Abstract
The latent HIV-1 reservoir in blood decays very slowly, even during prolonged suppression of viral replication by antiretroviral therapy (ART). Mechanisms for reservoir persistence include replenishment through low-level viral replication, longevity and homeostatic proliferation of memory T cells, and most recently appreciated, clonal expansion of HIV-infected cells. Clonally expanded cells make up a large and increasing fraction of the residual infected cell population on ART, and insertion of HIV proviruses into certain host cellular genes has been associated with this proliferation. That the vast majority of proviruses are defective clouds our assessment of the degree to which clonally expanded cells harbor infectious viruses, and thus the extent to which they contribute to reservoirs relevant to curing infection. This review summarizes past studies that have defined our current understanding and the gaps in our knowledge of the mechanisms by which proviral integration and clonal expansion sustain the HIV reservoir.
Keywords: HIV infected cell proliferation, clonal proliferation, integration sites, cancer genes, T-regulatory cells.
The potential for curing human immunodeficiency virus (HIV) infection is limited by an incomplete understanding of the mechanisms that allow HIV to persist during antiretroviral therapy (ART). In the 1990s, ART was predicted to completely inhibit HIV replication and, based on the decay of cell-free and cell-associated HIV in the blood, to result in clearance of HIV infection within approximately 3 years [1]. However, cells capable of producing infectious viruses were found to persist, mostly composed of memory CD4 T cells [2–7] with a half-life of 4.6–44 months [8, 9]. Then, long cellular lifespans and/or low levels of replication were suspected to sustain viral reservoirs [8]. More recently, homeostatic proliferation, central to survival of mature CD4 T cells [10] and proliferation of HIV-infected cells [11, 12] was also hypothesized to sustain infectious reservoirs. The finding that certain tissues have low, perhaps subinhibitory concentrations of antiretroviral drugs (ARVs), lent support to the concept of ongoing replication within certain tissues during ART [7, 13, 14]. However, analyses of viral sequences from many individuals with ART suppression for more than a decade have provided no evidence of virus evolution or selection of drug-resistant variants [15, 16]. This review will focus on the most recently recognized mechanism for sustaining virus reservoirs, that of infected cell proliferation.
PROLIFERATION OF HIV-INFECTED CELLS UNDER ART
Although approximately 75% of HIV infections result in the emergence of a single founder virus, and thus a nearly homogeneous virus population [17–21], stochastic as well as immune escape-driven viral evolution quickly results in virus diversification. Within a period of months, few identical viral sequences are found within more variable regions of the viral genome [22–25]. It was therefore surprising to observe that studies aimed at detecting “occult” HIV replication during ART [11, 12, 26] found that low-level viremias, between 50 and 250 c/mL during ART, were often composed of virions with identical env and pol sequences [12]. Often, these viral sequences were also identical to proviruses within infected cell DNA, leading to the hypothesis that clonal outgrowth of long-lived HIV-infected cells was occurring and that these cells could produce low levels of monotypic viremia [11, 12, 27]. The levels of viremia observed suggest that multiple cells from a clonal lineage were synchronously producing virions, possibly due to antigen-specific activation of memory cells, because single cells would not be capable of producing these quantities of virions.
The substantial proliferative potential of memory CD4+ T cells [28] and the demonstration that clonal viral sequences are found within these cells [7] led us to hypothesize that proliferating cells would form an increasing fraction of the residual HIV-infected population under ART [29]. This would not be the case during typical untreated infections because newly infected cells would be much more numerous than the persisting infected cell population. This would explain why despite many years of ART, the level of infected cells in the blood and the amount of infectious virus induced by global activators such as phytohemagglutinin (PHA) decline very slowly [8]. Our analysis [29] of 14 individuals over a median of 9.8 years of ART found that the proportion of all cell-associated viral DNA sequences corresponding to demonstrably clonal sequences increased at an average rate of 3.3% per year. Overall, infected cell numbers decreased over the study period, but the population of nonidentical viral sequences decreased by an average of 6.2% per year (95% confidence interval [CI] = 3.0–9.5%; P < .0001), whereas the clonal cell lineages experienced little if any decrease (mean = −1.0% per year; 95% CI = −0.2% to 2.3%; P > .10). Hence, although the number of infected cells decreases slowly under ART, the complexity of the virus population decreases more rapidly due to the increasing representation of what appeared to be clonally proliferating cells.
The aforementioned studies characterized viruses as “identical” using only short regions of the viral genome, usually 5%–10% of the entire virus, and in some cases, relatively highly conserved genes such as pol. In theory, if ARV levels are subtherapeutic during ART, an infected cell could transmit nearly identical virions cell-free, or more effectively through cell-to-cell contact, to vast numbers of susceptible cells. Thus, because analyses of short regions of HIV can overestimate the number of viruses thought to be emerging from proliferating cells [30], methods identifying HIV integration sites (HIV-ISs) are likely to be more accurate in identifying the source of monotypic virus populations.
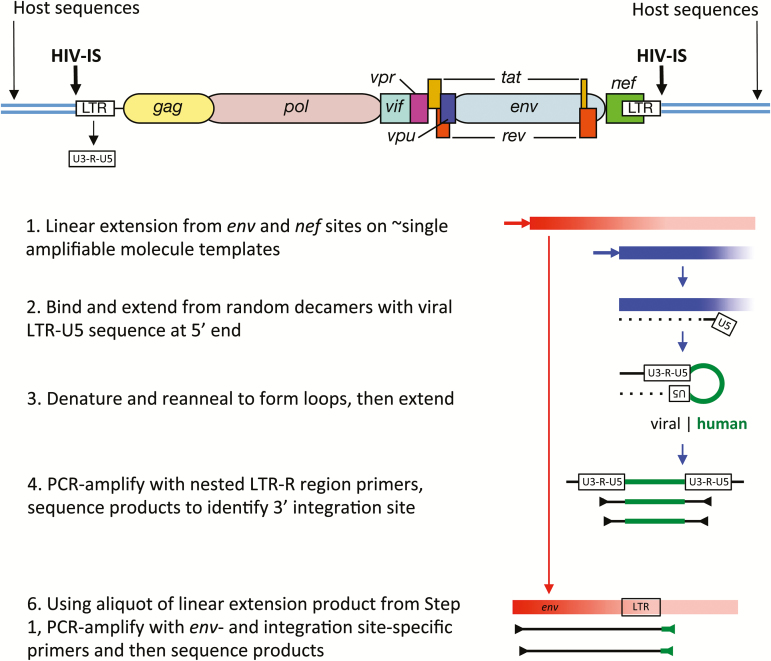
To formally prove that identical viral sequences were due to infected cell proliferation, we developed the integration site looping assay (ISLA), which amplifies a region of the viral genome that includes the variable regions within the gp120 Env coding sequence, the genomic region with the highest genetic diversity, through the 3’ long terminal repeat (LTR) and into the human cellular genome sequence flanking the viral integration site [31] (Figure 1). The utility of this approach stems from the extremely large number of possible HIV-ISs in the human genome and the extremely low likelihood that any 2 sites would be identical, as discussed below. Using the ISLA, we initially identified the host sequence at HIV-ISs plus the 3’ end of the viral genome (Figure 1, sections 1–4). Using primers specific to this host sequence, the viral sequences of the adjacent 2.8-kb region encompassing a portion of env-gp120, nef, and the remainder of the LTR were then recovered and sequenced (Figure 1, section 5) [31]. Using this method, each IS was linked to clonal provirus populations previously identified and to plasma HIV sequences. These studies provided a formal proof that identical proviral sequences arise from the proliferation of infected cells. Furthermore, in all cases evaluated to date (eg, Mullins JI, Hall B, Kim K, Deng W, Westfall DH, Bull M, Chen L, Aamer HA, Frenkel LM. 2015. Integration Sites & Propagation and Analysis of Individual Lineages of HIV-Infected Cells Persisting during ART. Banbury conference on HIV-1 and How to Kill a Killer: Attempts at Total or Functional Cure of HIV-1, Cold Spring Harbor Laboratory, October, 2015.) identical viral sequences were found to match the same ISs. And, identical whole genome sequences were obtained from several proviruses with identical ISs (James I. Mullins, Malika Aid, Hadega A. Aamer, Paul Edlefsen, Sherry McLaughlin, Marta E. Bull, Hannah Huang, Sheila Styrchak, Wenjie Deng, Thomas Sibley, Evan Silberman, Rafick Sekaly, and Lisa M. Frenkel, manuscript in preparation).
Figure 1.
Schematic summary of the Integration Site Looping Assay (ISLA). Essential aspects of this procedure are schematically shown here, whereas full details are provided in Supplementary Figure 1 from [31]. The structure of a full-length provirus and adjacent cellular sequences are shown at the top of the figure. Long terminal repeated sequences (LTRs) flank the provirus, with LTRs subdivided into U3, R, and U5 segments. These segment names derive from their origins in viral genomic RNA; they are found to be: unique to the 3’ end of the human immunodeficiency virus (HIV) genomic RNA (U3), repeated at each end of the HIV genomic RNA (R), and unique to the 5’ end of the HIV genomic RNA (U5). Using 2 virus-specific primers from the env and nef genes, linear amplification is carried out to increase the representation of these sequences in the cellular DNA preparation (Step 1). Products extending from the nef primer that reach the adjacent host sequences are much shorter (approximately 0.8kb vs approximately 2.8 kb) and hence are much more efficiently generated. An aliquot of this reaction is then used to extend DNA from random primer sequences containing the U5 region at the 5’ end (Step 2). This product is then denatured and reannealed to form a loop with the U5 region of the provirus hybridizing to its complement derived from extension products generated with random primers (Step 3). Such products are then selectively polymerase chain reaction (PCR)–amplified using R region primers and then sequenced to determine the host genome sequences immediately adjacent to the 3’ end of the provirus (ie, the integration site, Step 4). To link the integration site to a specific proviral genome, the sequence obtained needs to extend into the gp120 protein coding sequence of the viral env gene, the region of highest level of viral genetic variation. Hence, in step 5, integration site (IS)–specific primers are generated to provide the specificity to amplify the longer, less efficiently produced env IS products.
Ikeda and colleagues first demonstrated multiple identical HIV-ISs in longitudinal samples from individuals on ART, which suggested clonal expansion and long-term persistence of infected cell clones [32]. Subsequently, 2 studies firmly established the pervasiveness of clonal expansion, showing that such cells can make up a large fraction of the residual HIV population in individuals on ART. Using a high-throughput deep-sequencing approach to identify HIV-host sequence junction fragments, Maldarelli and colleagues [33] evaluated 2410 ISs in 5 individuals and found clonally expanded cells to comprise approximately 40% of all HIV-ISs. This approach has a higher throughput than the ISLA but does not permit linkage to variable segments of the viral genome to demonstrate the origin of monotypic viral sequences and identifies ISs at both ends of the provirus without being able to distinguish whether different sequences observed derive from the same or different cells. Using the ISLA, we [31] identified 534 ISs in 3 individuals over 11–13 years of infection. This longitudinal study confirmed that infected cell clones persisted and, importantly, showed an overall increase in representation during ART [31]. Subsequently, Cohn et al [34] used a deep sequencing approach that inferred identical integrations from unique sheared ends of sequences. They tallied 6719 HIV-ISs from 5 individuals before and after starting ART, as well as from 4 untreated viremic individuals and 4 viremic controllers (viral loads ranged 430–1070 copies of HIV RNA/mL plasma). As with our results, they found the representation of infected cell clones to increase with time on ART. However, their results also departed in multiple respects from prior studies. They found clonally expanded cells at high levels in all individuals irrespective of viremia, an unexpected finding given that identical HIV env sequences were rarely detected in the vast number of prior studies of untreated noncontroller individuals in chronic infection. They also reported 78%–90% of all infected cells as being derived from proliferating cells, versus 15%–65% in Wagner [31] and McLaughlin et al. [33], despite a longer period on ART, and ~40% in the Maldarelli study.
DISTRIBUTION OF INFECTED PROLIFERATING CELLS IN TISSUES
Bull et al [35] demonstrated that infected cell clones populate the blood and the female genital tract, and we have found identical HIV-ISs in >12 tissues examined from multiple individuals at autopsy, indicating that clones of infected cells spread throughout the body (Mullins et al, manuscript in preparation). Cohn [34], as well as Boritz et al [36], while studying viremic controllers, found infected cell clones in Central memory T-cells ( TCM), Transitional memory T-cells (TTM), and Effector memory T-cells (TEM)memory cell types. Boritz also found the same HIV-ISs in cells of multiple memory cell types, indicating that these cells were infected before undergoing differentiation into TEM cells.
INFECTED PROLIFERATING CELLS HARBOR DEFECTIVE AS WELL AS INFECTIOUS PROVIRUSES
The vast majority of infected cells that persist under ART harbor defective proviruses [37–42]. However, such proviruses are not necessarily innocuous: (1) Defective proviruses, including those from infected cell clones, are capable of producing viral antigens and viremias and populate tissues throughout the body [43–46]. This chronic antigenic stimulation could contribute to the inflammatory pathology observed during ART (Richard Fox, Brian Johnson, Atis Muehlenbachs, Paul T. Edlefsen, Dylan Westfall, Julie Elliott, Peter Anton, Lisa M. Frenkel and James I. Mullins, submitted). (2) Defective proviruses may undergo spontaneous partial repair in vivo [47], and immunodeficiency disease can develop following spontaneous repair in vivo of nef-deleted simian immunodeficiency virus (SIV) in macaques [48]. Data from the Sydney Blood Bank Cohort have shown some evidence of disease progression in people infected with viruses harboring defective nef genes [49–51]. (3) Infectious viruses can be rescued by recombination between 2 defective proviruses [52, 53] and following superinfection [54].
Clonally proliferating cells have been hypothesized to persist due to their genomes being noninfectious [31, 33, 34]. However, Anderson and colleagues have found that clonally produced plasma RNA can be recovered as virus from resting CD4+ T cells [55]. Simonetti, Bull, and their colleagues have separately shown that clonally proliferating cells can contain intact full-length proviruses and produce infectious viral genomes [44, 56]. Because >90% of proviruses examined under ART have been found to be defective [41, 42] (Breana M. Hall, Noelle P. Dahl, Katie B. Kim, Lennie Chen, Michael P. Dapp, Anne-Marie Rousseau, Tamara Babenko, Dylan H. Westfall, Hannah Huang, S. Styrchak, K. Wong, H. Zhao, Thomas R. Sibley, Evan J. Silberman, C. Alexander Hill, L.M. Frenkel and J.I. Mullins, manuscript in preparation), the extent to which clonally proliferating cells contribute to the residual infectious reservoir remains to be quantified.
FAVORED SITES OF HUMAN IMMUNODEFICIENCY VIRUS INTEGRATION
Integration of the HIV proviral genome involves duplication of a 4–5-basepair region of the host cellular DNA adjacent to the LTR at each end of the integrated viral genome [57]. The site at which the virus integrates into the host genome is not precise. If it were, we could not use identical integration sites to quantify the proliferation of infected cells. Pioneering and comprehensive studies by Bushman and colleagues [57, 58], as well as others [59], have established that there is no absolute chromosomal site or sequence specificity for virus integration within the host genome. Rather, HIV integration tends to favor repeated sequence elements in the host [34, 60, 61] and structural features such as bent or distorted DNA within nucleosomes [62, 63]. It is also clear that virus integration is favored within genes over intergenic regions of DNA in both T-cell and macrophage cultures and in vivo [23, 32, 64–67], especially within actively transcribed genes [64]. Thus, regions of open chromatin found during transcription, and potentially during cell division, serve as targets of HIV-ISs. Interactions between viral integration complexes and cellular proteins such as LEDGF/p75 enhance viral integration [68, 69] and tether viral preintegration complexes to specific regions [70, 71]. In addition, the close proximity of a gene to nuclear matrix attachment sequences [72, 73] and the nuclear periphery [74], especially nuclear pores [75], are also associated with increased frequencies of HIV-ISs, with depletion of nuclear pore proteins diminishing HIV-ISs in gene-dense regions [76, 77]. Lastly, transcription-associated histone and DNA modifications [78] are associated with increased HIV-IS frequencies. Interestingly, Cohn [34] reported that ART was associated with a decrease in cells with HIV-ISs in highly expressed genes.
GENES ASSOCIATED WITH HUMAN IMMUNODEFICIENCY VIRUS INTEGRATION SITES AND CLONAL PROLIFERATION
Our comparison of the patterns of HIV-ISs from individuals on suppressive ART to >44000 HIV-ISs found after acute infection of Jurkat or primary cells in vitro [78, 79] showed strong clustering of HIV-ISs in vivo but far less so in vitro and little evidence of clonal proliferation in vitro [31]. Similar results were found by Maldarelli and colleagues using their own cell culture HIV-IS data [33, 80]. Clonal proliferation would not necessarily be expected to be observed in recently infected cells in culture because of the limited time permitted for selective cell outgrowth. In contrast, the dramatic clonal cell proliferation observed in ART-treated individuals and the clustering of sites in certain genes, strongly suggests that these phenomena occur as a result of a selective process. Hence, we examined the distribution of HIV-ISs across genes [31]. Genes associated with cancer were substantially overrepresented among HIV-ISs during ART (P < .0001), but this bias was also observed in the HIV-ISs from in vitro infections (12.70% vs 11.14% of sites, respectively). Cells undergoing clonal proliferation, however, had an excess of HIV-ISs in genes associated with cancer (P = .008) and above that found for HIV-ISs in cell culture (P = .049). This cancer gene association was derived using gene sets compiled from several sources [31]. When gene ontology (GO) terms were used instead, uniquely detected HIV-ISs and HIV-ISs in infected cell clones were enriched in genes with hallmark cancer GO terms, as well those for cell proliferation and negative regulation of cell cycle (all with P < .0001). These data strongly support the hypothesis that HIV integration can promote cell proliferation and survival.
Ikeda and colleagues were also first to show clusters of HIV-ISs in a particular gene, the basic leucine zipper transcription factor 2 (BACH2), which they found in 2 individuals [32]. Human immunodeficiency virus integration sites have been found in this gene in a total of 9 of 33 (27%) individuals at 72 unique sites in the host, including 24 that have been shown to be from infected cell clonal populations [31–33, 41, 65, 81, 82] (the Cohn et al study [34] was not included because no precise HIV-IS mapping data was available at this writing). As previously noted [31], a striking feature of the HIV-ISs in BACH2 is that all are found in the same transcription orientation as the gene and in the 2 introns just upstream of the start of the coding sequence. It is also notable that integrations of murine leukemia virus into BACH2 have been associated with B-cell tumors [83]. BACH2 has been linked to a large number of functions in humans (eg, autoimmune and allergic diseases; activation of the tumor suppressor p53; regulation of a variety of immune cell functions, including CD4 T-cell senescence; immune activation; regulation of naive cells; generation of TEM cells; and formation and regulation of T regulatory cell (Treg) function [84–89]). It is therefore quite conceivable that BACH2 plays a role in HIV-infected cell survival and proliferation.
BACH2 is by no means the only gene selected for persistence and proliferation in association with HIV integration. Human immunodeficiency virus integration sites in the MKL1/myocardin-like-2 (MKL2; also referred to as megakaryoblastic leukemia 2) gene have been found in 4 individuals, at 21 different sites, including 11 in clonally proliferating cells [31–33]. Like BACH2, MKL2 is a nucleic acid binding protein and is known to be a transcription coactivator of serum response factor (which regulates cell growth) and has been associated with lipomas [90] and perhaps cancers [91]. Human immunodeficiency virus integration sites were also found in the same transcriptional orientation as the MKL2 gene at 20 of 21 sites. Human immunodeficiency virus integration sites within the related MKL1 gene has been found in 2 individuals, with HIV-ISs at total of 5 sites, including 2 in proliferating infected cell clones [31, 33].
Although many other genes are worth mentioning, the last we will discuss here is the signal transducer and transcription activator in JAK/STAT pathway 5B (STAT5B). This gene is involved in autoimmunity [92] and Treg maintenance [92, 93], T-cell receptor signaling, apoptosis, and different isoforms of the gene are associated with different cancers (eg, [94–97]). Human immunodeficiency virus integration sites in this gene have been found in 10 individuals, at 19 different sites, including 5 in clonally proliferating cells [31–33]. Unlike BACH2 and MKL2, however, no strong directional bias was observed in these ISs, with 12 of 19 in the same transcriptional orientation as the gene.
LATENCY AND THE PAUCITY OF T-CELL TUMORS
The site of virus integration has a strong influence on the level of expression of the provirus, and thus the development of latency, evident in both cell culture infections and HIV-ISs from patient specimens (ie, in vivo) [65, 98, 99]. Although HIV-ISs are preferentially found in highly expressed genes, elegant studies by Verdin and colleagues have shown that latency is readily induced after infection of cell lines and that the transcriptional activity of proviruses overall is inversely correlated with the transcriptional level of the gene [98–100] and is particularly low in proviruses found in heterochromatin [99] and gene deserts within euchromatin [100]. Furthermore, latency can be similarly and rapidly established after ex vivo infection of resting and activated human CD4+ T cells as well as tonsil and spleen cells [101]. DNA modifications such as cytosine methylation, histone methylation, and acetylation modifications also affect gene expression. Enzymes that modulate these modifications have become important targets of research and clinical studies directed to reversing viral latency (discussed in detail by Margolis in this supplement).
Human immunodeficiency virus may rarely cause insertional mutagenesis [102, 103], resulting in the development of tumors from infected cells in a few cases [80, 104, 105]. Notably, the infectious provirus found in clonally expanded cells reported by Simonetti was found in a person with a squamous cell cancer [106], and a prior report demonstrated clonality of HIV-ISs in association with a Kaposi’s sarcoma [107]. However, evidence for direct causation is lacking in the latter cases, as the virus was not shown to be in tumor cells. Kaposi’s sarcomas (KSs) and non-Hodgkin’s lymphomas accounted for approximately 90% of HIV-associated malignancies in the pre-ART era [108], and despite a sharp decline since the advent of effective ART [109], their attack rate remains much higher in HIV-infected individuals compared with the general population [110].
In contrast with HIV, infection with avian sarcoma/leukosis virus and murine (MLV) and feline leukemia viruses induce tumors within target cells by insertional mutagenesis [111] at relatively high frequencies. As noted herein, MLV integration within the BACH2 gene leads to B cell lymphomas in mice [83]. There are at least 4 potentially important differences between the host–virus interactions of these animal retroviruses and HIV that may explain differences in virally induced tumor frequencies: (1) HIV has a more complex genome structure and transcriptional control apparatus compared with the animal retroviruses (eg, avian, murine, feline). For example, HIV Tat is required for high-level transcription and Rev for transport of incompletely spliced mRNA to the cytoplasm for production of viral structural proteins. These regulatory features, plus the host’s local genetic and chromatin environment, typically lead to low levels of HIV gene activity and to latency. In contrast, little is known about animal retrovirus latency. (2) Animal retroviruses have a broader host cell range than HIV, and the infected cell burden in vivo is typically orders of magnitude higher than in HIV infection (eg, [112, 113]). (3) Murine leukemia virus integrations are often facilitated by interaction between the viral integrase and cellular BET proteins [114] and are found in association with promoter sequence elements and transcription start sites [115, 116]. Hence, MLV integrations are more likely to result in gene activation compared with the more random placement of HIV-ISs within genes. One MLV IS, found in association with B cell lymphomas, activated an alternative promoter, resulting in the production of a new BACH2 protein isoform [83]. Each of these host–virus interactions are likely to result in higher tumor frequencies in animals compared with HIV infections. (4) Lastly, MLV integrations that lead to BACH2-associated B-cell tumors and all HIV-ISs found to date in people have been found in the 2 introns just upstream of the start codon. However, all HIV-ISs were found in the same transcriptional orientation as the gene, whereas the overwhelming majority of the MLV-IS were in the opposite orientation. This suggests a differential effect on cellular gene and protein expression, although frequent HIV-ISs in other genes occur in both the reverse and same transcriptional orientations (eg, in STAT5B) within proliferating cells.
These observations, along with the finding that infected cell proliferation occurs with HIV-ISs in genes associated with immune control such as Tregs, suggest the possibility that clonally expanded HIV-infected cells may contribute to an increased susceptibility to cancers by contributing to an immunosuppressive cytokine milieu [107] in which cancer cells are better able to develop and persist.
PROLIFERATION OF INFECTED CELLS AND OTHER OBSTACLES TO CURE
Infected cell clones clearly resist depletion during ART; however, we do not know to what degree these cells will resist cure—as this will likely depend on the cure modality and the tissues in which the residual HIV reservoir is located. Although most studies of HIV reservoirs have focused on blood, it is highly likely that much more of the HIV reservoir is found in lymphoid tissues, the major site of viral replication early in infection [117, 118] when reservoirs are initially established [119, 120], or in organs with more pronounced physical barriers, such as the brain. Of particular interest are HIV-infected CD4+ T follicular helper (TFH) cells, which migrate into B-cell follicles and promote B-cell maturation and antibody production [121]. TFH cells within lymph nodes have been found to harbor a significant fraction of the (infectious) HIV reservoir under ART, and activated TFH cells, in particular the PD-1+CXCR3− subset, have been reported to make up a large fraction of the HIV reservoir in blood [122]. T follicular helper cells are also highly permissive for infection ex vivo and produce especially high levels of viral RNA found in lymph nodes [123, 124]. As will be discussed in more detail by Boritz and Douek in this supplement, a recent study of 14 natural HIV controllers (ie, <1000 copies of viral RNA/mL plasma) [36] showed that TEM and TTM cells in the blood were more likely to contain HIV DNA than TCM cells and much more than naive T cells (TN). However, when examining lymph nodes from 3 controllers, TFH cells within and outside germinal centers had higher levels than TCM and TEM cells, all of which were higher than levels found in blood CD4 cells. Also notable was that plasma HIV RNA levels and genotypes were more closely related to viral genomes from the lymph node than to those in blood CD4 cells, arguing that lymph node was an important source of viremia in these individuals and/or that infected cells within lymph nodes were more likely to survive immune depletion.
The persistence of HIV-infected cells harboring infectious as well as noninfectious proviruses begs the question as to why these cells are not eliminated by the immune system once viral antigens are produced. Latency is one factor. In addition, HIV-infected cells may persist in anatomic and cellular sanctuaries that restrict infected cell-targeted immune responses. Fukazawa and colleagues showed that SIV-infected viral RNA-expressing cells are not cleared from lymph node B-cell follicles [125], suggesting that these follicles may be a “sanctuary” for SIV and, perhaps HIV. infection. This also helps explain why TFH cells within lymph nodes harbor a significant fraction of the HIV reservoir under ART [36, 122, 126].
SUMMARY, CONCLUSIONS, AND SPECULATIONS
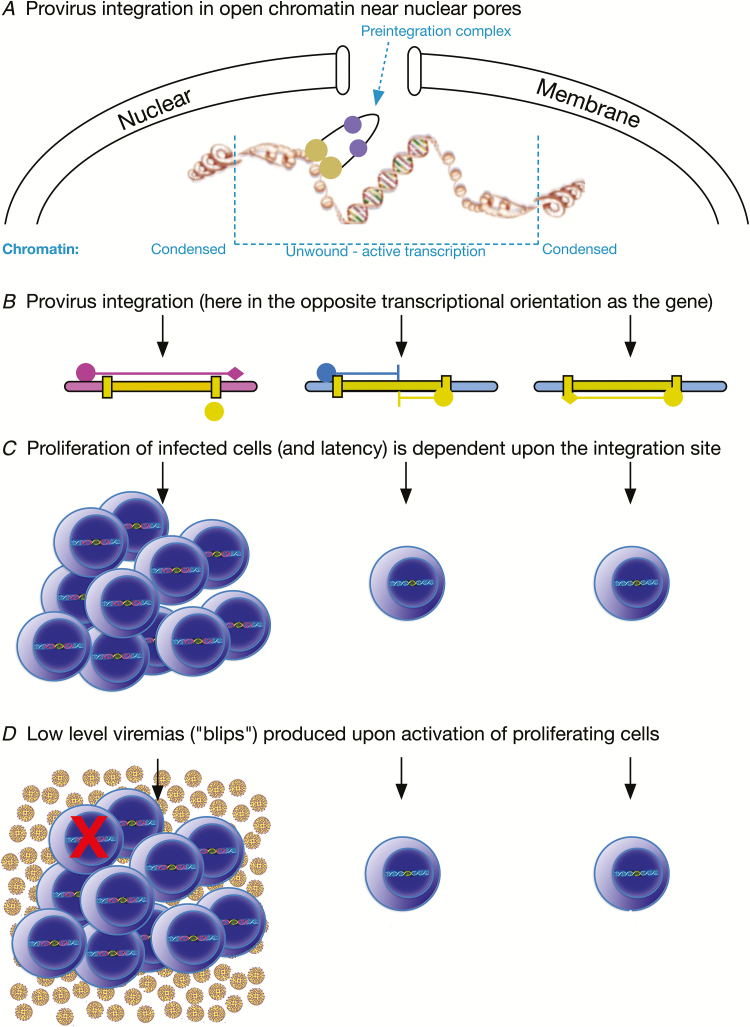
Human immunodeficiency virus integrates within the host genome preferentially in actively expressed genes and genes with spatial proximity to where the integration complex enters the nucleus and proximal to nuclear pores (Figure 2A). The site of viral integration in the host genome impacts viral latency. Although the mechanisms are unclear, integration into certain genes appears to favor persistence and proliferation of the cell and therefore is important to sustaining the HIV-infected cell population. Host gene transcription may interfere with viral transcription and vice versa (Figure 2B). Proliferating cells make up a substantial, if not a majority, component of the residual infected cell population under ART (Figure 2C) and have been shown to include infectious viruses in multiple studies. Because proliferating infected cells have memory phenotypes, cognate antigens may synchronously activate cell clones to produce the virions found in low-level viremias (or “blips”) under ART (Figure 2D). Additionally, proliferating cells may exert an immunosuppressive effect due to the provirus affecting transcription of Treg genes that impair their own elimination as well as elimination of other infected cells following activation (Figure 1D). Lastly, as individual cell purification methods are improved, proliferating infected cells, by virtue of their clonality, will provide a relatively rich source of material for study of the cellular environments that result in viral latency and recalcitrance to release from latency.
Figure 2.
Summary of findings and hypotheses discussed in this review. A, Provirus integration is favored in transcriptionally active genes near the nuclear membrane and pores. B, Human immunodeficiency virus (HIV) integration can result in the provirus and gene being in the same, or as shown here, the opposite transcriptional orientation. Subsequently, several scenarios for host gene and provirus transcription are possible, with 3 shown: Left, host gene transcription is initiated (pink circle) upstream of the provirus and travels through the provirus then terminates downstream (pink diamond), interfering with transcription of the proviral 5’ LTR (yellow circle). Middle, transcription is initiated both within the cellular gene and the provirus, leading to a transcriptional collision and incomplete expression of both transcription units. Right, transcription within the provirus is initiated and terminated correctly, yielding virions. C, Proliferation of infected cells as well as viral latency is associated with specific integration sites. D, Low-level viremias can result from proliferating infected cell populations, most likely from synchronous activation of a specific clonal cell population, perhaps by its cognate antigen. An immune-suppressive milieu may develop by HIV integrations affecting the function of regulatory T cells (Treg) cells that limit killing (red X) of virus-expressing cells, as well as cancerous cells (not depicted).
Notes
Financial support. This work was supported by US Public Health Services grants (AI111806, AI122361 and AI125026 to J. I. M. and HD36184, AI071212, AI091550 to L. M. F.)
Supplement sponsorship. This supplement was supported by grants from Merck & Co., Inc. and Gilead Sciences, Inc.
Potential conflicts of interest. Both authors certifies no potential conflicts of interest. Both authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Perelson AS, Essunger P, Cao Y, et al. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 1997; 387:188–91. [DOI] [PubMed] [Google Scholar]
- 2. Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med 1995; 1:1284–90. [DOI] [PubMed] [Google Scholar]
- 3. Finzi D, Hermankova M, Pierson T, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 1997; 278:1295–300. [DOI] [PubMed] [Google Scholar]
- 4. Chun TW, Carruth L, Finzi D, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 1997; 387:183–8. [DOI] [PubMed] [Google Scholar]
- 5. Chun TW, Stuyver L, Mizell SB, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A 1997; 94:13193–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wong JK, Hezareh M, Günthard HF, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 1997; 278:1291–5. [DOI] [PubMed] [Google Scholar]
- 7. Chomont N, El-Far M, Ancuta P, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 2009; 15:893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Siliciano JD, Kajdas J, Finzi D, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med 2003; 9:727–8. [DOI] [PubMed] [Google Scholar]
- 9. Chun TW, Justement JS, Moir S, et al. Decay of the HIV reservoir in patients receiving antiretroviral therapy for extended periods: implications for eradication of virus. J Infect Dis 2007; 195:1762–4. [DOI] [PubMed] [Google Scholar]
- 10. Boyman O, Purton JF, Surh CD, Sprent J. Cytokines and T-cell homeostasis. Curr Opin Immunol 2007; 19:320–6. [DOI] [PubMed] [Google Scholar]
- 11. Persaud D, Siberry GK, Ahonkhai A, et al. Continued production of drug-sensitive human immunodeficiency virus type 1 in children on combination antiretroviral therapy who have undetectable viral loads. J Virol 2004; 78:968–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tobin NH, Learn GH, Holte SE, et al. Evidence that low-level viremias during effective highly active antiretroviral therapy result from two processes: expression of archival virus and replication of virus. J Virol 2005; 79:9625–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fletcher CV, Staskus K, Wietgrefe SW, et al. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc Natl Acad Sci U S A 2014; 111:2307–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lorenzo-Redondo R, Fryer HR, Bedford T, et al. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature 2016; 530:51–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Evering TH, Mehandru S, Racz P, et al. Absence of HIV-1 evolution in the gut-associated lymphoid tissue from patients on combination antiviral therapy initiated during primary infection. PLoS Pathog 2012; 8:e1002506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kearney MF, Spindler J, Shao W, et al. Lack of detectable HIV-1 molecular evolution during suppressive antiretroviral therapy. PLoS Pathog 2014; 10:e1004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wolfs TF, Zwart G, Bakker M, Goudsmit J. HIV-1 genomic RNA diversification following sexual and parenteral virus transmission. Virology 1992; 189:103–10. [DOI] [PubMed] [Google Scholar]
- 18. Zhang LQ, MacKenzie P, Cleland A, Holmes EC, Brown AJ, Simmonds P. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J Virol 1993; 67:3345–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Delwart EL, Sheppard HW, Walker BD, Goudsmit J, Mullins JI. Human immunodeficiency virus type 1 evolution in vivo tracked by DNA heteroduplex mobility assays. J Virol 1994; 68:6672–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gottlieb GS, Heath L, Nickle DC, et al. HIV-1 variation before seroconversion in men who have sex with men: analysis of acute/early HIV infection in the multicenter AIDS cohort study. J Infect Dis 2008; 197:1011–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Keele BF, Giorgi EE, Salazar-Gonzalez JF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 2008; 105:7552–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shankarappa R, Margolick JB, Gange SJ, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol 1999; 73:10489–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu Y, McNevin J, Cao J, et al. Selection on the human immunodeficiency virus type 1 proteome following primary infection. J Virol 2006; 80:9519–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Herbeck JT, Rolland M, Liu Y, et al. Demographic processes affect HIV-1 evolution in primary infection before the onset of selective processes. J Virol 2011; 85:7523–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goonetilleke N, Liu MK, Salazar-Gonzalez JF, et al. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med 2009; 206:1253–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Frenkel LM, Wang Y, Learn GH, et al. Multiple viral genetic analyses detect low-level human immunodeficiency virus type 1 replication during effective highly active antiretroviral therapy. J Virol 2003; 77:5721–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bailey JR, Sedaghat AR, Kieffer T, et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol 2006; 80:6441–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pennock ND, White JT, Cross EW, Cheney EE, Tamburini BA, Kedl RM. T cell responses: naive to memory and everything in between. Adv Physiol Educ 2013; 37:273–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wagner TA, McKernan JL, Tobin NH, Tapia KA, Mullins JI, Frenkel LM. An increasing proportion of monotypic HIV-1 DNA sequences during antiretroviral treatment suggests proliferation of HIV-infected cells. J Virol 2013; 87:1770–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Laskey SB, Pohlmeyer CW, Bruner KM, Siliciano RF. Evaluating clonal expansion of HIV-infected cells: optimization of PCR strategies to predict clonality. PLoS Pathog 2016; 12:e1005689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wagner TA, McLaughlin S, Garg K, et al. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 2014; 345:570–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ikeda T, Shibata J, Yoshimura K, Koito A, Matsushita S. Recurrent HIV-1 integration at the BACH2 locus in resting CD4+ T cell populations during effective highly active antiretroviral therapy. J Infect Dis 2007; 195:716–25. [DOI] [PubMed] [Google Scholar]
- 33. Maldarelli F, Wu X, Su L, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 2014; 345:179–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cohn LB, Silva IT, Oliveira TY, et al. HIV-1 integration landscape during latent and active infection. Cell 2015; 160:420–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bull ME, Heath LM, McKernan-Mullin JL, et al. Human immunodeficiency viruses appear compartmentalized to the female genital tract in cross-sectional analyses but genital lineages do not persist over time. J Infect Dis 2013; 207:1206–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Boritz EA, Darko S, Swaszek L, et al. Multiple origins of virus persistence during natural control of HIV infection. Cell 2016; 166:1004–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Y, Kappes JC, Conway JA, Price RW, Shaw GM, Hahn BH. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J Virol 1991; 65:3973–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Iversen AK, Shpaer EG, Rodrigo AG, et al. Persistence of attenuated rev genes in a human immunodeficiency virus type 1-infected asymptomatic individual. J Virol 1995; 69:5743–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Deacon NJ, Tsykin A, Solomon A, et al. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science 1995; 270:988–91. [DOI] [PubMed] [Google Scholar]
- 40. Blankson JN, Bailey JR, Thayil S, et al. Isolation and characterization of replication-competent human immunodeficiency virus type 1 from a subset of elite suppressors. J Virol 2007; 81:2508–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ho YC, Shan L, Hosmane NN, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 2013; 155:540–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bruner KM, Murray AJ, Pollack RA, et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med 2016; 22:1043–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yunoki M, Maotani-Imai K, Kusuda H, et al. Production of infectious particles from defective human immunodeficiency virus type 1 (HIV-1)-producing cell clones by superinfection with infectious HIV-1. Arch Virol 1991; 116(1–4):143–58. [DOI] [PubMed] [Google Scholar]
- 44. Bull M, McLaughlin S, Mitchell C, et al. Low-level HIV viremias originate in part from infected proliferating cells. In: Program and abstracts of the 22nd Conference on Retroviruses and Opportunistic Infections (Seattle, Washington: ). [Google Scholar]
- 45. Trabaud MA, Cotte L, Saison J, et al. Persistent production of an integrase-deleted HIV-1 variant with no resistance mutation and wild-type proviral DNA in a treated patient. AIDS Res Hum Retroviruses 2015; 31:142–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Imamichi H, Dewar RL, Adelsberger JW, et al. Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proc Natl Acad Sci U S A 2016; 113:8783–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vicenzi E, Dimitrov DS, Engelman A, et al. An integration-defective U5 deletion mutant of human immunodeficiency virus type 1 reverts by eliminating additional long terminal repeat sequences. J Virol 1994; 68:7879–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Baba TW, Liska V, Khimani AH, et al. Live attenuated, multiply deleted simian immunodeficiency virus causes AIDS in infant and adult macaques. Nat Med 1999; 5:194–203. [DOI] [PubMed] [Google Scholar]
- 49. Birch MR, Learmont JC, Dyer WB, et al. An examination of signs of disease progression in survivors of the Sydney Blood Bank Cohort (SBBC). J Clin Virol 2001; 22:263–70. [DOI] [PubMed] [Google Scholar]
- 50. Ruprecht RM, Baba TW, Liska V. Attenuated HIV vaccine: caveats. Science 1996; 271:1790–2. [PubMed] [Google Scholar]
- 51. Deacon NJ, McPhee DA, Crowe S, Learmont J, Mills J. Response: attenuated HIV vaccine: caveats. Science 1996; 271:1791–2. [DOI] [PubMed] [Google Scholar]
- 52. Inoue M, Hoxie JA, Reddy MV, Srinivasan A, Reddy EP. Mechanisms associated with the generation of biologically active human immunodeficiency virus type 1 particles from defective proviruses. Proc Natl Acad Sci U S A 1991; 88:2278–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Boulerice F, Li XG, Lvovich A, Wainberg MA. Recovery of infectious human immunodeficiency virus type 1 after fusion of defectively infected clones of U-937 cells. J Virol 1991; 65:5589–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Quan Y, Xu H, Wainberg MA. Defective HIV-1 quasispecies in the form of multiply drug-resistant proviral DNA within cells can be rescued by superinfection with different subtype variants of HIV-1 and by HIV-2 and SIV. J Antimicrob Chemother 2014; 69:21–7. [DOI] [PubMed] [Google Scholar]
- 55. Anderson JA, Archin NM, Ince W, et al. Clonal sequences recovered from plasma from patients with residual HIV-1 viremia and on intensified antiretroviral therapy are identical to replicating viral RNAs recovered from circulating resting CD4+ T cells. J Virol 2011; 85:5220–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Simonetti F, Hill S, Wu X, et al. Residual viremia caused by clonally expanded tumor-infiltrating CD4+ cells. In: Program and abstracts of the 22nd Conference on Retroviruses and Opportunistic Infections (Seattle, Washington: ). [Google Scholar]
- 57. Craigie R, Bushman FD. HIV DNA integration. Cold Spring Harb Perspect Med 2012; 2:a006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bushman FD. Integration site selection by lentiviruses: biology and possible control. Curr Top Microbiol Immunol 2002; 261:165–77. [DOI] [PubMed] [Google Scholar]
- 59. Holman AG, Coffin JM. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proc Natl Acad Sci U S A 2005; 102:6103–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vincent KA, York-Higgins D, Quiroga M, Brown PO. Host sequences flanking the HIV provirus. Nucleic Acids Res 1990; 18:6045–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stevens SW, Griffith JD. Human immunodeficiency virus type 1 may preferentially integrate into chromatin occupied by L1Hs repetitive elements. Proc Natl Acad Sci U S A 1994; 91:5557–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Müller HP, Varmus HE. DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J 1994; 13:4704–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pruss D, Bushman FD, Wolffe AP. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc Natl Acad Sci U S A 1994; 91:5913–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schröder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 2002; 110:521–9. [DOI] [PubMed] [Google Scholar]
- 65. Han Y, Lassen K, Monie D, et al. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J Virol 2004; 78:6122–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Barr SD, Ciuffi A, Leipzig J, Shinn P, Ecker JR, Bushman FD. HIV integration site selection: targeting in macrophages and the effects of different routes of viral entry. Mol Ther 2006; 14:218–25. [DOI] [PubMed] [Google Scholar]
- 67. Liu H, Dow EC, Arora R, et al. Integration of human immunodeficiency virus type 1 in untreated infection occurs preferentially within genes. J Virol 2006; 80:7765–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shun MC, Raghavendra NK, Vandegraaff N, et al. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev 2007; 21:1767–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Marshall HM, Ronen K, Berry C, et al. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS One 2007; 2:e1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Llano M, Vanegas M, Fregoso O, et al. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J Virol 2004; 78:9524–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ciuffi A, Bushman FD. Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends Genet 2006; 22:388–95. [DOI] [PubMed] [Google Scholar]
- 72. Kulkarni A, Pavithra L, Rampalli S, et al. HIV-1 integration sites are flanked by potential MARs that alone can act as promoters. Biochem Biophys Res Commun 2004; 322:672–7. [DOI] [PubMed] [Google Scholar]
- 73. Johnson CN, Levy LS. Matrix attachment regions as targets for retroviral integration. Virol J 2005; 2:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Albanese A, Arosio D, Terreni M, Cereseto A. HIV-1 pre-integration complexes selectively target decondensed chromatin in the nuclear periphery. PLoS One 2008; 3:e2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Marini B, Kertesz-Farkas A, Ali H, et al. Nuclear architecture dictates HIV-1 integration site selection. Nature 2015; 521:227–31. [DOI] [PubMed] [Google Scholar]
- 76. Ocwieja KE, Brady TL, Ronen K, et al. HIV integration targeting: a pathway involving Transportin-3 and the nuclear pore protein RanBP2. PLoS Pathog 2011; 7:e1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. König R, Zhou Y, Elleder D, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 2008; 135:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res 2007; 17:1186–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sherrill-Mix S, Lewinski MK, Famiglietti M, et al. HIV latency and integration site placement in five cell-based models. Retrovirology 2013; 10:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shiramizu B, Herndier BG, McGrath MS. Identification of a common clonal human immunodeficiency virus integration site in human immunodeficiency virus-associated lymphomas. Cancer Res 1994; 54:2069–72. [PubMed] [Google Scholar]
- 81. Mack KD, Jin X, Yu S, et al. HIV insertions within and proximal to host cell genes are a common finding in tissues containing high levels of HIV DNA and macrophage-associated p24 antigen expression. J Acquir Immune Defic Syndr 2003; 33:308–20. [DOI] [PubMed] [Google Scholar]
- 82. Imamichi H, Natarajan V, Adelsberger JW, et al. Lifespan of effector memory CD4+ T cells determined by replication-incompetent integrated HIV-1 provirus. AIDS 2014; 28:1091–9. [DOI] [PubMed] [Google Scholar]
- 83. Liu J, Sørensen AB, Wang B, Wabl M, Nielsen AL, Pedersen FS. Identification of novel Bach2 transcripts and protein isoforms through tagging analysis of retroviral integrations in B-cell lymphomas. BMC Mol Biol 2009; 10:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tsukumo S, Unno M, Muto A, et al. Bach2 maintains T cells in a naive state by suppressing effector memory-related genes. Proc Natl Acad Sci U S A 2013; 110:10735–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kuwahara M, Suzuki J, Tofukuji S, et al. The Menin-Bach2 axis is critical for regulating CD4 T-cell senescence and cytokine homeostasis. Nat Commun 2014; 5:3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Roychoudhuri R, Hirahara K, Mousavi K, et al. BACH2 represses effector programs to stabilize T(reg)-mediated immune homeostasis. Nature 2013; 498:506–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Swaminathan S, Huang C, Geng H, et al. BACH2 mediates negative selection and p53-dependent tumor suppression at the pre-B cell receptor checkpoint. Nat Med 2013; 19:1014–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kallies A, Vasanthakumar A. Transcription factor Bach2 balances tolerance and immunity. Immunol Cell Biol 2013; 91:491–2. [DOI] [PubMed] [Google Scholar]
- 89. Kim EH, Gasper DJ, Lee SH, Plisch EH, Svaren J, Suresh M. Bach2 regulates homeostasis of Foxp3+ regulatory T cells and protects against fatal lung disease in mice. J Immunol 2014; 192:985–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Flucke U, Tops BB, de Saint Aubain Somerhausen N, et al. Presence of C11orf95-MKL2 fusion is a consistent finding in chondroid lipomas: a study of eight cases. Histopathology 2013; 62:925–30. [DOI] [PubMed] [Google Scholar]
- 91. Muehlich S, Hampl V, Khalid S, et al. The transcriptional coactivators megakaryoblastic leukemia ½ mediate the effects of loss of the tumor suppressor deleted in liver cancer 1. Oncogene 2012; 31:3913–23. [DOI] [PubMed] [Google Scholar]
- 92. Cohen AC, Nadeau KC, Tu W, et al. Cutting edge: decreased accumulation and regulatory function of CD4+ CD25(high) T cells in human STAT5b deficiency. J Immunol 2006; 177:2770–4. [DOI] [PubMed] [Google Scholar]
- 93. Jenks JA, Seki S, Kanai T, et al. Differentiating the roles of STAT5B and STAT5A in human CD4+ T cells. Clin Immunol 2013; 148:227–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Liang QC, Xiong H, Zhao ZW, et al. Inhibition of transcription factor STAT5b suppresses proliferation, induces G1 cell cycle arrest and reduces tumor cell invasion in human glioblastoma multiforme cells. Cancer Lett 2009; 273:164–71. [DOI] [PubMed] [Google Scholar]
- 95. Tang JZ, Zuo ZH, Kong XJ, et al. Signal transducer and activator of transcription (STAT)-5A and STAT5B differentially regulate human mammary carcinoma cell behavior. Endocrinology 2010; 151:43–55. [DOI] [PubMed] [Google Scholar]
- 96. Sobti RC, Singh N, Hussain S, Suri V, Bharadwaj M, Das BC. Deregulation of STAT-5 isoforms in the development of HPV-mediated cervical carcinogenesis. J Recept Signal Transduct Res 2010; 30:178–88. [DOI] [PubMed] [Google Scholar]
- 97. Du W, Wang YC, Hong J, et al. STAT5 isoforms regulate colorectal cancer cell apoptosis via reduction of mitochondrial membrane potential and generation of reactive oxygen species. J Cell Physiol 2012; 227:2421–9. [DOI] [PubMed] [Google Scholar]
- 98. Jordan A, Defechereux P, Verdin E. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J 2001; 20:1726–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Jordan A, Bisgrove D, Verdin E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J 2003; 22:1868–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lewinski MK, Bisgrove D, Shinn P, et al. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J Virol 2005; 79:6610–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Chavez L, Calvanese V, Verdin E. HIV latency is established directly and early in both resting and activated primary CD4 T cells. PLoS Pathog 2015; 11:e1004955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Hayward WS, Neel BG, Astrin SM. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature 1981; 290:475–80. [DOI] [PubMed] [Google Scholar]
- 103. Payne GS, Bishop JM, Varmus HE. Multiple arrangements of viral DNA and an activated host oncogene in bursal lymphomas. Nature 1982; 295:209–14. [DOI] [PubMed] [Google Scholar]
- 104. Herndier BG, Shiramizu BT, Jewett NE, Aldape KD, Reyes GR, McGrath MS. Acquired immunodeficiency syndrome-associated T-cell lymphoma: evidence for human immunodeficiency virus type 1-associated T-cell transformation. Blood 1992; 79:1768–74. [PubMed] [Google Scholar]
- 105. Katano H, Sato Y, Hoshino S, et al. Integration of HIV-1 caused STAT3-associated B cell lymphoma in an AIDS patient. Microbes Infect 2007; 9(14-15):1581–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Simonetti FR, Sobolewski MD, Fyne E, et al. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci U S A 2016; 113:1883–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. McGrath MS, Shiramizu BT, Herndier BG. Identification of a clonal form of HIV in early Kaposi’s sarcoma: evidence for a novel model of oncogenesis, “sequential neoplasia.” J Acquir Immune Defic Syndr Hum Retrovirol 1995; 8:379–85. [PubMed] [Google Scholar]
- 108. International Collaboration on HIV, Cancer. Highly active antiretroviral therapy and incidence of cancer in human immunodeficiency virus-infected adults. J Natl Cancer Inst 2000; 92:1823–30. [DOI] [PubMed] [Google Scholar]
- 109. Shiels MS, Engels EA. Evolving epidemiology of HIV-associated malignancies. Curr Opin HIV AIDS 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Goedert JJ. The epidemiology of acquired immunodeficiency syndrome malignancies. Semin Oncol 2000; 27:390–401. [PubMed] [Google Scholar]
- 111. Fan H, Johnson C. Insertional oncogenesis by non-acute retroviruses: implications for gene therapy. Viruses 2011; 3:398–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Jurriaans S, Dekker JT, de Ronde A. HIV-1 viral DNA load in peripheral blood mononuclear cells from seroconverters and long-term infected individuals. AIDS 1992; 6:635–41. [DOI] [PubMed] [Google Scholar]
- 113. Chan WT, Sherer NM, Uchil PD, Novak EK, Swank RT, Mothes W. Murine leukemia virus spreading in mice impaired in the biogenesis of secretory lysosomes and Ca2+-regulated exocytosis. PLoS One 2008; 3:e2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Sharma A, Larue RC, Plumb MR, et al. BET proteins promote efficient murine leukemia virus integration at transcription start sites. Proc Natl Acad Sci U S A 2013; 110:12036–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Mitchell RS, Beitzel BF, Schroder AR, et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol 2004; 2:E234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Felice B, Cattoglio C, Cittaro D, et al. Transcription factor binding sites are genetic determinants of retroviral integration in the human genome. PLoS One 2009; 4:e4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Pantaleo G, Graziosi C, Demarest JF, et al. HIV infection is active and progressive in lymphoid tissue during the clinically latent stage of disease. Nature 1993; 362:355–8. [DOI] [PubMed] [Google Scholar]
- 118. Embretson J, Zupancic M, Ribas JL, et al. Massive covert infection of helper T lymphocytes and macrophages by HIV during the incubation period of AIDS. Nature 1993; 362:359–62. [DOI] [PubMed] [Google Scholar]
- 119. Whitney JB, Hill AL, Sanisetty S, et al. Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature 2014; 512:74–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Sellier P, Mannioui A, Bourry O, et al. Antiretroviral treatment start-time during primary SIV(mac) infection in macaques exerts a different impact on early viral replication and dissemination. PLoS One 2010; 5:e10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity 2014; 41:529–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Perreau M, Savoye AL, De Crignis E, et al. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med 2013; 210:143–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Kohler SL, Pham MN, Folkvord JM, et al. Germinal center T follicular helper cells are highly permissive to HIV-1 and alter their phenotype during virus replication. J Immunol 2016; 196:2711–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Miles B, Connick E. TFH in HIV latency and as sources of replication-competent virus. Trends Microbiol 2016; 24:338–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Fukazawa Y, Lum R, Okoye AA, et al. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat Med 2015; 21:132–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Pallikkuth S, Sharkey M, Babic DZ, et al. Peripheral T follicular helper cells are the major HIV reservoir within central memory CD4 T cells in peripheral blood from chronically HIV-infected individuals on combination antiretroviral therapy. J Virol 2015; 90:2718–28. [DOI] [PMC free article] [PubMed] [Google Scholar]