Abstract
Protein synthesis, including the translation of specific messenger RNAs (mRNAs), is regulated by extracellular stimuli such as hormones and by the levels of certain nutrients within cells. This control involves several well-understood signaling pathways and protein kinases, which regulate the phosphorylation of proteins that control the translational machinery. These pathways include the mechanistic target of rapamycin complex 1 (mTORC1), its downstream effectors, and the mitogen-activated protein (MAP) kinase (extracellular ligand-regulated kinase [ERK]) signaling pathway. This review describes the regulatory mechanisms that control translation initiation and elongation factors, in particular the effects of phosphorylation on their interactions or activities. It also discusses current knowledge concerning the impact of these control systems on the translation of specific mRNAs or subsets of mRNAs, both in physiological processes and in diseases such as cancer.
The control of protein synthesis plays key roles in cell growth and proliferation and in many other processes, via shaping the cellular proteome. The importance of translational control is underscored, for example, by the lack of concordance between the transcriptome and the proteome—which, among other factors, reflects the differing efficiencies with which different messenger RNAs (mRNAs) are translated into polypeptides (Schwanhausser et al. 2011; Li et al. 2014)—and the role of dysregulation of translation in human diseases, including cancer (Robichaud et al. 2018a), virus infection (Stern-Ginossar et al. 2018), and many others (Tahmasebi et al. 2018). Furthermore, since mRNA translation consumes a substantial fraction of cellular metabolic energy and an even higher proportion of amino acids, it is crucial that cells match the rate of protein synthesis to the availability of nutrients.
Accordingly, sophisticated control mechanisms exist to allow extracellular stimuli (e.g., hormones, growth factors), intracellular metabolites (essential amino acids, nucleotides) and cues, such as energy status, to regulate protein synthesis. These mechanisms include control of the translation of specific mRNAs, via signaling pathways that impinge on components of the initiation or elongation machinery, which is mainly achieved through protein kinases that phosphorylate and thereby regulate translation initiation or elongation factors, and certain other proteins.
CAVEATS
Studies on the role of specific signaling pathways or their target phosphoproteins rely heavily on the use of small molecule inhibitors, especially of protein kinases, and on testing proteins in which a given phosphorylation site has been mutated to one that cannot be phosphorylated or a negatively charged one, which may mimic a phosphorylated residue. There are important caveats to both approaches. Compounds that inhibit protein kinases are often not specific for a single type of enzyme because the ATP-binding sites of many kinases are similar. For example, the compounds often used as inhibitors of the mitogen-activated protein (MAP) kinase-interacting kinases (MNKs, which phosphorylate eukaryotic initiation factor [eIF]4E), CGP57380 (Tschopp et al. 2000), and cercosporamide (Konicek et al. 2011), affect other protein kinases (Bain et al. 2007; Beggs et al. 2015), several of them more potently than their effects on MNK1 and MNK2. It is, therefore, important to test chemically distinct entities that affect the kinase of interest and, wherever possible, to complement such work with genetic studies using cells in which the enzyme of interest has been knocked out. Genetic studies have been made much more tractable by the advent of clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 genome editing, which can be applied to cell lines as well as to studies in animals.
Rapamycin is a macrolide compound that is a highly specific inhibitor of the protein kinase complex termed mechanistic (or mammalian) target of rapamycin complex 1 (mTORC1). Rapamycin is specific largely because it does not target the active site of mTOR, but instead binds (together with its partner, FKBP12, an immunophilin) to another domain, thereby partially occluding access to the active site for some protein substrates (Yang et al. 2013). Although in the short term its effects are limited to inhibiting mTORC1, longer-term exposure of cells to rapamycin also impairs mTORC2 signaling by interfering with assembly of this complex (Sarbassov et al. 2006).
The effects of mutating a phosphorylation site of interest must also be treated with caution. Nonphosphorylatable mutants are generally created by mutating the relevant serine or threonine to alanine, which, because it lacks a hydroxyl group, cannot accept a phosphate. However, alanine also differs from serine and threonine in being unable to form H-bonds. Conversely, phosphomimetic mutants of Ser or Thr to Glu or Asp may sometimes mimic the addition of a phosphate, but since the geometry (tetrahedral vs. planar) and charge (double vs. single) of phospho-Ser/Thr and Glu/Asp differ, phosphomimicry is by no means guaranteed.
mTORC1 SIGNALING
One major pathway that regulates the translational machinery in several ways is the mTOR pathway. mTOR forms two types of multisubunit complexes, mTORC1 and mTORC2. mTORC1 phosphorylates proteins that directly regulate the translational apparatus as well as protein kinases that phosphorylate and regulate translation factors (Saxton and Sabatini 2017).
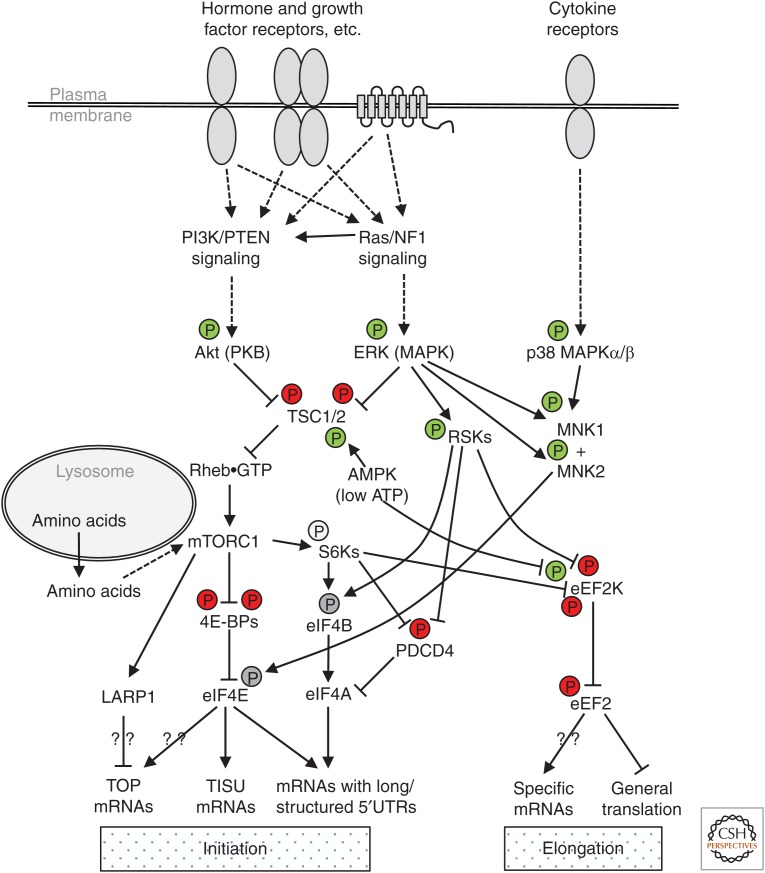
In addition to mTOR, mTORC1 contains Raptor, mLst8, and Rheb, a GTPase (Ben-Sahra and Manning 2017). Raptor and Rheb are found only in mTORC1, not in mTORC2. Raptor binds substrate proteins, conferring specificity, whereas Rheb activates mTORC1 when it is bound to GTP. Rheb provides a key link to extracellular stimuli through the phosphatidylinositide 3-kinase (PI3K) or classical MAP kinase (extracellular ligand-regulated kinase [ERK]) signaling pathways (Fig. 1) (Saxton and Sabatini 2017). Rheb is regulated by the tuberous sclerosis complex (TSC), which includes the proteins TSC1, TSC2, and TBC1D7 (Dibble et al. 2012). TSC2 acts as a GTPase activator protein for Rheb, promoting its conversion to its inactive GDP-bound state. Protein kinases within or downstream of these pathways, Akt (also called protein kinase B [PKB]), ERK, or the ERK-substrates termed RSKs, phosphorylate the TSC complex and impair its ability to promote GTP hydrolysis on Rheb. This allows stimuli that switch on PI3K or ERK signaling to enhance the levels of Rheb-GTP and thereby activate mTORC1.
Figure 1.
Major signaling connections to the translational machinery. This simplified schematic shows the main signaling links to the translational machinery. Solid lines depict direct links and dashed lines indirect ones (involving multiple components, for example). (P) indicates phosphorylation events, green indicates activation, red indicates an inhibitory phosphorylation, and gray denotes that the functional consequence is unclear. Not all the components or phosphorylation sites mentioned in the text are shown. Question marks denote some of the open questions described in the text.
Several components of the PI3K and ERK pathways undergo gain-of-function oncogenic mutations in cancers, whereas others are tumor suppressors whose function is lost in some cancers. These pathways, and hence mTORC1 signaling, are constitutively activated in a high percentage of human cancers, estimated at 70% or higher (Saxton and Sabatini 2017).
mTORC1 signaling is also activated by amino acids, at the surface of the lysosome (Fig. 1), by sophisticated sensing and signaling mechanisms involving a distinct set of GTPases, the Rags (Wolfson and Sabatini 2017). Interestingly, another nutrient-regulated signaling pathway, the AMP-activated protein kinase (AMPK), also impinges on mTORC1 at the lysosomal surface, where AMPK can be activated (Lin and Hardie 2017). This happens, for example, when cellular ATP levels fall and levels of ADP, or especially AMP, rise. Recent studies have identified an additional, novel link between intermediary metabolism and the control of AMPK, whereby levels of the glycolytic intermediate fructose 1,6-bisphosphate also control AMPK activity (Lin and Hardie 2017). When AMPK is switched on, it inactivates mTORC1 by phosphorylating TSC2 and/or Raptor, thereby restraining energy-using biosynthetic processes that are normally driven by mTORC1, including protein and ribosome biosynthesis (Ben-Sahra and Manning 2017).
mTORC1 PHOSPHORYLATES PROTEINS THAT REGULATE THE TRANSLATIONAL MACHINERY
Among the many substrates for mTORC1, two classes are particularly relevant for this review. The first class comprises the small phosphoproteins termed eIF4E-binding proteins (4E-BPs) that bind to and sequester eIF4E (Figs. 1 and 2) (Merrick and Pavitt 2018). (“eIF4E” is used here to mean eIF4E1; its paralogs bind to 4E-BPs less strongly [in the case of eIF4E2, also termed eIF4E-homologous protein, 4EHP] or not at all [eIF4E3]) (Joshi et al. 2004). The second class comprises the S6 kinases (S6Ks; Fig. 2), which are named for their ability to phosphorylate ribosomal protein (rp) S6, a component of the small ribosomal subunit.
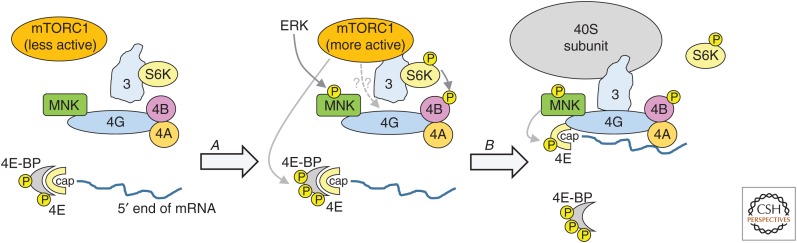
Figure 2.
Signaling links to the cap-binding initiation machinery. S6 kinase (S6K) indicates S6 kinases 1 and 2, and MAP kinase-interacting kinase (MNK) denotes MNK1/2. Not all phosphorylation events are depicted. Thin solid gray lines denote direct phosphorylation; the dashed gray line indicates that mechanistic target of rapamycin complex 1 (mTORC1) signaling can promote eukaryotic initiation factor (eIF)4G/eIF3 binding. Step A, stimulation of cells (e.g., with insulin or a growth factor), leads to the activation of kinases including mTORC1, MNKs, and S6Ks, leading to Step B where different protein:protein interactions occur and assembly of eIF4F, together with eIF3 and 40S ribosomal subunits. Please see the text for further information.
MAP KINASE SIGNALING
The MAP kinase (ERK) pathway regulates mTORC1, for example, through the phosphorylation of TSC1/2 (Huang and Manning 2008) or Raptor (Carriere et al. 2011). This pathway is activated by a range of extracellular stimuli and by oncogenic mutations (e.g., in the GTPase Ras or its GTPase-activator protein, neurofibromin 1). Thus, ERK signaling is activated in a high proportion of cancers.
ERK also phosphorylates and activates several other protein kinases, including the RSKs, whose original name (ribosomal S6 kinases) may cause confusion with the S6Ks (Fig. 1). They phosphorylate S6 at a subset of the sites targeted by S6Ks reflecting the fact that S6Ks and RSKs (as well as Akt) act at similar, but not identical, consensus recognition sequences. RSKs and S6Ks phosphorylate rpS6 on Ser235/236, whereas S6Ks additionally phosphorylate Ser240 and Ser244 (Pende et al. 2004), phosphorylation of which provides a useful readout of mTORC1 signaling. RSKs and S6Ks also phosphorylate the same sites in other proteins that regulate translation, including eIF4B (Shahbazian et al. 2006) and the eEF2 kinase (eEF2K) that regulates elongation (Wang et al. 2001).
ERKs are not the only members of the MAP kinase family relevant to control of the translational machinery; others include the p38 MAP kinases, which are turned on by distinct stimuli (such as cytokines) and include p38 MAPKs α and β, which can activate MNK1 (Fig. 1) (Waskiewicz et al. 1997; Wang et al. 1998). p38 MAPK δ (whose control is poorly understood) also phosphorylates eEF2K (see below and Dever et al. 2018; Fig. 3).
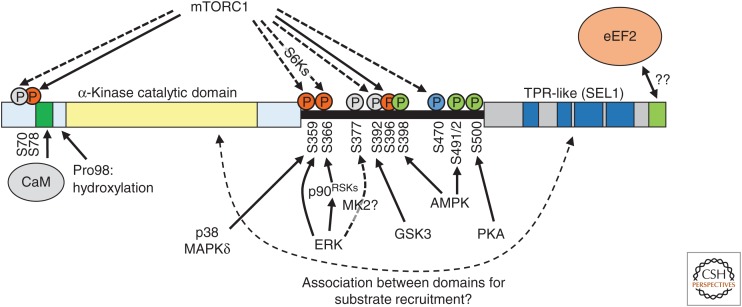
Figure 3.
Regulation of eukaryotic elongation factor 2 (eEF2) kinase. The principal domains of eEF2K are depicted schematically, not to scale, including the CaM-binding site and a region at the extreme carboxyl terminus, which is required for eEF2K to phosphorylate eEF2 and may interact directly with that protein. Known phosphorylation sites are shown; orange for inhibitory sites, green for one that activates eEF2K, and gray for ones whose function is not clear or do not affect activity. Solid lines indicate direct phosphorylation; dashed ones denote indirect or unclear connections.
PHOSPHORYLATION OF COMPONENTS OF THE INITIATION MACHINERY
Several eIFs are subject to regulatory phosphorylation. The first example to be discovered was the α subunit of the trimer eIF2. This phosphorylation event inhibits general translation and the inputs to this system are primarily intracellular stresses rather than extracellular stimuli (Wek 2018).
eIF2 is active only when bound (via its γ subunit) to GTP. The GTP is hydrolyzed during initiation. Because regeneration of active eIF2•GTP is slow, an additional factor is required, the GDP-dissociation stimulator protein (Williams et al. 2001), eIF2B, also referred to as a guanine nucleotide exchange factor (GEF). eIF2B is a decamer of five subunits (α2β2γ2δ2ε2) (Gordiyenko et al. 2014; Wortham et al. 2014). Its largest subunit, ε, which houses the catalytic domain (Gomez et al. 2002), is subject to phosphorylation at several sites. These include Ser540 (in human eIF2Bε), which is a substrate for GSK3 (Welsh et al. 1998), a kinase that is inactivated by stimuli such as insulin, which stimulate protein synthesis. Because phosphorylation of this site impairs eIF2B’s GEF function (Welsh et al. 1998), this link provides a potential mechanism by which agents such as insulin can activate translation initiation (alongside a number of other events discussed below).
mTORC1 can interact with eIF3 (Holz et al. 2005; Harris et al. 2006), as can its substrate S6K1 (Holz et al. 2005), which in turn phosphorylates components of initiation complexes, such as eIF4B (Figs. 1 and 2). mTORC1 signaling also leads to increased binding of eIF3 to eIF4G, which is induced by insulin (Harris et al. 2006). As eIF3 interacts with the 40S subunit, this interaction likely helps to promote recruitment of ribosomes to eIF4F and thus to mRNAs, although it remains unclear how mTOR modulates eIF3/eIF4G binding.
mTORC1 phosphorylates the 4E-BPs, impairing their ability to interact with eIF4E (Figs. 1 and 2). Phosphorylation occurs at multiple sites, Thr37, Thr46, Ser65, and Thr70, in human 4E-BP1. Phosphorylation of the first two sites promotes subsequent phosphorylation at the other sites, which in turn seem to be most important for the release of 4E-BP1 from eIF4E. mTORC1 can phosphorylate all four sites. The properties and regulation of 4E-BP2 and 3 are much less well studied than 4E-BP1. Binding to a 4E-BP prevents eIF4E from interacting with the scaffold protein eIF4G, which in turn interacts with several other proteins involved in translation initiation (Fig. 2) (Gingras et al. 1999). The binding sites on eIF4E for 4E-BPs and eIF4G overlap, and indeed these proteins possess similar linear sequence motifs involved in their binding to eIF4E (Mader et al. 1995). This mechanism therefore allows mTORC1 signaling to promote the formation of eIF4F complexes (Merrick and Pavitt 2018), and thus (through additional protein:protein interactions; Fig. 2) the recruitment of 40S subunits to the 5′-end of mRNAs.
MNK1 and MNK2 phosphorylate eIF4E on Ser209 (Fig. 1), which remains their only validated in vivo substrate (Ueda et al. 2004; Buxade et al. 2008); moreover, MNKs are the only kinases that act on this site. MNKs interact with eIF4G, bringing them close to their substrate eIF4E (Pyronnet et al. 1999).
It should be noted that eIF4E is not, as sometimes stated, always the least abundant initiation factor, although this varies between organisms and cell types (see, for example, Rau et al. 1996; von der Haar and McCarthy 2002; Schwanhausser et al. 2011); some others may occur at lower levels (Merrick and Pavitt 2018). When one copy of the eIF4E gene is deleted in mice, they show no adverse effects (Truitt et al. 2015). However, the 4E-BPs regulate eIF4E availability and it is this parameter, rather than the absolute levels of eIF4E, that can limit the factor’s activity.
Rapamycin generally has little effect on the phosphorylation of 4E-BP1, and therefore does not impair eIF4E:eIF4G binding (e.g., Choo et al. 2008; Huo et al. 2012). As one would expect, the phosphorylation of 4E-BP1 is efficiently blocked by compounds that interfere with the catalytic activity of mTOR (mTOR kinase inhibitors; mTOR-KIs). Thus, rapamycin is expected to have less impact on the control of translation initiation by 4E-BPs than mTOR-KIs do. The reason why rapamycin has little effect on the phosphorylation of 4E-BPs reflects its mode of action. Structural studies indicate that the FKBP12:rapamycin complex partially occludes but does not completely obstruct the active site cleft (Yang et al. 2013), thereby restricting substrate access. Detailed studies suggest that rapamycin inhibits the ability of mTOR to phosphorylate “weak” substrates such as the S6Ks mentioned above, but not “strong” substrates such as 4E-BPs (Kang et al. 2013).
IMPACT OF mTORC1 SIGNALING ON TRANSLATION
The fact that rapamycin only affects phosphorylation of some of the substrates of mTORC1 is important when evaluating the effects of rapamycin versus mTOR-KIs on overall protein synthesis and on the translation of specific mRNAs. Rapamycin has a lesser inhibitory effect compared to the much stronger effect of mTOR-KIs, and they have differential effects on the synthesis of specific proteins (Huo et al. 2012). One example is provided by the 5′-terminal oligopyrimidines (TOP) mRNAs, which possess a run of pyrimidine nucleotides at the extreme 5′ end and include those encoding all cytoplasmic ribosomal proteins and several translation elongation factors (Meyuhas and Kahan 2014). Synthesis of these proteins is impaired by rapamycin and more strongly inhibited by mTOR-KIs. This is presumably because mTOR-KIs inhibit mTORC1 signaling more strongly than rapamycin but one cannot formally rule out a contribution from mTORC2 signaling, which is also inhibited by mTOR-KIs. The regulation of TOP mRNA translation by mTORC1 provides a mechanism by which nutrient-activated signaling can also promote ribosome biogenesis, along with promoting rRNA transcription (Iadevaia et al. 2014) and nucleolar function (Chauvin et al. 2013) to increase the cellular capacity for protein synthesis. The control of this interesting and important set of mRNAs is discussed further below.
Several studies have examined the effect that interfering with mTOR signaling has on mRNA translation and protein synthesis. Such effects could be exerted through 4E-BP-mediated control of the availability of eIF4E; the associated effects on the recruitment of eIF4A (via eIF4G) to the 5′-end of mRNAs; the regulation of eIF4A by eIF4B, which is an indirect target of mTORC1 signaling (see later section); the effect of mTORC1 on the elongation machinery; or other mechanisms.
Larsson et al. (2012) used polysome profiling (i.e., sucrose gradient centrifugation) to assess which mRNAs were associated with larger polysomes. They studied the effects of treatment with rapamycin, PP242 (an mTOR-KI) or metformin, an agent that causes activation of AMPK and therefore inhibits mTORC1 signaling. Because AMPK has diverse substrates (Lin and Hardie 2017), it may impact translation in additional ways that do not occur in response to inhibition of mTOR. The data from their polysome profiling analysis revealed that the three agents they used decreased the association of almost 600 mRNAs with “large” polysomes; some were affected by all stimuli and others by only one or two of them. The great majority of mRNAs affected by PP242 were also influenced by rapamycin (with some exceptions), and two-thirds of them were also affected by metformin, although, interestingly, almost half the mRNAs whose polysomal association was decreased by metformin were not affected by mTOR inhibition, consistent with additional actions of this drug, likely through other AMPK targets. Gene ontology (GO) analysis revealed distinct functional categories of mRNAs that were affected by the different treatments. For example, those selectively affected by PP242 and by metformin, but not rapamycin, included many involved in cell-cycle control, whereas the set affected only by PP242 was enriched in mRNAs encoding proteins involved in mitochondrial functions and mRNA translation; those affected only by metformin included many involved in RNA processing.
Thoreen et al. (2012) and Hsieh et al. (2012) used ribosome profiling technology (Ingolia et al. 2018) to assess which mRNAs were associated with ribosomes under different conditions, in combination with pharmacological inhibition of mTOR/mTORC1 using mTOR-KIs or rapamycin. These studies concluded that mTOR inhibition predominantly reduces ribosome footprints on TOP mRNAs. Hsieh et al. (2012) also identified several mRNAs for proteins involved in metastasis or invasion as being regulated by mTOR, including Y-box protein 1 (YB1), a posttranscriptional regulator of genes involved in these processes. A distinct feature, a pyrimidine-rich translational element distinct in make-up and position from TOP elements, was present in many mTOR-regulated mRNAs. The importance of this feature for translation initiation, and its mode of action, remain unknown.
Many of the differences in the conclusions drawn by the ribosome profiling studies as compared to the polysome profiling work (Larsson et al. 2012) likely arise for methodological reasons (Gandin et al. 2016). Ribosome profiling provides data on the positions of ribosomes on mRNAs and, as performed by Thoreen et al. (2012) and Hsieh et al. (2012), detects big shifts in ribosome occupancy on highly expressed mRNAs, whereas polysome profiling can detect more modest shifts of mRNAs from larger to smaller polysomes. TOP mRNAs are abundant, and as discussed in Masvidal et al. (2017), next-generation sequencing (NGS) reads corresponding to them therefore tend to make up a high proportion of the data from such ribosome-profiling experiments. High-read “depth” is necessary to obtain quantifiable data for less abundant mRNAs.
Gandin et al. (2016) extended polysome-profiling studies to examine the impacts both of inhibiting mTOR and depleting the helicase eIF4A on the set of mRNAs found in larger polysomes and coupled this with analysis of transcription start sites. This study identified two distinct subsets of “mTOR-sensitive” mRNAs, with quite distinct features in their 5′ untranslated regions (UTRs) (Fig. 1). One subset has long 5′UTRs and includes mRNAs for proteins involved in cell survival (such as that for Mcl1 or survivin [Birc5]) or proliferation (e.g., cyclin D3, ODC). This subset was anticipated given that translation of such mRNAs is expected to require eIF4A’s helicase function and thus assembly of the eIF4F complex on them. Consistent with this, the polysomal association of members of this set is also impaired by depleting cells of eIF4A1. The effect of mTOR signaling on the translation of these mRNAs very likely reflects the control of eIF4F assembly by mTOR (through 4E-BPs) and perhaps also the input from mTORC1 via S6Ks to eIF4B (Figs. 1 and 2).
In contrast, mRNAs in the second subset of mTOR-sensitive transcripts possess very short, unstructured 5′UTRs and, presumably as a consequence, their translation is not impaired by depletion or chemical inhibition of eIF4A (Gandin et al. 2016). These mRNAs have been described as containing translation initiator of short 5′UTR (TISU) elements (Elfakess et al. 2011; Kwan and Thompson 2018), although the brevity of the 5′UTR rather than a specific sequence element may be the key feature for their translational control. Translation of these mRNAs requires eIF4E-eIF4G binding and is regulated by 4E-BPs (Fig. 1) (Morita et al. 2013; Sinvani et al. 2015). Many such mRNAs encode proteins involved in mitochondrial function, the vast majority of mitochondrial proteins being encoded by nuclear genes.
The ability of mTORC1 signaling to promote the translation of mRNAs for mitochondrial proteins likely represents part of a concerted program through which this pathway upregulates mitochondrial biogenesis. This program also involves the transcription factor PGC-1α, which drives the transcription of many genes involved in mitochondrial function or biogenesis (Cunningham et al. 2007; Duvel et al. 2010). This mechanism may serve to increase the mitochondrial capacity for ATP production to support the many anabolic processes that are turned on by mTORC1 signaling, such as protein synthesis and ribosome biogenesis.
A LIMITATION OF RIBOSOME AND POLYSOME PROFILING TECHNIQUES
A limitation of both ribosome profiling and polysome profiling approaches concerns the control of elongation. mRNAs whose rate of elongation is slower under a given condition will shift into larger polysomes as ribosomes build up on them, so that in both analyses these mRNAs will appear to be “more translated,” whereas in fact the opposite is true. Methods such as bioorthogonal noncanonical amino acid tagging (BONCAT), involving the labeling of newly synthesized polypeptides with chemical and mass tags (Dieterich et al. 2006), report the rate of accumulation of new polypeptides and thus the rate of synthesis of proteins rather than the association of their mRNAs with ribosomes/polysomes. These approaches have been used to study global proteome dynamics (Schwanhausser et al. 2011), the role of eEF2K in the synthesis of new proteins in primary neurons (Kenney et al. 2016), and the impact of mTOR signaling on the synthesis of specific proteins (Huo et al. 2012). The latter study concluded that mTOR-KIs and rapamycin have differing effects on the synthesis of many proteins. In particular, in agreement with other studies, rapamycin inhibits the synthesis of proteins encoded by 5′TOP mRNAs rather less strongly than mTOR-KIs. Interestingly, the synthesis of certain other proteins shows a similar pattern of inhibition; the mRNAs for some of them may belong to the TOP class although others evidently do not. The fact that the annotation of the 5′ end of mRNAs is often incorrect makes interpretation more difficult, an issue that is addressed by Gandin et al. (2016).
REGULATION OF THE TRANSLATION OF TOP mRNAs
Although the translation (polysomal association) of TOP mRNAs has been known to be promoted by mTORC1 signaling for almost three decades, and despite extensive work in several laboratories, the mechanisms that link mTORC1 signaling to control of TOP mRNA translation still remain to be definitively clarified (Meyuhas and Kahan 2014). At one time, S6K-catalyzed phosphorylation of S6 was thought to be involved but there is now unequivocal evidence against this idea, for example, from data obtained using cells in which S6Ks have been knocked out or the phosphorylation sites in S6 have been mutated (reviewed in Meyuhas and Kahan 2014). The phosphorylation of S6 is discussed in more detail below.
As mTORC1-regulated repressors of translation initiation, the 4E-BPs appear well-placed to regulate TOP mRNA translation, and data show that they do indeed play such a role in response to pharmacological inhibition of mTOR. Thoreen et al. (2012) concluded that, by interfering with eIF4E-eIF4G binding, mTOR inhibitors cause a greater dissociation of eIF4E from TOP mRNAs than from other mRNAs, thereby reducing the translation of the TOP mRNAs. However, several lines of evidence indicate that 4E-BPs are not the key regulators of TOP mRNA translation under conditions such as amino acid deprivation or hypoxia, both of which impair mTORC1 signaling (Meyuhas and Kahan 2014; Miloslavski et al. 2014).
Recent studies have identified a new potential regulator of TOP mRNA translation, La-related protein 1 (LARP1) (Fig. 1), although the data for the role of LARP1 in controlling TOP mRNA translation are conflicting. Tcherkezian et al. (2014) used a proteomic screen to identify proteins that associate with the 5′ cap of mRNAs in an mTORC1-dependent manner; one of them was LARP1, which also binds poly(A)-binding protein (PABP) and mTORC1 itself. Their data indicate that LARP1 is a positive regulator of mRNA translation based on the observation that knockdown of LARP1 reduced polysome levels, suggesting impaired translation initiation. The degree of polysome size reduction of TOP mRNAs was greater than that of mRNAs in general, although the effect was clearly not restricted to TOP messages.
Fonseca et al. (2015) agree that LARP1 interacts with mTORC1 but provide evidence that it acts as a repressor of TOP mRNA translation and interacts with TOP elements. Their data show a “preferential” association of LARP1 with TOP mRNAs and that such association is increased in response to rapamycin or Torin1, an mTOR-KI. In particular, the repression of TOP mRNA translation caused by amino acid starvation is attenuated when LARP1 is knocked down, consistent with LARP1 being a repressor rather than an activator of TOP mRNA translation. It should be noted that the effect of amino acid starvation on TOP mRNA translation also involves the RNA-binding proteins T-cell intracellular antigen 1 (TIA1) and TIA-R (Damgaard and Lykke-Andersen 2011), although, again, this appears not to be a general mechanism for their control.
Further studies have provided additional evidence that LARP1 acts as a repressor of TOP mRNA translation, although the mechanisms proposed differ. Two studies reported that LARP1 directly binds the cap of TOP mRNAs, thus impeding access for eIF4E (Lahr et al. 2017; Philippe et al. 2017). Lahr et al. solved structures of the carboxy-terminal DM15 domain of LARP1, including structures of co-complexes with nucleotides 4-11 of the 5′ end of the TOP mRNA encoding rpS6 (i.e., without the initial cytosine and the next two nucleotides) and m7GTP-C (analog of the extreme 5′ end of a TOP mRNA). This revealed binding pockets in the DM15 domain of LARP1 for the 7-methylated GTP, the first C, and a TOP. Philippe et al. (2017) also reported that LARP1 binds the cap and TOP sequence, that LARP1 represses TOP mRNA translation in vitro, and that this ability is enhanced using LARP1 from cells pretreated with Torin1. A recent report shows that LARP1 is phosphorylated by mTORC1; this causes LARP1 to dissociate from the 5′UTRs of TOP mRNAs and relieves its inhibitory effect on their translation (Hong et al. 2017). Puzzlingly, however, the binding sites for LARP1 on these mRNAs generally did not match the TOP elements at their 5′-termini.
Overall, these data suggest a model whereby (1) LARP1 is regulated by mTORC1 such that its ability to bind TOP mRNAs is somehow enhanced when mTORC1 signaling is inhibited, and (2) association of LARP1 with the cap and 5′ end of a TOP mRNA blocks access for eIF4E and thus ribosomes, leading to impairment of initiation on such mRNAs. It remains unclear precisely how mTORC1 regulates LARP1—presumably by phosphorylation—and given that LARP1 binds Raptor, perhaps direct phosphorylation by mTORC1. LARP1 contains numerous phosphorylation sites, but it is not yet known which are regulated by mTORC1 signaling and which control LARP1’s ability to bind TOP mRNAs. It should be noted that overexpression of eIF4E failed to outcompete a repressor activity that represses the translation of TOP mRNAs (Shama et al. 1995), which appears to argue against roles for 4E-BPs or LARP1.
eIF4E PHOSPHORYLATION BY THE MNKs
Phosphorylation of eIF4E was discovered early in studies on translation factor modifications (Duncan et al. 1987) but the function of this regulated modification remains obscure. Phosphorylation occurs at a single site, Ser209 (Flynn and Proud 1995; Joshi et al. 1995), and is not required for animal viability, as mice carrying a homozygous Ser209Ala mutation are viable (Furic et al. 2010). Furthermore, mice lacking both copies of both genes for the relevant kinases, the MNKs, are also viable and fertile (Ueda et al. 2004). However, as described below, there are now a number of examples of the control of the translation of mRNAs requiring phosphorylation of eIF4E and/or MNK function. eIF4E2/4EHP and eIF4E3 do not contain an equivalent of Ser209.
In humans (but apparently not mice), each MNK gene gives rise to two distinct mRNA species caused by alternative splicing (Buxade et al. 2008); this results in two protein isoforms each for MNK1 and MNK2 termed “a” and “b,” which differ at their carboxyl termini. All isoforms contain the complete kinase domain and an amino-terminal polybasic region that interacts with eIF4G, which also serves as a nuclear localization signal (Parra-Palau et al. 2003; Scheper et al. 2003). MNK1a also contains a nuclear export signal and is therefore mainly cytoplasmic; MNK2a does not have this feature and is largely nuclear. MNK1a and MNK2a each contain a carboxy-terminal sequence that interacts with MAP kinases; this motif is absent from the shorter MNK1b and MNK2b isoforms. The MAP kinase-binding sequences differ however, such that although MNK1a interacts with ERK and p38 MAP kinase αβ, MNK2a only binds ERK (Waskiewicz et al. 1997). Indeed, unlike MNK1a, MNK2a can bind active phosphorylated ERK, which likely explains its high basal activity. In contrast, in unstimulated cells, MNK1a activity is low but can be markedly increased by stimuli that switch on ERK signaling (e.g., serum, some growth factors, various receptor agonists) or p38 MAP kinase α/β (some cytokines, certain “stress” conditions). The control of MNK1b and MNK2b, which lack the MAP kinase interaction site, is not well understood (see O’Loghlen et al. 2007; Buxade et al. 2008).
MNK signaling intersects with mTORC1 signaling in several ways. First, as noted, MNKs interact with eIF4G, facilitating the phosphorylation of eIF4E (Pyronnet et al. 1999). A consequence of this is that agents that promote the phosphorylation of 4E-BPs and their release from eIF4E, allowing increased eIF4E-eIF4G binding, should facilitate eIF4E phosphorylation. Second, rapamycin treatment can paradoxically enhance eIF4E phosphorylation and the activity of MNK2 (rather than MNK1 [Stead and Proud 2013]). Third, MNKs were recently reported to stimulate signaling through mTORC1 by phosphorylating TELO2, a partner for mTORC1 that promotes interactions between mTORC1 and its substrates (Brown and Gromeier 2017).
WHAT ROLE DOES eIF4E PHOSPHORYLATION PLAY IN CONTROLLING mRNA TRANSLATION?
To date, no comprehensive datasets have been published for the effects of inhibiting phosphorylation of eIF4E on the translation of specific mRNAs of the kind published for mTOR or eIF4A (e.g., Hsieh et al. 2012; Thoreen et al. 2012; Gandin et al. 2016). However, a set of data was reported by Furic et al. (2010) for embryonic fibroblasts from wild-type and eIF4ESer209Ala knock-in mice; this polysome-profiling study identified 35 mRNAs whose association with polysomes was reduced in the latter cells, implying that eIF4E phosphorylation promotes the initiation of their translation.
It remains unclear how phosphorylation of eIF4E influences the translation of specific transcripts, for example, whether a feature of these mRNAs (and, if so, which feature) confers sensitivity to eIF4E phosphorylation. Several studies, using compounds that inhibit MNKs, MNK-knockout (KO) cells, or cells where the eIF4E phosphorylation site has been changed (Ser209Ala), show that MNKs and/or the phosphorylation of eIF4E positively regulate the translation of mRNAs for proteins involved in a range of processes, including migration/invasion, the epithelial–mesenchymal transition (EMT), cell proliferation and survival, immune responses, and circadian rhythms. A handful of other MNK substrates has been reported, two of which, heterogeneous nuclear ribonucleoprotein (hnRNP) A1 and polypyrimidine tract-binding protein-associated splicing factor (PSF) (Buxade et al. 2008), are nucleic acid-binding proteins. Their phosphorylation may affect gene expression, although it remains to be shown that they are indeed substrates for the MNKs in vivo.
Structural data for eIF4E reveal that Ser209 is located near the “channel” through which the mRNA exits the cap-binding site on eIF4E (Marcotrigiano et al. 1997; Matsuo et al. 1997); it is therefore not close to this binding site, or indeed to the region of the surface of eIF4E for 4E-BPs/eIF4G. Biophysical studies (Scheper et al. 2002; Zuberek et al. 2004; Slepenkov et al. 2006) indicate that phosphorylation of eIF4E decreases its affinity for a cap analog or short capped RNAs. The consequences for translational control of the reduced affinity and lower affinity of the eIF4E-cap interaction when eIF4E is phosphorylated remain unclear.
Reflecting both the ability of eIF4E overexpression to transform cells (Lazaris-Karatzas et al. 1990) and the fact that MNKs are activated by the oncogenic Ras/Raf/ERK pathway, initial studies focused on the role of the MNKs and phosphorylation of eIF4E in cancers. This work and other studies revealed requirements for the MNKs and/or Ser209 in eIF4E for cell transformation and tumor progression (Wendel et al. 2004, 2007; Furic et al. 2010; Ueda et al. 2010; Proud 2015). Key to understanding the mechanistic basis of these observations will be to identify which mRNAs’ translation is regulated by MNK-mediated phosphorylation of eIF4E. A modest number of candidates has been identified as being positively controlled in this way, including those for Mcl1 and other prosurvival proteins, certain cyclins, chemokines, matrix metalloproteinases, and vascular endothelial growth factor C (VEGF C), a proangiogenic factor (Wendel et al. 2007; Furic et al. 2010, and references therein; Proud 2015; Robichaud et al. 2018b).
PHOSPHORYLATION OF eIF4E CAN INDIRECTLY INFLUENCE THE TRANSCRIPTIONAL MACHINERY
Lim et al. (2013) discovered that eIF4E phosphorylation favors signaling through the Wnt/β-catenin pathway in blast crisis chronic myeloid leukemia. Of particular relevance here is that eIF4E and its phosphorylation promote translation of the mRNA for β-catenin and also, by an unknown mechanism, the translocation of β-catenin into the nucleus where it mediates Wnt regulation of gene transcription.
eIF4E phosphorylation regulates the translation of mRNAs for regulators of transcription in cells of the immune system. For example, the translation of the mRNA for IκBα (inhibitor of κBα), a negative regulator of the key transcription factor NF-κB, is reduced in cells that homozygously express the nonphosphorylatable eIF4ESer209Ala mutant (Herdy et al. 2012), allowing greater activation of NF-κB and enhanced production of interferon β. In other words, reduced eIF4E phosphorylation can enhance antiviral responses, a device that may be used in virus-infected cells to limit viral spread. However, the regulation of eIF4E phosphorylation during infection differs between viruses (Cuesta et al. 2000; Walsh and Mohr 2004; Stern-Ginossar et al. 2018). The control of the IκBα/NF-κB system by the MNKs may also be important in inflammatory responses (Joshi and Platanias 2014; Bao et al. 2017).
MNKs and/or eIF4E phosphorylation also promote the translation of positive regulators of transcription in immune cells (i.e., interferon regulatory factor [IRF]-8; Xu et al. 2012), which supports the induction of genes involved in host defenses against pathogen infection and the development of lymphoid and myeloid cell lineages. MNKs may also regulate cytokine expression through translational control of the expression of RANTES factor of late-activated T lymphocytes-1 (RFLAT-1), which in turn drives production of the cytokine RANTES/CCL5 (Nikolcheva et al. 2002).
Taken together, these and other data (Buxade et al. 2005; Su et al. 2015; Brace et al. 2017) suggest that the MNKs play multiple, and to some extent opposing, effects in immune and inflammatory responses by modulating the translation of mRNAs for transcriptional regulators.
ROLES FOR eIF4E PHOSPHORYLATION IN NEUROLOGICAL PROCESSES
In neurons of the suprachiasmatic nucleus (SCN), translational control of proteins such as period 1 and 2 (Per1/2) and cryptochrome (Cry) plays a key role in establishing circadian rhythms as part of sophisticated and highly conserved feedback mechanisms. Cao et al. (2015) showed that brief exposure to light activates MNK signaling and promotes the translation of the mRNAs for Per1 and Per2. Interestingly, 4E-BP1 regulates the translation of another mRNA, Vip1, in the SCN, where its product, vasoactive intestinal peptide, plays a key role in the SCN in maintaining circadian synchrony (Cao et al. 2013).
The protein Arc plays a crucial role in long-term synaptic plasticity and memory formation. Its levels are subject to rapid regulation via translational control mechanisms that require eIF4E phosphorylation (and therefore MNK1, the main MNK in neurons) as well as other events (Panja et al. 2009, 2014). Although brain-derived neurotrophic factor (BDNF) normally promotes eIF4E-eIF4G binding in synaptoneurosome preparations from cells of the dentate gyrus (part of the hippocampal formation that plays a key role in memory), it fails to do so when MNK1 has been knocked out (Panja and Bramham 2014). BDNF normally causes the release from eIF4E of another partner, cytoplasmic fragile X mental retardation protein (FMRP)-interacting protein 1 (CYFIP1), but does not do so when eIF4E cannot be phosphorylated (e.g., in MNK1-KO cells; see Panja and Bramham 2014). CYFIP1 also binds FMRP mutations, which are the most frequent cause of inherited autism (Hagerman et al. 2017). FMRP binds subsets of mRNAs to repress their translation; thus, the lack of eIF4G-eIF4E binding and the sustained eIF4E-CYFIP1-FMRP association likely work together to prevent engagement of the proper translational program in MNK1-KO cells. In primary neurons from MNK1-KO mice, BDNF fails to activate protein synthesis and, in particular, the synthesis of proteins involved in neurotransmission and synaptic plasticity is impaired. Many of the corresponding mRNAs interact with FMRP (Genheden et al. 2015).
ROLES FOR eIF4E PHOSPHORYLATION IN EMT AND TUMOR METASTASIS
FMRP also binds mRNAs for proteins that are involved in the EMT, a process that is critical for tumor cells to migrate, invade, and metastasize (Luca et al. 2013). Consistent with this, the phosphorylation of eIF4E augments the translation of the mRNAs for SNAIL and matrix metalloproteinase-3 (MMP3), proteins important for EMT and metastasis (Furic et al. 2010; Robichaud et al. 2015). MNKs also positively regulate cell migration and the translation of the mRNA for vimentin, a protein that is characteristic of mesenchymal cells (Beggs et al. 2015).
Two major questions yet to be addressed are (1) how does the phosphorylation of eIF4E, which occurs at a location in eIF4E adjacent only to the extreme 5′ nucleotides of eIF4E-associated mRNAs, regulate the translation of specific transcripts; and (2) which other mRNAs are translationally regulated through phosphorylation of eIF4E? Published studies have generally focused on only single mRNAs or small numbers of messages.
PHOSPHORYLATION OF OTHER TRANSLATION INITIATION FACTORS
In mammalian cells, several other initiation factors are phosphorylated, including eIF4G, eIF4B and several subunits of eIF3, some of which are modified at high stoichiometry, suggesting that at least some may play roles in the control of translation (Andaya et al. 2014). However, information on the consequences of the phosphorylation of these translation factors on protein synthesis remains sparse, and discussion here focuses on examples where there is some information on the functional consequences of phosphorylation of specific sites.
Mammalian eIF3 consists of 13 different subunits, some of which form a “core” complex (Merrick and Pavitt 2018). The phosphorylation of eIF3f at Ser46 by cyclin-dependent kinase 11 (CDK11p46) (Shi et al. 2009) is reported to enhance its association with the core complex. However, the consequences of this for translation are unclear. Phosphorylation of Ser183 in eIF3h is implicated in oncogenesis; overexpressing eIF4h is oncogenic, whereas expressing a Ser-to-Ala mutant of this phosphorylation site is not. In contrast, when Ser183 is mutated to Asp, eIF3H retains its oncogenic properties (Zhang et al. 2008).
The genomes of yeast (Saccharomyces cerevisiae) and mammals encode two different forms of eIF4G; here, the mammalian proteins are termed eIF4GI and eIF4GII (rather confusingly, and they are curated as eIF4G1 and eIF4G3). Human eIF4GI contains more than a dozen phosphorylated residues (Andaya et al. 2014), although the functional significance of most of them is not known. Protein kinase Cα phosphorylates eIF4GI at Thr1186 in the linker region between two of eIF4GI's domains and this phosphorylation promotes binding of eIF4GI to MNK1 even though the MNK1-binding site is at the far carboxyl terminus of eIF4GI (i.e., not adjacent to Thr1186; Dobrikov et al. 2011). This effect would be expected to promote the phosphorylation of eIF4E by MNK1. The linker region also contains phosphorylation sites that are stimulated by serum in a rapamycin-sensitive manner (Raught et al. 2000); their function is unclear.
eIF4GI is also phosphorylated on Ser896 by a stress-activated protein kinase termed p21-activated kinase 2 (Pak2), and this modification impairs cap-dependent translation (Ling et al. 2005).
eIF4GII undergoes phosphorylation during mitosis and eIF4GII hyperphosphorylation is associated with its reduced association with eIF4E (Pyronnet et al. 2001). This effect may contribute to the impairment of cap-dependent mRNA translation, which occurs during mitosis. In budding yeast, both isoforms of eIF4G are phosphorylated during glucose starvation by the protein kinase Ksp1 (Chang and Huh 2018). Interestingly, data obtained using a mutant of Tif4631 (encoding eIF4GI in yeast) indicate a role for phosphorylation of eIF4GI in modulating the stability of mRNAs encoding enzymes of glycolysis when cells are deprived of glucose (Chang and Huh 2018).
Further studies are clearly needed to determine the regulation and functional consequences of other known phosphorylation events in eIF4G isoforms, eIF3 subunits, and other translation factors.
PHOSPHORYLATION OF ELONGATION FACTORS
In mammalian cells, there are two main translation elongation factors, eEF1A and eEF2. A third multimeric factor, eEF1B, acts as a GEF for eEF1A, to regenerate active eEF1A•GTP, the form of this factor that is competent to bind aminoacyl-tRNAs and recruit them to the ribosomal A site (Dever et al. 2018). eEF2 is required for the translocation step of elongation where the ribosome moves by one codon relative to the mRNA and the tRNA carrying the nascent chain shifts from the A site to the P site.
eEF1A and all four subunits of eEF1B are subject to phosphorylation, with CDKs phosphorylating eIF1Bβ and δ (Sasikumar et al. 2012). However, the role of phosphorylation in regulating the activities of eEF1A and eEF1B and in modulating translation elongation remains to be established.
eEF2 is subject to phosphorylation at Thr56 (residue 57 of its encoded sequence, before the initial methionine is removed [Price et al. 1991; Redpath et al. 1993]). This impairs its binding to ribosomes and thus its activity, but in a nondominant way; phosphorylated eEF2 is inactive and removed from the active pool of this factor but it does not interfere with the function of nonphosphorylated eEF2. Thus, phosphorylation of eEF2 slows elongation, as shown by “transit time” measurements (Redpath et al. 1996a); this should conserve energy (the equivalent of at least four ATPs being needed to add each amino acid to a polypeptide) and, of course, amino acids.
eEF2K IS AN ATYPICAL PROTEIN KINASE AND IS SUBJECT TO DIVERSE REGULATORY INPUTS
Phosphorylation of Thr56 in eEF2 is catalyzed by the specific and unusual kinase, eEF2K (Ryazanov et al. 1988; Mitsui et al. 1993; Redpath et al. 1996b). Such phosphorylation is entirely lost in cells lacking eEF2K, showing it is the only enzyme involved (although AMPK had previously been reported to phosphorylate Thr56 [Hong-Brown et al. 2007]). AMPK can phosphorylate and activate eEF2K (Browne et al. 2004; Johanns et al. 2017), providing a mechanism by which low energy levels can slow down rates of elongation and thus reduce the consumption of ATP/GTP.
eEF2K is not a member of the main protein kinase superfamily comprising almost 500 genes but is one of only six α-kinases present in mammals (Ryazanov et al. 1997). The catalytic domains of α-kinases show no sequence homology with the main superfamily (Drennan and Ryazanov 2004), although there are similarities at the 3D structural level (Yamaguchi et al. 2001; Drennan and Ryazanov 2004; Ye et al. 2010; Galan et al. 2014). eEF2K activity is normally dependent on Ca2+-ions and calmodulin (CaM), and it is activated in contracting muscle where Ca2+-ions trigger contraction. Because contraction uses a great deal of ATP, coordinating eEF2K activity with contraction could help ensure that contraction is not affected by reduced ATP levels caused by protein synthesis. Consistent with this model, eEF2 phosphorylation rises very quickly in muscle after the onset of exercise (Rose et al. 2005).
eEF2K is also subjected to other regulatory inputs linked to nutrients, extracellular stimuli, or other cues (Fig. 3) (Liu and Proud 2016). In brief, eEF2K is switched off by signaling through mTORC1, which promotes the phosphorylation of eEF2K at several sites including one (Ser78) near its CaM-binding site, which inhibits activation by CaM (Browne and Proud 2004). Another inhibitory site, Ser366 (Wang et al. 2001), is a target for S6Ks (which are activated by mTORC1). Ser78 and also Ser396 are direct substrates for mTORC1, whereas other sites appear to be regulated indirectly by this pathway. Signaling through the MAP kinase pathway also inhibits eEF2K, via direct phosphorylation by ERK at Ser359 and by the RSKs at Ser366 (Fig. 3) (Wang et al. 2001, 2014).
The CaM-binding site also acts as a locus for regulation of eEF2K by hypoxia, because a neighboring proline is subject to oxygen-dependent hydroxylation that impairs eEF2K activity (Fig. 3) (Moore et al. 2015). This binding site contains several histidines, whose protonation at pH values associated with acidosis enhances CaM binding and thus eEF2K activity (Xie et al. 2015).
Taken together, these inputs allow eEF2K to be activated under conditions where energy requirements are high or energy is depleted, where amino acids are in short supply, or where levels of oxygen (needed to efficiently generate ATP in many cell types) are inadequate (Fig. 1). Inhibiting elongation rather than initiation when nutrients are scarce could have two advantages: (1) by preserving polysomes, rather than causing their disaggregation, it will allow translation to resume quickly once conditions improve; and (2) the continued association of mRNAs with polysomes may protect them against degradation. Conversely, under favorable conditions such as the presence of anabolic hormones or growth factors, eEF2K is switched off to remove a potential limitation on elongation rates.
WHAT IS THE ROLE OF CONTROL OF ELONGATION?
Early studies indicated that chemical inhibition of elongation could, paradoxically, enhance the synthesis of certain proteins (Fig. 1) (Walden and Thach 1986; Scheetz et al. 2000). eEF2K regulates the synthesis of certain proteins in neurons (Kenney et al. 2016), where it undergoes transient activation in response to stimulation of cells with a neurotransmitter receptor antagonist and controls the synthesis of microtubule-associated proteins. It remains unclear how slowing elongation promotes translation of some mRNAs; one suggestion is that for “weak” mRNAs (ones for which initiation is inefficient and thus limiting), slowing elongation results in initiation no longer being the limiting event, enabling these transcripts to compete better in recruiting ribosomes. Two aspects of this model are puzzling: (1) slow moving ribosomes near the beginning of the coding region would be expected to impede access for initiating ribosomes, and (2) it also requires an adequate supply of free (“spare”) ribosomes that could be recruited onto these mRNAs when elongation is slowed. Conversely, the levels of certain short-lived proteins, such as some antiapoptotic regulators, may drop when elongation is inhibited; this mechanism is suggested to explain the role of eEF2K in ensuring oocyte quality by promoting the apoptotic elimination of defective cells (Chu et al. 2014).
Several lines of evidence indicate that eEF2K helps to protect cancer cells against nutrient deprivation or other insults including some chemotherapeutic agents (Kenney et al. 2014). Mice lacking eEF2K or bearing a “kinase-dead” mutation in its gene are viable and fertile under vivarium conditions, indicating that eEF2K is not an essential gene (Chu et al. 2014; Moore et al. 2015). Thus, eEF2K appears to be an attractive target for novel anticancer agents. However, the picture is clouded by the observation that eEF2K actually restrains the development of colon tumors in a well-characterized mouse model (Faller et al. 2014), raising the prospect that inhibiting it might promote development of some tumors. This “double-edged sword” situation also applies to other pathways that are considered or used as targets in anticancer therapy, such as autophagy (White and DiPaola 2009) and indeed mTORC1 (Palm et al. 2015), inhibition of which can exert both pro- and antitumor effects depending on the stage in tumor development or the setting. Inhibition of eEF2K may be most effective for treating established solid tumors that are poorly vascularized. Reported connections between eEF2K and other diseases have recently been reviewed (Liu and Proud 2016).
PHOSPHORYLATION OF RIBOSOMAL PROTEIN S6
Several ribosomal proteins are subject to phosphorylation but by far the most attention has focused on S6; that said, the function of its phosphorylation is still not understood. As mentioned above, S6Ks phosphorylate several sites in the carboxyl terminus of S6, whereas RSKs act on two of them. Increased S6 phosphorylation is seen under conditions where cell growth and/or proliferation are enhanced, giving rise to the early idea that S6 phosphorylation promoted these outcomes. Indeed, mice lacking S6K1 (Shima et al. 1998) or fruit-flies lacking the Drosophila ortholog (Montagne et al. 1999) are smaller than control animals, revealing a role for S6K signaling in cell and organismal growth.
One possible explanation for these observations would be that phosphorylation of S6 promotes protein synthesis and/or the translation of specific mRNAs whose products are important for cell growth. One such class of mRNAs, once thought to be controlled by S6Ks, is the TOP mRNAs, which are well placed to control cell growth as they encode components of the translational machinery (Meyuhas and Kahan 2014). However, their association with active polysomes is still reduced by rapamycin in S6K-KO cells (Pende et al. 2004) and in cells where the phosphorylation sites in S6 have been mutated to nonphosphorylatable alanine residues (Ruvinsky et al. 2005).
Further studies on cells in which the phosphorylation sites in S6 are mutated revealed faster rates of cell division and protein synthesis, and smaller cell size, which was not further decreased by rapamycin (Ruvinsky et al. 2005). Thus, S6 phosphorylation appears to be a positive regulator of cell size but not of general protein synthesis or TOP mRNA translation. Intriguingly, pancreatic β cells from mice expressing nonphosphorylatable S6 are resistant to transformation by active Akt (which, among the effects, turns on mTORC1 signaling) and also to genotoxic or “proteotoxic” stress (Wittenberg et al. 2016). Probing the molecular mechanisms that underlie these effects should reveal much-needed insights into the function(s) of S6 phosphorylation.
OTHER S6K SUBSTRATES RELEVANT TO TRANSLATION
S6Ks phosphorylate a number of other substrates relevant to mRNA translation. These include eEF2K (Figs. 1 and 3) and eIF4B, which interacts with eIF4F and eIF4G and promotes the helicase activity of its eIF4A subunit (Fig. 2) (Raught et al. 2004; Shahbazian et al. 2006). The S6K site in eIF4B, Ser422, is also phosphorylated by the RSKs, downstream of MAP kinase (ERK) signaling (Shahbazian et al. 2006); it, and a second site Ser406, are also targets for the related kinase Akt (van Gorp et al. 2009). The effects of phosphorylation of eIF4B at this site, Ser422, are unclear, with one report concluding that phosphorylation impairs translation initiation (Raught et al. 2004) and a second that it enhances the association of eIF4B with eIF3 (Shahbazian et al. 2006). Mutation of either Ser406 or Ser422 to alanine abolished the ability of eIF4B to promote translation initiation when overexpressed (van Gorp et al. 2009), suggesting that both sites positively regulate eIF4B function (Shahbazian et al. 2006; van Gorp et al. 2009).
S6Ks also phosphorylate the programmed cell death 4 protein (PDCD4), promoting its release from eIF4A (and its degradation), thereby allowing eIF4A to bind eIF4G (Yang et al. 2003; Dorrello et al. 2006). The availability of small-molecule inhibitors of eIF4A has permitted studies on the impact of eIF4A function on the translation of specific mRNAs in mammalian cells (Rubio et al. 2014; Wolfe et al. 2014), revealing roles in tumor biology.
Both eIF4B (Figs. 1 and 2) and PDCD4 are also phosphorylated by RSKs (Shahbazian et al. 2006; Galan et al. 2014). S6Ks also regulate other proteins involved in RNA biology, including SKAR (S6K1 Aly/REF-like substrate), which plays a role in splicing and whose phosphorylation enhances the translation of newly made, spliced mRNAs, perhaps by recruiting S6K (Ma et al. 2008).
S6Ks also help promote the transcriptional program of ribosome biogenesis, an effect that seems to require phosphorylation of S6; again, the mechanism is not known (Chauvin et al. 2013). This role of the S6Ks in the function of the nucleolus, where ribosomes are assembled, may well be related to the observations linking S6Ks with cell size, as ribosome biogenesis is a key factor controlling cell growth (Rudra and Warner 2004).
WHAT DON’T WE KNOW?
Recent years have seen substantial progress is identifying the links between signaling pathways and the translational machinery, such as phosphorylation events. Further examples remain to be discovered. Our understanding of the impact of signaling pathways on the translation of specific mRNAs is greatly helped by the advent of new technologies such as ribosome profiling and stable-isotope labeling methods to monitor new protein synthesis. However, further work is clearly needed to elucidate the molecular mechanisms by which events such as the phosphorylation of eIF4E, or the control of elongation, modulate the synthesis of specific proteins. Conversely, it is unclear how the features of subsets of mRNAs, such as TISU elements, confer translational control. mTOR signaling modulates translation of many mRNAs that are not TOP messages; how does it do this?
One study has indicated that the MNKs negatively modulate the expression of eIF4E3 and that loss of MNK function is associated with enhanced eIF4E3-dependent translation of a subset of mRNAs (Landon et al. 2014). Although this is just a single report, it does raise the possibility that signaling pathways modulate the use of isoforms of translation factors. For example, the roles of eIF4E2, which has been reported to be involved both in repressing and promoting translation of different subsets of mRNAs (Cho et al. 2005; Uniacke et al. 2012; Chapat et al. 2017; Jafarnejad et al. 2018), and of eIF4E3 remain to be fully elucidated. What role do signaling events play in modulating their effects on gene expression?
Control of translation under different conditions almost certainly involves the combined effects of several signaling pathways and control of multiple translation factors. How do these inputs work together to modulate protein synthesis?
LARP1 appears to provide an example of an RNA-binding protein that is regulated by phosphorylation and controls the translation of a specific subset of mRNAs (TOP mRNAs). Do other RNA-binding proteins serve similar roles, linking signaling pathways to regulation of the translation of certain mRNAs?
Lastly, the signaling pathways that control the translational machinery are dysregulated in diseases such as cancer. How can modulating these pathways or the translational components they control be exploited to prevent, manage, or treat human disease?
ACKNOWLEDGMENTS
Research in the author’s laboratory is funded by the National Health & Medical Research Council and the South Australian Health & Medical Research Institute (SAHMRI). There are no competing interests to disclose. I thank Oded Meyuhas (Jerusalem) and Kirk Jensen, Xuemin Wang, and Jianling Xie (SAHMRI) for their helpful comments.
Footnotes
Editors: Michael B. Mathews, Nahum Sonenberg, and John W.B. Hershey
Additional Perspectives on Translation Mechanisms and Control available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Andaya A, Villa N, Jia W, Fraser CS, Leary JA. 2014. Phosphorylation stoichiometries of human eukaryotic initiation factors. Int J Mol Sci 15: 11523–11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P. 2007. The selectivity of protein kinase inhibitors: A further update. Biochem J 408: 297–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Wu X, Chen J, Hu X, Zeng F, Cheng J, Jin H, Lin X, Chen LF. 2017. Brd4 modulates the innate immune response through Mnk2-eIF4E pathway-dependent translational control of IκBα. Proc Natl Acad Sci 114: E3993–E4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs JE, Tian S, Jones GG, Xie J, Iadevaia V, Jenei V, Thomas GJ, Proud CG. 2015. The MAP kinase-interacting kinases regulate cell migration, vimentin expression and eIF4E/CYFIP1 binding. Biochem J 467: 63–76. [DOI] [PubMed] [Google Scholar]
- Ben-Sahra I, Manning BD. 2017. mTORC1 signaling and the metabolic control of cell growth. Curr Opin Cell Biol 45: 72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brace PT, Tezera LB, Bielecka MK, Mellows T, Garay D, Tian S, Rand L, Green J, Jogai S, Steele AJ, et al. 2017. Mycobacterium tuberculosis subverts negative regulatory pathways in human macrophages to drive immunopathology. PLoS Pathog 13: e1006367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MC, Gromeier M. 2017. MNK controls mTORC1:Substrate association through regulation of TELO2 binding with mTORC1. Cell Rep 18: 1444–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne GJ, Proud CG. 2004. A novel mTOR-regulated phosphorylation site in elongation factor 2 kinase modulates the activity of the kinase and its binding to calmodulin. Mol Cell Biol 24: 2986–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne GJ, Finn SG, Proud CG. 2004. Stimulation of the AMP-activated protein kinase leads to activation of eukaryotic elongation factor 2 kinase and to its phosphorylation at a novel site, serine 398. J Biol Chem 279: 12220–12231. [DOI] [PubMed] [Google Scholar]
- Buxade M, Parra JL, Rousseau S, Shpiro N, Marquez R, Morrice N, Bain J, Espel E, Proud CG. 2005. The Mnks are novel components in the control of TNFα biosynthesis and phosphorylate and regulate hnRNP A1. Immunity 23: 177–189. [DOI] [PubMed] [Google Scholar]
- Buxade M, Parra-Palau JL, Proud CG. 2008. The Mnks: MAP kinase-interacting kinases (MAP kinase signal-integrating kinases). Front Biosci 13: 5359–5373. [DOI] [PubMed] [Google Scholar]
- Cao R, Robinson B, Xu H, Gkogkas C, Khoutorsky A, Alain T, Yanagiya A, Nevarko T, Liu AC, Amir S, et al. 2013. Translational control of entrainment and synchrony of the suprachiasmatic circadian clock by mTOR/4E-BP1 signaling. Neuron 79: 712–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Gkogkas CG, de ZN, Blum ID, Yanagiya A, Tsukumo Y, Xu H, Lee C, Storch KF, Liu AC, et al. 2015. Light-regulated translational control of circadian behavior by eIF4E phosphorylation. Nat Neurosci 18: 855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carriere A, Romeo Y, Acosta-Jaquez HA, Moreau J, Bonneil E, Thibault P, Fingar DC, Roux PP. 2011. ERK1/2 phosphorylate Raptor to promote Ras-dependent activation of mTOR complex 1 (mTORC1). J Biol Chem 286: 567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Huh WK. 2018. Ksp1-dependent phosphorylation of eIF4G modulates post-transcriptional regulation of specific mRNAs under glucose deprivation conditions. Nucleic Acids Res 10.1093/nar/gky1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapat C, Jafarnejad SM, Matta-Camacho E, Hesketh GG, Gelbart IA, Attig J, Gkogkas CG, Alain T, Stern-Ginossar N, Fabian MR, et al. 2017. Cap-binding protein 4EHP effects translation silencing by microRNAs. Proc Natl Acad Sci 114: 5425–5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvin C, Koka V, Nouschi A, Mieulet V, Hoareau-Aveilla C, Dreazen A, Cagnard N, Carpentier W, Kiss T, Meyuhas O, et al. 2013. Ribosomal protein S6 kinase activity controls the ribosome biogenesis transcriptional program. Oncogene 33: 474–483. [DOI] [PubMed] [Google Scholar]
- Cho PF, Poulin F, Cho-Park YA, Cho-Park IB, Chicoine JD, Lasko P, Sonenberg N. 2005. A new paradigm for translational control: Inhibition via 5′-3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell 121: 411–423. [DOI] [PubMed] [Google Scholar]
- Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. 2008. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci 105: 17414–17419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu HP, Liao Y, Novak JS, Hu Z, Merkin JJ, Shymkiv Y, Braeckman BP, Dorovkov MV, Nguyen A, Clifford PM, et al. 2014. Germline quality control: eEF2K stands guard to eliminate defective oocytes. Dev Cell 28: 561–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta R, Xi Q, Schneider RJ. 2000. Adenovirus-specific translation by displacement of kinase Mnk1 from cap-initiation complex eIF4F. EMBO J 19: 3465–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. 2007. mTOR controls mitochondrial oxidative function through a YY1-PGC-1α transcriptional complex. Nature 450: 736–740. [DOI] [PubMed] [Google Scholar]
- Damgaard CK, Lykke-Andersen J. 2011. Translational coregulation of 5′TOP mRNAs by TIA-1 and TIAR. Genes Dev 25: 2057–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Dever TE, Dinman JD, Green R. 2018. Translation elongation and recoding in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble CC, Elis W, Menon S, Qin W, Klekota J, Asara JM, Finan PM, Kwiatkowski DJ, Murphy LO, Manning BD. 2012. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell 47: 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. 2006. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc Natl Acad Sci 103: 9482–9487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrikov M, Dobrikova E, Shveygert M, Gromeier M. 2011. Phosphorylation of eukaryotic translation initiation factor 4G1 (eIF4G1) by protein kinase Cα regulates eIF4G1 binding to Mnk1. Mol Cell Biol 31: 2947–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. 2006. S6K1- and βTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science 314: 467–471. [DOI] [PubMed] [Google Scholar]
- Drennan D, Ryazanov AG. 2004. α-Kinases: Analysis of the family and comparison with conventional protein kinases. Prog Biophys Mol Biol 85: 1–32. [DOI] [PubMed] [Google Scholar]
- Duncan R, Milburn SC, Hershey JW. 1987. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J Biol Chem 262: 380–388. [PubMed] [Google Scholar]
- Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, et al. 2010. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell 39: 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfakess R, Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Dikstein R. 2011. Unique translation initiation of mRNAs-containing TISU element. Nucleic Acids Res 39: 7598–7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faller WJ, Jackson TJ, Knight JR, Ridgway RA, Jamieson T, Karim SA, Jones C, Radulescu S, Huels DJ, Myant KB, et al. 2014. mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature 517: 497–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn A, Proud CG. 1995. Serine 209, not serine 53, is the major site of phosphorylation in initiation factor eIF-4E in serum-treated Chinese hamster ovary cells. J Biol Chem 270: 21684–21688. [DOI] [PubMed] [Google Scholar]
- Fonseca BD, Zakaria C, Jia JJ, Graber TE, Svitkin Y, Tahmasebi S, Healy D, Hoang HD, Jensen JM, Diao IT, et al. 2015. La-related protein 1 (LARP1) represses terminal oligopyrimidine (TOP) mRNA translation downstream of mTOR complex 1 (mTORC1). J Biol Chem 290: 15996–16020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furic L, Rong L, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, Petroulakis E, Robichaud N, Pollak M, Gaboury LA, et al. 2010. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc Natl Acad Sci 107: 14134–14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JA, Geraghty KM, Lavoie G, Kanshin E, Tcherkezian J, Calabrese V, Jeschke GR, Turk BE, Ballif BA, Blenis J, et al. 2014. Phosphoproteomic analysis identifies the tumor suppressor PDCD4 as a RSK substrate negatively regulated by 14–3-3. Proc Natl Acad Sci 111: E2918–E2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin V, Masvidal L, Hulea L, Gravel SP, Cargnello M, McLaughlan S, Cai Y, Balanathan P, Morita M, Rajakumar A, et al. 2016. nanoCAGE reveals 5′ UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Genome Res 26: 636–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genheden M, Kenney JW, Johnston HE, Manousopoulou A, Garbis SD, Proud CG. 2015. BDNF Stimulation of protein synthesis in cortical neurons requires the MAP kinase-interacting kinase MNK1. J Neurosci 35: 972–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A-C, Raught B, Sonenberg N. 1999. eIF4 translation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 68: 913–963. [DOI] [PubMed] [Google Scholar]
- Gomez E, Mohammad SS, Pavitt GD. 2002. Characterization of the minimal catalytic domain within eIF2B: The guanine-nucleotide exchange factor for translation initiation. EMBO J 21: 5292–5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordiyenko Y, Schmidt C, Jennings MD, Matak-Vinkovic D, Pavitt GD, Robinson CV. 2014. eIF2B is a decameric guanine nucleotide exchange factor with a γ2ɛ2 tetrameric core. Nat Commun 5: 3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman RJ, Berry-Kravis E, Hazlett HC, Bailey DB Jr, Moine H, Kooy RF, Tassone F, Gantois I, Sonenberg N, Mandel JL, et al. 2017. Fragile X syndrome. Nat Rev Dis Primers 3: 17065. [DOI] [PubMed] [Google Scholar]
- Harris TE, Chi A, Shabanowitz J, Hunt DF, Rhoads RE, Lawrence JC Jr. 2006. mTOR-dependent stimulation of the association of eIF4G and eIF3 by insulin. EMBO J 25: 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdy B, Jaramillo M, Svitkin YV, Rosenfeld AB, Kobayashi M, Walsh D, Alain T, Sean P, Robichaud N, Topisirovic I, et al. 2012. Translational control of the activation of transcription factor NF-κB and production of type I interferon by phosphorylation of the translation factor eIF4E. Nat Immunol 13: 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz MK, Ballif BA, Gygi SP, Blenis J. 2005. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 123: 569–580. [DOI] [PubMed] [Google Scholar]
- Hong-Brown LQ, Brown CR, Huber DS, Lang CH. 2007. Alcohol regulates eukaryotic elongation factor 2 phosphorylation via an AMP-activated protein kinase-dependent mechanism in C2C12 skeletal myocytes. J Biol Chem 282: 3702–3712. [DOI] [PubMed] [Google Scholar]
- Hong S, Freeberg MA, Han T, Kamath A, Yao Y, Fukuda T, Suzuki T, Kim JK, Inoki K. 2017. LARP1 functions as a molecular switch for mTORC1-mediated translation of an essential class of mRNAs. eLife 6: e25237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al. 2012. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485: 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Manning BD. 2008. The TSC1-TSC2 complex: A molecular switchboard controlling cell growth. Biochem J 412: 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y, Iadevaia V, Yao Z, Kelly I, Cosulich S, Guichard S, Foster LJ, Proud CG. 2012. Stable isotope-labelling analysis of the impact of inhibition of the mammalian target of rapamycin on protein synthesis. Biochem J 444: 141–151. [DOI] [PubMed] [Google Scholar]
- Iadevaia V, Liu R, Proud CG. 2014. mTORC1 signaling controls multiple steps in ribosome biogenesis. Semin Cell Dev Biol 36: 113–120. [DOI] [PubMed] [Google Scholar]
- *.Ingolia NT, Hussmann JA, Weissman JS. 2018. Ribosome profiling: Global views of translation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafarnejad SM, Chapat C, Matta-Camacho E, Gelbart IA, Hesketh GG, Arguello M, Garzia A, Kim SH, Attig J, Shapiro M, et al. 2018. Translational control of ERK signaling through miRNA/4EHP-directed silencing. eLife 7: e35034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanns M, Pyr Dit Ruys S, Houddane A, Vertommen D, Herinckx G, Hue L, Proud CG, Rider MH. 2017. Direct and indirect activation of eukaryotic elongation factor 2 kinase by AMP-activated protein kinase. Cell Signal 36: 212–221. [DOI] [PubMed] [Google Scholar]
- Joshi S, Platanias LC. 2014. Mnk kinase pathway: Cellular functions and biological outcomes. World J Biol Chem 5: 321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi B, Cai AL, Keiper BD, Minich WB, Mendez R, Beach CM, Stolarski R, Darzynkiewicz E, Rhoads RE. 1995. Phosphorylation of eukaryotic protein synthesis initiation factor 4E at serine 209. J Biol Chem 270: 14597–14603. [DOI] [PubMed] [Google Scholar]
- Joshi B, Cameron A, Jagus R. 2004. Characterization of mammalian eIF4E-family members. Eur J Biochem 271: 2189–2203. [DOI] [PubMed] [Google Scholar]
- Kang SA, Pacold ME, Cervantes CL, Lim D, Lou HJ, Ottina K, Gray NS, Turk BE, Yaffe MB, Sabatini DM. 2013. mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science 341: 1236566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney JW, Moore CE, Wang X, Proud CG. 2014. Eukaryotic elongation factor 2 kinase, an unusual enzyme with multiple roles. Adv Biol Regul 55: 15–27. [DOI] [PubMed] [Google Scholar]
- Kenney JW, Genheden M, Moon KM, Wang X, Foster LJ, Proud CG. 2016. Eukaryotic elongation factor 2 kinase regulates the synthesis of microtubule-related proteins in neurons. J Neurochem 136: 276–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konicek BW, Stephens JR, McNulty AM, Robichaud N, Peery RB, Dumstorf CA, Dowless MS, Iversen PW, Parsons S, Ellis KE, et al. 2011. Therapeutic inhibition of MAP kinase interacting kinase blocks eukaryotic initiation factor 4E phosphorylation and suppresses outgrowth of experimental lung metastases. Cancer Res 71: 1849–1857. [DOI] [PubMed] [Google Scholar]
- *.Kwan T, Thompson SR. 2018. Noncanonical translation initiation in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahr RM, Fonseca BD, Ciotti GE, Al-Ashtal HA, Jia JJ, Niklaus MR, Blagden SP, Alain T, Berman AJ. 2017. La-related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs. eLife 6: e24146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landon AL, Muniandy PA, Shetty AC, Lehrmann E, Volpon L, Houng S, Zhang Y, Dai B, Peroutka R, Mazan-Mamczarz K, et al. 2014. MNKs act as a regulatory switch for eIF4E1 and eIF4E3 driven mRNA translation in DLBCL. Nat Commun 5: 5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson O, Morita M, Topisirovic I, Alain T, Blouin MJ, Pollak M, Sonenberg N. 2012. Distinct perturbation of the translatome by the antidiabetic drug metformin. Proc Natl Acad Sci 109: 8977–8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaris-Karatzas A, Montine KS, Sonenberg N. 1990. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 345: 544–547. [DOI] [PubMed] [Google Scholar]
- Li JJ, Bickel PJ, Biggin MD. 2014. System wide analyses have underestimated protein abundances and the importance of transcription in mammals. Peer J 2: e270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Saw TY, Zhang M, Janes MR, Nacro K, Hill J, Lim AQ, Chang CT, Fruman DA, Rizzieri DA, et al. 2013. Targeting of the MNK-eIF4E axis in blast crisis chronic myeloid leukemia inhibits leukemia stem cell function. Proc Natl Acad Sci 110: E2298–E2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Hardie DG. 2017. AMPK: Sensing glucose as well as cellular energy status. Cell Metab 27: 299–313. [DOI] [PubMed] [Google Scholar]
- Ling J, Morley SJ, Traugh JA. 2005. Inhibition of cap-dependent translation via phosphorylation of eIF4G by protein kinase Pak2. EMBO J 24: 4094–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Proud CG. 2016. Eukaryotic elongation factor 2 kinase as a drug target in cancer, and in cardiovascular and neurodegenerative diseases. Acta Pharmacol Sin 37: 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca R, Averna M, Zalfa F, Vecchi M, Bianchi F, La FG, Del NF, Nardacci R, Bianchi M, Nuciforo P, et al. 2013. The fragile X protein binds mRNAs involved in cancer progression and modulates metastasis formation. EMBO Mol Med 5: 1523–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Yoon SO, Richardson CJ, Julich K, Blenis J. 2008. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell 133: 303–313. [DOI] [PubMed] [Google Scholar]
- Mader S, Lee H, Pause A, Sonenberg N. 1995. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4γ and the translational repressors 4E-binding proteins. Mol Cell Biol 15: 4990–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras A-C, Sonenberg N, Burley SK. 1997. Co-crystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89: 951–961. [DOI] [PubMed] [Google Scholar]
- Masvidal L, Hulea L, Furic L, Topisirovic I, Larsson O. 2017. mTOR-sensitive translation: Cleared fog reveals more trees. RNA Biol 14: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo H, Li HJ, McGuire AM, Fletcher CM, Gingras A-C, Sonenberg N, Wagner G. 1997. Structure of translation factor eIF4E bound to m(7)GDP and interaction with 4E-binding protein. Nat Struct Biol 4: 717–724. [DOI] [PubMed] [Google Scholar]
- *.Merrick WC, Pavitt GD. 2018. Protein synthesis initiation in eukaryotic cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyuhas O, Kahan T. 2014. The race to decipher the top secrets of TOP mRNAs. Biochim Biophys Acta 1849: 801–811. [DOI] [PubMed] [Google Scholar]
- Miloslavski R, Cohen E, Avraham A, Iluz Y, Hayouka Z, Kasir J, Mudhasani R, Jones SN, Cybulski N, Ruegg MA, et al. 2014. Oxygen sufficiency controls TOP mRNA translation via the TSC-Rheb-mTOR pathway in a 4E-BP-independent manner. J Mol Cell Biol 6: 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Brady M, Palfrey HC, Nairn AC. 1993. Purification and characterization of calmodulin-dependent protein kinase III from rabbit reticulocytes and rat pancreas. J Biol Chem 268: 13422–13433. [PubMed] [Google Scholar]
- Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, Thomas G. 1999. Drosophila S6 kinase: A regulator of cell size. Science 285: 2126–2129. [DOI] [PubMed] [Google Scholar]
- Moore CE, Mikolajek H, Regufe da MS, Wang X, Kenney JW, Werner JM, Proud CG. 2015. Elongation factor 2 kinase is regulated by proline hydroxylation and protects cells during hypoxia. Mol Cell Biol 35: 1788–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Gravel SP, Chenard V, Sikstrom K, Zheng L, Alain T, Gandin V, Avizonis D, Arguello M, Zakaria C, et al. 2013. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab 18: 698–711. [DOI] [PubMed] [Google Scholar]
- Nikolcheva T, Pyronnet S, Chou SY, Sonenberg N, Song A, Clayberger C, Krensky AM. 2002. A translational rheostat for RFLAT-1 regulates RANTES expression in T lymphocytes. J Clin Invest 110: 119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Loghlen A, Gonzalez VM, Jurado T, Salinas M, Martin ME. 2007. Characterization of the activity of human MAP kinase-interacting kinase Mnk1b. Biochim Biophys Acta 1773: 1416–1427. [DOI] [PubMed] [Google Scholar]
- Palm W, Park Y, Wright K, Pavlova NN, Tuveson DA, Thompson CB. 2015. The utilization of extracellular proteins as nutrients is suppressed by mTORC1. Cell 162: 259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panja D, Bramham CR. 2014. BDNF mechanisms in late LTP formation: A synthesis and breakdown. Neuropharmacology 76: 664–676. [DOI] [PubMed] [Google Scholar]
- Panja D, Dagyte G, Bidinosti M, Wibrand K, Kristiansen AM, Sonenberg N, Bramham CR. 2009. Novel translational control in Arc-dependent long term potentiation consolidation in vivo. J Biol Chem 284: 31498–31511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panja D, Kenney JW, D’Andrea L, Zalfa F, Vedeler A, Wibrand K, Fukunaga R, Bagni C, Proud CG, Bramham CR. 2014. Two-stage translational control of dentate gyrus LTP consolidation is mediated by sustained BDNF-TrkB signaling to MNK. Cell Rep 9: 1430–1445. [DOI] [PubMed] [Google Scholar]
- Parra-Palau JL, Scheper GC, Wilson ML, Proud CG. 2003. Features in the N and C termini of the MAPK-interacting kinase Mnk1 mediate its nucleocytoplasmic shuttling. J Biol Chem 278: 44197–44204. [DOI] [PubMed] [Google Scholar]
- Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, Mueller M, Fumagalli S, Kozma SC, Thomas G. 2004. S6K1−/−/S6K2−/− mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol 24: 3112–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe L, Vasseur JJ, Debart F, Thoreen CC. 2017. La-related protein 1 (LARP1) repression of TOP mRNA translation is mediated through its cap-binding domain and controlled by an adjacent regulatory region. Nucleic Acids Res 16: 1457–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price NT, Redpath NT, Severinov KV, Campbell DG, Russell JM, Proud CG. 1991. Identification of the phosphorylation sites in elongation factor-2 from rabbit reticulocytes. FEBS Lett 282: 253–258. [DOI] [PubMed] [Google Scholar]
- Proud CG. 2015. Mnks, eIF4E phosphorylation and cancer. Biochim Biophys Acta 1849: 766–773. [DOI] [PubMed] [Google Scholar]
- Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T, Sonenberg N. 1999. Human eukaryotic translation initiation factor 4G (eIF4G) recruits Mnk1 to phosphorylate eIF4E. EMBO J 18: 270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyronnet S, Dostie J, Sonenberg N. 2001. Suppression of cap-dependent translation in mitosis. Genes Dev 15: 2083–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau M, Ohlmann T, Morley SJ, Pain VM. 1996. A re-evaluation of the cap-binding translation factor, eIF4E, as a rate-limiting factor for initiation of translation in reticulocyte lysate. J Biol Chem 271: 8983–8990. [DOI] [PubMed] [Google Scholar]
- Raught B, Gingras AC, Gygi SP, Imataka H, Morino S, Gradi A, Aebersold R, Sonenberg N. 2000. Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. EMBO J 19: 434–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raught B, Peiretti F, Gingras AC, Livingstone M, Shahbazian D, Mayeur GL, Polakiewicz RD, Sonenberg N, Hershey JW. 2004. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J 23: 1761–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redpath NT, Price NT, Severinov KV, Proud CG. 1993. Regulation of elongation factor-2 by multisite phosphorylation. Eur J Biochem 213: 689–699. [DOI] [PubMed] [Google Scholar]
- Redpath NT, Foulstone EJ, Proud CG. 1996a. Regulation of translation elongation factor-2 by insulin via a rapamycin-sensitive signalling pathway. EMBO J 15: 2291–2297. [PMC free article] [PubMed] [Google Scholar]
- Redpath NT, Price NT, Proud CG. 1996b. Cloning and expression of cDNA encoding protein synthesis elongation factor-2 kinase. J Biol Chem 271: 17547–17554. [PubMed] [Google Scholar]
- Robichaud N, del Rincon SV, Huor B, Alain T, Petruccelli LA, Hearnden J, Goncalves C, Grotegut S, Spruck CH, Furic L, et al. 2015. Phosphorylation of eIF4E promotes EMT and metastasis via translational control of SNAIL and MMP-3. Oncogene 34: 2032–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Robichaud N, Sonenberg N, Ruggero D, Schneider RJ. 2018a. Translational control in cancer. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robichaud N, Hsu BE, Istomine R, Alvarez F, Blagih J, Ma EH, Morales SV, Dai DL, Li G, Souleimanova M, et al. 2018b. Translational control in the tumor microenvironment promotes lung metastasis: Phosphorylation of eIF4E in neutrophils. Proc Natl Acad Sci 115: E2202–E2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AJ, Broholm C, Kiillerich K, Finn SG, Proud CG, Rider MH, Richter EA, Kiens B. 2005. Exercise rapidly increases eukaryotic elongation factor 2 phosphorylation in skeletal muscle of men. J Physiol 569: 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio CA, Weisburd B, Holderfield M, Arias C, Fang E, DeRisi JL, Fanidi A. 2014. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol 15: 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra D, Warner JR. 2004. What better measure than ribosome synthesis? Genes Dev 18: 2431–2436. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T, Dor Y, Zisman P, Meyuhas O. 2005. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev 19: 2199–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryazanov AG, Natapov PG, Shestakova EA, Severin FF, Spirin AS. 1988. Phosphorylation of the elongation factor 2: The fifth Ca2+/calmodulin-dependent system of protein phosphorylation. Biochimie 70: 619–626. [DOI] [PubMed] [Google Scholar]
- Ryazanov AG, Ward MD, Mendola CE, Pavur KS, Dorovkov MV, Wiedmann M, Erdjument-Bromage H, Tempst P, Parmer TG, Prostko CR, et al. 1997. Identification of a new class of protein kinases represented by eukaryotic elongation factor-2 kinase. Proc Natl Acad Sci 94: 4884–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov dD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. 2006. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell 22: 159–168. [DOI] [PubMed] [Google Scholar]
- Sasikumar AN, Perez WB, Kinzy TG. 2012. The many roles of the eukaryotic elongation factor 1 complex. Wiley Interdiscip Rev RNA 3: 543–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton RA, Sabatini DM. 2017. mTOR signaling in growth, metabolism, and disease. Cell 169: 361–371. [DOI] [PubMed] [Google Scholar]
- Scheetz AJ, Nairn AC, Constantine-Paton M. 2000. NMDA-mediated control of protein synthesis at developing synapses. Nat Neurosci 3: 1–7. [DOI] [PubMed] [Google Scholar]
- Scheper GC, van Kollenburg B, Hu J, Luo Y, Goss DJ, Proud CG. 2002. Phosphorylation of eukaryotic initiation factor 4E markedly reduces its affinity for capped mRNA. J Biol Chem 277: 3303–3309. [DOI] [PubMed] [Google Scholar]
- Scheper GC, Parra JL, Wilson M, van KB, Vertegaal AC, Han ZG, Proud CG. 2003. The N and C termini of the splice variants of the human mitogen-activated protein kinase-interacting kinase Mnk2 determine activity and localization. Mol Cell Biol 23: 5692–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473: 337–342. [DOI] [PubMed] [Google Scholar]
- Shahbazian D, Roux PP, Mieulet V, Cohen MS, Raught B, Taunton J, Hershey JW, Blenis J, Pende M, Sonenberg N. 2006. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J 25: 2781–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shama S, Avni D, Frederickson RM, Sonenberg N, Meyuhas O. 1995. Overexpression of initiation factor eIF-4E does not relieve the translational repression of ribosomal protein mRNAs in quiescent cells. Gene Expr 4: 241–252. [PMC free article] [PubMed] [Google Scholar]
- Shi J, Hershey JW, Nelson MA. 2009. Phosphorylation of the eukaryotic initiation factor 3f by cyclin-dependent kinase 11 during apoptosis. FEBS Lett 583: 971–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima H, Pende M, Chen Y, Fumagalli S, Thomas G, Kozma SC. 1998. Disruption of the p70S6k/p85S6k gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J 17: 6649–6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Tamarkin-Ben-Harush A, Viollet B, Dikstein R. 2015. Translational tolerance of mitochondrial genes to metabolic energy stress involves TISU and eIF1-eIF4GI cooperation in start codon selection. Cell Metab 21: 479–492. [DOI] [PubMed] [Google Scholar]
- Slepenkov SV, Darzynkiewicz E, Rhoads RE. 2006. Stopped-flow kinetic analysis of eIF4E and phosphorylated eIF4E binding to cap analogs and capped oligoribonucleotides: Evidence for a one-step binding mechanism. J Biol Chem 281: 14927–14938. [DOI] [PubMed] [Google Scholar]
- Stead RL, Proud CG. 2013. Rapamycin enhances eIF4E phosphorylation by activating MAP kinase-interacting kinase 2a (Mnk2a). FEBS Lett 587: 2623–2628. [DOI] [PubMed] [Google Scholar]
- *.Stern-Ginossar N, Thompson SR, Mathews MB, Mohr I. 2018. Translational control in virus-infected cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Yu Y, Zhong Y, Giannopoulou EG, Hu X, Liu H, Cross JR, Ratsch G, Rice CM, Ivashkiv LB. 2015. Interferon-γ regulates cellular metabolism and mRNA translation to potentiate macrophage activation. Nat Immunol 16: 838–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahmasebi S, Khoutorsky A, Mathews MB, Sonenberg N. 2018. Translation deregulation in human disease. Nature Rev Mol Cell Biol (in press). [DOI] [PubMed] [Google Scholar]
- Tcherkezian J, Cargnello M, Romeo Y, Huttlin EL, Lavoie G, Gygi SP, Roux PP. 2014. Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5′TOP mRNA translation. Genes Dev 28: 357–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. 2012. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485: 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truitt ML, Conn CS, Shi Z, Pang X, Tokuyasu T, Coady AM, Seo Y, Barna M, Ruggero D. 2015. Differential requirements for eIF4E dose in normal development and cancer. Cell 162: 59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp C, Knauf U, Brauchle M, Zurini M, Ramage P, Glueck D, New L, Han J, Gram H. 2000. Phosphorylation of eIF-4E on Ser 209 in response to mitogenic and inflammatory stimuli is faithfully detected by specific antibodies. Mol Cell Biol Res Commun 3: 205–211. [DOI] [PubMed] [Google Scholar]
- Ueda T, Watanabe-Fukunaga R, Fukuyama H, Nagata S, Fukunaga R. 2004. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol Cell Biol 24: 6539–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Sasaki M, Elia AJ, Chio II, Hamada K, Fukunaga R, Mak TW. 2010. Combined deficiency for MAP kinase-interacting kinase 1 and 2 (Mnk1 and Mnk2) delays tumor development. Proc Natl Acad Sci 107: 13984–13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uniacke J, Holterman CE, Lachance G, Franovic A, Jacob MD, Fabian MR, Payette J, Holcik M, Pause A, Lee S. 2012. An oxygen-regulated switch in the protein synthesis machinery. Nature 486: 126–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gorp AG, van der Vos KE, Brenkman AB, Bremer A, van den Broek N, Zwartkruis F, Hershey JW, Burgering BM, Calkhoven CF, Coffer PJ. 2009. AGC kinases regulate phosphorylation and activation of eukaryotic translation initiation factor 4B. Oncogene 28: 95–106. [DOI] [PubMed] [Google Scholar]
- von der Haar T, McCarthy JE. 2002. Intracellular translation initiation factor levels in Saccharomyces cerevisiae and their role in cap-complex function. Mol Microbiol 46: 531–544. [DOI] [PubMed] [Google Scholar]
- Walden WE, Thach RE. 1986. Translational control of gene expression in a normal fibroblast. Characterization of a subclass of mRNAs with unusual kinetic properties. Biochemistry 25: 2033–2041. [DOI] [PubMed] [Google Scholar]
- Walsh D, Mohr I. 2004. Phosphorylation of eIF4E by Mnk-1 enhances HSV-1 translation and replication in quiescent cells. Genes Dev 18: 660–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Flynn A, Waskiewicz AJ, Webb BLJ, Vries RG, Baines IA, Cooper J, Proud CG. 1998. The phosphorylation of eukaryotic initiation factor eIF4E in response to phorbol esters, cell stresses and cytokines is mediated by distinct MAP kinase pathways. J Biol Chem 273: 9373–9377. [DOI] [PubMed] [Google Scholar]
- Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG. 2001. Regulation of elongation factor 2 kinase by p90RSK1 and p70 S6 kinase. EMBO J 20: 4370–4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Regufe da MS, Liu R, Moore CE, Xie J, Lanucara F, Agarwala U, Pyr Dit RS, Vertommen D, Rider MH, et al. 2014. Eukaryotic elongation factor 2 kinase activity is controlled by multiple inputs from oncogenic signaling. Mol Cell Biol 34: 4088–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz AJ, Flynn A, Proud CG, Cooper JA. 1997. Mitogen-activated kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J 16: 1909–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Wek RC. 2018. Role of elF2α kinases in translational control and adaptation to cellular stresses. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh GI, Miller CM, Loughlin AJ, Price NT, Proud CG. 1998. Regulation of eukaryotic initiation factor eIF2B: Glycogen synthase kinase-3 phosphorylates a conserved serine which undergoes dephosphorylation in response to insulin. FEBS Lett 421: 125–130. [DOI] [PubMed] [Google Scholar]
- Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, Cordon-Cardo C, Pelletier J, Lowe SW. 2004. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature 428: 332–337. [DOI] [PubMed] [Google Scholar]
- Wendel HG, Silva RL, Malina A, Mills JR, Zhu H, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Teruya-Feldstein J, Pelletier J, et al. 2007. Dissecting eIF4E action in tumorigenesis. Genes Dev 21: 3232–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E, DiPaola RS. 2009. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res 15: 5308–5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DD, Price NT, Loughlin AJ, Proud CG. 2001. Characterisation of the initiation factor eIF2B complex from mammalian cells as a GDP-dissociation stimulator protein. J Biol Chem 276: 24697–24703. [DOI] [PubMed] [Google Scholar]
- Wittenberg AD, Azar S, Klochendler A, Stolovich-Rain M, Avraham S, Birnbaum L, Binder Gallimidi A, Katz M, Dor Y, Meyuhas O. 2016. Phosphorylated ribosomal protein S6 is required for Akt-driven hyperplasia and malignant transformation, but not for hypertrophy, aneuploidy and hyperfunction of pancreatic β-cells. PLoS ONE 11: e0149995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AL, Singh K, Zhong Y, Drewe P, Rajasekhar VK, Sanghvi VR, Mavrakis KJ, Jiang M, Roderick JE, Van der Meulen J, et al. 2014. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature 513: 65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson RL, Sabatini DM. 2017. The dawn of the age of amino acid sensors for the mTORC1 pathway. Cell Metab 26: 301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortham NC, Martinez M, Gordiyenko Y, Robinson CV, Proud CG. 2014. Analysis of the subunit organization of the eIF2B complex reveals new insights into its structure and regulation. FASEB J 28: 2225–2237. [DOI] [PubMed] [Google Scholar]
- Xie J, Mikolajek H, Pigott CR, Hooper KJ, Mellows T, Moore CE, Mohammed H, Werner JM, Thomas GJ, Proud CG. 2015. Molecular mechanism for the control of eukaryotic elongation factor 2 kinase by pH: Role in cancer cell survival. Mol Cell Biol 35: 1805–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Zhu J, Smith S, Foldi J, Zhao B, Chung AY, Outtz H, Kitajewski J, Shi C, Weber S, et al. 2012. Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nat Immunol 13: 642–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Matsushita M, Nairn AC, Kuriyan J. 2001. Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity. Mol Cell 7: 1047–1057. [DOI] [PubMed] [Google Scholar]
- Yang HS, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S, Lockett SJ, Sonenberg N, Colburn NH. 2003. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol 23: 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Rudge DG, Koos JD, Vaidialingam B, Yang HJ, Pavletich NP. 2013. mTOR kinase structure, mechanism and regulation. Nature 497: 217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Crawley SW, Yang Y, Cote GP, Jia Z. 2010. Crystal structure of the α-kinase domain of Dictyostelium myosin heavy chain kinase A. Sci Signal 3: ra17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Smit-McBride Z, Pan X, Rheinhardt J, Hershey JW. 2008. An oncogenic role for the phosphorylated h-subunit of human translation initiation factor eIF3. J Biol Chem 283: 24047–24060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuberek J, Jemielity J, Jablonowska A, Stepinski J, Dadlez M, Stolarski R, Darzynkiewicz E. 2004. Influence of electric charge variation at residues 209 and 159 on the interaction of eIF4E with the mRNA 5′ terminus. Biochemistry 43: 5370–5379. [DOI] [PubMed] [Google Scholar]