and fungal cultures provided negative results after culturing for 2 weeks.
Figure 3.

Histopathological examination with hematoxylin and eosin revealing an inflammatory reaction, including a massive granuloma with central necrosis, lymphocytic infiltration and a small amount of neutrophil infiltration. Magnification, ×200.
The mNGS process was utilized to clarify the pathogenic source. DNA was extracted from synovial fluid collected intra-operatively through the above-mentioned method. The extracted DNA was sonicated to generate 200–300 bp fragments. DNA libraries were constructed according to the standard protocol of the BGISEQ-500 sequencing platform (BGI-Tianjin). The quantified libraries were sequenced with the BGISEQ-500 platform.
The raw sequencing data were analyzed by a Bioinformatics pipeline developed by BGI that included the following steps: i) Clean reads of high-quality sequencing data were generated by filtering out the short, low-quality and low-complexity reads. ii) Human host sequences were eliminated by mapping to the human reference genome (hg19) with Burrows-Wheeler alignment (http://bio-bwa.sourceforge.net). iii) The remaining sequencing data were aligned to the Microbial Genome Database (an in-house database built by BGI, with no plan to publish publicly), which contains the genomic sequences of 2,700 viruses, 1,494 bacteria, 73 fungi and 48 parasites that are all associated with human diseases. The reference genomes in the database were downloaded from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/).
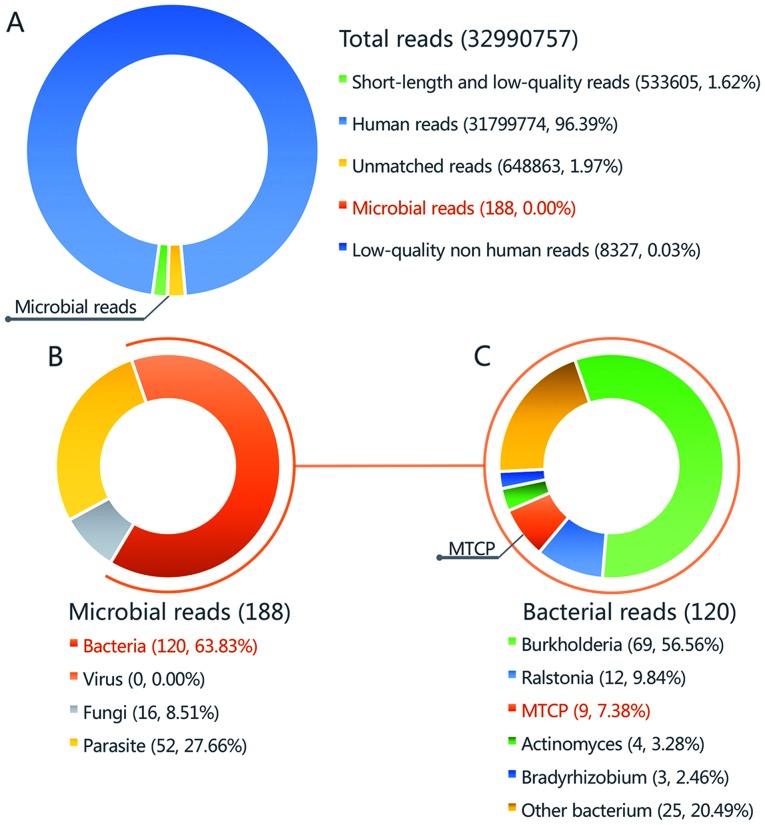
The number of microbial reads was 260 (0.000801% of total clean reads). Burkholderia accounted for the highest number of microbial reads with 31 (11.92% of the total microbial reads). This species was observed to be the most common contaminant according to this sequencing method (data not shown). A total of 6 unique reads (2.31% of the total microbial reads) of the Mycobacterium tuberculosis complex group (MTCP) were obtained (Fig. 4). As this had never been validated as a contaminant in previous experiments and analyses in the same lab during past 3 months, MTCP was regarded as the pathogenic organism.
Figure 4.
Taxonomic classification of sequencing reads from synovial fluid by a bioinformatics pipeline. (A) A total of 32,990,757 reads were detected by mNGS, including 188 microbial reads. (B) A total of 120 reads of bacteria were detected in the synovial fluid (63.83% of bacterial reads). (C) MTCP with 9 reads were detected in the synovial fluid (7.38% of bacterial reads). MTCP, Mycobacterium tuberculosis complex group.
To verify the mNGS test results, the intra-operative synovial fluid was examined using a variety of validated molecular diagnostic methods for detecting MTB. i) The results of nested PCR targeting the IS6110 gene were positive. ii) PCR and hybridization using the MTB Drug Resistance Detection Array kit (CapitalBio) indicated no rpoB, katG or inhA mutations, indicating that the strain is sensitive to rifampicin and isoniazid. iii) Results of the GeneXpert MTB/RIF test (Cepheid) suggested the presence of MTB and no resistance to rifampin.
Due to the consistent results of several different molecular diagnostic methods, the patient was finally diagnosed with right knee tuberculosis. Anti-tuberculosis treatments included isoniazid, rifampicin, ethambutol and pyrazinamide, which were started immediately after the surgery. The patient's incision site healed within 2 weeks of surgery.
MTB culture of synovial fluid and periprosthetic tissues on modified Lowenstein-Jensen medium were positive for tuberculosis at 5 weeks post-operatively. Drug resistance tests indicated that the strain was sensitive to isoniazid, rifampicin and ethambutol, but resistant to streptomycin. During the 3 months of follow-up, the patient reported no fever and fatigue, no swelling or pain in his knee, and being able to walk without a cane. The patient's ESR and CRP values returned normal. The patient is presently undergoing oral anti-tuberculosis chemotherapy, including isoniazid, rifampicin, ethambutol and pyrazinamide. Until now, no signs of recurrence of infection have been noted.
Discussion
Approximately 2.2–4.7% of tuberculosis cases affect the bone and joint systems and are referred to as OAT (7,8). OAT most frequently occurs in the spine, followed by the knee, hip and elbow. If not treated properly, OAT may lead to limb and spine deformities, spinal cord compression and even irreversible disability (1,2). Early OAT is frequently localized and may be successfully treated by anti-tuberculosis chemotherapy. However, OAT is difficult to diagnose in its early stage due to its lack of acute manifestations of infection. Early OAT is difficult to differentiate from bacterial or non-tuberculosis mycobacterial infections, immunological arthritis or spondylitis, and even degenerative diseases (8). Due to this difficulty, the treatment is frequently delayed, leading to poor outcomes. Although MTB culture remains the ‘gold standard’ for diagnosing OAT, existing methods have numerous drawbacks, including long culture times (3–8 weeks) and low sensitivity (30–40%) (9,10).
Molecular diagnostic methods have been applied to improve the diagnosis of OAT. Primers or probes targeting specific genes or drug resistance-associated genes of MTB are used for PCR detection, including fluorescent probe quantitative PCR, GeneXpert MTB/RIF, loop-mediated isothermal amplification and microarrays. These methods have advantages over culture, including higher sensitivity, faster detection and the ability to detect drug resistance spontaneously. However, the currently available commercial kits are unable to identify non-MTB bacteria or other atypical pathogens simultaneously. Clinically, mixed infection is common in tuberculosis patients due to compromised immunity and side effects of injections or surgery. Traditional anti-tuberculosis chemotherapy drugs are frequently ineffective against bacterial infections, which may lead to failure of treatment (11–14).
Use of the mNGS method to detect pathogens in clinical samples has been applied for the diagnosis of meningitis, bacteremia and orthopedic implant-associated infections (15–17). Using high-throughput sequencing and subsequent Bioinformatics processing, mNGS may simultaneously align thousands of sequences of pathogenic microorganisms, including bacteria, fungi, mycoplasma and even parasites, to obtain unbiased information on the species and abundance of all microorganisms in the sample. The mNGS method has been demonstrated to accurately detect MTB in sputum specimens from patients with suspected pulmonary tuberculosis (12). However, few studies have described the diagnosis of OAT using mNGS.
In the present case, MTCP was identified by mNGS of synovial fluid. The result was successfully verified by other methods, including culture and PCR. The patient's symptoms were relieved through surgery and chemotherapy, and inflammatory biomarker levels returned to normal, which confirmed the diagnosis.
mNGS is a type of sequencing with no preferences and allows for identification of microorganisms in an unbiased manner. The ability of mNGS to identify multiple pathogens with a single detection method has great potential for uncovering mixed infections. The case of the present study received multiple intra-articular injections and therefore had a risk of exogenous infection. The mNGS results did not indicate the presence of other pathogenic microorganisms after excluding common background microorganisms during mNGS procedures (e.g., Burkholderia, Ralstonia, Actinomyces and Bradyrhizobium) (18,19). Consequently, only anti-tuberculosis drugs were prescribed. To date, no signs of recurrence of infection have been noted, which further verifies the mNGS result. The present study demonstrated the superiority of mNGS over other molecular diagnostic methods (17).
In the present study, the number of reads uniquely aligned to the reference genome of MTCP was insufficient; therefore, it was impossible to differentiate MTB from MTCP to obtain resistance-associated information. This may be due to the small amount of nucleic acid extracted from the MTB in the specimen. The major causes of this effect may be the low burden of planktonic bacteria in the synovial fluid, difficulty in lysing the tenacious cell wall of MTB and interference of overwhelming host sequences during sequencing. More robust and comprehensive lysis, removal of human DNA (18) and utilization of a single-cell whole-genome analysis method may be adopted to fix these issues (20).
In conclusion, the present case study reported on a patient with inflammatory pain and swelling of the knee. Using mNGS, the presence of MTB was detected and mixed infection was excluded. Culture, specific PCR and GeneXpert MTB/RIF were used to verify the results. The patient's symptoms were relieved through surgical treatment and chemotherapy, and functional recovery was observed. The successful course of diagnosis in the present case indicates that mNGS has great potential for diagnosing OAT and excluding mixed bacterial infection in a single assay. This method provides strong support to guide physicians in selecting the appropriate pharmacotherapy and surgical treatment.
Acknowledgements
Not applicable.
Funding
The current study was supported by the Fujian Education and Scientific Research Projects for Young Teachers (grant no. JAT170241), the National Science Foundation for Young Scientists of China (grant no. 81702168), and the Natural Science Foundation of Fujian Province (grant nos. 2018I0006 and 2018Y4003).
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ZH and ChoZ analyzed and interpreted the data, wrote the manuscript and organized the figures. QijW and ML performed the 16 s ribosome RNA gene polymerase chain reaction analysis. QiqW, DH, KS and WL performed the surgery. BY performed the microbial culture. ChaZ and WZ designed the study, edited and reviewed the manuscript, and approved the version to be published. XL, LH and NX performed the metagenomic next-generation sequencing. All authors reviewed the final version of the manuscript.
Ethics approval and consent to participate
This study was approved by the Institutional Review Board of the First Affiliated Hospital of Fujian Medical University (Fuzhou, China; process no. 2014-047).
Patient consent for publication
Informed consent for publication of data and images was obtained from the patient included in the study.
Competing interest
The authors have no competing interests to declare.
References
- 1.Pigrau-Serrallach C, Rodríguez-Pardo D. Bone and joint tuberculosis. Eur Spine J. 2013;22(Suppl 4):S556–S566. doi: 10.1007/s00586-012-2331-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Magnussen A, Dinneen A, Ramesh P. Osteoarticular tuberculosis: Increasing incidence of a difficult clinical diagnosis. Br J Gen Pract. 2013;63:385–386. doi: 10.3399/bjgp13X669383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tarashi S, Fateh A, Mirsaeidi M, Siadat SD, Vaziri F. Mixed infections in tuberculosis: The missing part in a puzzle. Tuberculosis (Edinb) 2017;107:168–174. doi: 10.1016/j.tube.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Brown AC, Bryant JM, Einer-Jensen K, Holdstock J, Houniet DT, Chan JZ, Depledge DP, Nikolayevskyy V, Broda A, Stone MJ, et al. Rapid whole-genome sequencing of mycobacterium tuberculosis isolates directly from clinical samples. J Clin Microbiol. 2015;53:2230–2237. doi: 10.1128/JCM.00486-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang Z, Wu Q, Fang X, Li W, Zhang C, Zeng H, Wang Q, Lin J, Zhang W. Comparison of culture and broad-range polymerase chain reaction methods for diagnosing periprosthetic joint infection: Analysis of joint fluid, periprosthetic tissue, and sonicated fluid. Int Orthop. 2018;42:2035–2040. doi: 10.1007/s00264-018-3827-9. [DOI] [PubMed] [Google Scholar]
- 6.Bovonratwet P, Nelson SJ, Bellamkonda K, Ondeck NT, Shultz BN, Medvecky MJ, Grauer JN. Similar 30-day complications for septic knee arthritis treated with arthrotomy or arthroscopy: An American college of surgeons National surgical quality improvement program analysis. Arthroscopy. 2018;34:213–219. doi: 10.1016/j.arthro.2017.06.046. [DOI] [PubMed] [Google Scholar]
- 7.Global tuberculosis report 2017, corp-author. World Health Organization; Geneva: 2017. [Feb 11;2019 ]. [Google Scholar]
- 8.García-Elorriaga G, Martínez-Elizondo O, Del Rey-Pineda G, González-Bonilla C. Clinical, radiological and molecular diagnosis correlation in serum samples from patients with osteoarticular tuberculosis. Asian Pac J Trop Biomed. 2014;4:581–585. doi: 10.12980/APJTB.4.201414B112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lakhanpal VP, Tuli SM, Singh H, Sen PC. The value of histology, culture and guinea pig inoculation examination in osteo-articular tuberculosis. Acta Orthop Scand. 1974;45:36–42. doi: 10.3109/17453677408989119. [DOI] [PubMed] [Google Scholar]
- 10.Chen ST, Zhao LP, Dong WJ, Gu YT, Li YX, Dong LL, Ma YF, Qin SB, Huang HR. The clinical features and bacteriological characterizations of bone and joint tuberculosis in China. Sci Rep. 2015;5:11084. doi: 10.1038/srep11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pérez-Osorio AC, Boyle DS, Ingham ZK, Ostash A, Gautom RK, Colombel C, Houze Y, Leader BT. Rapid identification of mycobacteria and drug-resistant mycobacterium tuberculosis by use of a single multiplex PCR and DNA sequencing. J Clin Microbiol. 2012;50:326–336. doi: 10.1128/JCM.05570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.N'guessan K, Horo K, Coulibaly I, Adegbele J, Kouame-Adjei N, Seck-Angu H, Guei A, Kouakou J, Dosso M. Rapid detection of mycobacterium tuberculosis complex in sputum samples using PURE TB-LAMP assay. Int J Mycobacteriol. 2016;5(Suppl 1):S164–S165. doi: 10.1016/j.ijmyco.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 13.Bojang AL, Mendy FS, Tientcheu LD, Otu J, Antonio M, Kampmann B, Agbla S, Sutherland JS. Comparison of TB-LAMP, GeneXpert MTB/RIF and culture for diagnosis of pulmonary tuberculosis in The Gambia. J Infect. 2016;72:332–337. doi: 10.1016/j.jinf.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 14.Cheng XH, Bian SN, Zhang YQ, Zhang LF, Shi XC, Yang B, Zhang FC, Liu XQ. Diagnostic value of t-cell interferon-γ release assays on synovial fluid for articular tuberculosis: A pilot study. Chin Med J (Engl) 2016;129:1171–1178. doi: 10.4103/0366-6999.181958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grumaz S, Stevens P, Grumaz C, Decker SO, Weigand MA, Hofer S, Brenner T, von Haeseler A, Sohn K. Next-generation sequencing diagnostics of bacteremia in septic patients. Genome Med. 2016;8:73. doi: 10.1186/s13073-016-0326-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan H, Shen A, Lv X, Yang X, Ren H, Zhao Y, Zhang Y, Gong Y, Ni P, Wu H, et al. Detection of virus in CSF from the cases with meningoencephalitis by next-generation sequencing. J Neurovirol. 2016;22:240–245. doi: 10.1007/s13365-015-0390-7. [DOI] [PubMed] [Google Scholar]
- 17.Lecuit M, Eloit M. The diagnosis of infectious diseases by whole genome next generation sequencing: A new era is opening. Front Cell Infect Microbiol. 2014;4:25. doi: 10.3389/fcimb.2014.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bukowska-Ośko I, Perlejewski K, Nakamura S, Motooka D, Stokowy T, Kosińska J, Popiel M, Płoski R, Horban A, Lipowski D, et al. Sensitivity of next-generation sequencing metagenomic analysis for detection of RNA and DNA viruses in cerebrospinal fluid: The confounding effect of background contamination. Adv Exp Med Biol. 2016 Jul 13; doi: 10.1007/5584_2016_42. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 19.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang L, Ma F, Chapman A, Lu S, Xie XS. Single-cell whole-genome amplification and sequencing: Methodology and applications. Annu Rev Genomics Hum Genet. 2015;16:79–102. doi: 10.1146/annurev-genom-090413-025352. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.