Abstract
The translation of messenger RNAs (mRNAs) into proteins is a key event in the regulation of gene expression. This is especially true in the cancer setting, as many oncogenes and transforming events are regulated at this level. Cancer-promoting factors that are translationally regulated include cyclins, antiapoptotic factors, proangiogenic factors, regulators of cell metabolism, prometastatic factors, immune modulators, and proteins involved in DNA repair. This review discusses the diverse means by which cancer cells deregulate and reprogram translation, and the resulting oncogenic impacts, providing insights into the complexity of translational control in cancer and its targeting for cancer therapy.
ON THE IMPORTANCE OF TRANSLATIONAL CONTROL IN CANCER
Considerable resources are dedicated to messenger RNA (mRNA) translation in normal proliferating cells. Up to 20% of cellular energy is used for protein synthesis, as compared with 15% for transcription and DNA replication, and 20% for various cation pumps (Buttgereit and Brand 1995). Moreover, the majority of transcription is directed to the synthesis of ribosomal RNA (rRNA) and mRNAs encoding ribosomal proteins, further increasing the dedication of cellular energy to mRNA translation, making it the most energy-demanding cellular process (Rolfe and Brown 1997). The rapid and continuous proliferation of highly malignant cancers requires continuous protein synthesis and increased ribosome content, further increasing the energy consumption directed to protein synthesis (Silvera et al. 2010). Most tumor cells are under physiological stresses such as hypoxia and nutritional deprivation that down-regulate mRNA translation in normal cells but become uncoupled from regulation as a part of the transformation process, further stressing the cell.
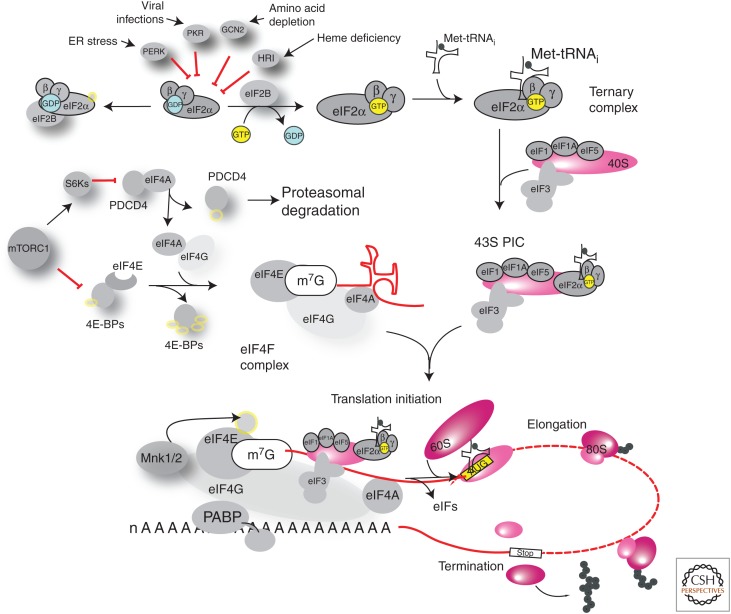
In this article, we review how cancer cells hijack the translational machinery for their sustained proliferation, survival, and metastasis (spread) to distant tissue sites, and how the changes in activity and expression of distinct translation factors confer cancer-specific translation of mRNAs. A brief overview of the scanning mechanism of translation initiation and the factors involved in this process is presented in Figure 1, as discussed at length elsewhere (Kwan and Thompson 2018; Merrick and Pavitt 2018).
Figure 1.
Overview of translation initiation. Initiation proceeds via a scanning mechanism, whereby the 40S ribosomal subunit is recruited to the 5′ extremity of the messenger RNA (mRNA) and scans the 5′ untranslated region (UTR) of the mRNA toward its 3′ end. When the anticodon of the initiator methionyl-transfer RNA (tRNA) (Met-tRNAi) base-pairs with the start codon, the 60S subunit is recruited and elongation begins with the sequential addition of amino acids until a stop codon is reached and termination occurs. (Top) Formation of the ternary complex (TC): eukaryotic initiation factor (eIF)2—composed of α, β, and γ subunits—GTP and Met-tRNAi. eIF2α can be phosphorylated by protein kinase R (PKR), PERK, GCN2, or HRI, responding to different stresses such as double-stranded RNA, misfolded proteins, amino acid deficiency, and heme deficiency, respectively. Phosphorylation of eIF2α leads to stabilization of the GDP-loaded complex with the guanine nucleotide exchange factor eIF2B and reduced cycling to the active, GTP-bound TC, resulting in inhibition of global protein synthesis and active translation of upstream open reading frame (uORF)-containing mRNAs such as ATF4. TCs associate with the 40S ribosome and other initiation factors, forming the preinitiation complex (PIC). (Middle) The eIF4F complex consists of the cap-binding protein eIF4E, the scaffolding protein eIF4G, and the eIF4A helicase. The eIF4E-binding proteins (4E-BP1/2/3) sequester eIF4E and prevent its binding to eIF4G. Similarly, PDCD4 sequesters eIF4A. Phosphorylation downstream from mammalian target of rapamycin (mTOR) alleviates the inhibitory activity of PDCD4 and 4E-BPs, allowing for eIF4F complex formation, recruitment of the 43S PIC and the initiation of translation. In addition, eIF4E can be phosphorylated by the MNKs.
MECHANISMS OF DEREGULATED AND SELECTIVE mRNA TRANSLATION IN CANCER
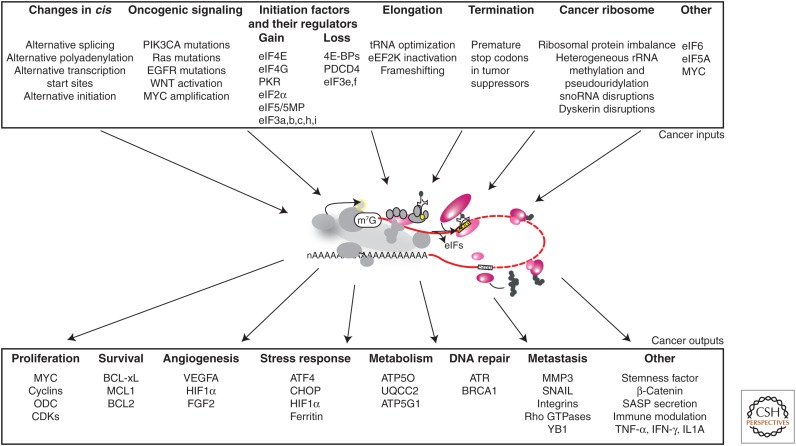
Mathematical modeling has predicted that, given constant rates of ribosome elongation on mRNAs and limited translation initiation factors, the translation of specific mRNAs for which ribosome recruitment is inefficient will be disproportionately affected by changes in the level or activity of initiation factors (Lodish 1974). In principle, this basic model helps explain a number of aspects of translational deregulation in cancer cells discussed here, including how changes in factors involved in the translation of most mRNAs can yield specific advantages. This model predicts that cancer cells are particularly well equipped to promote angiogenesis, survival, and proliferation, even in times of high physiological stress. This can be achieved through a variety of complex molecular alterations that increase selective translation of poorly translated mRNAs, including increased expression or availability of certain translation initiation factors, and increased activity of signaling pathways regulating them (reviewed in Silvera et al. 2010; Ruggero 2013; Bhat et al. 2015; Truitt and Ruggero 2016; de la Parra et al. 2017). Some of the many ways by which this can be achieved are described below and summarized in Figure 2.
Figure 2.
Cancer inputs and outputs. Summarized view of the oncogenic lesions feeding into the translational machinery (cancer inputs, top) and of the resulting advantages conferred by aberrant translation, with examples of regulated mRNAs (cancer outputs, bottom). (Center) Schematic of translation, as shown in Figure 1.
Gain/Loss of Initiation Factors
Aberrant expression of translation initiation factors was the first mechanism to be identified by which cancer cells deregulate translation, shown originally by the ability of overexpressed eukaryotic initiation factor (eIF)4E to transform NIH 3T3 cells in vitro (Lazaris-Karatzas et al. 1990). Several initiation factors were subsequently found to be overexpressed in human cancers, as discussed below and shown in Figure 2.
eIF4F Complex Formation
Ribosomes are recruited to the 5′ end of the mRNA via the eIF4F complex, which consists of eIF4E, eIF4G, and eIF4A (Fig. 1). The enzymatic component, eIF4A, provides the helicase activity required to unwind secondary structures present in mRNA 5′ untranslated regions (UTRs), a process that is vastly enhanced by binding to eIF4G and eIF4E (Feoktistova et al. 2013). Considering that oncogenic mRNAs tend to have long and stable 5′UTRs, they are particularly sensitive to the activity of eIF4A and the formation of eIF4F (Kozak 1987; Chu and Pelletier 2015; Gandin et al. 2016). All three eIF4F subunits can be deregulated in cancer cells, their genomic loci have all been shown to be amplified in human tumors, and they are all targets of the MYC oncoprotein (Silvera et al. 2010; Ruggero 2013; Bhat et al. 2015). eIF4E and eIF4G act as classical oncogenes, their overexpression resulting in transformation in vitro (in cell culture) and in vivo (in animals) (Lazaris-Karatzas et al. 1990; Fukuchi-Shimogori et al. 1997; Ruggero et al. 2004; Silvera et al. 2009). Importantly, it was recently shown by using a mouse model of haploinsufficient levels of eIF4E (loss of one allele) that a 50% reduction of eIF4E levels does not limit general protein synthesis and embryonic development in the eIF4E+/− mouse, which is quite remarkably viable. However, eIF4E+/− cells and mice are highly resistant to cellular transformation and tumorigenicity, even when driven by a powerful oncogene such as Hras-V12 (Truitt et al. 2015). eIF4E overexpression in cancer cells is therefore highly specific for oncogenic transformation.
Translation initiation in cancer cells can also be regulated by phosphorylation of eIF4F components. For example, eIF4E phosphorylation by MNK1 and MNK2 kinases has been shown to promote tumor development and dissemination (Topisirovic et al. 2004; Wendel et al. 2007; Furic et al. 2010; Robichaud et al. 2015; Proud 2018), and is elevated in human lung, breast, and prostate cancers, among others (Fan et al. 2009; Graff et al. 2009; Ramon y Cajal et al. 2014). Multiple phosphorylation sites exist on eIF4G, not all of which have known functions, although Ser1186 phosphorylation is thought to regulate MNK recruitment, and thus phosphorylation of eIF4E (Raught et al. 2000; Dobrikov et al. 2011). MNK-mediated phosphorylation of eIF4E has also been shown to be involved in translational reprogramming that drives resistance to tamoxifen in estrogen-receptor-positive breast cancer (Geter et al. 2017). The mechanism underlying translational regulation by eIF4E phosphorylation is poorly understood, but is thought to involve initiation factor recycling (Scheper and Proud 2002; Proud 2018). This hypothesis is based on the requirement of eIF4G and eIF3 for MNK recruitment and the decreased affinity of eIF4E for the cap resulting from its phosphorylation (Pyronnet et al. 1999; Scheper et al. 2002; Slepenkov et al. 2006; Walsh and Mohr 2014).
Another regulatory mechanism involves sequestration of initiation factors to prevent eIF4F complex formation. Thus, eIF4A can be sequestered by the tumor suppressor programmed cell death 4 (PDCD4), the loss of which is associated with cancer cell invasion and poor patient survival in some cancers (Proud 2018). The exact role of PDCD4 in cancer remains to be established and better characterized (Yang et al. 2003, 2004; Wang et al. 2008; Meric-Bernstam et al. 2012; Modelska et al. 2015). Using a similar mechanism, the 4E-BPs, which compete with eIF4G for binding to eIF4E, are thought to act as tumor suppressors by inhibiting cap-dependent translation (Alain et al. 2012). 4E-BP expression can be lost, as in pancreatic cancer, or its function impaired by inhibitory phosphorylation (Martineau et al. 2014). In contrast, 4E-BP expression is increased in the setting of stage III nonmetastatic esophageal, breast, and prostate cancers, in which it is proposed to oppose metastasis, but lead to the development of large locally advanced tumors (Salehi and Mashayekhi 2006; Braunstein et al. 2007; Coleman et al. 2009; Graff et al. 2009). Thus, the role of the 4E-BPs in cancer may be more complex than originally thought.
Ternary Complex Formation
The ternary complex (TC) is composed of eIF2, GTP, and the initiator methionine transfer RNA (tRNA) (Fig. 1). Deregulated TC formation in cancer cells is a complex issue that has led to different, occasionally conflicting findings regarding the role of eIF2α phosphorylation (Koromilas 2015). On the one hand, it is generally thought that increased eIF2α phosphorylation grants cancer cells a heightened ability to respond to stress conditions encountered along the path to malignancy, by promoting the translation of upstream open reading frame (uORF)-containing stress-response mRNAs such as ATF4 (Robichaud and Sonenberg 2017; Sendoel et al. 2017; Wek 2018). Accordingly, overexpression of eIF2α or one of its kinases, protein kinase R (PKR), has been shown to promote transformation in some contexts, although the mechanism remains unclear (Wang et al. 1999; Rosenwald et al. 2001; Kim et al. 2002; Rosenwald et al. 2003; Ye et al. 2010). On the other hand, long-term eIF2α phosphorylation promotes apoptosis, and has prompted research into the development of cancer therapies that promote the activity of eIF2α kinases or the inhibition of eIF2α phosphatases (Schewe and Aguirre-Ghiso 2009; Denoyelle et al. 2012; Hamamura et al. 2014). These results indicate that the outcome of eIF2α phosphorylation in cancer cells is highly context specific, perhaps related to disease site or underlying driver mutations, and may change over time (Silvera et al. 2010).
Additional ways to modulate TC activity in cancer cells have been less explored but include overexpression of eIF5 or its mimic proteins (MPs), 5MP1 and 5MP2. When present in excess, these proteins can bind to eIF2 and sequester it from the 40S ribosome (Singh et al. 2006, 2011). Similar to eIF2α phosphorylation, eIF2 binding by eIF5 or the 5MPs reduces global protein synthesis but enhances translation of uORF-containing mRNAs, including ATF4 (Kozel et al. 2016). This mechanism appears to be important for the malignant properties of some cancer types such as fibrosarcoma and salivary mucoepidermoid carcinoma (Li et al. 2009).
eIF3, Connecting eIF4F and Preinitiation Complexes
eIF3 is a multisubunit complex that binds directly to eIF4G, bridging it to the preinitiation complex (PIC) (Fig. 1), thus connecting mRNAs with the 40S ribosomal subunit and allowing scanning to occur (Hinnebusch et al. 2016; Merrick and Pavitt 2018). Increased eIF3 levels should promote bridging mRNA to the PIC, and therefore increase the rate of translation initiation. This appears to occur when the a, b, and c subunits of eIF3 are overexpressed, resulting in increased levels of whole eIF3 complex, increased global protein synthesis, and increased translation of oncogenic transcripts (Zhang et al. 2007). However, studies on eIF3 present a more complex picture. Indeed, when overexpressed or silenced in immortalized cells, certain individual subunits of eIF3 display oncogenic properties, while other subunits behave oppositely, as tumor suppressors (Hershey 2015). Such confusing results may arise from the nontranslational roles of certain eIF3 subunits, such as eIF3a, which has been reported to bind to components of the cytoskeleton (MacDonald et al. 1999; Lin et al. 2001), or eIF3f and eIF3i, which have been proposed to regulate signal transduction pathways (Wang et al. 2013; Lee et al. 2016). New roles for eIF3 in translation have also been reported. These include binding to mRNA structures in the 5′UTR of such cancer-relevant mRNAs as c-Jun and Btg1 (Lee et al. 2015), or even directly to the cap of the c-Jun mRNA (Lee et al. 2016), opening new avenues of research on eIF3-dependent translation in cancer.
Translation Elongation and Termination
Although much of the scientific literature has been focused on translation initiation, an understanding of oncogenic changes in elongation and termination is emerging as well. For example, a dominant role for the loss of inhibitory regulation of elongation via eukaryotic elongation factor 2 phosphorylation by its kinase (eEF2K) has been shown for intestinal tumor formation (Faller et al. 2015; Proud 2018). Furthermore, the increased availability of specific tRNA isoaccepting species in cancer cells appears to play a role in tumorigenesis (Gingold et al. 2014). Indeed, the speed of amino acid incorporation during the elongation phase is dependent on the availability of the corresponding charged tRNA (Novoa and Ribas de Pouplana 2012). Several studies have reported distinct translation programs in which proliferating undifferentiated cells and cancer cells express tRNAs optimized to correspond to the codon usage of pro-proliferative mRNAs (Pavon-Eternod et al. 2009; Gingold et al. 2014; Topisirovic and Sonenberg 2014). Hence, in cancer cells, the repertoire of available tRNAs is thought to be reprogrammed such that the species required for the translation of oncogenic mRNAs are present at sufficient levels. In addition, elongation can be deregulated in cancer via programmed -1 ribosomal frameshifting (-1 RPF), a process by which sequence elements force elongating ribosomes back by one base, leading to frameshifts, premature stop codons, and nonsense-mediated mRNA decay (NMD) (Dever et al. 2018; Karousis and Mühlemann 2018). This mechanism may explain the oncogenic role of, for example, silent mutations inducing frameshifting in tumor suppressors (Sulima et al. 2017).
An area that is underexplored is aberrant or altered regulation of termination in the cancer setting. However, termination at premature stop codons can be a cancer driver if it occurs as a result of somatic mutations in tumor-suppressor genes (Bordeira-Carrico et al. 2012), resulting in NMD of the corresponding transcript (Karousis and Mühlemann 2018). NMD can be prevented by using aminoglycosides or small molecule drugs that promote readthrough of premature stop codons. Clinical introduction of such a small molecule inhibitor for the treatment of Duchenne muscular dystrophy, known as Translarna (ataluren) (Welch et al. 2007; Finkel et al. 2013) could be useful in the oncology setting, although its level of efficacy remains to be established.
Two initiation factors with confusing, multiple roles in mRNA translation are also associated with altered translational regulation in cancer cells. One is eIF6, a ribosomal subunit anti-association factor that prevents aberrant interactions between the 40S and 60S ribosomal subunits. eIF6 must be displaced from the ribosome for the final step of 60S ribosome biosynthesis in the nucleolus, and it can promote 80S ribosome disassembly in the cytosol by preventing the reassociation of post-termination 60S ribosomes, thereby impairing further rounds of initiation with prolonged sequestration (Ceci et al. 2003; Brina et al. 2015). Thus, eIF6 may play multiple roles in deregulating translation. Aberrant eIF6 expression has been observed in colorectal, and head and neck cancers, in which it accumulates in the nucleolus (Sanvito et al. 2000; Rosso et al. 2004). In contrast, reduced eIF6 levels have been shown to prevent oncogene-induced transformation and delay lymphomagenesis (Gandin et al. 2008; Miluzio et al. 2011).
The second translation factor that has multiple oncogenic activities is eIF5A. eIF5A was originally described as an initiation factor important for the formation of the first peptide bond during elongation, but has since been found to play a role in the elongation of poorly translated tripeptide regions in mRNAs: prolines, glycines, and/or basic residues (Benne et al. 1978; Gutierrez et al. 2013; Mathews and Hershey 2015; Pelechano and Alepuz 2017; Dever et al. 2018). More general roles in elongation and translation termination have also been proposed based on the accumulation of stalled ribosomes in cells lacking eIF5A in yeast (Pelechano and Alepuz 2017; Schuller et al. 2017). There is considerable interest in eIF5A, as both of its isoforms are overexpressed in a variety of cancers, including pancreatic, hepatic, colon, lung, and ovarian, and have been linked to the metastatic capacity of cancer cells (reviewed in Mathews and Hershey 2015). As the only known mammalian protein containing a hypusine modification, eIF5A is an attractive target, since its activity can be abrogated by inhibiting the enzymes catalyzing the hypusination (Nakanishi and Cleveland 2016).
Changes in 5′ and 3′UTR Length and Composition in Cancer Cells
Sequence and structural motifs present in mRNAs determine their intrinsic translational efficiency and their ability to be regulated by trans-acting factors such as microRNAs (miRNAs), RNA-binding proteins, and initiation factors (Truitt and Ruggero 2016; Duchaine and Fabian 2018). These motifs tend to be overrepresented in oncogenic mRNAs, conferring tight translational regulation (Kozak 1987). Furthermore, mutations in these noncoding motifs have been found to significantly modulate the expression of proto-oncogenes (Diederichs et al. 2016). Increased secondary structure in the 5′UTR was one of the first cis-acting elements to be identified that affect the rate or efficiency of cap-dependent mRNA translation initiation (Sonenberg et al. 1981). Subsequently, studies have shown that oncogenic mRNAs typically possess stable 5′UTR structures and, accordingly, show greater dependence on eIF4F. These include mRNAs encoding MYC, ornithine decarboxylase (ODC), a number of cyclins and cyclin-dependent kinases, among others (Darveau et al. 1985; Grens and Scheffler 1990; Rosenwald et al. 1993).
mRNA sequence elements found in the 5′ and 3′UTRs other than length and stability can also regulate the efficiency of translation. For example, enhanced dependence on eIF4E, but not eIF4A, was shown for the translation initiator of the short 5′UTR (TISU) element found in certain mRNAs (Elfakess et al. 2011; Kwan and Thompson 2018). This is in contrast to mRNAs containing internal ribosome entry sites (IRESs) that are cap-independent, but more highly dependent on eIF4G and eIF4A (Komar and Hatzoglou 2011). mRNAs can contain alternative initiation codons and inhibitory ORFs upstream of the canonical initiating AUG, which can severely hamper normal start site identification by the 43S PIC. These sequence elements are enriched in oncogenic transcripts (Kozak 1987). However, under periods of stress, including oncogenic stress (hypoxia and nutritional deprivation), some mRNAs with uORFs show selectively increased translation resulting from increased eIF2α phosphorylation (Hinnebusch et al. 2016; Wek 2018). Additionally, structural or sequence motifs in some 5′UTRs mediate the recruitment of RNA-binding proteins that modulate mRNA translation. One well-investigated example is that of the transforming growth factor β (TGF-β)-activated translation (BAT) element, which regulates the translation of certain mRNAs involved in the epithelial-to-mesenchymal transition (EMT) that promotes cell migration (Chaudhury et al. 2010). Finally, binding sites for miRNAs are particularly common motifs that affect translation, in addition to mRNA stability, as are AU-rich motifs. All of these elements have been thoroughly reviewed elsewhere (Komar and Hatzoglou 2011; Jonas and Izaurralde 2015; Wurth and Gebauer 2015; Hinnebusch et al. 2016; Truitt and Ruggero 2016) and are described by Duchaine and Fabian (2018).
The majority of identified cis-acting elements reduce mRNA translation efficiency, and are found in various combinations in individual oncogenic mRNAs, probably to govern and attenuate translation of mRNAs that have the capacity to transform cells when overexpressed (Mayr and Bartel 2009; Dieudonne et al. 2015). Nevertheless, cancer cells often develop mechanisms that bypass this stringent control. For example, the genome-wide shortening of 5′ and/or 3′UTRs is quite common in cellular transformation, which eliminates the suppressive RNA motifs (Mayr and Bartel 2009; Dieudonne et al. 2015). Alternative transcription start sites downstream from the 5′UTR translation regulatory element are one such reported mechanism. For example, initiating transcription downstream from two inhibitory uORFs in MDM2 results in increased translation of the MDM2 mRNA and inhibition of p53 (Landers et al. 1997). Similarly, translation inhibitory elements in the 3′UTR are sometimes eliminated by alternative polyadenylation in cancer cells, producing shorter 3′UTRs lacking, for instance, miRNA-binding sites and AU-rich elements that promote rapid mRNA decay (Mayr 2016).
Oncogenic Signaling
Most physiological signals, including stresses, nutritional and growth factor stimulation, metabolic functions, and endocrine factors, among others, are integrated through the translational machinery (Robichaud and Sonenberg 2017). The mammalian target of rapamycin (mTOR) in particular plays a crucial role in translation regulatory signaling (Fig. 1) by phosphorylating the 4E-BPs to allow eIF4F complex formation, and ribosomal protein S6 kinase (S6K), which, in turn, regulates eIF4A via PDCD4 and eIF4B, as well as eEF2K, to alleviate inhibition of elongation (Raught et al. 2004; Dorrello et al. 2006; Faller et al. 2015; Proud 2018). Many of the most commonly mutated genes across many cancer types encode key proteins that regulate signaling pathways impinging on translation, including PIK3CA, KRAS, PTEN, APC, and EGFR, among others (Kandoth et al. 2013). Particularly interesting is the case of the MYC oncogene, which promotes almost all aspects of translation (reviewed in van Riggelen et al. 2010). Although mRNA translation is far from the only effector of these signaling pathways, its importance is shown by the central role for “oncogenic translation” activity required to initiate and maintain the transformed phenotype (Truitt and Ruggero 2016). For example, rapid inhibition of translation following inhibition of upstream kinases such as AKT and KRAS occurs before any transcriptional changes in models of glioblastoma (Rajasekhar et al. 2003). Consequently, deregulation of the translation machinery in response to cancer drug treatment may well be an early driver of drug resistance as a means of maintaining and/or reprogramming cancer cell protein synthesis (Ilic et al. 2011; Zindy et al. 2011; Alain et al. 2012; Boussemart et al. 2014; Cope et al. 2014; Musa et al. 2016).
Is There a Cancer Ribosome?
It has long been discussed whether there are cancer-specific modifications in ribosomes themselves that could promote cancer-specific mRNA translational reprogramming. Discovery of “ribosomopathies,” a family of syndromes caused by inherited mutations in genes encoding ribosomal proteins and their regulators that are characterized by initial defects in hematopoiesis, followed by increased cancer susceptibility, supports this hypothesis (Sulima et al. 2017). However, the mechanism underlying the oncogenic effects of these mutations is unclear.
It is noteworthy that the stoichiometry of ribosomal proteins and rRNA modifications, such as methylation and pseudouridylation, varies in cancer cells (Truitt and Ruggero 2016). This suggests that individual ribosomes may possess unique modifications that alter their ability to translate certain mRNAs with respect to others. Whether such “cancer ribosomes” could promote the selective translation of oncogenic mRNAs and/or restrict the synthesis of tumor suppressors remains to be established. However, in support of this model is the finding that disruption of dyskerin, the enzyme-catalyzing pseudouridylation, or of small nucleolar RNAs (snoRNAs) that guide dyskerin to rRNA sites, are common in many cancers and can impair the translation of mRNAs encoding critical tumor suppressors, such as p53 and p27 (Truitt and Ruggero 2016). Most recently, the existence of ribosome heterogeneity has recently been shown in embryonic stem cells, in which a distinct subset of mRNAs is preferentially translated by different ribosome pools (Shi et al. 2017).
Alternatively, the link between ribosomal proteins, ribosomopathies, and cancer has been attributed to nontranslational roles of components of the translation machinery, mainly p53 stabilization (Gentilella et al. 2015; Zhou et al. 2015b). Thus, a subribosomal complex composed of the 5S rRNA and ribosomal proteins RPL5 and RPL11, binds to and sequesters MDM2, resulting in p53 stabilization and cell-cycle arrest (Gentilella et al. 2015). The complex forms when deregulated ribosome biogenesis leads to the imbalance of ribosomal components, as is the case in ribosomopathies (Gentilella et al. 2015). It is thought that somatic loss of p53 signaling allows hematopoietic cells to escape from the cell cycle arrest caused by defective ribosome biosynthesis, resulting in the cancer predisposition associated with ribosomopathies (Sulima et al. 2017).
SELECTIVE ONCOGENIC ADVANTAGES OF DEREGULATED TRANSLATION
Proliferation and Apoptosis
A large body of research has shown translational regulation of antiapoptotic factors, cyclins, and cyclin-dependent kinases (Sonenberg 1994; Silvera et al. 2010; Ruggero 2013; Teng et al. 2013; de la Parra et al. 2017). Although the role of translation in cell proliferation and survival has historically been the focus of much of the research in this field, other hallmarks of cancer are also regulated at the level of translation, as delineated below (Fig. 2).
Angiogenesis
Tumor angiogenesis is an ongoing process of continuous remodeling to accommodate tumor growth and is promoted by a variety of translational mechanisms. The mRNAs encoding two major regulators of angiogenesis, VEGFA and HIF1α, are translated via a variety of mechanisms that ensure cancer cells’ ability to adapt to hypoxia. Thus, the translation of VEGFA and HIF1α mRNAs can be promoted by both cap-dependent and cap-independent mechanisms, via the use of IRESs, uORFs, and possibly other noncanonical regulatory elements (Lang et al. 2002; Braunstein et al. 2007; Bastide et al. 2008; Arcondeguy et al. 2013). Although their translation is associated with increased eIF4E expression in human tumors (Nathan et al. 1997; Scott et al. 1998; Dodd et al. 2015), the complex translational regulation of VEGFA and HIF1α mRNAs allows for their translation to be maintained even in profound hypoxia and nutrient deprivation. Interestingly, HIF1α binds to the EIF4E promoter to promote its transcription, suggesting the possibility that the response to hypoxia could switch from an initial cap-independent mechanism to a cap-dependent one (Yi et al. 2013).
Stress Responses
In addition to hypoxia, cancer cells must modulate translation in the face of a variety of other stresses (Young and Wek 2016; Robichaud and Sonenberg 2017). Interestingly, the responses to diverse stressors share common regulatory mechanisms. Thus, up to 49% of the transcriptome, and essentially all mRNAs translated under stress conditions, are regulated by eIF2α phosphorylation, as they have been reported to include uORFs. These mRNAs disproportionately encode proteins involved in pathways that allow cancer cells to adapt to their environment (Calvo et al. 2009; Andreev et al. 2015; Young and Wek 2016; Wek 2018). Furthermore, IRESs, mRNA methylation, and a variety of noncanonical mechanisms of translation initiation maintain protein synthesis in the face of various stresses that inhibit cap-dependent translation (Meyer et al. 2015; Zhou et al. 2015a; Lacerda et al. 2017; Robichaud and Sonenberg 2017). How specific subsets of mRNAs are selectively translated in response to each stress is not well understood. Another issue requiring further investigation relates to the fact that inhibition of general translation associated with eIF2α phosphorylation, if persistent, eventually causes cell death (Young and Wek 2016; Robichaud and Sonenberg 2017). Cancer cells may partially escape apoptosis because of the fact that eIF2α phosphorylation promotes the translation of factors promoting its dephosphorylation, resulting in a feedback inhibitory loop (Andreev et al. 2015).
Emerging Oncogenic Advantages of Deregulated Translation
Considering that most cancer deaths are caused by metastatic dissemination, a key emerging concept is the ability of cancer cells to deregulate the translation of prometastatic factors such as matrix metalloproteases, integrins, transcription factors involved in the EMT, and GTPases involved in migration (Silvera et al. 2009; Nasr et al. 2013; Fujimura et al. 2015; Pinzaglia et al. 2015; Robichaud et al. 2015). The importance of cancer-cell-specific translation in the maintenance of cellular energy balance is also becoming clearer, as energy status and protein synthesis are regulated reciprocally to achieve an equilibrium (Proud 2006; Morita et al. 2013; Gandin et al. 2016; Miluzio et al. 2016; Robichaud and Sonenberg 2017). In addition, the interplay between translation and reactive oxygen species (ROS) in cancer cells has recently been revealed. Thus, components of the translation machinery are particularly sensitive to cysteine oxidation by ROS (Chio et al. 2016), whereas the mRNAs encoding key antioxidant proteins possess a motif termed cytosine-enriched regulator of translation (CERT) that confers translation regulation in response to increased eIF4E expression levels (Truitt et al. 2015). Finally, deregulated translation can also promote the expression of proteins involved in DNA repair such as BRCA1, thus enabling the escape from oncogene-induced senescence and resistance to DNA-damaging agents (Badura et al. 2012; Avdulov et al. 2015; Musa et al. 2016). Protein synthesis thus provides a crucial means for cancer cells to disrupt a variety of processes important for all steps of tumor biology.
CONSIDERING HETEROGENEOUS CELL POPULATIONS IN TUMORS
Cancer Stem Cells
Translational regulation in stem cells has emerged as an important new area of study, especially given the low transcriptional activity of these cells in embryonic and hematopoietic systems (see Teixeira and Lehmann 2018). Thus, the 4E-BPs are required to limit translation and ensure the maintenance of hematopoietic and embryonic stem cells (Signer et al. 2016; Tahmasebi et al. 2016), whereas eIF2α phosphorylation promotes the maintenance of muscle stem cells (Zismanov et al. 2016). In the cancer setting, tumor-initiating cells in a mouse skin cancer model display reduced protein synthesis and aberrant uORF translation, linking their stem-like state to eIF2α phosphorylation (Blanco et al. 2016). Phosphorylation of eIF4E has also been implicated in the stem-like phenotype by promoting the synthesis of stem cell maintenance factors such as β-catenin (Altman et al. 2013; Lim et al. 2013; Bell et al. 2016). These studies suggest that cancer stem-like cells may be characterized by low protein synthesis rates compared with most cancer cells caused by hypophosphorylation of 4E-BPs and/or hyperphosphorylation of eIF2α. The spectrum of inhibitors of translation to which cancer stem cells respond may therefore be different than that of bulk tumor cells, which must be taken into consideration for potential cancer therapies.
Immune Cells
The importance of translational regulation in immune cell populations is evident from the immune-suppressive effects of inhibitors such as rapamycin (Martel et al. 1977; So et al. 2016). Indeed, mTOR activity in T cells regulates translation, metabolic reprogramming, differentiation, lineage determination, and activation (Bjur et al. 2013; Araki et al. 2017; Linke et al. 2017; Yoo et al. 2017). The above-cited studies notwithstanding, little is known regarding translation regulatory events specific to the many different immune cell populations, particularly in the cancer-stroma context. eIF4E phosphorylation appears to be important for the synthesis of cancer-relevant cytokines and chemokines, such as tumor necrosis factor α (TNF-α) and interferon γ (IFN-γ) (Andersson and Sundler 2006; Rowlett et al. 2008; Herdy et al. 2012; Salvador-Bernáldez et al. 2017), and for the development of experimental autoimmune encephalomyelitis (Gorentla et al. 2013). In neutrophils, eIF4E phosphorylation promotes survival and metastatic dissemination in a mouse model of breast cancer (Robichaud et al. 2018). Thus, translation in a variety of immune cell populations can regulate various aspects of tumor biology.
CONCLUDING REMARKS
As we ponder the remarkable and numerous ways in which translational control can be usurped in cancer biology, we are left to discover exciting and promising paths to therapeutic interventions. The possibility that benefits can be derived from targeting translation in both the cancer and immune compartments, as appears to be the case for MNK inhibitors, should improve patient outcome. These findings are particularly meaningful considering the clinical development of MNK inhibitors in immune-oncology (ClinicalTrials.gov identifiers: NCT03258398, NCT02439346). Whether other means of inhibiting translation may be similarly useful or even more potent is an intriguing direction that remains to be investigated (Chu and Pelletier 2018). Indeed, studying the effect of newly developed translation-targeting drugs on the tumor microenvironment should help better predict their clinical benefit, and identify useful therapeutic combinations.
ACKNOWLEDGMENTS
This work is by supported by grants to N.S. from The Susan G. Komen Breast Cancer Foundation (IIR12224057) and CIHR (MOP-7214 and FND-148423). N.S. is a Howard Hughes Medical Institute Senior International Scholar. N.R. is supported by the Vanier Canada Graduate Scholarship (267839) and Rosalind Goodman Commemorative Scholarship (GCRC). R.J.S. is supported by the National Institutes of Health (1RO1CA178509, 1R01CA207893), the New York State Department of Health (DOH01-ROWLEY-2015-00035), and the Breast Cancer Research Foundation (BCRF-16-143). D.R. is supported by the National Institutes of Health (R01 CA140456, R01 CA184624, R01 CA154916, and R01NS089868).
Footnotes
Editors: Michael B. Mathews, Nahum Sonenberg, and John W.B. Hershey
Additional Perspectives on Translation Mechanisms and Control available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Alain T, Morita M, Fonseca BD, Yanagiya A, Siddiqui N, Bhat M, Zammit D, Marcus V, Metrakos P, Voyer LA, et al. 2012. eIF4E/4E-BP ratio predicts the efficacy of mTOR targeted therapies. Cancer Res 72: 6468–6476. [DOI] [PubMed] [Google Scholar]
- Altman JK, Szilard A, Konicek BW, Iversen PW, Kroczynska B, Glaser H, Sassano A, Vakana E, Graff JR, Platanias LC. 2013. Inhibition of Mnk kinase activity by cercosporamide and suppressive effects on acute myeloid leukemia precursors. Blood 121: 3675–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson K, Sundler R. 2006. Posttranscriptional regulation of TNFα expression via eukaryotic initiation factor 4E (eIF4E) phosphorylation in mouse macrophages. Cytokine 33: 52–57. [DOI] [PubMed] [Google Scholar]
- Andreev DE, O’Connor PB, Fahey C, Kenny EM, Terenin IM, Dmitriev SE, Cormican P, Morris DW, Shatsky IN, Baranov PV. 2015. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. eLife 4: e03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, Morita M, Bederman AG, Konieczny BT, Kissick HT, Sonenberg N, Ahmed R. 2017. Translation is actively regulated during the differentiation of CD8+ effector T cells. Nat Immunol 18: 1046–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcondeguy T, Lacazette E, Millevoi S, Prats H, Touriol C. 2013. VEGF-A mRNA processing, stability and translation: A paradigm for intricate regulation of gene expression at the post-transcriptional level. Nucleic Acids Res 41: 7997–8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avdulov S, Herrera J, Smith K, Peterson M, Gomez-Garcia JR, Beadnell TC, Schwertfeger KL, Benyumov AO, Manivel JC, Li S, et al. 2015. eIF4E threshold levels differ in governing normal and neoplastic expansion of mammary stem and luminal progenitor cells. Cancer Res 75: 687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badura M, Braunstein S, Zavadil J, Schneider RJ. 2012. DNA damage and eIF4G1 in breast cancer cells reprogram translation for survival and DNA repair mRNAs. Proc Natl Acad Sci 109: 18767–18772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastide A, Karaa Z, Bornes S, Hieblot C, Lacazette E, Prats H, Touriol C. 2008. An upstream open reading frame within an IRES controls expression of a specific VEGF-A isoform. Nucleic Acids Res 36: 2434–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JB, Eckerdt FD, Alley K, Magnusson LP, Hussain H, Bi Y, Arslan AD, Clymer J, Alvarez AA, Goldman S, et al. 2016. MNK inhibition disrupts mesenchymal glioma stem cells and prolongs survival in a mouse model of glioblastoma. Mol Cancer Res 14: 984–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benne R, Brown-Luedi ML, Hershey JW. 1978. Purification and characterization of protein synthesis initiation factors eIF-1, eIF-4C, eIF-4D, and eIF-5 from rabbit reticulocytes. J Biol Chem 253: 3070–3077. [PubMed] [Google Scholar]
- Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. 2015. Targeting the translation machinery in cancer. Nat Rev Drug Discov 14: 261–278. [DOI] [PubMed] [Google Scholar]
- Bjur E, Larsson O, Yurchenko E, Zheng L, Gandin V, Topisirovic I, Li S, Wagner CR, Sonenberg N, Piccirillo CA. 2013. Distinct translational control in CD4+ T cell subsets. PLoS Genet 9: e1003494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, Sajini A, Tanna H, Cortes-Garrido R, Gkatza N, et al. 2016. Stem cell function and stress response are controlled by protein synthesis. Nature 534: 335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeira-Carrico R, Pego AP, Santos M, Oliveira C. 2012. Cancer syndromes and therapy by stop-codon readthrough. Trends Mol Med 18: 667–678. [DOI] [PubMed] [Google Scholar]
- Boussemart L, Malka-Mahieu H, Girault I, Allard D, Hemmingsson O, Tomasic G, Thomas M, Basmadjian C, Ribeiro N, Thuaud F, et al. 2014. eIF4F is a nexus of resistance to anti-BRAF and anti-MEK cancer therapies. Nature 513: 105–109. [DOI] [PubMed] [Google Scholar]
- Braunstein S, Karpisheva K, Pola C, Goldberg J, Hochman T, Yee H, Cangiarella J, Arju R, Formenti SC, Schneider RJ. 2007. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol Cell 28: 501–512. [DOI] [PubMed] [Google Scholar]
- Brina D, Miluzio A, Ricciardi S, Biffo S. 2015. eIF6 anti-association activity is required for ribosome biogenesis, translational control and tumor progression. Biochim Biophys Acta 1849: 830–835. [DOI] [PubMed] [Google Scholar]
- Buttgereit F, Brand MD. 1995. A hierarchy of ATP-consuming processes in mammalian cells. Biochem J 312: 163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Pagliarini DJ, Mootha VK. 2009. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci 106: 7507–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceci M, Gaviraghi C, Gorrini C, Sala LA, Offenhauser N, Marchisio PC, Biffo S. 2003. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature 426: 579–584. [DOI] [PubMed] [Google Scholar]
- Chaudhury A, Hussey GS, Ray PS, Jin G, Fox PL, Howe PH. 2010. TGF-β-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat Cell Biol 12: 286–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chio II, Jafarnejad SM, Ponz-Sarvise M, Park Y, Rivera K, Palm W, Wilson J, Sangar V, Hao Y, Ohlund D, et al. 2016. NRF2 Promotes tumor maintenance by modulating mRNA translation in pancreatic cancer. Cell 166: 963–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J, Pelletier J. 2015. Targeting the eIF4A RNA helicase as an anti-neoplastic approach. Biochim Biophys Acta 1849: 781–791. [DOI] [PubMed] [Google Scholar]
- *.Chu J, Pelletier J. 2018. Translating therapeutics. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman LJ, Peter MB, Teall TJ, Brannan RA, Hanby AM, Honarpisheh H, Shaaban AM, Smith L, Speirs V, Verghese ET, et al. 2009. Combined analysis of eIF4E and 4E-binding protein expression predicts breast cancer survival and estimates eIF4E activity. Br J Cancer 100: 1393–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope CL, Gilley R, Balmanno K, Sale MJ, Howarth KD, Hampson M, Smith PD, Guichard SM, Cook SJ. 2014. Adaptation to mTOR kinase inhibitors by amplification of eIF4E to maintain cap-dependent translation. J Cell Sci 127: 788–800. [DOI] [PubMed] [Google Scholar]
- Darveau A, Pelletier J, Sonenberg N. 1985. Differential efficiencies of in vitro translation of mouse c-myc transcripts differing in the 5′ untranslated region. Proc Natl Acad Sci 82: 2315–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Parra C, Walters BA, Geter P, Schneider RJ. 2017. Translation initiation factors and their relevance in cancer. Curr Opin Genet Dev 48: 82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoyelle S, Chen T, Chen L, Wang Y, Klosi E, Halperin JA, Aktas BH, Chorev M. 2012. In vitro inhibition of translation initiation by N,N′-diarylureas—Potential anti-cancer agents. Bioorg Med Chem Lett 22: 402–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Dever TE, Dinman JD, Green R. 2018. Translation elongation and recoding in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S, Bartsch L, Berkmann JC, Frose K, Heitmann J, Hoppe C, Iggena D, Jazmati D, Karschnia P, Linsenmeier M, et al. 2016. The dark matter of the cancer genome: Aberrations in regulatory elements, untranslated regions, splice sites, non-coding RNA and synonymous mutations. EMBO Mol Med 8: 442–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieudonne FX, O’Connor PB, Gubler-Jaquier P, Yasrebi H, Conne B, Nikolaev S, Antonarakis S, Baranov PV, Curran J. 2015. The effect of heterogeneous transcription start sites (TSS) on the translatome: Implications for the mammalian cellular phenotype. BMC Genomics 16: 986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrikov M, Dobrikova E, Shveygert M, Gromeier M. 2011. Phosphorylation of eukaryotic translation initiation factor 4G1 (eIF4G1) by protein kinase Cα regulates eIF4G1 binding to Mnk1. Mol Cell Biol 31: 2947–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd KM, Yang J, Shen MH, Sampson JR, Tee AR. 2015. mTORC1 drives HIF-1α and VEGF-A signalling via multiple mechanisms involving 4E-BP1, S6K1 and STAT3. Oncogene 34: 2239–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. 2006. S6K1- and βTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science 314: 467–471. [DOI] [PubMed] [Google Scholar]
- *.Duchaine TF, Fabian MR. 2018. Mechanistic insights into microRNA-mediated gene silencing. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfakess R, Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Dikstein R. 2011. Unique translation initiation of mRNAs-containing TISU element. Nucleic Acids Res 39: 7598–7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faller WJ, Jackson TJ, Knight JR, Ridgway RA, Jamieson T, Karim SA, Jones C, Radulescu S, Huels DJ, Myant KB, et al. 2015. mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature 517: 497–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S, Ramalingam SS, Kauh J, Xu Z, Khuri FR, Sun SY. 2009. Phosphorylated eukaryotic translation initiation factor 4 (eIF4E) is elevated in human cancer tissues. Cancer Biol Ther 8: 1463–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feoktistova K, Tuvshintogs E, Do A, Fraser CS. 2013. Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc Natl Acad Sci 110: 13339–13344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel RS, Flanigan KM, Wong B, Bonnemann C, Sampson J, Sweeney HL, Reha A, Northcutt VJ, Elfring G, Barth J, et al. 2013. Phase 2a study of ataluren-mediated dystrophin production in patients with nonsense mutation Duchenne muscular dystrophy. PLoS ONE 8: e81302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K, Choi S, Wyse M, Strnadel J, Wright T, Klemke R. 2015. Eukaryotic translation initiation factor 5A (EIF5A) regulates pancreatic cancer metastasis by modulating RhoA and Rho-associated kinase (ROCK) protein expression levels. J Biol Chem 290: 29907–29919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi-Shimogori T, Ishii I, Kashiwagi K, Mashiba H, Ekimoto H, Igarashi K. 1997. Malignant transformation by overproduction of translation initiation factor eIF4G. Cancer Res 57: 5041–5044. [PubMed] [Google Scholar]
- Furic L, Rong L, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, Petroulakis E, Robichaud N, Pollak M, Gaboury LA, et al. 2010. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc Natl Acad Sci 107: 14134–14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin V, Miluzio A, Barbieri AM, Beugnet A, Kiyokawa H, Marchisio PC, Biffo S. 2008. Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation. Nature 455: 684–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin V, Masvidal L, Hulea L, Gravel SP, Cargnello M, McLaughlan S, Cai Y, Balanathan P, Morita M, Rajakumar A, et al. 2016. nanoCAGE reveals 5′ UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Genome Res 26: 636–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentilella A, Kozma SC, Thomas G. 2015. A liaison between mTOR signaling, ribosome biogenesis and cancer. Biochim Biophys Acta 1849: 812–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geter PA, Ernlund AW, Bakogianni S, Alard A, Arju R, Giashuddin S, Gadi A, Bromberg J, Schneider RJ. 2017. Hyperactive mTOR and MNK1 phosphorylation of eIF4E confer tamoxifen resistance and estrogen independence through selective mRNA translation reprogramming. Genes Dev 31: 2235–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, Kooistra SM, Christophersen NS, Christensen LL, Borre M, Sorensen KD, et al. 2014. A dual program for translation regulation in cellular proliferation and differentiation. Cell 158: 1281–1292. [DOI] [PubMed] [Google Scholar]
- Gorentla BK, Krishna S, Shin J, Inoue M, Shinohara ML, Grayson JM, Fukunaga R, Zhong XP. 2013. Mnk1 and 2 are dispensable for T cell development and activation but important for the pathogenesis of experimental autoimmune encephalomyelitis. J Immunol 190: 1026–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff JR, Konicek BW, Lynch RL, Dumstorf CA, Dowless MS, McNulty AM, Parsons SH, Brail LH, Colligan BM, Koop JW, et al. 2009. eIF4E activation is commonly elevated in advanced human prostate cancers and significantly related to reduced patient survival. Cancer Res 69: 3866–3873. [DOI] [PubMed] [Google Scholar]
- Grens A, Scheffler IE. 1990. The 5′- and 3′-untranslated regions of ornithine decarboxylase mRNA affect the translational efficiency. J Biol Chem 265: 11810–11816. [PubMed] [Google Scholar]
- Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, Dever TE. 2013. eIF5A promotes translation of polyproline motifs. Mol Cell 51: 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamura K, Minami K, Tanjung N, Wan Q, Koizumi M, Matsuura N, Na S, Yokota H. 2014. Attenuation of malignant phenotypes of breast cancer cells through eIF2α-mediated downregulation of Rac1 signaling. Intl J Oncol 44: 1980–1988. [DOI] [PubMed] [Google Scholar]
- Herdy B, Jaramillo M, Svitkin YV, Rosenfeld AB, Kobayashi M, Walsh D, Alain T, Sean P, Robichaud N, Topisirovic I, et al. 2012. Translational control of the activation of transcription factor NF-κB and production of type I interferon by phosphorylation of the translation factor eIF4E. Nat Immunol 13: 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey JW. 2015. The role of eIF3 and its individual subunits in cancer. Biochim Biophys Acta 1849: 792–800. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Ivanov IP, Sonenberg N. 2016. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 352: 1413–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic N, Utermark T, Widlund HR, Roberts TM. 2011. PI3K-targeted therapy can be evaded by gene amplification along the MYC-eukaryotic translation initiation factor 4E (eIF4E) axis. Proc Natl Acad Sci 108: E699–E708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas S, Izaurralde E. 2015. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 16: 421–433. [DOI] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. 2013. Mutational landscape and significance across 12 major cancer types. Nature 502: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Karousis ED, Mühlemann O. 2018. Nonsense-mediated mRNA decay begins where translation ends. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Gunnery S, Choe JK, Mathews MB. 2002. Neoplastic progression in melanoma and colon cancer is associated with increased expression and activity of the interferon-inducible protein kinase, PKR. Oncogene 21: 8741–8748. [DOI] [PubMed] [Google Scholar]
- Komar AA, Hatzoglou M. 2011. Cellular IRES-mediated translation: The war of ITAFs in pathophysiological states. Cell Cycle 10: 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koromilas AE. 2015. Roles of the translation initiation factor eIF2α serine 51 phosphorylation in cancer formation and treatment. Biochim Biophys Acta 1849: 871–880. [DOI] [PubMed] [Google Scholar]
- Kozak M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15: 8125–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozel C, Thompson B, Hustak S, Moore C, Nakashima A, Singh CR, Reid M, Cox C, Papadopoulos E, Luna RE, et al. 2016. Overexpression of eIF5 or its protein mimic 5MP perturbs eIF2 function and induces ATF4 translation through delayed re-initiation. Nucleic Acids Res 44: 8704–8713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Kwan T, Thompson SR. 2018. Noncanonical translation initiation in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacerda R, Menezes J, Romao L. 2017. More than just scanning: The importance of cap-independent mRNA translation initiation for cellular stress response and cancer. Cell Mol Life Sci 74: 1659–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landers JE, Cassel SL, George DL. 1997. Translational enhancement of mdm2 oncogene expression in human tumor cells containing a stabilized wild-type p53 protein. Cancer Res 57: 3562–3568. [PubMed] [Google Scholar]
- Lang KJ, Kappel A, Goodall GJ. 2002. Hypoxia-inducible factor-1α mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Mol Biol Cell 13: 1792–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaris-Karatzas A, Montine KS, Sonenberg N. 1990. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 345: 544–547. [DOI] [PubMed] [Google Scholar]
- Lee AS, Kranzusch PJ, Cate JH. 2015. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature 522: 111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS, Kranzusch PJ, Doudna JA, Cate JH. 2016. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 536: 96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Chai Z, Li Y, Liu D, Bai Z, Situ Z. 2009. BZW1, a novel proliferation regulator that promotes growth of salivary muocepodermoid carcinoma. Cancer Lett 284: 86–94. [DOI] [PubMed] [Google Scholar]
- Lim S, Saw TY, Zhang M, Janes MR, Nacro K, Hill J, Lim AQ, Chang CT, Fruman DA, Rizzieri DA, et al. 2013. Targeting of the MNK-eIF4E axis in blast crisis chronic myeloid leukemia inhibits leukemia stem cell function. Proc Natl Acad Sci 110: E2298–E2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Holbro T, Alonso G, Gerosa D, Burger MM. 2001. Molecular interaction between human tumor marker protein p150, the largest subunit of eIF3, and intermediate filament protein K7. J Cell Biochem 80: 483–490. [DOI] [PubMed] [Google Scholar]
- Linke M, Fritsch SD, Sukhbaatar N, Hengstschlager M, Weichhart T. 2017. mTORC1 and mTORC2 as regulators of cell metabolism in immunity. FEBS Lett 591: 3089–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish HF. 1974. Model for the regulation of mRNA translation applied to haemoglobin synthesis. Nature 251: 385–388. [DOI] [PubMed] [Google Scholar]
- MacDonald JI, Verdi JM, Meakin SO. 1999. Activity-dependent interaction of the intracellular domain of rat trkA with intermediate filament proteins, the β-6 proteasomal subunit, Ras-GRF1, and the p162 subunit of eIF3. J Mol Neurosci 13: 141–158. [DOI] [PubMed] [Google Scholar]
- Martel RR, Klicius J, Galet S. 1977. Inhibition of the immune response by rapamycin, a new antifungal antibiotic. Can J Physiol Pharmacol 55: 48–51. [DOI] [PubMed] [Google Scholar]
- Martineau Y, Azar R, Muller D, Lasfargues C, El Khawand S, Anesia R, Pelletier J, Bousquet C, Pyronnet S. 2014. Pancreatic tumours escape from translational control through 4E-BP1 loss. Oncogene 33: 1367–1374. [DOI] [PubMed] [Google Scholar]
- Mathews MB, Hershey JW. 2015. The translation factor eIF5A and human cancer. Biochim Biophys Acta 1849: 836–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C. 2016. Evolution and biological roles of alternative 3′UTRs. Trends Cell Biol 26: 227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meric-Bernstam F, Chen H, Akcakanat A, Do KA, Lluch A, Hennessy BT, Hortobagyi GN, Mills GB, Gonzalez-Angulo AM. 2012. Aberrations in translational regulation are associated with poor prognosis in hormone receptor-positive breast cancer. Breast Cancer Res 14: R138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Merrick WC, Pavitt GD. 2018. Protein synthesis initiation in eukaryotic cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 2015. 5′ UTR m6A promotes cap-independent translation. Cell 163: 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miluzio A, Beugnet A, Grosso S, Brina D, Mancino M, Campaner S, Amati B, de Marco A, Biffo S. 2011. Impairment of cytoplasmic eIF6 activity restricts lymphomagenesis and tumor progression without affecting normal growth. Cancer Cell 19: 765–775. [DOI] [PubMed] [Google Scholar]
- Miluzio A, Ricciardi S, Manfrini N, Alfieri R, Oliveto S, Brina D, Biffo S. 2016. Translational control by mTOR-independent routes: How eIF6 organizes metabolism. Biochem Soc Trans 44: 1667–1673. [DOI] [PubMed] [Google Scholar]
- Modelska A, Turro E, Russell R, Beaton J, Sbarrato T, Spriggs K, Miller J, Graf S, Provenzano E, Blows F, et al. 2015. The malignant phenotype in breast cancer is driven by eIF4A1-mediated changes in the translational landscape. Cell Death Dis 6: e1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Gravel SP, Chenard V, Sikstrom K, Zheng L, Alain T, Gandin V, Avizonis D, Arguello M, Zakaria C, et al. 2013. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab 18: 698–711. [DOI] [PubMed] [Google Scholar]
- Musa F, Alard A, David-West G, Curtin JP, Blank SV, Schneider RJ. 2016. Dual mTORC1/2 Inhibition as a novel strategy for the resensitization and treatment of platinum-resistant ovarian cancer. Mol Cancer Ther 15: 1557–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi S, Cleveland JL. 2016. Targeting the polyamine-hypusine circuit for the prevention and treatment of cancer. Amino Acids 48: 2353–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasr Z, Robert F, Porco JA Jr, Muller WJ, Pelletier J. 2013. eIF4F suppression in breast cancer affects maintenance and progression. Oncogene 32: 861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan CO, Carter P, Liu L, Li BD, Abreo F, Tudor A, Zimmer SG, De Benedetti A. 1997. Elevated expression of eIF4E and FGF-2 isoforms during vascularization of breast carcinomas. Oncogene 15: 1087–1094. [DOI] [PubMed] [Google Scholar]
- Novoa EM, Ribas de Pouplana L. 2012. Speeding with control: Codon usage, tRNAs, and ribosomes. Trends Genet 28: 574–581. [DOI] [PubMed] [Google Scholar]
- Pavon-Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T. 2009. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res 37: 7268–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Alepuz P. 2017. eIF5A facilitates translation termination globally and promotes the elongation of many non polyproline-specific tripeptide sequences. Nucleic Acids Res 45: 7326–7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzaglia M, Montaldo C, Polinari D, Simone M, La Teana A, Tripodi M, Mancone C, Londei P, Benelli D. 2015. eIF6 over-expression increases the motility and invasiveness of cancer cells by modulating the expression of a critical subset of membrane-bound proteins. BMC Cancer 15: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud CG. 2006. Regulation of protein synthesis by insulin. Biochem Soc Trans 34: 213–216. [DOI] [PubMed] [Google Scholar]
- *.Proud CG. 2018. Phosphorylation and signal transduction pathways in translational control. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T, Sonenberg N. 1999. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J 18: 270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekhar VK, Viale A, Socci ND, Wiedmann M, Hu X, Holland EC. 2003. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol Cell 12: 889–901. [DOI] [PubMed] [Google Scholar]
- Ramon y Cajal S, De Mattos-Arruda L, Sonenberg N, Cortes J, Peg V. 2014. The intra-tumor heterogeneity of cell signaling factors in breast cancer: p4E–BP1 and peIF4E are diffusely expressed and are real potential targets. Clin Transl Oncol 16: 937–941. [DOI] [PubMed] [Google Scholar]
- Raught B, Gingras AC, Gygi SP, Imataka H, Morino S, Gradi A, Aebersold R, Sonenberg N. 2000. Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. EMBO J 19: 434–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raught B, Peiretti F, Gingras AC, Livingstone M, Shahbazian D, Mayeur GL, Polakiewicz RD, Sonenberg N, Hershey JW. 2004. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J 23: 1761–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robichaud N, Sonenberg N. 2017. Translational control and the cancer cell response to stress. Curr Opin Cell Biol 45: 102–109. [DOI] [PubMed] [Google Scholar]
- Robichaud N, del Rincon SV, Huor B, Alain T, Petruccelli LA, Hearnden J, Goncalves C, Grotegut S, Spruck CH, Furic L, et al. 2015. Phosphorylation of eIF4E promotes EMT and metastasis via translational control of SNAIL and MMP-3. Oncogene 34: 2032–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robichaud N, Hsu BE, Istomine R, Alvarez F, Blagih J, Ma EH, Morales SV, Dai DL, Li G, Souleimanova M, et al. 2018. Translational control in the tumor microenvironment promotes lung metastasis: Phosphorylation of eIF4E in neutrophils. Proc Natl Acad Sci 16: 937–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfe DF, Brown GC. 1997. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol Rev 77: 731–758. [DOI] [PubMed] [Google Scholar]
- Rosenwald IB, Lazaris-Karatzas A, Sonenberg N, Schmidt EV. 1993. Elevated levels of cyclin D1 protein in response to increased expression of eukaryotic initiation factor 4E. Mol Cell Biol 13: 7358–7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwald IB, Hutzler MJ, Wang S, Savas L, Fraire AE. 2001. Expression of eukaryotic translation initiation factors 4E and 2α is increased frequently in bronchioloalveolar but not in squamous cell carcinomas of the lung. Cancer 92: 2164–2171. [DOI] [PubMed] [Google Scholar]
- Rosenwald IB, Wang S, Savas L, Woda B, Pullman J. 2003. Expression of translation initiation factor eIF-2α is increased in benign and malignant melanocytic and colonic epithelial neoplasms. Cancer 98: 1080–1088. [DOI] [PubMed] [Google Scholar]
- Rosso P, Cortesina G, Sanvito F, Donadini A, Di Benedetto B, Biffo S, Marchisio PC. 2004. Overexpression of p27BBP in head and neck carcinomas and their lymph node metastases. Head Neck 26: 408–417. [DOI] [PubMed] [Google Scholar]
- Rowlett RM, Chrestensen CA, Nyce M, Harp MG, Pelo JW, Cominelli F, Ernst PB, Pizarro TT, Sturgill TW, Worthington MT. 2008. MNK kinases regulate multiple TLR pathways and innate proinflammatory cytokines in macrophages. Am J Physiol Gastrointest Liver Physiol 294: G452–G459. [DOI] [PubMed] [Google Scholar]
- Ruggero D. 2013. Translational control in cancer etiology. Cold Spring Harb Perspect Biol 5: a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, Pandolfi PP. 2004. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med 10: 484–486. [DOI] [PubMed] [Google Scholar]
- Salehi Z, Mashayekhi F. 2006. Expression of the eukaryotic translation initiation factor 4E (eIF4E) and 4E-BP1 in esophageal cancer. Clin Biochem 39: 404–409. [DOI] [PubMed] [Google Scholar]
- Salvador-Bernáldez M, Mateus SB, Del Barco Barrantes I, Arthur SC, Martínez-A C, Nebreda AR, Salvador JM. 2017. p38α regulates cytokine-induced IFNγ secretion via the Mnk1/eIF4E pathway in Th1 cells. Immunol Cell Biol 95: 814–823. [DOI] [PubMed] [Google Scholar]
- Sanvito F, Vivoli F, Gambini S, Santambrogio G, Catena M, Viale E, Veglia F, Donadini A, Biffo S, Marchisio PC. 2000. Expression of a highly conserved protein, p27BBP, during the progression of human colorectal cancer. Cancer Res 60: 510–516. [PubMed] [Google Scholar]
- Scheper GC, Proud CG. 2002. Does phosphorylation of the cap-binding protein eIF4E play a role in translation initiation? Eur J Biochem 269: 5350–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheper GC, van Kollenburg B, Hu J, Luo Y, Goss DJ, Proud CG. 2002. Phosphorylation of eukaryotic initiation factor 4E markedly reduces its affinity for capped mRNA. J Biol Chem 277: 3303–3309. [DOI] [PubMed] [Google Scholar]
- Schewe DM, Aguirre-Ghiso JA. 2009. Inhibition of eIF2α dephosphorylation maximizes bortezomib efficiency and eliminates quiescent multiple myeloma cells surviving proteasome inhibitor therapy. Cancer Res 69: 1545–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller AP, Wu CC, Dever TE, Buskirk AR, Green R. 2017. eIF5A functions globally in translation elongation and termination. Mol Cell 66: 194–205.e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott PA, Smith K, Poulsom R, De Benedetti A, Bicknell R, Harris AL. 1998. Differential expression of vascular endothelial growth factor mRNA vs protein isoform expression in human breast cancer and relationship to eIF-4E. Br J Cancer 77: 2120–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendoel A, Dunn JG, Rodriguez EH, Naik S, Gomez NC, Hurwitz B, Levorse J, Dill BD, Schramek D, Molina H, et al. 2017. Translation from unconventional 5′ start sites drives tumour initiation. Nature 541: 494–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Fujii K, Kovary KM, Genuth NR, Rost HL, Teruel MN, Barna M. 2017. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol Cell 67: 71–83.e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer RA, Qi L, Zhao Z, Thompson D, Sigova AA, Fan ZP, DeMartino GN, Young RA, Sonenberg N, Morrison SJ. 2016. The rate of protein synthesis in hematopoietic stem cells is limited partly by 4E-BPs. Genes Dev 30: 1698–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvera D, Arju R, Darvishian F, Levine PH, Zolfaghari L, Goldberg J, Hochman T, Formenti SC, Schneider RJ. 2009. Essential role for eIF4GI overexpression in the pathogenesis of inflammatory breast cancer. Nat Cell Biol 11: 903–908. [DOI] [PubMed] [Google Scholar]
- Silvera D, Formenti SC, Schneider RJ. 2010. Translational control in cancer. Nat Rev Cancer 10: 254–266. [DOI] [PubMed] [Google Scholar]
- Singh CR, Lee B, Udagawa T, Mohammad-Qureshi SS, Yamamoto Y, Pavitt GD, Asano K. 2006. An eIF5/eIF2 complex antagonizes guanine nucleotide exchange by eIF2B during translation initiation. EMBO J 25: 4537–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh CR, Watanabe R, Zhou D, Jennings MD, Fukao A, Lee B, Ikeda Y, Chiorini JA, Campbell SG, Ashe MP, et al. 2011. Mechanisms of translational regulation by a human eIF5-mimic protein. Nucleic Acids Res 39: 8314–8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slepenkov SV, Darzynkiewicz E, Rhoads RE. 2006. Stopped-flow kinetic analysis of eIF4E and phosphorylated eIF4E binding to cap analogs and capped oligoribonucleotides: Evidence for a one-step binding mechanism. J Biol Chem 281: 14927–14938. [DOI] [PubMed] [Google Scholar]
- So L, Lee J, Palafox M, Mallya S, Woxland CG, Arguello M, Truitt ML, Sonenberg N, Ruggero D, Fruman DA. 2016. The 4E-BP-eIF4E axis promotes rapamycin-sensitive growth and proliferation in lymphocytes. Sci Signal 9: ra57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N. 1994. Regulation of translation and cell growth by eIF-4E. Biochimie 76: 839–846. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Guertin D, Cleveland D, Trachsel H. 1981. Probing the function of the eucaryotic 5′ cap structure by using a monoclonal antibody directed against cap-binding proteins. Cell 27: 563–572. [DOI] [PubMed] [Google Scholar]
- Sulima SO, Hofman IJF, De Keersmaecker K, Dinman JD. 2017. How ribosomes translate cancer. Cancer Discov 7: 1069–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahmasebi S, Jafarnejad SM, Tam IS, Gonatopoulos-Pournatzis T, Matta-Camacho E, Tsukumo Y, Yanagiya A, Li W, Atlasi Y, Caron M, et al. 2016. Control of embryonic stem cell self-renewal and differentiation via coordinated alternative splicing and translation of YY2. Proc Natl Acad Sci 113: 12360–12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Teixeira FK, Lehmann R. 2018. Translational control during developmental transitions. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng T, Thomas G, Mercer CA. 2013. Growth control and ribosomopathies. Curr Opin Genet Dev 23: 63–71. [DOI] [PubMed] [Google Scholar]
- Topisirovic I, Sonenberg N. 2014. Distinctive tRNA repertoires in proliferating versus differentiating cells. Cell 158: 1238–1239. [DOI] [PubMed] [Google Scholar]
- Topisirovic I, Ruiz-Gutierrez M, Borden KL. 2004. Phosphorylation of the eukaryotic translation initiation factor eIF4E contributes to its transformation and mRNA transport activities. Cancer Res 64: 8639–8642. [DOI] [PubMed] [Google Scholar]
- Truitt ML, Ruggero D. 2016. New frontiers in translational control of the cancer genome. Nat Rev Cancer 16: 288–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truitt ML, Conn CS, Shi Z, Pang X, Tokuyasu T, Coady AM, Seo Y, Barna M, Ruggero D. 2015. Differential requirements for eIF4E dose in normal development and cancer. Cell 162: 59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riggelen J, Yetil A, Felsher DW. 2010. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 10: 301–309. [DOI] [PubMed] [Google Scholar]
- Walsh D, Mohr I. 2014. Coupling 40S ribosome recruitment to modification of a cap-binding initiation factor by eIF3 subunit e. Genes Dev 28: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Rosenwald IB, Hutzler MJ, Pihan GA, Savas L, Chen JJ, Woda BA. 1999. Expression of the eukaryotic translation initiation factors 4E and 2α in non-Hodgkin’s lymphomas. Am J Pathol 155: 247–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wei Z, Gao F, Zhang X, Zhou C, Zhu F, Wang Q, Gao Q, Ma C, Sun W, et al. 2008. Expression and prognostic significance of PDCD4 in human epithelial ovarian carcinoma. Anticancer Res 28: 2991–2996. [PubMed] [Google Scholar]
- Wang YW, Lin KT, Chen SC, Gu DL, Chen CF, Tu PH, Jou YS. 2013. Overexpressed-eIF3I interacted and activated oncogenic Akt1 is a theranostic target in human hepatocellular carcinoma. Hepatology 58: 239–250. [DOI] [PubMed] [Google Scholar]
- *.Wek RC. 2018. Role of eIF2α kinases in translational control and adaptation to cellular stress. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch EM, Barton ER, Zhuo J, Tomizawa Y, Friesen WJ, Trifillis P, Paushkin S, Patel M, Trotta CR, Hwang S, et al. 2007. PTC124 targets genetic disorders caused by nonsense mutations. Nature 447: 87–91. [DOI] [PubMed] [Google Scholar]
- Wendel HG, Silva RL, Malina A, Mills JR, Zhu H, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Teruya-Feldstein J, Pelletier J, et al. 2007. Dissecting eIF4E action in tumorigenesis. Genes Dev 21: 3232–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurth L, Gebauer F. 2015. RNA-binding proteins, multifaceted translational regulators in cancer. Biochim Biophys Acta 1849: 881–886. [DOI] [PubMed] [Google Scholar]
- Yang HS, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S, Lockett SJ, Sonenberg N, Colburn NH. 2003. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol 23: 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HS, Cho MH, Zakowicz H, Hegamyer G, Sonenberg N, Colburn NH. 2004. A novel function of the MA-3 domains in transformation and translation suppressor Pdcd4 is essential for its binding to eukaryotic translation initiation factor 4A. Mol Cell Biol 24: 3894–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, Bobrovnikova-Marjon E, Diehl JA, Ron D, Koumenis C. 2010. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J 29: 2082–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi T, Papadopoulos E, Hagner PR, Wagner G. 2013. Hypoxia-inducible factor-1α (HIF-1α) promotes cap-dependent translation of selective mRNAs through up-regulating initiation factor eIF4E1 in breast cancer cells under hypoxia conditions. J Biol Chem 288: 18732–18742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HS, Lee K, Na K, Zhang YX, Lim HJ, Yi T, Song SU, Jeon MS. 2017. Mesenchymal stromal cells inhibit CD25 expression via the mTOR pathway to potentiate T-cell suppression. Cell Death Dis 8: e2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SK, Wek RC. 2016. Upstream open reading frames differentially regulate gene-specific translation in the integrated stress response. J Biol Chem 291: 16927–16935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Pan X, Hershey JW. 2007. Individual overexpression of five subunits of human translation initiation factor eIF3 promotes malignant transformation of immortal fibroblast cells. J Biol Chem 282: 5790–5800. [DOI] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. 2015a. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526: 591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Liao WJ, Liao JM, Liao P, Lu H. 2015b. Ribosomal proteins: Functions beyond the ribosome. J Mol Cell Biol 7: 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy P, Berge Y, Allal B, Filleron T, Pierredon S, Cammas A, Beck S, Mhamdi L, Fan L, Favre G, et al. 2011. Formation of the eIF4F translation-initiation complex determines sensitivity to anticancer drugs targeting the EGFR and HER2 receptors. Cancer Res 71: 4068–4073. [DOI] [PubMed] [Google Scholar]
- Zismanov V, Chichkov V, Colangelo V, Jamet S, Wang S, Syme A, Koromilas AE, Crist C. 2016. Phosphorylation of eIF2α is a translational control mechanism regulating muscle stem cell quiescence and self-renewal. Cell Stem Cell 18: 79–90. [DOI] [PubMed] [Google Scholar]