Abstract
Non-small-cell lung cancer (NSCLC) is the leading type of cancer worldwide and sex determining region Y-box 2 (SOX2) has been implicated as an oncogene in various types of cancer. In the present study, SOX2 was positively associated with NSCLC stage and lymph node metastasis. Wound healing and Transwell assays demonstrated that knockdown of SOX2 inhibited A549 and H1299 cell migration. Furthermore, it was identified that knockdown of SOX2 inhibited epithelial-to-mesenchymal transition of NSCLC cells, which was demonstrated by increased expression of epithelial-cadherin and decreased expression of vimentin, zinc finger protein SNAI1 and zinc finger protein SNAI2. It was then demonstrated that SOX2 may be targeted by microRNA (miR)-590-5p, which indicated a potential therapeutic strategy for NSCLC focusing on the miR-590-5p/SOX2 axis.
Keywords: sex determining region Y-box 2, non-small-cell lung cancer, microRNA-590-5p, epithelial-to-mesenchymal transition, metastasis
Introduction
SOX family proteins are a group of transcriptional factors that recognize and bind DNA via the high-mobility group domain (1). Sex determining region Y-box 2 (SOX2) is located on chromosome 3q26.3 and belongs to the SOXB1 group (2–4). An increasing number of studies have implicated SOX2 in the tumorigenesis of gastric, lung and breast cancer (5,6).
Lung cancer, which includes non-small-cell lung cancer (NSCLC) and small-cell lung cancer, is one of the most common causes of cancer-associated mortality worldwide, among which NSCLC accounts for ~85%. Currently, NSCLC is a non-curable disease with marked genetic complexity (1). A fluorescence in situ hybridization analysis performed in 447 resected NSCLC tissue samples indicated that amplification of SOX2 was positively associated with increased copy number of the FGFR1 and PI3KCA genes (1). SOX2 has been demonstrated to promote cellular proliferation via evading apoptotic signals in NSCLC (7,8). However, the role of SOX2 in the metastasis and epithelial-to-mesenchymal transition (EMT) regulation of NSCLC remains poorly understood.
MicroRNAs (miRNAs) comprise a large family of regulatory RNAs that may repress expression of target mRNAs in a sequence-specific manner. miRNAs participate in diverse biological processes, including cell development, differentiation, proliferation and apoptosis. In addition, aberrant miRNA expression patterns have been identified in a number of types of human cancer, including lung cancer. According to a previous study, the expression of miR-590-5p significantly suppressed the proliferation, migration and invasion of NSCLC cells (9), and bioinformatics analysis using TargetScan also demonstrated that SOX2 is targeted by miRNA-590-5p (10). The aim of the present study was to investigate the role of SOX2 in the regulation of NSCLC metastasis and EMT, and determine whether miR-590-5p is a regulator of SOX2 in NSCLC.
Materials and methods
Patients and specimens
Between June 2014 and March 2016, 33 paraffin-embedded tumor specimens and adjacent normal specimens from patients with NSCLC were collected from The Second Affiliated Hospital of Zhejiang University (Zhejiang, China). The patients were aged between 32–60 years old with a median age of 48 years; the cohort contained 25 females and 8 males. The specimens were definitively diagnosed by experienced pathologists. The pathological Tumor-Node-Metastasis classification stage was determined by the 2009 staging system of the Union for International Cancer Control (11). Patients were separated into high or low expression groups based on the median expression value of mRNA for miR-590-5p (cut-off value, 0.036) or SOX2 (cut-off value, 0.0469). Written informed consent was obtained from all patients regarding the use of their tissues for research purposes. All the procedures were performed in accordance with the guidelines of the Ethics Committee of The Second Affiliated Hospital of Zhejiang University.
Computational target prediction
For the identification of miR-590-5p targets, TargetScan release 7.2 (http://www.targetscan.org/vert_72/) was used with the default parameters.
Plasmids
The SOX2 over-expression plasmid (SOX2-OE), SOX2 3untranslated region (UTR) and SOX2 3UTR-mut were purchased from GenePharma (Shanghai, China). Briefly, the SOX2 cDNA was inserted into the pcDNA3.1 vector (cat. no. 20011; Addgene, Inc.), which harbors a FLAG epitope sequence to generate the SOX2-OE plasmid. The complete SOX2 3UTR was inserted into the pmirGLO Dual-luciferase plasmid (Promega Corporation) to generate the SOX2 3UTR plasmid. To generate SOX2 3UTR-mut plasmid, the predicted miR-590-5p binding site in the SOX2 3UTR was mutated (mut) and inserted into the pmirGLO plasmid to generate SOX2 3UTR-mut plasmid.
Cell culture and transfection
The lung cancer A549 and H1299 cell lines (American Type Culture Collection) were cultured in RPMI-1640 medium (Life Technologies; Thermo Fisher Scientific, Inc.) containing 10% FBS (Hyclone; GE Healthcare), 100 U/ml penicillin and 100 U/ml streptomycin. The cells were cultured in an incubator with 5% CO2 at 37°C. According to the manufacturer's instructions, cells were seeded in 12-well plates for 24 h and then transfected with 100 pmol short interfering (si)RNAs and 10 nM miR mimics with Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) for 48 h. The SOX2 siRNA sequence was 5′-ACCTCCGGGACATGATCAGCA-3′ (Shanghai GenePharma Co., Ltd.). Scrambled siRNA (siRNA NC; 5′-TTCTCCGAACGTGTCACGT-3′; Shanghai GenePharma Co., Ltd.) was used as a negative control. miR-590-5p mimic (5′-GAGCUUAUUCAUAAAAUGCAG-3′) and miRNA NC (5′-GUCCAGUGAAUUCCCAG-3′) were obtained from Guangzhou RiboBio Co., Ltd.
Luciferase activity assay
A total of 1×105 A549 cells were seeded in 24-well plates for 24 h. Then the mixture of 1 µg SOX2 3UTR or SOX2 3UTR-mut, 0.1 µg of Renilla luciferase plasmids (Promega Corporation) and 25 nM miR-590-5p mimic or miRNA NC were incubated with Lipofectamine 2000 at room temperature for 15 min and added to each well. After 48 h, the cells were lysed and the luciferase activity was detected using the Dual Luciferase Reporter Assay System (Promega Corporation) on an LD400 luminometer (Beckman Coulter, Inc.). The firefly luciferase activity was normalized to the Renilla luciferase activity.
RNA extraction and reverse transcription quantitative polymerase chain reaction (RT-qPCR) assay
TRIzol® reagent (Thermo Fisher Scientific, Inc.) was used to extract total RNA, and the cDNA library was generated using a cDNA kit (Thermo Fisher Scientific, Inc.). According to the manufacturer's protocol, SYBR Green Master mix (Thermo Fisher Scientific) was used to perform RT-qPCR in triplicate with the following thermocycler conditions: Denaturation of DNA at 95°C for 5 min, followed by 40 cycles of amplification: 95°C for 60 sec, 55°C for 60 sec and 72°C for 60 sec. To analyze the relative expression of mRNAs or miRNA, the 2−ΔΔCq method (12) was used and normalized by GAPDH or U6 expression. The primer sequences were as follows: SOX2 forward, 5′-CTCGTGCAGTTCTACTCGTCG-3′; SOX2 reverse, 5′-AGCTCTCGGTCAGGTCCTTT-3′; GAPDH forward, 5′-CTCCTCCTGTTCGACAGTCAGC-3′; GAPDH reverse, 5′-CCCAATACGACCAAATCCGTT-3′.
Cell migration assay
To analyze the cell migration ability, a wound healing assay was performed. The transfected A549 and H1299 cells were plated into 12-well plates at a density of 5×104 for 24 h, followed by creation of a wound in the cell monolayer using a sterile 100 µl micropipette tip, washed with PBS twice and then cultured in serum-free medium for an additional 24 h. A phase-contrast inverted microscope (Olympus IX71; Olympus Corporation) at a magnification of ×100 was used to measure and capture images of the wound width.
Transwell invasion assay
To analyze the cell invasive ability, a Transwell assay was performed. The upper chamber was pre-coated with 1 mg/ml Matrigel (Growth Factor Reduced BD Matrigel™ Matrix; BD Biosciences) for 1 h in an incubator with 5% CO2 at 37°C, followed by seeding cells (5×104) in the upper chamber with RPMI-1640 medium without FBS (8 µm pore size membrane; EMD Millipore). Complete medium (600 µl) was added to the lower chamber and the cells were incubated for 48 h. The non-invading cells in the top chamber were removed with a cotton swab, and the cells on the lower surface of the membrane were fixed with 4% formaldehyde for 15 min at room temperature and stained with 0.1% crystal violet solution (both Sigma-Aldrich; Merck KGaA) for 20 min at room temperature. Images were captured using an IX71 microscope and cells were counted at a magnification of ×200.
Western blot analysis
Radioimmunoprecipitation assay lysis buffer was used to obtain the cell lysates, protein was quantified by a bicinchoninic acid protein assay (Pierce; Thermo Fisher Scientific, Inc.) and the protein expression was analyzed via standard western blot analysis. Briefly, equal amounts of protein (50 µg/lane) were subjected to 10% SDS-PAGE and subsequently transferred to polyvinylidene fluoride membrane (EMD Millipore). Following blocking with 5% non-fat milk for 1.5 h at room temperature, the membrane was incubated with the primary antibodies [anti-SOX2 (cat. no. ab75485), anti-epithelial cadherin (E-cadherin; cat. no. ab15148), anti-vimentin (cat. no. ab92547), anti-zinc finger protein SNAI1 (Snail; cat. no. ab216347), anti-zinc finger protein SNAI2 (Slug; cat. no. ab106077) and anti-GAPDH (cat. no. ab9484; all 1:500; Abcam)] at 4°C overnight, and then rinsed twice with PBS, followed by incubation with the horseradish peroxidase-conjugated secondary anti-rabbit antibody (cat. no. ab150077; 1:2,000; Abcam) for 1 h at room temperature. The membrane was subsequently incubated with an ECL Chemiluminescent Substrate kit (Thermo Fisher Scientific, Inc.) and exposed to X-ray film. ImageJ version 1.41 software (National Institutes of Health, Bethesda, MD, USA) was used for the quantitative analysis of band densities.
Statistical analysis
All statistical analyses were performed using the GraphPad software 8 (GraphPad Software, Inc.). A t-test was used to compare data between two groups. For the comparison of multiple groups, a one-way analysis of variance followed by Tukey's post-hoc test was performed. P<0.05 was considered to indicate a statistically significant difference.
Results
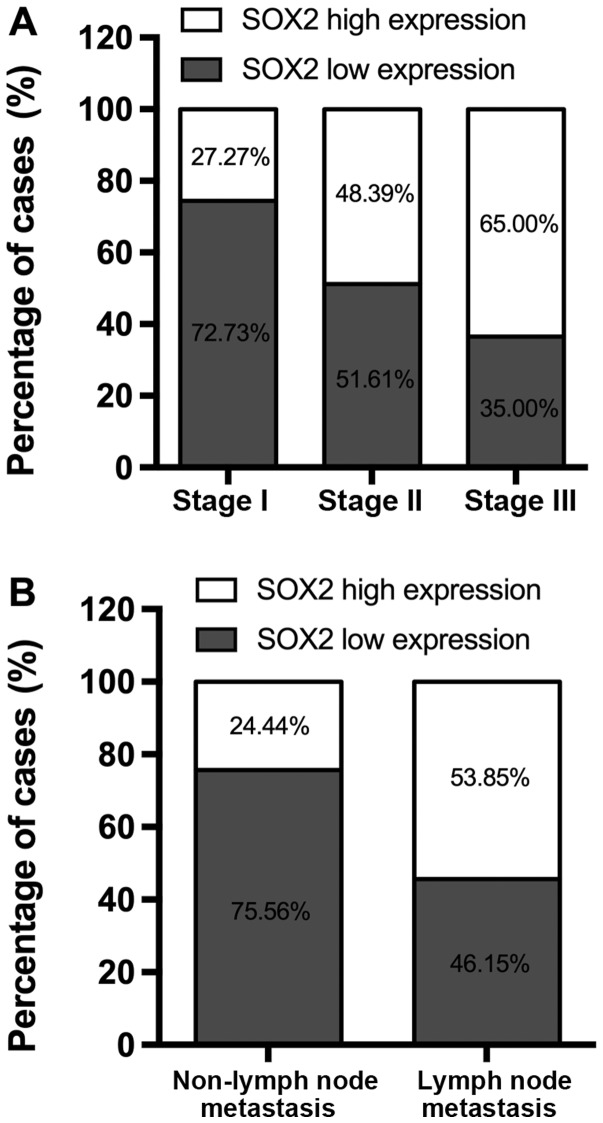
SOX2 is upregulated in NSCLC tissues and is associated with pathological stage and lymph node metastasis
Of the 33 NSCLC tissue samples at pathological stage I, 24 (72.73%) exhibited low expression of SOX2, while 16/31 (51.61%) stage II samples and 7/20 (35.00%) stage III cases exhibited low SOX2 expression rates (Fig. 1A). Changes in SOX2 expression were also significantly associated with lymph node metastasis. Of the 39 cases of NSCLC with lymph node metastasis, 21 (53.85%) exhibited high expression of SOX2. By contrast, of the 45 cases without lymph node metastasis, only 11 (24.44%) exhibited high SOX2 expression (Fig. 1B).
Figure 1.
Expression of SOX2 is associated with non-small-cell lung cancer clinicopathological characteristics including (A) pathological stage and (B) lymph node metastasis. SOX2, sex determining region Y-box 2.
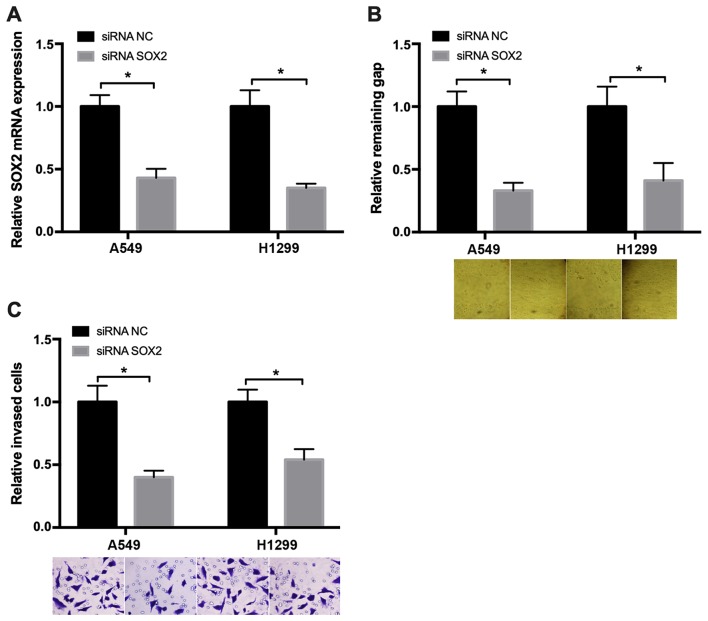
Knockdown of SOX2 suppresses NSCLC cell migration
To examine the role of SOX2 in regulating NSCLC cell migration, wound healing and Transwell assays were performed. The SOX2 mRNA expression was measured to evaluate the small interfering RNA (siRNA) knockdown efficiency in cancer cells, and the results are presented in Fig. 2A. The wound healing assay results revealed that, after 48 h, the wound in the SOX2 downregulated group was markedly narrower compared with that in the negative control group (Fig. 2B). The Transwell assay demonstrated that, at 48 h after transfection with SOX2 siRNA, the number of migrated cells was decreased compared with that in the negative control group (Fig. 2C). These results demonstrated that knockdown of SOX2 may suppress NSCLC cell migration.
Figure 2.
SOX2 promotes NSCLC cell migration. (A) The SOX2 mRNA expression was measured to evaluate the siRNA knockdown efficiency in A549 and H1299 cells. (B) Knockdown of SOX2 significantly inhibited the migration of A549 and H1299 cells. (C) Knockdown of SOX2 decreased the migratory ability of A549 and H1299 cells. *P<0.05. SOX2, sex determining region Y-box 2; NSCLC, non-small-cell lung cancer; siRNA, small interfering RNA; NC, negative control.
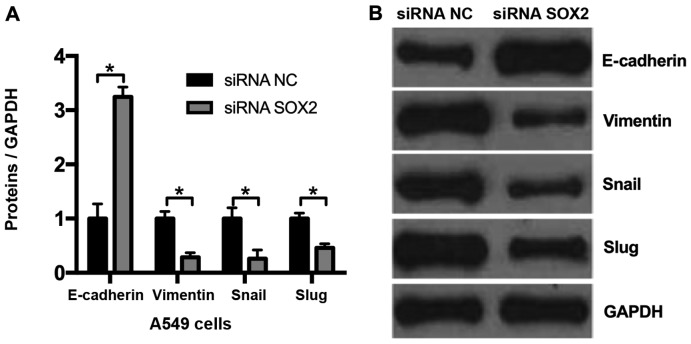
Knockdown of SOX2 suppresses EMT in NSCLC cells
As knockdown of SOX2 may decrease cell migration, it was hypothesized that SOX2 may affect EMT. Compared with the negative control (siRNA NC), it was observed that the expression of the epithelial marker E-cadherin was markedly upregulated and that of the mesenchymal marker vimentin was downregulated in A549 cells transfected with SOX2 siRNA (Fig. 3). The 2 EMT-associated genes, Snail and Slug, were additionally assessed by western blot analysis, and the results demonstrated that knockdown of SOX2 decreased Snail and Slug expression. These results indicate that knockdown of SOX2 may suppress EMT in NSCLC cells.
Figure 3.
Knockdown of SOX2 inhibits epithelial-to-mesenchymal transition in NSCLC cells. (A) Densitometric analysis of western blot analysis data of E-cadherin, vimentin, Snail and Slug expression. (B) Representative western blot analysis gel performed in A549 cells transfected with SOX2 siRNA. *P<0.05. SOX2, sex determining region Y-box 2; NSCLC, non-small-cell lung cancer; E-cadherin, epithelial cadherin; Snail, zinc finger protein SNAI1; Slug, zinc finger protein SNAI2; siRNA, small interfering RNA; NC, negative control.
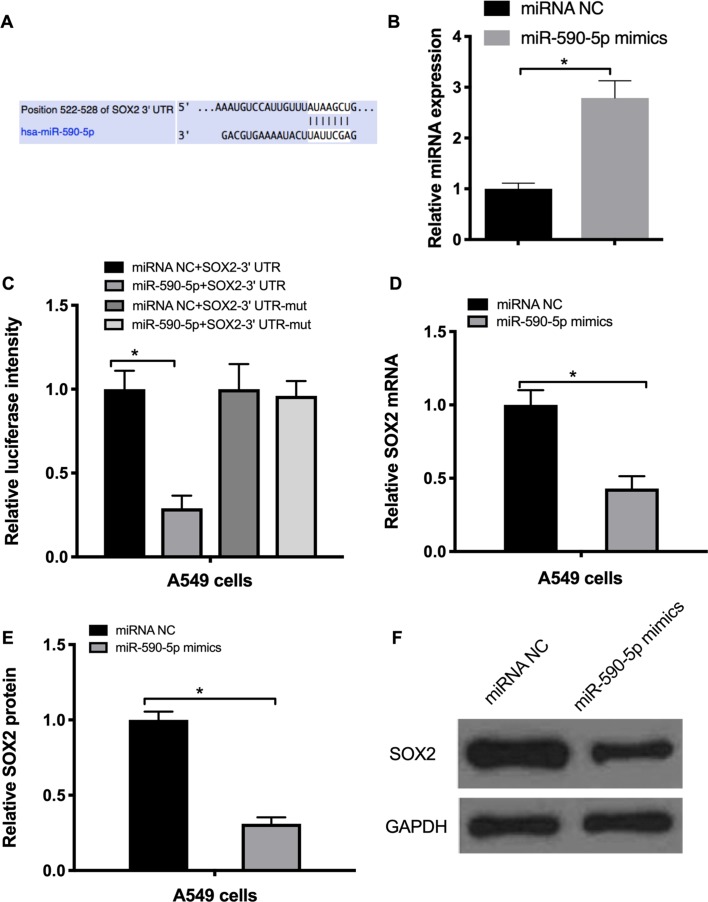
miR-590-5p targets SOX2 and suppresses EMT in NSCLC cells
In order to explore the factors that may regulate SOX2 expression, TargetScan software was used to identify miRNAs targeting SOX2. The bioinformatics analysis demonstrated that SOX2 may be targeted by miR-590-5p (Fig. 4A). RT-qPCR data indicated that miR-590-5p mimics markedly increased its expression by 2.8-fold (Fig. 4B). To confirm whether miR-590-5p directly targeted SOX2, a 3′-untranslated region (UTR) luciferase reporter assay was performed. The results revealed that miR-590-5p mimics repressed the luciferase activity of the wild-type SOX2 3′UTR, but not that of the mutated SOX2 3′UTR (Fig. 4C). To additionally confirm these results, A549 cells were transfected with the miR-590-5p mimics and their negative control. The results of RT-qPCR and western blot analysis demonstrated that miR-590-5p mimics markedly decreased the mRNA and protein levels of SOX2 (Fig. 4D and E).
Figure 4.
SOX2 is a target of miR-590-5p. (A) Bioinformatics analysis predicted a miR-590-5p seed region in the 3′ UTR of SOX2. (B) Transfection efficiency of miR-590-5p mimics detected by RT-qPCR. (C) Effect of miR-590-5p mimics on the luciferase activity of SOX2-3′ UTR and SOX2-3′ UTR-mut. (D) RT-qPCR analysis of SOX2 mRNA in A549 cells transfected with miR-590-5p mimics. (E and F) Western blot analysis of SOX2 protein expression in A549 cells transfected with miR-590-5p mimics. *P<0.05. SOX2, sex determining region Y-box 2; UTR, untranslated region; mut, mutant; RT-qPCR, reverse transcription quantitative polymerase chain reaction; miR, microRNA; NC, negative control; hsa, Homo sapiens.
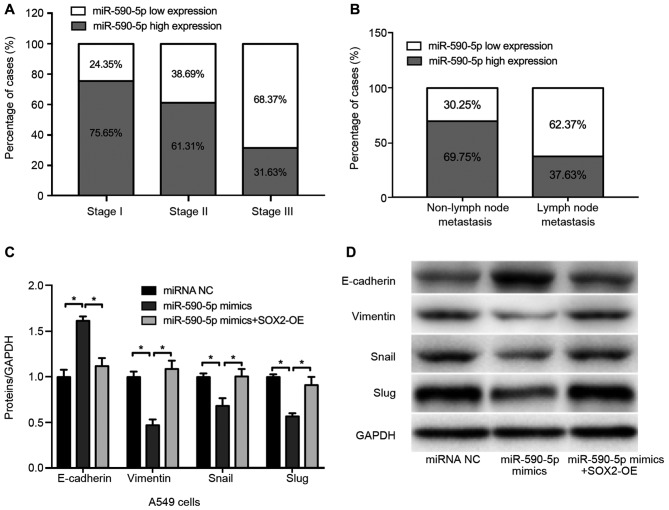
To determine the expression pattern of miR-590-5p in patients with NSCLC, the expression of miR-590-5p was measured in 33 NSCLC tissue specimens. The results revealed that, among the cases at pathological stage I, 75.65% exhibited high expression of miR-590-5p, 61.31% of pathological stage II and 31.63% of pathological stage III cases exhibited high miR-590-5p expression (Fig. 5A). Changes in miR-590-5p expression were also significantly associated with lymph node metastasis. Among the 39 cases of NSCLC without lymph node metastasis, 69.75% exhibited high expression of miR-590-5p. By contrast, of the 45 cases without lymph node metastasis, only 37.63% exhibited high miR-590-5p expression (Fig. 5B).
Figure 5.
Expression of miR-590-5p in NSCLC tissues and effect of miR-590-5p on cell migration and epithelial-to-mesenchymal transition-associated protein expression. (A) The expression of miR-590-5p was significantly associated with pathological stage in 33 patients. (B) The expression of miR-590-5p was associated with lymph node metastasis in NSCLC. (C and D) Densitometric analysis of western blot analysis data of E-cadherin, vimentin, Snail and Slug expression. (D) Representative western blot analysis gel performed in A549 cells transfected with miRNA NC, miR-590-5p mimics, miR-590-5p mimics + SOX2 overexpression. *P<0.05. SOX2, sex determining region Y-box 2; NSCLC, non-small-cell lung cancer; miRNA/miR, microRNA; OE, overexpression; E-cadherin, epithelial cadherin; Snail, zinc finger protein SNAI1; Slug, zinc finger protein SNAI2; NC, negative control.
The effect of miR-590-5p on cell migration and EMT-associated protein expression was also investigated in the present study. The results demonstrated that, compared with the negative control (miRNA NC), the expression of the epithelial marker E-cadherin was markedly upregulated and that of the mesenchymal marker vimentin was downregulated in A549 cells transfected with miR-590-5p mimics, whereas restoring SOX2 expression reversed these effects (Fig. 5C and D).
Discussion
SOX2 has been demonstrated to be an oncogene regulating the transcription of downstream target genes. It has also been revealed that the alteration of SOX2 expression may affect the progression and prognosis of multiple tumors via regulating various cell signaling pathways. SOX2 enhances tumor growth via the RAC-alpha serine/threonine-protein kinase signaling pathway in esophageal squamous cell carcinoma (13); in hepatocellular cancer, SOX2 enhances cell invasion through activating Slug (14), whereas in lung cancer, cell apoptosis may be inhibited by SOX2 through the mitogen-activated protein kinase 4/survivin signaling pathway (15). In addition, SOX2 is involved in the epidermal growth factor receptor, Bcl-2 like 1 and Wnt signaling pathways. The emerging role of SOX2 in cell proliferation and survival and its crosstalk with oncogenic signaling was previously described in lung cancer (16–18). In the present study, the expression of SOX2 was evaluated in NSCLC tissues. The specimens were collected from patients with stage I–III NSCLC, and the SOX2 mRNA expression in the tissues was measured using RT-qPCR. The results revealed that an increased level of SOX2 expression was markedly associated with a more advanced pathological stage and lymph node metastasis. Due to the limitation of clinical specimen collection, the sample size of the present study was not very large. In the future, more specimens must be collected to validate the results.
EMT enhances tumor cell migration and invasion ability, eventually promoting the dissemination of tumor cells to various parts of the body (19). The process involves downregulation of E-cadherin and upregulation of vimentin, Snail and Slug expression (20). Functional associations have been established between the SOX protein family and key effectors of EMT occurring in the context of carcinogenesis and embryonic development. In the present study, it was observed that the knockdown of SOX2 increased the expression of the epithelial marker E-cadherin, while the mesenchymal markers vimentin, Snail and Slug and their associated genes were downregulated, indicating that SOX2 may promote EMT in NSCLC.
miR-590-5p appears to exhibit different functions in various types of cancer. In cervical cancer cells, miR-590-5p was demonstrated to facilitate cell growth due to its close homology with the major capsid protein L1 (21). However, miR-590-5p has the opposite function in glioblastoma multiforme cells (22), suggesting that the role of miR-590-5p is context-dependent. A recent study demonstrated that ectopic expression of miR-590-5p significantly suppressed the proliferation, migration and invasion of NSCLC cells (9). Evidence has demonstrated that miRNAs may regulate multiple cellular functions, including EMT, through modulating the expression of their target genes in NSCLC. Using bioinformatics analysis, SOX2 was targeted by miR-590-5p, and the 3′UTR luciferase assay confirmed that SOX2 expression is regulated by miR-590-5p. It was also observed that the expression of the epithelial marker E-cadherin was markedly upregulated and that of the mesenchymal marker vimentin downregulated in A549 cells transfected with miR-590-5p mimics, whereas restoring SOX2 expression reversed these effects. The results of the present study suggested that the miR-590-5p/SOX2 axis may serve as an EMT regulator in NSCLC, which may be useful as a target for NSCLC treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Ethics approval and consent to participate
Written informed consent was obtained from all the patients regarding the use of their tissues for research purposes. All the procedures were performed in accordance with the guidelines of the Ethics Committee of Zhejiang Medical University.
Patient consent for publication
Written informed consent was obtained from all the patients regarding the use of their tissues for research purposes.
Availability of data and materials
All the datasets generated/analyzed in the present study are included in the published manuscript.
Authors' contributions
ZBC is the only contributor of this work, who analyzed and interpreted the patient data, performed the experiments, and wrote and approved the final manuscript.
Competing interests
The author declares that they have no competing interests.
References
- 1.Sarkar A, Hochedlinger K. The sox family of transcription factors: Versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12:15–30. doi: 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schepers GE, Teasdale RD, Koopman P. Twenty pairs of sox: Extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev Cell. 2002;3:167–170. doi: 10.1016/S1534-5807(02)00223-X. [DOI] [PubMed] [Google Scholar]
- 3.Stevanovic M, Zuffardi O, Collignon J, Lovell-Badge R, Goodfellow P. The cDNA sequence and chromosomal location of the human SOX2 gene. Mamm Genome. 1994;5:640–642. doi: 10.1007/BF00411460. [DOI] [PubMed] [Google Scholar]
- 4.Collignon J, Sockanathan S, Hacker A, Cohen-Tannoudji M, Norris D, Rastan S, Stevanovic M, Goodfellow PN, Lovell-Badge R. A comparison of the properties of Sox-3 with Sry and two related genes, Sox-1 and Sox-2. Development. 1996;122:509–520. doi: 10.1242/dev.122.2.509. [DOI] [PubMed] [Google Scholar]
- 5.Lengerke C, Fehm T, Kurth R, Neubauer H, Scheble V, Müller F, Schneider F, Petersen K, Wallwiener D, Kanz L, et al. Expression of the embryonic stem cell marker SOX2 in early-stage breast carcinoma. BMC Cancer. 2011;11:42. doi: 10.1186/1471-2407-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boumahdi S, Driessens G, Lapouge G, Rorive S, Nassar D, Le Mercier M, Delatte B, Caauwe A, Lenglez S, Nkusi E, et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014;511:246–250. doi: 10.1038/nature13305. [DOI] [PubMed] [Google Scholar]
- 7.Xie C, Han Y, Liu Y, Han L, Liu J. miRNA-124 down-regulates SOX8 expression and suppresses cell proliferation in non-small cell lung cancer. Int J Clin Exp Pathol. 2014;7:6534–6542. [PMC free article] [PubMed] [Google Scholar]
- 8.Toschi L, Finocchiaro G, Nguyen TT, Skokan MC, Giordano L, Gianoncelli L, Perrino M, Siracusano L, Di Tommaso L, Infante M, et al. Increased SOX2 gene copy number is associated with FGFR1 and PIK3CA gene gain in non-small cell lung cancer and predicts improved survival in early stage disease. PLoS One. 2014;9:e95303. doi: 10.1371/journal.pone.0095303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang FF, Wang S, Xue WH, Cheng JL. microRNA-590 suppresses the tumorigenesis and invasiveness of non-small cell lung cancer cells by targeting ADAM9. Mol Cell Biochem. 2016;423:29–37. doi: 10.1007/s11010-016-2822-y. [DOI] [PubMed] [Google Scholar]
- 10.Zhou L, Zhao LC, Jiang N, Wang XL, Zhou XN, Luo XL, Ren J. MicroRNA miR-590-5p inhibits breast cancer cell stemness and metastasis by targeting SOX2. Eur Rev Med Pharmacol Sci. 2017;21:87–94. [PubMed] [Google Scholar]
- 11.Edge SB, Compton CC. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Gen Y, Yasui K, Nishikawa T, Yoshikawa T. SOX2 promotes tumor growth of esophageal squamous cell carcinoma through the AKT/mammalian target of rapamycin complex 1 signaling pathway. Cancer Sci. 2013;104:810–816. doi: 10.1111/cas.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun C, Sun L, Li Y, Kang X, Zhang S, Liu Y. Sox2 expression predicts poor survival of hepatocellular carcinoma patients and it promotes liver cancer cell invasion by activating Slug. Med Oncol. 2013;30:503. doi: 10.1007/s12032-013-0503-1. [DOI] [PubMed] [Google Scholar]
- 15.Chen S, Li X, Lu D, Xu Y, Mou W, Wang L, Chen Y, Liu Y, Li X, Li LY, et al. SOX2 regulates apoptosis through MAP4K4-survivin signaling pathway in human lung cancer cells. Carcinogenesis. 2014;35:613–623. doi: 10.1093/carcin/bgt371. [DOI] [PubMed] [Google Scholar]
- 16.Chou YT, Lee CC, Hsiao SH, Lin SE, Lin SC, Chung CH, Chung CH, Kao YR, Wang YH, Chen CT, et al. The emerging role of SOX2 in cell proliferation and survival and its crosstalk with oncogenic signaling in lung cancer. Stem Cells. 2013;31:2607–2619. doi: 10.1002/stem.1518. [DOI] [PubMed] [Google Scholar]
- 17.Stewart DJ. Wnt signaling pathway in non-small cell lung cancer. J Natl Cancer Inst. 2014;106:djt356. doi: 10.1093/jnci/djt356. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Xu Y, Chen Y, Chen S, Jia X, Sun T, Liu Y, Li X, Xiang R, Li N. SOX2 promotes tumor metastasis by stimulating epithelial-to-mesenchymal transition via regulation of WNT/β-catenin signal network. Cancer Lett. 2013;336:379–389. doi: 10.1016/j.canlet.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 19.Yuan XW, Wang DM, Hu Y, Tang YN, Shi WW, Guo XJ, Song JG. Hepatocyte nuclear factor 6 suppresses the migration and invasive growth of lung cancer cells through p53 and the inhibition of epithelial-mesenchymal transition. J Biol Chem. 2013;288:31206–31216. doi: 10.1074/jbc.M113.480285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitamura K, Seike M, Okano T, Matsuda K, Miyanaga A, Mizutani H, Noro R, Minegishi Y, Kubota K, Gemma A. MiR-134/487b/655 cluster regulates TGF-β-induced epithelial-mesenchymal transition and drug resistance to gefitinib by targeting MAGI2 in lung adenocarcinoma cells. Mol Cancer Ther. 2014;13:444–453. doi: 10.1158/1535-7163.MCT-13-0448. [DOI] [PubMed] [Google Scholar]
- 21.Chu Y, Ouyang Y, Wang F, Zheng A, Bai L, Han L, Chen Y, Wang H. MicroRNA-590 promotes cervical cancer cell growth and invasion by targeting CHL1. J Cell Biochem. 2014;115:847–853. doi: 10.1002/jcb.24726. [DOI] [PubMed] [Google Scholar]
- 22.Pang H, Zheng Y, Zhao Y, Xiu X, Wang J. miR-590-3p suppresses cancer cell migration, invasion and epithelial-mesenchymal transition in glioblastoma multiforme by targeting ZEB1 and ZEB2. Biochem Biophys Res Commun. 2015;468:739–745. doi: 10.1016/j.bbrc.2015.11.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the datasets generated/analyzed in the present study are included in the published manuscript.