Abstract
Long non-coding (lnc)RNA small nucleolar RNA host gene 12 (SNHG12) has an oncogenic role in various common human cancer types, including colorectal cancer (CRC). However, the detailed regulatory mechanisms of SNHG12 in CRC cells have remained largely elusive, and the investigation thereof was the purpose of the present study. Polymerase chain reaction analysis was performed to examine the expression of lncRNA and microRNA (miR). Cell Counting Kit-8 and Transwell assays were used to assess cell proliferation and invasion. A luciferase reporter assay was performed to confirm a predicted targeting association between lncRNA and miR. It was observed that SNHG12 was markedly upregulated in CRC tissues when compared with that in adjacent non-tumour tissues, and its high expression was associated with CRC progression, as well as poor prognosis of patients. In addition, the expression of SNHG12 was higher in CRC cell lines when compared with that in a normal intestinal epithelial cell line. Knockdown of SNHG12 significantly inhibited CRC cell proliferation and invasion, while ectopic overexpression of SNHG12 had the opposite effect. A Bioinformatics analysis predicted that SNHG12 and miR-16 have complementary binding sites, which was confirmed by a luciferase reporter gene assay. The expression levels of miR-16 were markedly decreased in CRC tissues and cell lines compared with those in normal tissues or cells, and were inversely correlated with the expression levels of SNHG12 in CRC tissues. Furthermore, silencing of miR-16 eliminated the suppressive effects of SNHG12 knockdown on CRC cell proliferation and invasion. In conclusion, the present study demonstrated that SNHG12 promotes CRC cell proliferation and invasion, at least in part, by acting as a molecular sponge of miR-16, suggesting that SNHG12 may be a promising therapeutic target for CRC.
Keywords: long non-coding RNA, microRNA, proliferation, invasion, colorectal cancer, small nucleolar RNA host gene 12, microRNA16
Introduction
Colorectal cancer (CRC) is one of the most common types of human malignancy worldwide (1). In recent decades, the combined treatment for CRC has improved, which includes surgical resection, radiotherapy and chemoradiotherapy; however, the prognosis of patients with advanced CRC remains poor (2). It has been demonstrated that CRC is driven by a series of successive genetic and epigenetic changes, which cause malignant transformation and metastasis (3–5). Non-coding RNAs are important epigenetic factors associated with cancer development (6–10), but the functions of various non-coding RNAs in CRC remain to be fully elucidated.
Long non-coding RNAs (lncRNAs), a class of non-coding RNAs longer than 200 nucleotides, are 10-fold lower in abundance than mRNAs (11). It has been demonstrated that there are thousands of lncRNA small nucleolar RNA host gene 12 (SNHG12) sequences in mammals and that the majority of lncRNAs are likely to be functional; however, only a small proportion of lncRNAs have been studied (12). Through regulation of gene expression at transcriptional, post-transcriptional and epigenetic levels, lncRNAs have important roles in various physiological and pathological processes, including cell growth, proliferation, differentiation, apoptosis and tumourigenesis (13,14). Numerous lncRNAs have been reported to function as promoters during cancer development and progression (15,16). Therefore, these lncRNAs may become promising biomarkers and/or therapeutic targets for various cancer types.
Recently, several studies have indicated that the lncRNA SNHG12 functions as an oncogene in certain, common cancer types, including osteosarcoma, hepatocellular carcinoma, breast cancer, lung cancer, gastric cancer, glioma, cervical cancer, papillary thyroid carcinoma and CRC (17–25). For instance, Lei et al (20) reported that SNHG12 promoted the proliferation and migration of glioma cells by binding to ELAV like RNA binding protein 1. Ding et al (21) indicated that SNHG12 enhanced the proliferation and metastasis of papillary thyroid carcinoma cells by regulating the Wnt/β-catenin signalling pathway. Furthermore, Wang et al (17) reported that SNHG12 promotes the proliferation and inhibits apoptosis of CRC cells. However, the regulatory mechanisms of how SNHG12 affects CRC growth and metastasis have remained elusive.
MicroRNAs (miRs), a class of small non-coding RNAs of 22–25 nt in length, are key regulators of gene expression (26). They directly bind to the 3′-untranslated region (3′-UTR) of their target mRNAs to thereby cause translational repression or mRNA degradation (26). A large number of studies have reported that miRs have key roles in the development and progression of human cancer types, including CRC (27,28). In recent years, Qian et al (29) reported that miR-16 was significantly downregulated in CRC tissues compared with that in normal colonic mucosa, and miR-16 expression was associated with tumour differentiation, lymph node metastasis, TNM stage and tumour recurrence in CRC. Furthermore, low miR-16 expression is an independent factor predicting a poor prognosis for CRC patients (29). In addition, miR-16 has been indicated to inhibit CRC cell growth in vitro by regulating the p53-survivin signalling pathway (30). However, the association between SNHG12 and miR-16 in CRC has remained elusive.
Therefore, the present study was performed to explore the association of SNHG12 and miR-16 in CRC and the underlying molecular mechanisms in CRC cells in vitro.
Materials and methods
Clinical tissue samples
A total of 53 CRC tissues and their matched adjacent non-tumour tissues were obtained from primary CRC patients at the Second Xiangya Hospital, Central South University (Changsha, China) between April 2011 and June 2012. These 53 CRC patients included 31 males and 22 females aged between 34 to 71 years (mean age, 53.7 years). The tissue samples were immediately snap-frozen in liquid nitrogen after surgical resection and stored until use.
Cell culture
Normal intestinal epithelial NCM460 cells and CRC cell lines, including HT-29, SW480, SW620 and LOVO, were obtained from the Cell Bank of Central South University and were cultured in Dulbecco's modified Eagle's medium (Thermo Fisher Scientific, Inc.) with 10% foetal bovine serum (FBS; HyClone; GE Healthcare) in a humidified atmosphere with 5% CO2 at 37°C.
Cell transfection
For cell transfection, SW620 and HT-29 cells (1×106 per well) were seeded into 6-well plates, cultured overnight and transfected with 100 nM non-specific short interfering (si)RNA (NC siRNA; cat. no. 12935300; Thermo Fisher Scientific, Inc.), 100 nM SNHG12-specific siRNA (SNHG12 siRNA; cat. no. AM16708; Thermo Fisher Scientific, Inc.), 100 nM pcDNA3.1 vector (cat. no. V00008; Yearthbio, Inc.), 100 nM pcDNA-SNHG12 expression plasmid (cat. no. P00951; Yearthbio, Inc.), 100 nM miR-16 inhibitor (cat. no. 4464084; Thermo Fisher Scientific, Inc.) and/or 100 nM negative control (NC) inhibitor (cat. no. 4464076; Thermo Fisher Scientific, Inc.) by using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The subsequent experiments were performed a 48 h after transfection.
Reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted from tissues and cells using TRIzol reagent (Thermo Fisher Scientific, Inc). To detect the expression of miR-16 or SNHG12, RNA was reverse-transcribed into complementary (c)DNA using the PrimeScript™ II 1st Strand cDNA Synthesis kit (Takara Bio, Inc.) according to the manufacturer's protocol. Subsequently, qPCR was performed using the SYBR Premix kit (Takara Bio, Inc.) or the SYBR Prime Script miRNA RT-PCR kit (Takara Bio, Inc.). U6 and GAPDH were used as internal references. The relative expression was determined using the 2−∆∆Cq method (31). The following primers were used: GAPDH forward, 5′-CTGGGCTACACTGAGCACC-3′ and reverse, 5′-AAGTGGTCGTTGAGGGCAATG-3′; and SNHG16 forward, 5′-GTGCCTCAGGAAGTCTCTTGCC-3′ and reverse, 5′-ATCCAAACAAGTTATCACACAGCAC-3′. Primers for U6 (cat. no. HmiRQP9001) and miR-16 (cat. no. HmiRQP0226) were obtained from Guangzhou FulenGen, Co., Ltd.
Bioinformatics analysis and luciferase reporter gene assay
TargetScan (www.targetscan.org) software predicted that SNHG12 was a potential target gene of miR-16. SW620 and HT-29 cells were transfected with a wild-type (WT) or a mutant (MT) SNHG12 pMIR-REPORT™ miRNA Expression Reporter Vector (Ambion; Thermo Fisher Scientific, Inc.) containing a 4-bp mutation at the predicted miR-16 binding site within SNHG12, along with miR-16 mimics (cat. no. 4464066; Thermo Fisher Scientific, Inc.) or miR-NC mimics (cat. no. 4464058; Thermo Fisher Scientific, Inc.) using Lipofectamine 2000. After transfection for 48 h, a luciferase reporter gene assay was performed using the Dual-Luciferase® Reporter Assay System (Promega Corporation). The activity of firefly luciferase was normalized to the activity of Renilla luciferase.
Cell Counting Kit (CCK)-8 assay
The transfected cells (2,000 cells/well) were seeded in 96-well plates and cultured at 37°C with 5% CO2 for 0, 24, 48, 72 and 96 h, respectively. The cell proliferation was then assessed using a CCK-8 (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The optical density value was read at a wavelength of 450 nm using a microplate reader (Bio-Rad Laboratories, Inc.).
Cell invasion assay
Cell invasion was assessed using Transwell chambers pre-coated with Matrigel® (BD Biosciences). The transfected cells (1×105 per chamber) were re-suspended in 300 µl serum-free DMEM and added to the upper chamber, and the lower chamber was filled with medium containing 10% FBS. After incubation at 37°C for 48 h, cells that had transgressed through the membrane were fixed with 37% methanol at room temperature for 30 min, and stained with 0.5% crystal violet at room temperature for 10 min. The invaded cells were counted under a microscope.
Statistical analysis
Values are expressed as the mean ± standard deviation. SPSS 19.0 (IBM Corp.) was used for statistical analysis. An unpaired or paired Student's t-test was used for comparison between two groups. One-way analysis of variance followed by Tukey's post-hoc test was used for comparisons of more than two groups. A chi-square test was used to analyse the association between SNHG12 expression and clinicopathological characteristics in patients with CRC. Pearson correlation analysis was used to determine the association between miR-16 and SNHG12 expression in CRC tissues. The Kaplan-Meier method and a log-rank test were used to analyse the overall survival of patients with CRC. P<0.05 was considered to indicate statistical significance.
Results
Upregulation of SNHG12 in CRC tissues and cell lines
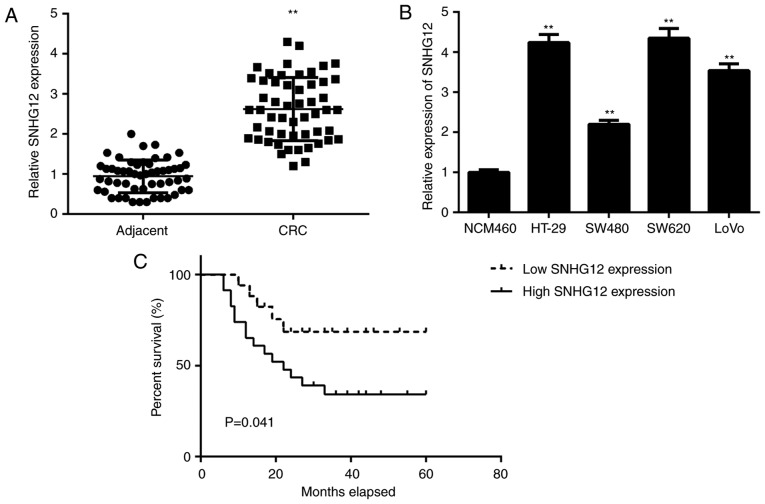
First, the expression of lncRNA SNHG12 in CRC and adjacent non-tumour tissue samples was examined using RT-qPCR. As indicated in Fig. 1, the levels of SNHG12 were significantly higher in CRC tissues compared with those in adjacent non-tumour tissue samples. Consistently, the expression levels of SNHG12 were also markedly increased in CRC cell lines, including HT-29, SW480, SW620 and LOVO, when compared with those in NCM460 cells (Fig. 1B).
Figure 1.
SNHG12 is upregulated in CRC. (A) SNHG12 is upregulated in CRC tissues compared with that in adjacent non-tumour tissues. **P<0.01 vs. Adjacent. (B) SNHG12 is upregulated in CRC cell lines compared with that in NCM460 cells. **P<0.01 vs. NCM460 cells. (C) Kaplan-Meier analysis indicated that CRC patients with high SNHG12 expression had a significantly poorer overall survival when compared with that of the group with low SNHG12 expression. SNHG12, small nucleolar RNA host gene 12; CRC, colorectal cancer.
To further assess the clinical significance of SNHG12 expression in CRC, the CRC patients were divided into high and low SNHG12 expression groups using the mean expression level of SNHG12 as the cut-off value. As presented in Table I, high expression of SNHG12 was significantly associated with lymph node metastasis and advanced clinical stage. Furthermore, the CRC patients with high SNHG12 expression had a poorer overall survival when compared with that in the group with low SNHG12 expression (Fig. 1C), suggesting that high SNHG12 expression may predict poor prognosis for CRC patients.
Table I.
Association between SNHG12 expression and clinicopathological characteristics in patients with colorectal cancer (n=53).
| SNHG12 expression | |||||
|---|---|---|---|---|---|
| Variable | N | Low (n=27) | High (n=26) | Chi-square value | P-value |
| Age (years) | 0.930 | 0.335 | |||
| ≤55 | 27 | 12 | 15 | ||
| >55 | 26 | 15 | 11 | ||
| Sex | 0.195 | 0.442 | |||
| Male | 31 | 15 | 16 | ||
| Female | 22 | 12 | 10 | ||
| Differentiation | 2.163 | 0.141 | |||
| Well and moderate | 43 | 24 | 19 | ||
| Poor | 10 | 3 | 7 | ||
| Lymph node metastasis | 2.419 | 0.016 | |||
| Present | 18 | 5 | 13 | ||
| Absent | 35 | 22 | 13 | ||
| Tumor, nodes and metastasis stage | 2.419 | 0.016 | |||
| I–II | 35 | 22 | 13 | ||
| III–IV | 18 | 5 | 13 | ||
SNHG12, small nucleolar RNA host gene 12.
Function of SNHG12 in CRC cell proliferation and invasion
Next, the function of SNHG12 in the proliferation and invasion of CRC cells was examined. As the above results had indicated that SNHG12 was upregulated in CRC, SW620 and HT-29 cells were transfected with SNHG12 siRNA to reduce its expression. After transfection, the expression of SNHG12 was markedly downregulated in the siSNHG12 group when compared with that in the siNC group (Fig. 2A). A CKK-8 assay indicated that the proliferation of SW620 and HT-29 cells was significantly decreased in the siSNHG12 group compared with that in the siNC group, indicating that knockdown of SNHG12 expression inhibits CRC cell proliferation (Fig. 2B and C). Similarly, the Transwell assay suggested that the invasive capacity of SW620 and HT-29 cells was markedly decreased after silencing of SNHG12 expression (Fig. 2D).
Figure 2.
Inhibition of SNHG12 inhibits colorectal cancer cell proliferation and invasion. (A-D) SW620 and HT-29 cells were transfected with SNHG12 siRNA or NC siRNA, respectively. (A) RT-qPCR was used to examine the levels of SNHG12. CCK-8 assay using (B) SW620 and (C) HT-29 cells, and (D) a Transwell assay (magnification, ×200) were performed to assess cell proliferation and invasion, respectively. For A-D, **P<0.01 vs. NC siRNA. (E-H) Next, SW620 and HT-29 cells were transfected with SNHG12 expression plasmid or a blank vector, respectively. (E) RT-qPCR was used to determine the levels of SNHG12. CCK-8 assay using (F) SW620 and (G) HT-29 cells, and (H) a Transwell assay (magnification, ×200) were performed to study cell proliferation and invasion. For E-H, **P<0.01 vs. Blank. Groups: siNC, cells transfected with NC siRNA; siSNHG12, cells transfected with siRNA targeting SNHG12; SNHG12, cells transfected with SNHG12 overexpression vector; Blank, cells transfected with empty vector. SNHG12, small nucleolar RNA host gene 12; CKK-8, Cell Counting Kit 8; siRNA, small interfering RNA; NC, negative control; OD, optical density; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
To further confirm the above results, the effects of SNHG12 overexpression on the proliferation and invasion of CRC cells were then assessed. SW620 and HT-29 cells were transfected with SNHG12 plasmid or blank vector. After transfection, SNHG12 was significantly upregulated in the SNHG12 group when compared with that in the blank vector group (Fig. 2E). Furthermore, as indicated in Fig. 2F-H, the proliferation and invasion of SW620 and HT-29 cells was significantly increased in the SNHG12 group compared with that in the blank vector group. These results suggest that SNHG12 may have a role in promoting CRC growth and metastasis.
Targeting association between SNHG12 and miR-16 in CRC cells
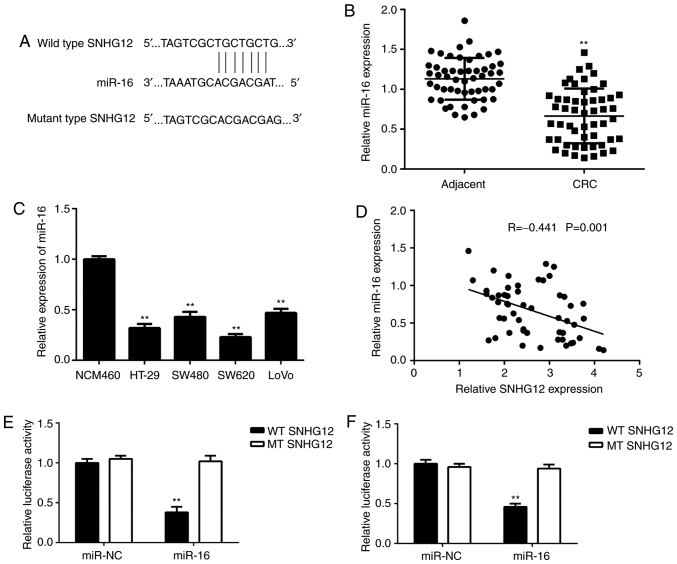
Next, a Bioinformatics analysis was performed to identify miRs targeted by SNHG12, and it was indicated that miR-16 and SNHG12 had complementary binding sites (Fig. 3A). Therefore, the expression levels of miR-16 in CRC tissue and cell lines were examined. As indicated in Fig. 3B and C, the expression levels of miR-16 were markedly reduced in CRC tissues and cell lines when compared with those in adjacent non-tumour tissues and NCM460 cells, respectively. Furthermore, a significant inverse correlation was observed between SNHG12 and miR-16 expression in CRC tissues (Fig. 3D). To further confirm the association between SNHG12 and miR-16 in CRC, luciferase reporter plasmids containing WT or MT miR-16 binding sites of SNHG12 were constructed (Fig. 3E). The luciferase reporter assay indicated that transfection with miR-16 mimics markedly decreased the luciferase activity of the vector containing WT SNHG12 but had no effect on that in the vector carrying MT SNHG12 in SW620 and HT-29 cells (Fig. 3F-G), suggesting that SNHG12 acts as a sponge for miR-16 in CRC cells.
Figure 3.
Targeting association between SNHG12 and miR-16 in CRC cells. (A) miR-16 was predicted to bind to SNHG12 by a Bioinformatics analysis. (B) miR-16 is downregulated in CRC tissues compared with that in adjacent non-tumour tissues. **P<0.01 vs. Adjacent. (C) miR-16 is downregulated in CRC cell lines compared with that in NCM460 cells. **P<0.01 vs. NCM460 cells. (D) Inverse correlation between SNHG12 and miR-16 expression in CRC tissue. (E) A luciferase reporter plasmid containing WT or MT miR-16 binding sites of SNHG12 was constructed. A luciferase reporter gene assay was performed in (E) SW620 and (F) HT-29 cells by co-transfection of the respective reporter plasmids and miR-16 mimics or miR-NC. WT, wild-type; MT, mutant type; miR, microRNA; SNHG12, small nucleolar RNA host gene 12; NC, negative control; CRC, colorectal cancer.
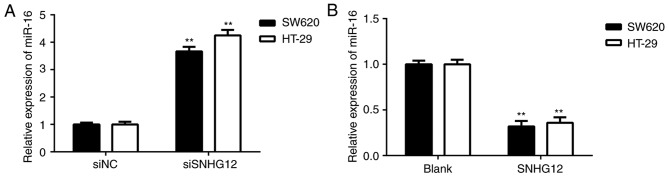
SNHG12 inhibits miR-16 expression in CRC cells
The effects of SNHG12 on miR-16 expression in CRC cells were then further studied. As presented in Fig. 4A, silencing of SNHG12 increased the expression of miR-16 in SW620 and HT-29 cells. Furthermore, overexpression of SNHG12 markedly reduced the expression of miR-16 in CRC cells (Fig. 4B). It was therefore indicated that SNHG12 negatively affects the miR-16 levels in CRC cells.
Figure 4.
SNHG12 inhibits miR-16 expression in CRC cells. (A) Silencing of SNHG12 increased the expression of miR-16 in SW620 and HT-29 cells. **P<0.01 vs. NC siRNA. (B) SW620 and HT-29 cells were transfected with SNHG12 expression plasmid or a blank vector, respectively. The expression of miR-16 was then examined using reverse transcription-quantitative polymerase chain reaction. **P<0.01 vs. Blank. Groups: siNC, cells transfected with NC siRNA; siSNHG12, cells transfected with siRNA targeting SNHG12; SNHG12, cells transfected with SNHG12 overexpression vector; Blank, cells transfected with empty vector. SNHG12, small nucleolar RNA host gene 12; siRNA, small interfering RNA; NC, negative control; miR, microRNA.
miR-16 is involved in SNHG12-induced CRC cell proliferation and invasion
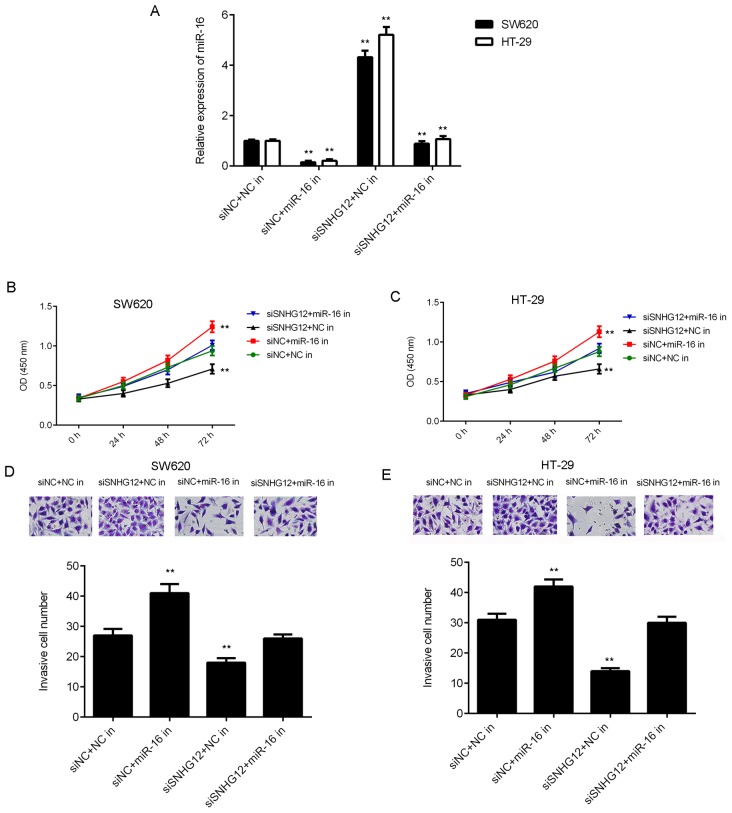
Based on the above results, it was speculated that SNHG12 promotes CRC cell proliferation and invasion by acting as a sponge of miR-16. To validate this speculation, SW620 and HT-29 cells were co-transfected with NC siRNA and NC inhibitor (siNC+NC in group), NC siRNA and miR-16 inhibitor (siNC+miR-16 in group), SNHG12 siRNA and NC inhibitor (siSNHG12+NC in group), or with SNHG12 siRNA and miR-16 inhibitor (siSNHG12+miR-16 in group). As presented in Fig. 5A, the expression levels of miR-16 were markedly lower in the siNC+miR-16 in group, while they were higher in the siSNHG12+NC group, when compared with those in the siNC+NC in group. Furthermore, the expression levels of miR-16 were lower in the siSNHG12+miR-16 in group when compared with those in the siSNHG12+NC in group. CKK-8 assay and Transwell assays further indicated that the proliferation and invasion of CRC cells were significantly reduced in the siSNHG12+NC in group when compared with those in the siNC+NC in group, while these effects were rescued in the siSNHG12+miR-16 in group (Fig. 5B-E). These results suggested that knockdown of SNHG12 inhibited CRC cell proliferation and invasion by upregulating miR-16.
Figure 5.
miR-16 is involved in SNHG12-mediated colorectal cancer cell proliferation and invasion. SW620 and HT-29 cells were co-transfected with NC siRNA and NC inhibitor (siNC+NC in), NC siRNA and miR-16 inhibitor (siNC+miR-16 in), SNHG12 siRNA and NC inhibitor (siSNHG12+NC in), or with SNHG12 siRNA and miR-16 inhibitor (siSNHG12+miR-16 in). After transfection, (A) reverse transcription-quantitative polymerase chain reaction was used to examine the levels of miR-16. CCK-8 assay using (B) SW620 and (C) HT-29 cells, and (D and E) a Transwell assay (magnification, ×200) were performed to examine cell proliferation and invasion, respectively. **P<0.01 vs. siSNHG12+NC. siSNHG12, siRNA targeting SNHG12; in, inhibitor; SNHG12, small nucleolar RNA host gene 12; siRNA, small interfering RNA; NC, negative control; OD, optical density; miR, microRNA.
Discussion
The regulatory mechanism of SNHG12 in CRC cells has remained largely elusive. In the present study, SNHG12 was identified to be markedly upregulated in CRC tissue when compared with adjacent non-tumour tissues, and its upregulation was markedly associated with CRC progression, as well as poor prognosis of patients. In addition, SNHG12 expression was higher in CRC cell lines when compared with that in normal intestinal epithelial cells. Knockdown of SNHG12 significantly repressed CRC cell proliferation and invasion, while SNHG12 overexpression significantly increased the proliferation and invasion of CRC cells. A Bioinformatics analysis indicated that SNHG12 and miR-16 had complementary binding sites, which was confirmed by a luciferase reporter gene assay. The expression levels of miR-16 were markedly downregulated in CRC tissues and cell lines, and were inversely correlated with the expression of SNHG12 in CRC tissues. In addition, silencing of miR-16 eliminated the suppressive effects of SNHG12 downregulation on CRC cell proliferation and invasion.
It has been reported that SNHG12 has a role in promoting certain common human cancer types. For instance, SNHG12 has been indicated to promote cell proliferation and migration by increasing the expression of angiomotin in osteosarcoma cells (19). It was also reported to promote tumourigenesis and metastasis by targeting miR-199a/b-5p in hepatocellular carcinoma (18). Furthermore, SNHG12 contributes to multidrug resistance by activating the MAPK/Slug pathway by sponging miR-181a in non-small-cell lung cancer (32). In addition, C-MYC-induced upregulation of lncRNA SNHG12 promoted cell proliferation and migration, while inhibiting cell apoptosis in triple-negative breast cancer (25). Thus, SNHG12 has been suggested as a promising therapeutic target for specific cancer types (18,32,33). Furthermore, knockdown of SNHG12 was demonstrated to inhibit non-small cell lung cancer cell growth and induce apoptosis by upregulating miR-138 (33). Recently, Wang et al (17) reported that SNHG12 promotes the proliferation and inhibits apoptosis of CRC cells. However, the underlying mechanisms of the regulatory function of SNHG12 during CRC growth and metastasis has remained elusive. The present study confirmed that the expression of SNHG12 was markedly upregulated in CRC tissues and cell lines when compared with that in adjacent normal tissues and normal intestinal epithelial cells, respectively. Consistent with the present results, Wang et al (17) also observed an increased expression of SNHG12 in CRC tissues and cell lines. Furthermore, the present study revealed that high expression of SNHG12 was associated with lymph node metastasis, advanced clinical stage and poor overall survival of CRC patients.
Next, the exact roles of SNHG12 to regulate CRC cell proliferation and invasion were investigated. It was demonstrated that inhibition of SNHG12 expression led to a significant reduction in CRC cell proliferation and invasion. Consistent with these results, Wang et al (17) also reported that SNHG12 promoted CRC cell proliferation and that overexpression of SNHG12 enhanced the cell cycle progression of SW480 CRC cells, while knockdown of SNHG12 repressed the cell cycle progression of HT29 CRC cells. Furthermore, they demonstrated that SNHG12 also inhibited CRC cell apoptosis by increasing the expression of cell cycle-associated proteins and suppressing the expression of caspase 3 (17). These previous and the present results indicated that SNHG12 may have a role in promoting CRC proliferation and metastasis.
Next, the underlying mechanisms of the regulatory effect of SNHG12 on CRC cell proliferation and invasion were assessed in vitro. TargetScan software predicted that miR-16 and SNHG12 had complementary binding sites. To confirm this predicted binding interaction, the expression of miR-16 was examined in CRC, revealing that miR-16 was markedly downregulated in CRC tissues and cell lines. Furthermore, an inverse correlation was observed between miR-16 and SNHG12 in CRC tissues. To further clarify the association between miR-16 and SNHG12 in CRC, a luciferase reporter gene assay was performed, and the results indicated that transfection with miR-16 mimics markedly reduced the luciferase activity of WT SNHG12 reporter vector but did not affect the luciferase activity of MT SNHG12 in SW620 and HT-29 cells. These results suggest that SNHG12 may sponge miR-16 in CRC cells. As it was further observed that miR-16 expression was negatively regulated by SNHG12 in CRC cells, it was speculated that miR-16 is involved in SNHG12-mediated CRC cell proliferation and invasion, which was further confirmed by the result that inhibition of miR-16 expression abrogated the suppressive effects of SNHG12 downregulation on CRC cell proliferation and invasion.
In summary, the present study demonstrated that SNHG12 is upregulated in CRC and has a role in promoting CRC cell proliferation and invasion by acting as a sponge of miR-16. Thus, SNHG12 may be a promising therapeutic target for CRC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analysed during the present study are included in this published article.
Authors' contributions
FR designed the study and wrote the manuscript. YL and J-YZ collected clinical tissues. SW, YS, J-PZ and FR performed the in-vitro experiments and statistical analysis.
Ethics approval and consent to participate
The present study was approved by the Research Ethics Committee of the Second Xiangya Hospital, Central South University (Changsha, China) and written informed consent had been obtained from all subjects.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Li JW, Huang CZ, Li JH, Yuan JH, Chen QH, Zhang WF, Xu ZS, Liu YP, Li Y, Zhan MX, Lu LG. Knockdown of metadherin inhibits cell proliferation and migration in colorectal cancer. Oncol Rep. 2018;40:2215–2223. doi: 10.3892/or.2018.6581. [DOI] [PubMed] [Google Scholar]
- 4.Cen C, Li J, Liu J, Yang M, Zhang T, Zuo Y, Lin C, Li X. Long noncoding RNA LINC01510 promotes the growth of colorectal cancer cells by modulating MET expression. Cancer Cell Int. 2018;18:45. doi: 10.1186/s12935-018-0503-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Price TJ, Tang M, Gibbs P, Haller DG, Peeters M, Arnold D, Segelov E, Roy A, Tebbutt N, Pavlakis N, et al. Targeted therapy for metastatic colorectal cancer. Expert Rev Anticancer Ther. 2018;18:991–1006. doi: 10.1080/14737140.2018.1502664. [DOI] [PubMed] [Google Scholar]
- 6.Islam F, Gopalan V, Vider J, Wahab R, Ebrahimi F, Lu CT, Kasem K, Lam AKY. MicroRNA-186-5p overexpression modulates colon cancer growth by repressing the expression of the FAM134B tumour inhibitor. Exp Cell Res. 2017;357:260–270. doi: 10.1016/j.yexcr.2017.05.021. [DOI] [PubMed] [Google Scholar]
- 7.Xiao Z, Qu Z, Chen Z, Fang Z, Zhou K, Huang Z, Guo X, Zhang Y. LncRNA HOTAIR is a prognostic biomarker for the proliferation and chemoresistance of colorectal cancer via MiR-203a-3p-mediated Wnt/β-catenin signaling pathway. Cell Physiol Biochem. 2018;46:1275–1285. doi: 10.1159/000489110. [DOI] [PubMed] [Google Scholar]
- 8.Xie B, Deng Z, Pan Y, Fu C, Fan S, Tao Y, Zhou J, Xiao D. Post-transcriptional regulation DPC4 gene by miR-190 in colorectal cancer cells. J Cancer Res Ther. 2018;14:838–843. doi: 10.4103/jcrt.JCRT_577_17. [DOI] [PubMed] [Google Scholar]
- 9.Liu K, Yao H, Wen Y, Zhao H, Zhou N, Lei S, Xiong L. Functional role of a long non-coding RNA LIFR-AS1/miR-29a/TNFAIP3 axis in colorectal cancer resistance to pohotodynamic therapy. Biochim Biophys Acta. 2018;1864:2871–2880. doi: 10.1016/j.bbadis.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 10.Liao D, Li T, Ye C, Zeng L, Li H, Pu X, Ding C, He Z, Huang GL. miR-221 inhibits autophagy and targets TP53INP1 in colorectal cancer cells. Exp Ther Med. 2018;15:1712–1717. doi: 10.3892/etm.2017.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yunusov D, Anderson L, DaSilva LF, Wysocka J, Ezashi T, Roberts RM, Verjovski-Almeida S. HIPSTR and thousands of lncRNAs are heterogeneously expressed in human embryos, primordial germ cells and stable cell lines. Sci Rep. 2016;6:32753. doi: 10.1038/srep32753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Y, Chen Y, Ding W, Hua Z, Wang L, Zhu Y, Qian H, Dai T. LncRNA UCA1 impacts cell proliferation, invasion, and migration of pancreatic cancer through regulating miR-96/FOXO3. IUBMB Life. 2018;70:276–290. doi: 10.1002/iub.1699. [DOI] [PubMed] [Google Scholar]
- 14.Peng Z, Liu C, Wu M. New insights into long noncoding RNAs and their roles in glioma. Mol Cancer. 2018;17:61. doi: 10.1186/s12943-018-0812-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smolle MA, Pichler M. The role of long non-coding RNAs in osteosarcoma. Noncoding RNA. 2018;4:E7. doi: 10.3390/ncrna4010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu S, Kong D, Chen Q, Ping Y, Pang D. Oncogenic long noncoding RNA landscape in breast cancer. Mol Cancer. 2017;16:129. doi: 10.1186/s12943-017-0696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang JZ, Xu CL, Wu H, Shen SJ. LncRNA SNHG12 promotes cell growth and inhibits cell apoptosis in colorectal cancer cells. Braz J Med Biol Res. 2017;50:e6079. doi: 10.1590/1414-431x20176079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lan T, Ma W, Hong Z, Wu L, Chen X, Yuan Y. Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes tumorigenesis and metastasis by targeting miR-199a/b-5p in hepatocellular carcinoma. J Exp Clin Cancer Res. 2017;36:11. doi: 10.1186/s13046-016-0486-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruan W, Wang P, Feng S, Xue Y, Li Y. Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes cell proliferation and migration by upregulating angiomotin gene expression in human osteosarcoma cells. Tumour Biol. 2016;37:4065–4073. doi: 10.1007/s13277-015-4256-7. [DOI] [PubMed] [Google Scholar]
- 20.Lei W, Wang ZL, Feng HJ, Lin XD, Li CZ, Fan D. Long non-coding RNA SNHG12promotes the proliferation and migration of glioma cells by binding to HuR. Int J Oncol. 2018;53:1374–1384. doi: 10.3892/ijo.2018.4478. [DOI] [PubMed] [Google Scholar]
- 21.Ding S, Qu W, Jiao Y, Zhang J, Zhang C, Dang S. LncRNA SNHG12 promotes the proliferation and metastasis of papillary thyroid carcinoma cells through regulating wnt/beta-catenin signaling pathway. Cancer Biomark. 2018;22:217–226. doi: 10.3233/CBM-170777. [DOI] [PubMed] [Google Scholar]
- 22.Yang BF, Cai W, Chen B. LncRNA SNHG12 regulated the proliferation of gastric carcinoma cell BGC-823 by targeting microRNA-199a/b-5p. Eur Rev Med Pharmacol Sci. 2018;22:1297–1306. doi: 10.26355/eurrev_201803_14471. [DOI] [PubMed] [Google Scholar]
- 23.Dong J, Wang Q, Li L, Xiao-Jin Z. Upregulation of long non-coding RNA small nucleolar RNA host gene 12 contributes to cell growth and invasion in cervical cancer by acting as a sponge for MiR-424-5p. Cell Physiol Biochem. 2018;45:2086–2094. doi: 10.1159/000488045. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H, Lu W. LncRNA SNHG12 regulates gastric cancer progression by acting as a molecular sponge of miR320. Mol Med Rep. 2018;17:2743–2749. doi: 10.3892/mmr.2017.8143. [DOI] [PubMed] [Google Scholar]
- 25.Wang O, Yang F, Liu Y, Lv L, Ma R, Chen C, Wang J, Tan Q, Cheng Y, Xia E, et al. C-MYC-induced upregulation of lncRNA SNHG12 regulates cell proliferation, apoptosis and migration in triple-negative breast cancer. Am J Transl Res. 2017;9:533–545. [PMC free article] [PubMed] [Google Scholar]
- 26.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Sun Z, Liu B, Shan Y, Zhao L, Jia L. Tumor-suppressive miR-26a and miR-26b inhibit cell aggressiveness by regulating FUT4 in colorectal cancer. Cell Death Dis. 2017;8:e2892. doi: 10.1038/cddis.2017.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lv H, Zhang Z, Wang Y, Li C, Gong W, Wang X. MicroRNA-92a promotes colorectal cancer cell growth and migration by inhibiting KLF4. Oncol Res. 2016;23:283–290. doi: 10.3727/096504016X14562725373833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qian J, Jiang B, Li M, Chen J, Fang M. Prognostic significance of microRNA-16 expression in human colorectal cancer. World J Surg. 2013;37:2944–2949. doi: 10.1007/s00268-013-2205-4. [DOI] [PubMed] [Google Scholar]
- 30.Ma Q, Wang X, Li Z, Li B, Ma F, Peng L, Zhang Y, Xu A, Jiang B. microRNA-16 represses colorectal cancer cell growth in vitro by regulating the p53/survivin signaling pathway. Oncol Rep. 2013;29:1652–1658. doi: 10.3892/or.2013.2262. [DOI] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Wang P, Chen D, Ma H, Li Y. LncRNA SNHG12 contributes to multidrug resistance through activating the MAPK/Slug pathway by sponging miR-181a in non-small cell lung cancer. Oncotarget. 2017;8:84086–84101. doi: 10.18632/oncotarget.20475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Qi G, Zhang J, Wu J, Zhou N, Li L, Ma J. Knockdown of long noncoding RNA small nucleolar RNA host gene 12 inhibits cell growth and induces apoptosis by upregulating miR-138 in nonsmall cell lung cancer. DNA Cell Biol. 2017;36:892–900. doi: 10.1089/dna.2017.3830. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during the present study are included in this published article.