Here, Kearse et al. used start codon-specific reporters and ribosome profiling to characterize how translation from non-AUG start codons responds to protein synthesis inhibitors in human cells. Their data indicate that ribosome queuing enables mRNAs with poor initiation context—namely, those with non-AUG start codons—to be resistant to pharmacological translation inhibitors at concentrations that robustly inhibit global translation.
Keywords: near-cognate, start codon, translation initiation, translational control, cycloheximide, RAN translation
Abstract
Aberrant translation initiation at non-AUG start codons is associated with multiple cancers and neurodegenerative diseases. Nevertheless, how non-AUG translation may be regulated differently from canonical translation is poorly understood. Here, we used start codon-specific reporters and ribosome profiling to characterize how translation from non-AUG start codons responds to protein synthesis inhibitors in human cells. These analyses surprisingly revealed that translation of multiple non-AUG-encoded reporters and the endogenous GUG-encoded DAP5 (eIF4G2/p97) mRNA is resistant to cycloheximide (CHX), a translation inhibitor that severely slows but does not completely abrogate elongation. Our data suggest that slowly elongating ribosomes can lead to queuing/stacking of scanning preinitiation complexes (PICs), preferentially enhancing recognition of weak non-AUG start codons. Consistent with this model, limiting PIC formation or scanning sensitizes non-AUG translation to CHX. We further found that non-AUG translation is resistant to other inhibitors that target ribosomes within the coding sequence but not those targeting newly initiated ribosomes. Together, these data indicate that ribosome queuing enables mRNAs with poor initiation context—namely, those with non-AUG start codons—to be resistant to pharmacological translation inhibitors at concentrations that robustly inhibit global translation.
Canonical eukaryotic translation follows a scanning model of initiation in which the 7-methylguanosine (m7G) cap at the 5′ end of the mRNA is first bound by the eIF4F complex, which recruits the 43S preinitiation complex [PIC; comprised of the 40S small ribosomal subunit, eIF2•GTP•Met-tRNAiMet (methionyl-tRNA) ternary complex (TC), eIF1, eIF1A, eIF3, and eIF5]. The resulting 48S PIC then scans 5′ to 3′ in an ATP-dependent manner (with help from the eIF4A helicase) until the charged initiator Met-tRNAiMet bound by eIF2 base-pairs with an AUG start codon (for reviews, see Hinnebusch 2017; Shirokikh and Preiss 2018). A number of initiation factors are then displaced to allow for joining of the 60S large ribosomal subunit to form the complete 80S ribosome. Due to its perfect complementarity with the anticodon of tRNAiMet, AUG codons are preferred for initiation, but near-cognate codons—i.e., those that differ from AUG by a single nucleotide, such as CUG, GUG, ACG—can support low levels of initiation (for reviews, see Touriol et al. 2003; Kearse and Wilusz 2017).
Ribosome profiling has revealed that the use of non-AUG start codons for translation initiation (referred to here as non-AUG translation) is far more frequent in eukaryotic cells than predicted previously (Ingolia et al. 2009, 2011). Moreover, proteins produced from non-AUG translation can have critical biological functions, especially in cancer and neurodegenerative diseases (for review, see Kearse and Wilusz 2017). For example, fibroblast growth factor 2 (FGF2) translation can be initiated from a canonical AUG start codon or from one of four upstream in-frame CUG codons (Bugler et al. 1991; Arnaud et al. 1999). Interestingly, the CUG-encoded FGF2 isoforms are uniquely observed in transformed cells, and their ectopic expression is sufficient to cause cell immortalization in culture and increased tumorigenic phenotypes in mice (Couderc et al. 1991; Quarto et al. 1991). The oncoprotein c-myc can likewise be synthesized from different start codons: c-myc 2 (p64) is generated from a canonical AUG start codon, whereas c-myc 1 (p67) is made from an upstream in-frame CUG codon (Hann et al. 1992). Overexpression of the CUG-encoded c-myc 1 isoform (but not the AUG-encoded c-myc 2 isoform) inhibits growth of cultured cells (Hann 1994), and inactivation of c-myc 1 is observed in some tumors, including human Burkitt's lymphomas (Hann et al. 1988). Recently, translation of upstream ORFs (uORFs) from non-AUG codons on oncoprotein-encoding mRNAs has been shown to promote expression of the downstream ORFs and drive initiation of squamous cell carcinomas (Sendoel et al. 2017).
Aberrant non-AUG translation has further been associated with at least seven neurodegenerative diseases that are caused by nucleotide repeat expansions, including the fragile X disorders, Huntington's disease, and amyotrophic lateral sclerosis (ALS) (for reviews, see Green et al. 2016; Cleary and Ranum 2017). In repeat-associated non-AUG (RAN) translation, non-AUG start codons upstream of the expanded repeat are used to initiate translation in multiple reading frames, resulting in the synthesis of toxic homopolymeric proteins from the expanded nucleotide repeat itself. Recent work indicates that RAN translation primarily uses a cap- and scanning-dependent initiation mechanism (Kearse et al. 2016; Green et al. 2017; Tabet et al. 2018), although cap-independent mechanisms may also play a role in some cases (Cheng et al. 2018).
Start codon selection is regulated by nearly all of the canonical initiation factors (eIFs), including eIF1, eIF2, and eIF5 (for review, see Hinnebusch 2017). At most non-AUG start codons, it appears that Met-tRNAiMet is used for initiation due to the high specificity of eIF2; other tRNAs are rarely loaded by eIF2 into the P site of the small ribosomal subunit (Peabody 1989; Kolitz and Lorsch 2010; Sellier et al. 2017). Nevertheless, alternative factors that control initiation from some non-AUG start codons have been identified, including eIF2A and eIF2D, but they remain poorly understood. eIF2A can use Leu-tRNACUG for initiation at CUG codons (Komar et al. 2005; Kim et al. 2011; Starck et al. 2012), whereas eIF2D (sometimes referred to as ligatin [LGTN]) can regulate non-AUG initiation in a somewhat selective manner in vitro (Dmitriev et al. 2010; Skabkin et al. 2010). Given that start codon selection patterns change with cellular state (especially in human diseases), targeting non-AUG translation may be a promising therapeutic strategy. However, exact mechanistic differences that may distinguish canonical translation from non-AUG translation remain poorly understood.
In the present study, we generated a series of reporters to characterize in detail how canonical translation and non-AUG translation are differentially regulated in human cells. Canonical translation was sensitive to many well-characterized protein synthesis inhibitors, but non-AUG translation was strikingly resistant to cycloheximide (CHX) treatment. This was observed with multiple ORFs and in RAN translation. Using ribosome profiling and polysome analysis to probe endogenous translation patterns, we found that synthesis of the GUG-encoded DAP5 (eIF4G2/p97) protein and the CUG-encoded BAG1 N-terminally extended protein is likewise resistant to CHX. CHX is a well-characterized and potent elongation inhibitor (Ennis and Lubin 1964; Schneider-Poetsch et al. 2010), but we found that high doses of CHX do not completely block elongation in human cells. Notably, non-AUG translation becomes sensitive to CHX when PIC formation or scanning is blocked. We thus propose that the observed resistance of non-AUG translation to CHX treatment is caused by a ribosome queuing-based mechanism. Slowly elongating ribosomes induce queuing/stacking of scanning PICs that can become positioned over an otherwise poorly recognized non-AUG start codon, resulting in increased initiation. Consistent with this model, we show that non-AUG translation is also resistant to other elongation inhibitors that are expected to generate ribosome queues over the non-AUG start codon. In total, these data reveal how ribosome queuing enables non-AUG translation events to be resistant to ribosome-binding protein synthesis inhibitors.
Results
Non-AUG translation reporters use the canonical TC for initiation
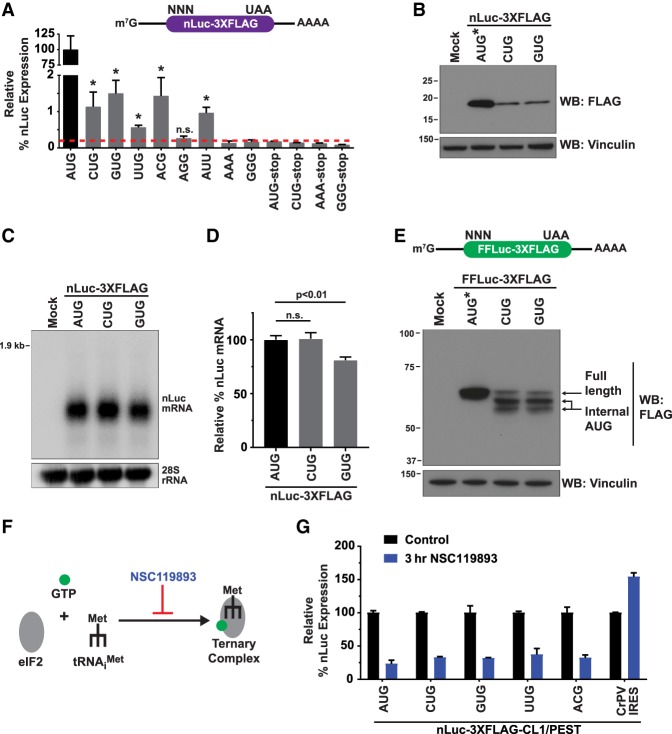
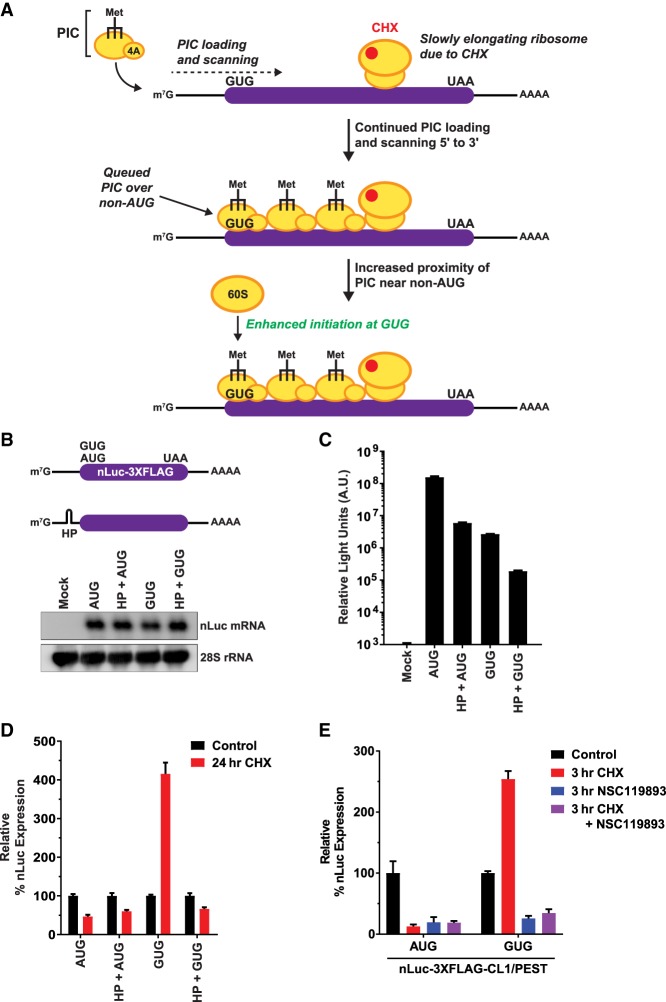
To characterize how translation from non-AUG start codons is regulated differently from canonical AUG-initiated translation in human cells, we generated a series of nanoLuciferase (nLuc) reporters that have the same 5′ leader, C-terminal 3XFlag tag, and polyadenylation signal but a different initiation codon (Fig. 1A, top). Mutating the nLuc AUG start codon to a near-cognate CUG, GUG, ACG, or AUU codon drastically reduced the nLuc signal to ∼1%–2% of that observed with an AUG start codon in HeLa cells (Fig. 1A, bottom). Nevertheless, this represents a true nLuc signal, as it was substantially above the level of signal obtained with reporters that (1) begin with AAA or GGG codons that are known to not support initiation (Kearse et al. 2016; Ivanov et al. 2018) or (2) harbor a stop codon immediately downstream from the start codon (e.g., AUG-stop or CUG-stop) (Fig. 1A, bottom). We confirmed by Western blotting that the single expected nLuc protein product (19 kDa) accumulated in cells (Fig. 1B) and that the steady-state nLuc mRNA levels were largely similar across reporters (Fig. 1C,D). These results strongly suggest that the differences in nLuc protein levels are primarily due to differences in translation initiation efficiencies.
Figure 1.
Development of reporters that are specific for non-AUG translation. (A) HeLa cells were transfected with nLuc reporters harboring the indicated start codons and a C-terminal 3XFlag tag. Twenty-four hours after transfection, the nLuc luminescence was quantified and normalized to a cotransfected firefly luciferase (FFLuc; pGL4.13) reporter. Signal from the AUG-nLuc reporter (black) was then used to determine the relative expression levels of the other nLuc reporters (gray). The red dashed line indicates expression above the negative control reporters. Data are shown as mean ± SD. n = 3. Signals from the non-AUG-nLuc reporters were compared with the GGG-nLuc negative control using a two-tailed unpaired t-test with Welch's correction. (*) P < 0.05. (B) Western blotting (WB) using an anti-Flag antibody was used to examine expression of the nLuc-3XFlag reporters 24 h after transfection. Vinculin was used as a loading control. AUG* denotes that 1/20th of the AUG-nLuc-3XFlag plasmid was transfected to avoid overexposure during film development. (C) Northern blotting was used to examine expression of the nLuc reporter mRNAs 24 h after transfection. 28S ribosomal RNA was used as a loading control. (D) RT-qPCR was used to quantify nLuc reporter mRNA levels 24 h after transfection. mRNA levels were first normalized to the cotransfected FFLuc (pGL4.13) reporter, and AUG-nLuc (black) was then used to determine the relative expression levels of the other nLuc reporters (gray). Data are shown as mean ± SD. n = 3. A two-tailed unpaired t-test with Welch's correction was used. (n.s.) Not significant. (E) HeLa cells were transfected with FFLuc-3XFlag reporters harboring the indicated start codons. Total protein was isolated 24 h after transfection and analyzed using Western blotting. Vinculin was used as a loading control. AUG* denotes that 1/20th of the AUG-FFLuc-3XFlag plasmid was transfected to avoid overexposure during film development. (F) NSC119893 inhibits formation of the canonical TC. (G) HeLa cells were transfected with the destabilized nLuc-3XFlag-CL1/PEST reporters for 24 h and then either collected (control) or treated for 3 h with 200 µM NSC119893. Luminescence signals for each reporter were quantified and set relative to the associated control samples. The CrPV IRES does not require the TC for initiation. Data are shown as mean ± SD. n = 3.
Similar low levels of non-AUG translation were observed with reporters that generate destabilized nLuc proteins (Supplemental Fig. S1), indicating that the long half-life of the canonical nLuc protein does not exaggerate or influence these phenotypes. For example, addition of C-terminal degron motifs decreased the nLuc protein half-life from 24 h (Fig. 2C) to ∼1 h (nLuc-PEST) or <1 h (nLuc-CL1/PEST) (Supplemental Fig. S1A,B), but translation from the non-AUG start codons was still ∼1% as efficient as an AUG start codon (Supplemental Fig. S1C). In contrast, CUG- and GUG-encoded firefly luciferase (FFLuc) reporters produced multiple protein products, including truncated variants that initiated from internal AUG codons (Fig. 1E; Supplemental Fig. S2). We thus focused most of our subsequent characterization efforts on the well-behaved nLuc reporters.
Figure 2.
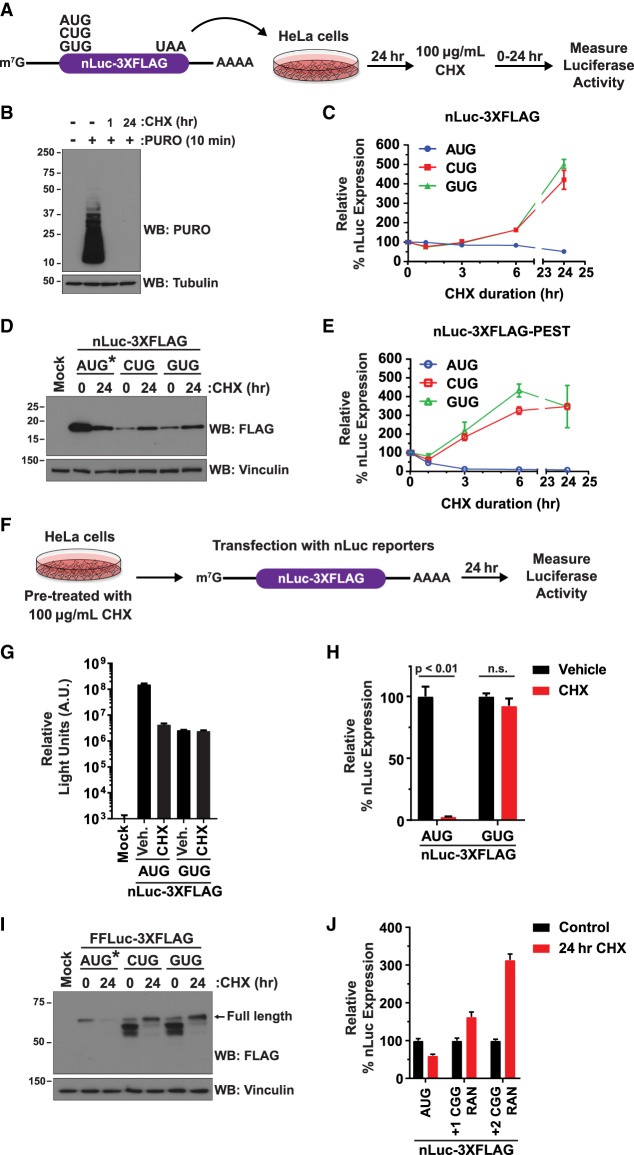
Non-AUG translation reporters are resistant to inhibition by CHX. (A) HeLa cells were transfected with the nLuc-3XFlag reporters followed by treatment with 100 µg/mL CHX and quantification of luminescence signals. (B) After 1 or 24 h of treatment with 100 µg/mL CHX, HeLa cells were subjected to puromycin (PURO) labeling and Western blotting (WB) to confirm global translation inhibition. Tubulin was used as a loading control. (C) Luminescence from the nLuc-3XFlag reporters was measured at the indicated time points after 100 µg/mL CHX treatment. Data for each reporter were set relative to the 0-h time point and are shown as mean ± SD. n = 3. (D) Western blotting using an anti-Flag antibody was used to examine expression of the nLuc-3XFlag reporters before and after 24 h of 100 µg/mL CHX treatment. Vinculin was used as a loading control. AUG* denotes that 1/20th of the AUG-nLuc-3XFlag plasmid was transfected to avoid overexposure during film development. (E) Luminescence from the destabilized nLuc-3XFlag-PEST reporters was measured at the indicated time points after CHX treatment. Data for each reporter were set relative to the 0-h time point and are shown as mean ± SD. n = 3. (F) HeLa cells were pretreated (15 min) with vehicle (Veh.; 0.1% DMSO) or 100 µg/mL CHX followed by transfection of the nLuc-3XFlag reporters. Luminescence was measured after 24 h. (G) Raw luciferase values of nLuc-3XFlag reporters 24 h after transfection. Data are shown as mean ± SD. n = 3. (H) Luminescence signals for each reporter were quantified and set relative to the associated vehicle-treated samples. Data are shown as mean ± SD. n = 3. A two-tailed unpaired t-test with Welch's correction was used. (I) Western blotting was used to examine expression of the FFLuc-3XFlag reporters before and after 24 h of 100 µg/mL CHX treatment. Vinculin was used as a loading control. AUG* denotes that 1/20th of the AUG-FFLuc-3XFlag plasmid was transfected to avoid overexposure during film development. (J) HeLa cells were transfected with expanded (CGG)100 repeat RAN translation reporters for 24 h and then collected (control) or treated for 24 h with 100 µg/mL CHX. “+1” and “+2” refer to the reading frame of the (CGG)100 repeat in relation to the nLuc-coding sequence. Luminescence signals for each reporter were quantified and set relative to the associated control samples. Data are shown as mean ± SD. n = 3.
In the canonical scanning model of eukaryotic translation initiation, the eIF2•GTP•Met-tRNAiMet TC enables methionine to be used as the first amino acid in protein synthesis. However, alternative initiation factors, including eIF2A and eIF2D, have been reported (Komar et al. 2005; Dmitriev et al. 2010). To test whether the nLuc reporters use the canonical TC for initiation, we treated HeLa cells with the pharmacological inhibitor NSC119893, which impairs TC formation (Fig. 1F; Robert et al. 2006). Both the AUG and non-AUG nLuc reporters were inhibited by NSC119893 to similar extents (Fig. 1G), unlike a reporter containing the cricket paralysis virus intergenic region internal ribosome entry site (CrPV IRES) that initiates translation in a manner independent of eIFs or Met-tRNAiMet (Wilson et al. 2000; Pestova and Hellen 2003). This strongly suggests that the canonical Met-tRNAiMet-containing TC is used to initiate non-AUG translation from the nLuc reporters.
Non-AUG translation reporters are resistant to CHX
We next assessed the response of these reporters to treatment with CHX (Fig. 2A), which inhibits translation elongation by blocking eEF2-mediated translocation via binding to the E site of the 60S ribosomal subunit (Schneider-Poetsch et al. 2010). HeLa cells were treated with a standard high dose of CHX (100 µg/mL; 355 µM) (Ennis and Lubin 1964; Warner et al. 1966; Schneider-Poetsch et al. 2010) for 1 or 24 h, and puromycin (PURO) labeling was then used to confirm that global translation was potently inhibited (Fig. 2B). Upon measuring the responses of the nLuc reporters, a surprising dichotomous effect was observed. Expression of the AUG-nLuc reporter decreased after addition of CHX (Fig. 2C, blue), consistent with inhibition of new translation coupled to turnover of the pre-existing nLuc protein over time. In stark contrast, translation of the non-AUG nLuc reporters was strikingly resistant to CHX and, in fact, increased over time (Fig. 2C, red and green). Similar increases in nLuc expression were observed regardless of the near-cognate start codon (CUG, GUG, ACG, or AUU) tested in both HeLa and HEK293T cells (Fig. 2C; Supplemental Fig. S3A). Moreover, Western blotting confirmed the results from the luminescence assays (Fig. 2D).
To verify that the observed increases in non-AUG nLuc reporter expression were due to new protein synthesis, we first measured the responses of the destabilized nLuc constructs to CHX treatment. The destabilized nLuc proteins rapidly turn over (Supplemental Fig. S1), suggesting that increases in their protein levels likely are due to increases in translation efficiency (TE). Expression of the destabilized AUG-nLuc reporters (Fig. 2E, blue; Supplemental Fig. S3B, blue) rapidly decreased after CHX treatment. In contrast, but consistent with the above results, expression of the destabilized CUG-nLuc and GUG-nLuc reporters (Fig. 2E, red and green, respectively; Supplemental Fig. S3B, red and green, respectively) increased over time, including after 3 or 6 h of CHX treatment. These increases in non-AUG translation upon CHX treatment were not due to the preferential accumulation of the non-AUG reporter mRNAs (Supplemental Fig. S3C) and occurred in a dose-dependent manner, with the largest effects observed upon treatment with 10 or 100 µg/mL CHX in HeLa cells (Supplemental Fig. S3B,D). A similar dose response was observed in normal human fibroblasts (IMR90), although these cells overall were more sensitive to CHX, and the largest effects were observed with 1 µg/mL CHX (Supplemental Fig. S3E).
It should be noted that in all experiments described to this point, cells were first transfected with the reporter plasmids for 24 h and then CHX was added. Therefore, to definitively prove that new translation can occur in the presence of CHX, HeLa cells were pretreated with 100 µg/mL CHX and then transfected with the AUG-nLuc or GUG-nLuc reporter plasmids (Fig. 2F). Again, CHX significantly decreased translation of the AUG-nLuc reporter (to ∼3% of that observed in vehicle-treated cells), whereas translation of the GUG-nLuc reporter was resistant (Fig. 2G,H). In contrast to when CHX was added 24 h after transfection (Fig. 2A–E), we found that GUG-nLuc expression was steady (not increased) when CHX was already present on the cells (Fig. 2G,H). This appears to be due to decreased transfection efficiency when CHX is present, as mRNA levels for the nLuc reporters as well as a cotransfected control FFLuc reporter (pGL4.13) were decreased compared with when these reporters were transfected into vehicle-treated cells (Supplemental Fig. S4). It is also important to stress here that the high dose of CHX did not result in a complete block to elongation and that even the AUG-nLuc luminescence signal was orders of magnitude higher than the background signal observed in mock transfected cells (Fig. 2G). Translation elongation thus still occurs but is greatly slowed by CHX (Hussmann et al. 2015).
CHX-resistant non-AUG translation was further observed with a different ORF (FFLuc) (Fig. 2I) as well as with reporters for RAN translation of the CGG repeats in the fragile X mRNA (Fig. 2J; Kearse et al. 2016). These data thus indicate that translation from non-AUG start codons is resistant to CHX in multiple sequence contexts.
Increases in non-AUG translation could be observed within a few hours of CHX addition (Fig. 2E; Supplemental Fig. S3B), but we nevertheless wanted to determine whether this phenotype was due to overall cell dysfunction or a cell death response. Cell viability (Supplemental Fig. S5A) and metabolism (Supplemental Fig. S5B) were thus assayed in cells treated with vehicle (0.1% DMSO), 100 µg/mL CHX, 50 µg/mL PURO (a polypeptide chain terminator that inhibits cytoplasmic and mitochondrial translation), or a dilute solution of Triton X-100 (which permeabilizes cell membranes). Vehicle-treated cells doubled after 24 h (consistent with the known HeLa cell cycle length) (Boisvert et al. 2012), while CHX-treated cells remained attached but not actively proliferating (Supplemental Fig. S5A). This was in stark contrast to the widespread cell death observed after treatment with PURO or Triton X-100. The metabolic activities of the cells (as determined by their reducing potential) mirrored these trends, with CHX-treated cells retaining ∼80% metabolic activity after 24 h (Supplemental Fig. S5B). Therefore, even after 24 h of CHX treatment, HeLa cells remained viable and metabolically active, indicating that the observed increases in non-AUG translation are likely not due to widespread cell death responses.
Endogenous non-AUG translation is likewise resistant to CHX
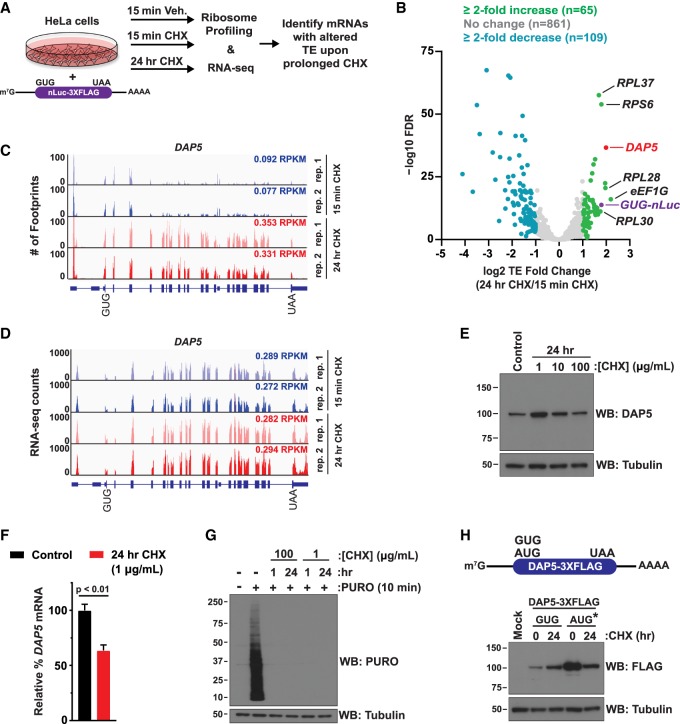
To identify endogenous mRNAs that might be similarly resistant to CHX, HeLa cells were treated with vehicle for 15 min or 100 µg/mL CHX for 15 min or 24 h followed by ribosome profiling and RNA sequencing (RNA-seq) analysis (Fig. 3A). As CHX stabilizes monosomes and polysomes (Low et al. 2005; Schneider-Poetsch et al. 2010), we used 15 min of CHX treatment (when cellular translation should be maximally inhibited) (Warner et al. 1966) as the control condition in order to account for these stabilizing effects. More than 1000 mRNAs had sufficient sequencing read coverage in both biological replicates to allow accurate determinations of their TEs (ratio of ribosome footprint RPKM [reads per kilobase per million mapped reads] to RNA-seq RPKM) (Supplemental Fig. S6). As expected, compared with cells treated with vehicle or CHX for 15 min, the vast majority (∼85%) of mRNAs exhibited a less than twofold change in TE after prolonged CHX treatment (Fig. 3B, gray; Supplemental Table S1).
Figure 3.
Translation of endogenous DAP5 (eIF4G2/p97) is resistant to CHX. (A) HeLa cells were transfected with the GUG-nLuc-3XFlag reporter. After 24 h, cells were treated with vehicle (Veh., 0.1% DMSO) or 100 µg/mL CHX and then incubated for 15 min or 24 h before being subjected to ribosome profiling and RNA-seq. (B) A volcano plot showing changes in TE of endogenous mRNAs (24 h of CHX vs. 15 min of CHX) compared with the associated false discovery rates (FDRs). FDRs for each mRNA are listed in Supplemental Table S1. (C,D) Ribosome footprints (C) and RNA-seq fragments (D) that mapped to the GUG-encoded DAP5 locus in cells that had been treated with CHX for 15 min (blue) or 24 h (red). Raw counts are depicted, and the ribosome footprint and RNA-seq RPKMs (which account for library size and ORF length) of the DAP5-coding sequence are also given for each replicate and time point. (E) Western blotting was used to assess endogenous DAP5 protein levels in HeLa cells before (control) and after 24 h of treatment with 1, 10, or 100 µg/mL CHX. Tubulin was used as a loading control. (F) RT-qPCR was used to assess endogenous DAP5 mRNA levels before (control) and after 24 h of 1 µg/mL CHX treatment. FH and NPTX1 mRNAs (whose levels did not change during CHX treatment as determined by RNA-seq) were used as dual internal references. Data are shown as mean ± SD. n = 3. A two-tailed unpaired t-test with Welch's correction was used. (G) After 1 or 24 h of treatment with 100 or 1 µg/mL CHX, HeLa cells were subjected to PURO labeling and Western blotting to confirm global translation inhibition. Tubulin was used as a loading control. (H) Western blotting was used to examine expression of the GUG- and AUG-encoded DAP5-3XFlag reporters before and after 24 h of 100 µg/mL CHX treatment. Tubulin was used as a loading control. AUG* denotes that one-fourth of the AUG-DAP5-3XFlag plasmid was transfected to avoid overexposure during film development.
Sixty-five mRNAs had a twofold or greater increase in TE after prolonged CHX treatment, including the GUG-nLuc reporter (Fig. 3B, purple; Supplemental Table S1). Remarkably, DAP5 (eIF4G2/p97) mRNA (Fig. 3B, red; Supplemental Table S1), which encodes an N-terminally truncated eIF4G1 homolog and has been shown previously to solely use a GUG start codon (Imataka et al. 1997; Levy-Strumpf et al. 1997; Takahashi et al. 2005), had nearly the largest increase in TE. More ribosome footprints were detected throughout the DAP5 ORF after prolonged CHX treatment (Fig. 3C) without substantial changes in mRNA levels as determined by RNA-seq (Fig. 3D; Supplemental Fig. S7A) or RT-qPCR (Supplemental Fig. S7B). This suggests that the observed increase in TE is not simply due to an accumulation of 80S ribosomes that were inhibited soon after initiation but rather that the DAP5 ORF was being actively translated in a manner that is resistant to CHX. Indeed, DAP5 protein levels increased with prolonged CHX treatment, albeit most noticeably with a lower CHX dose (1 µg/mL) than was used for the ribosome profiling (Fig. 3E). Interestingly, DAP5 mRNA levels somewhat decreased at this lower CHX dose (Fig. 3F), confirming that the increase in protein levels is driven primarily by increased translation. We further verified that this lower dose of CHX robustly inhibits global translation (Fig. 3G; Schneider-Poetsch et al. 2010). An increase in DAP5 protein levels was also observed upon prolonged treatment with CHX when we measured translation from a GUG-encoded DAP5-3XFlag expression construct (Fig. 3H). This increase was dependent on the non-AUG start codon because mutating the GUG start codon to an AUG start codon rendered DAP5-3XFlag expression sensitive to CHX (Fig. 3H).
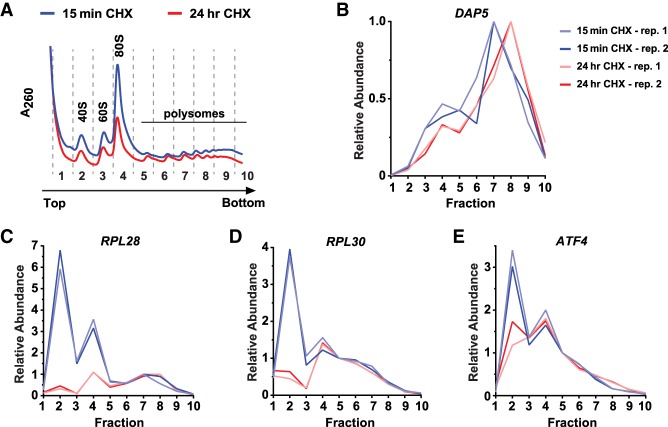
As an additional independent approach to verify that DAP5 translation increases upon CHX treatment, we confirmed that endogenous DAP5 mRNA shifted to heavier polysomes on sucrose gradients (Fig. 4A,B). In general, the polysome profiles showed that the levels of 40S and 60S ribosomal subunits as well as the 80S monosomes were decreased after 24 h of CHX treatment (Fig. 4A), which may be partially due to turnover of a short-lived rDNA-specific transcription factor (Gokal et al. 1986).
Figure 4.
Endogenous DAP5 mRNA cosediments with larger polysomes upon CHX treatment. (A) Polysome profiles of HeLa cells that had been treated with 100 µg/mL CHX for 15 min (blue) or 24 h (red). (B–E) RT-qPCR was used to determine the distribution of DAP5 (B), RPL28 (C), RPL30 (D), and ATF4 (E) mRNAs across 10%–50% sucrose gradients after 15 min (blue) or 24 h (red) of 100 µg/mL CHX treatment. Two biological replicates for each treatment are shown.
Upon examining the other mRNAs besides DAP5 with a greater than or equal to twofold increase in TE upon prolonged CHX treatment (Supplemental Table S1), we noted that 23 of the top 30 transcripts encode ribosomal proteins or translation factors (including eEF1G, which had the largest TE increase), all of which are thought to use a canonical AUG start codon. mRNAs encoding ribosomal proteins and translation factors are typically poorly translated (Supplemental Fig. S8A; Supplemental Table S1; Wang et al. 2004; Gismondi et al. 2014; Liu et al. 2014). We confirmed that the bulk of ribosomal protein mRNAs was not found on actively translating ribosomes in control conditions (15 min of CHX) but instead in the lighter sucrose gradient fractions. This is exemplified by the RPL28 (Fig. 4C) and RPL30 mRNAs (Fig. 4D) and was true for the other ribosomal protein mRNAs examined (Supplemental Fig. S8B). Ribosome profiling revealed no substantial change in footprint accumulation across these coding sequences (Supplemental Fig. S9), and, in fact, significant decreases in their mRNA levels were detected by RNA-seq (Supplemental Figs. S7A, S9; Supplemental Table S1) and confirmed by RT-qPCR (Supplemental Fig. S7B). By definition, calculation of the TE should account for such changes in mRNA levels, but we reasoned that the TE could become artificially inflated if untranslated mRNAs were degraded preferentially during the prolonged CHX treatment. Indeed, a clear reduction in the amounts of ribosomal protein mRNAs present in the lighter fractions was observed after 24 h of CHX treatment (Fig. 4C,D; Supplemental Fig. S8B).
Recent work has revealed that ribosomal protein mRNAs are preferentially degraded when not bound by free 40S ribosomal subunits (Gentilella et al. 2017). It thus appears that CHX treatment leads to the accumulation of slowly elongating ribosomes (Schneider-Poetsch et al. 2010), thereby limiting the amount of both subunits entering the free pool and causing decay of these particular mRNAs. As further evidence that the TE of these genes did not truly increase with prolonged CHX treatment, we found that the distribution of these mRNAs in the polysome fractions did not shift toward heavier polysomes (Fig. 4C,D). Similar patterns were observed for ATF4 mRNA (Fig. 4E), a well-established poorly translated mRNA that harbors inhibitory upstream and overlapping ORFs in its 5′ leader (Harding et al. 2000), as well as for other non-translation-related mRNAs that appeared to have a greater than twofold increased TE by ribosome profiling (Supplemental Fig. S8C). Together, these data indicate that a number of nontranslated mRNAs are selectively degraded over prolonged CHX treatment, whereas translation of the GUG-encoded DAP5 (eIF4G2/p97) mRNA appears to be truly resistant to CHX (Figs. 3, 4B).
We then examined translation from other annotated non-AUG start codons, most of which encode uORFs or N-terminal extensions (Touriol et al. 2003; Ivanov et al. 2011). This analysis is complicated, as ribosome profiling usually cannot decipher which start codon was used for footprints that map within a coding sequence. Furthermore, because non-AUG start codons are very inefficient (Fig. 1A), one would predict that most ribosome footprints on an mRNA with a non-AUG N-terminal extension would be derived from the AUG start codon. Nevertheless, it does appear that non-AUG translation from the BAG1 mRNA is also resistant to CHX (Supplemental Fig. S10). Translation of BAG1 mRNA, which encodes an antiapoptotic oncoprotein, can initiate at a CUG start codon upstream of at least two AUG start codons (Packham et al. 1997), and we observed a 2.6-fold increase in TE upon prolonged CHX treatment (Supplemental Fig. S10A,B). Note that BAG1 is not included in Figure 3B, as it did not meet the stringent coverage thresholds originally used (due to low ribosome footprints in the 15-min vehicle samples). Upon prolonged CHX treatment, increased ribosome footprints were detected across the BAG1-coding sequence (Supplemental Fig. S10A), with minimal changes in mRNA levels as detected by RNA-seq (Supplemental Fig. S10B) and confirmed by RT-qPCR (Supplemental Fig. S10C). Endogenous BAG1 mRNA was further detected on heavier polysomes upon prolonged CHX treatment, consistent with increased translation (Supplemental Fig. S10D). Upon cloning the BAG1 5′ untranslated region (UTR) upstream of an nLuc reporter, we observed that the reporter was resistant to CHX when it was translated using the natural BAG1 CUG start codon but was inhibited by CHX when the CUG was mutated to an AUG start codon (Supplemental Fig. S10E). These results indicate that translation from multiple endogenous non-AUG start codons, including in the DAP5 (eIF4G2/p97) and BAG1 mRNAs, is resistant to CHX.
CHX likely induces ribosome queuing to promote non-AUG translation
How, then, is non-AUG translation specifically resistant to CHX? Initiation at non-AUG start codons has long been known to be influenced by downstream mRNA secondary structures that block scanning PICs and position them optimally over the non-AUG start codon (Kozak 1990). This blockage biases the scanning PIC to recognize the nonoptimal start codon, thereby stimulating initiation. Ribosome queuing (or stacking of ribosomes) can likewise increase non-AUG translation when PICs accumulate upstream of a stalled 80S ribosome (Ivanov et al. 2018). We thus hypothesized that CHX may similarly stimulate formation of a ribosome queue: Once CHX slows a ribosome within the coding sequence, a stalling point for the upstream PICs would be generated (Fig. 5A). New PICs would continue to be loaded and scan 5′ to 3′ (as CHX targets only elongating 80S ribosomes) (Schneider-Poetsch et al. 2010; Garreau de Loubresse et al. 2014), usually passing weak non-AUG start codons and ultimately generating a queue/stack of PICs (perhaps with an occasional elongating ribosome also included). This queue/stack could then allow PICs to be positioned over the non-AUG start codon for a longer period of time, allowing for increased initiation. Given that CHX treatment does not cause a complete block to translation elongation (Fig. 2F–H), this model predicts that the expression of proteins from non-AUG codons (but not from canonical AUG codons that are efficiently recognized by scanning PICs) should increase in the presence of CHX.
Figure 5.
Ribosome queuing likely occurs upon CHX treatment. (A) A model of CHX-resistant translation via ribosome queuing. Scanning PICs inefficiently recognize non-AUG start codons, and thus most PICs continue to scan downstream in search of a start codon. When an elongating 80S ribosome is slowed by CHX (top), PICs continue to be loaded and ultimately start to queue (or stack) upstream of the slowed 80S ribosome (middle). (Bottom) This places a PIC near a non-AUG start codon for an extended period of time, allowing for increased non-AUG translation. (4A) eIF4A. (B) A stable hairpin (HP) was inserted within the 5′ leader of the AUG-nLuc-3XFlag and GUG-nLuc-3XFlag reporters. Northern blotting was used to examine expression of the nLuc reporter mRNAs in HeLa cells 24 h after transfection. 28S ribosomal RNA was used as a loading control. (C) Raw luciferase values of nLuc-3XFlag reporters with and without the hairpin (HP) 24 h after transfection. Data are shown as mean ± SD. n = 3. (D) HeLa cells were transfected with the indicated nLuc-3XFlag reporters for 24 h and then either collected (control) or treated for 24 h with 100 µg/mL CHX. Luminescence signals for each reporter were quantified and set relative to the associated control samples. Data are shown as mean ± SD. n = 3. (E) HeLa cells were transfected with the destabilized nLuc-3XFlag-CL1/PEST reporters for 24 h and then collected (control) or treated for 3 h with 100 µg/mL CHX, 200 µM NSC119893, or 100 µg/mL CHX + 200 µM NSC119893. Luminescence signals for each reporter were quantified and set relative to the associated control samples. Data are shown as mean ± SD. n = 3.
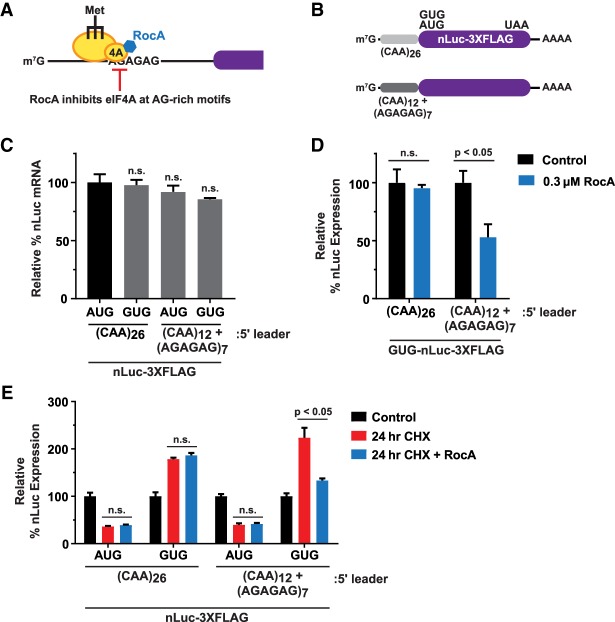
If this inhibitor-induced ribosome queuing model is correct, we reasoned that impairing PIC formation or scanning should block formation of the queue (Ivanov et al. 2018) and cause non-AUG translation to now be sensitive to CHX. We thus used three distinct strategies to impair PIC formation or scanning and then tested their respective effects on the non-AUG nLuc reporters. First, we sterically inhibited scanning PICs by inserting a 40-nucleotide (nt) hairpin upstream of the nLuc-coding sequence (Fig. 5B, top). In alignment with prior reports that stable secondary structures in 5′ leaders limit downstream start codon recognition (Kozak 1989), insertion of the hairpin reduced the basal translational output of the AUG-nLuc and GUG-nLuc reporters by ∼20 fold and ∼10-fold, respectively (Fig. 5C), without decreasing steady-state mRNA levels (Fig. 5B, bottom). Notably, insertion of the hairpin was sufficient to cause the GUG-nLuc reporter to be inhibited by CHX (Fig. 5D). This is likely because the ribosome queue was generated upstream of the hairpin, not over the non-AUG start codon. Second, we limited PIC formation by disrupting the eIF2•GTP•Met-tRNAiMet TC by treating HeLa cells with NSC119893 (Fig. 1F). This treatment blocked CHX-resistant translation of the GUG-nLuc reporter (Fig. 5E), consistent with a requirement for PIC loading for a queue to form. Third, we used the pharmacological inhibitor rocaglamide A (RocA), which inhibits the eIF4A helicase and stalls scanning PICs at AGAGAG-rich motifs (Fig. 6A; Iwasaki et al. 2016). nLuc reporters that lack a (AGAGAG)7 motif in their 5′ leader were unaffected by RocA, as expected (Fig. 6B–D). In contrast, when the (AGAGAG)7 motif was present, the GUG-nLuc reporter was inhibited by RocA (Fig. 6D). Furthermore, in the presence of CHX, RocA significantly decreased the expression of the (AGAGAG)7-containing GUG-nLuc reporter (Fig. 6E), likely by stalling PICs upstream of the non-AUG start codon. These three sets of data thus are all consistent with a model in which CHX generates a ribosome queue that positions PICs and allows for increased initiation at a non-AUG start codon.
Figure 6.
Inhibition of scanning PICs by RocA blocks CHX-resistant translation. (A) RocA inhibits scanning PICs by causing the eIF4A helicase to clamp onto AGAGAG-rich sequence motifs. (B) nLuc-3XFlag reporters harboring either a (CAA)26 5′ leader, which is insensitive to RocA, or a (CAA)12 + (AGAGAG)7 5′ leader, which is sensitive to RocA. (C) RT-qPCR was used to quantify nLuc reporter mRNA levels 24 h after transfection. mRNA levels were first normalized to the cotransfected FFLuc (pGL4.13) reporter. (CAA)26 5′ leader AUG-nLuc (black) was then used to determine the relative expression levels of the other nLuc reporters (gray). Data are shown as mean ± SD. n = 3. A two-tailed unpaired t-test with Welch's correction was used. (D) HeLa cells were transfected with the indicated nLuc-3XFlag reporters and treated with 0.1% DMSO (control) or 0.3 µM RocA for 24 h. Luminescence signals for each reporter were quantified and set relative to the associated control samples. Data are shown as mean ± SD. n = 3. A two-tailed unpaired t-test with Welch's correction was used. (E) HeLa cells were transfected with the indicated nLuc-3XFlag reporters for 24 h and then collected (control) or treated for 24 h with 100 µg/mL CHX ± 0.3 µM RocA. Luminescence signals for each reporter were quantified and set relative to the associated control samples. Data are shown as mean ± SD. n = 3. A two-tailed unpaired t-test with Welch's correction was used.
Non-AUG translation is also resistant to other elongation inhibitors that are predicted to induce ribosome queuing
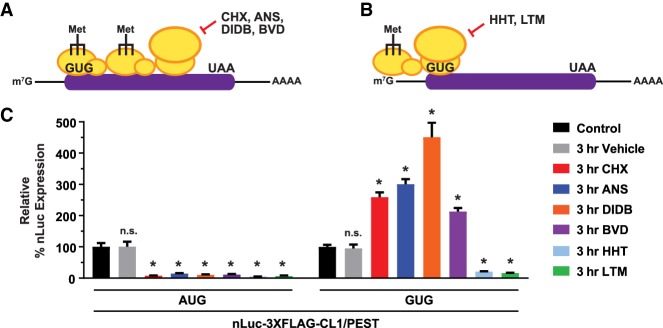
Last, we hypothesized that other elongation inhibitors that can stall ribosomes within the coding sequence may also generate queues and stimulate non-AUG translation. Anisomycin (ANS), didemnin B (DIDB), and bouvardin (BVD) inhibit different steps of elongation (Supplemental Table S2), but each should result in stalled or slowed 80S ribosomes downstream from the start codon (Fig. 7A). When HeLa cells were treated with these inhibitors at concentrations that robustly inhibited translation of the AUG-nLuc reporter, ANS, DIDB, and BVD each caused increased translation of the GUG-nLuc reporter (Fig. 7C). In contrast, the early elongation inhibitors homoharringtonine (HHT) and lactimidomycin (LTM) have a high preference to stall 80S ribosomes at the start codon after initiation (Supplemental Table S2). This would lead to a PIC queue being generated upstream of the start codon, with no scanning PIC able to access the non-AUG start codon (Fig. 7B). We thus predicted that no stimulation of the non-AUG start codon itself should be observed. Indeed, HHT and LTM inhibited the AUG-nLuc and GUG-nLuc reporters (Fig. 7C; Supplemental Fig. S11A,B). These data thus suggest that any method that slows translation elongation (but does not fully block it) downstream from a non-AUG start codon may be able to generate a PIC/ribosome queue that increases non-AUG translation. In fact, it was shown recently that translation of a non-AUG uORF within the antizyme inhibitor 1 (AZIN1) mRNA is regulated in this manner by a downstream ribosome pause sequence (Ivanov et al. 2018).
Figure 7.
Slowing elongation within coding sequences stimulates non-AUG translation. (A) Multiple translation inhibitors, including CHX, ANS, DIDB, and BVD, that can inhibit elongating ribosomes within the coding sequence are predicted to generate a PIC queue over the non-AUG-start codon. (B) HHT and LTM preferentially inhibit 80S ribosomes shortly after initiation and thus should generate a PIC queue upstream of the non-AUG start codon. (C) HeLa cells were transfected with the destabilized nLuc-3XFlag-CL1/PEST reporters for 24 h and then collected (control) or treated for 3 h with vehicle (0.1% DMSO) or one of the following protein synthesis inhibitors: 100 µg/mL CHX, 0.5 µg/mL ANS, 1 µM DIDB, 1 µM BVD, 1 µg/mL HHT, or 5 µM LTM. Luminescence signals for each reporter were quantified and set relative to the associated control samples. Data are shown as mean ± SD. n = 3. A two-tailed unpaired t-test with Welch's correction was used to compare expression levels with the control treatment samples. (*) P < 0.01.
Discussion
Canonical eukaryotic translation initiates at an AUG start codon, but it is becoming increasingly clear that near-cognate start codons can also be used. Such initiation events are typically not efficient but can (1) provide a layer of translational control when encoding uORFs, (2) be used to generate functional proteins (including DAP5 and proteins with N-terminal extensions), and/or (3) drive human disease progression (for review, see Kearse and Wilusz 2017). It has further long been known that treating eukaryotic cells with a high dose (100 µg/mL; 355 µM) of the elongation inhibitor CHX robustly blocks cytoplasmic translation (Fig. 2B; Ennis and Lubin 1964; Schneider-Poetsch et al. 2010). In the present study, we used highly validated reporters and ribosome profiling to surprisingly demonstrate that non-AUG translation is resistant to high doses of CHX and other elongation inhibitors that robustly inhibit global translation. We further provide multiple lines of evidence that are consistent with a model in which ribosome queuing drives this increase in translation from non-AUG start codons (Fig. 5A). This study thus reveals a previously unappreciated way in which the efficiency of non-AUG translation can increase in cellular conditions that strongly inhibit global translation. Additionally, this study also provides important new insights into how translation inhibitors can influence ribosome profiling data (Gerashchenko and Gladyshev 2014; Jackson and Standart 2015; Andreev et al. 2017), revealing that inhibitors can induce PIC queuing to increase non-AUG initiation as well as decrease the amounts of untranslated mRNAs.
In order to study the mechanism of non-AUG translation, it was critical in the present study to first generate and stringently validate new nLuc-based reporters (Fig. 1A; Supplemental Fig. S1). nLuc is ∼100-fold brighter than other luciferase reporters (Hall et al. 2012), thereby allowing a better signal:noise ratio, which is especially critical when measuring low-efficiency non-AUG translation events. Furthermore, we found that commonly used FFLuc-based non-AUG translation reporters produce multiple polypeptides when expressed in human cells, including at least two truncated versions that initiate from internal AUG codons (Fig. 1E; Supplemental Fig. S2). These truncated FFLuc protein species may or may not directly contribute to the measured luminescence but accumulate to higher levels than the full-length protein (Fig. 1E). This complicates the analysis of FFLuc protein levels by Western blotting or immunofluorescence as well as the analysis of TEs by ribosome profiling or polysome profiling. In contrast, the nLuc-based reporters had none of these issues.
When the non-AUG nLuc reporters were expressed in human cells, we found that they became more efficiently translated in the presence of a high dose (100 µg/mL) of CHX (Fig. 2C–E). Likewise, ribosome profiling and polysome analysis revealed that synthesis of the GUG-encoded DAP5 (eIF4G2/p97) and the CUG-encoded BAG1 proteins are resistant to CHX (Figs. 3, 4A,B; Supplemental Fig. S10). It had been shown previously that very low concentrations (0.02–0.5 µg/mL) of CHX can stimulate the translation of poorly translated mRNAs (Walden et al. 1981). At these low levels of inhibition, highly translated mRNAs likely spend more time bound by slowly elongating 80S ribosomes and less time bound by rate-limiting initiation factors, thus allowing poorly translated mRNAs more access to initiation factors (Walden et al. 1981). However, as the CHX concentration was increased to ≥0.5 µg/mL, these stimulatory effects on poorly translated mRNAs were no longer observed (Walden et al. 1981), as elongation of these mRNAs should now be more robustly inhibited. If this competition-based model explained the CHX-resistant translation phenotype that is observed in the present study, we would have expected that reducing the TE of a canonically translated mRNA would result in resistance to CHX. However, our experiments strongly indicate that low TE is not sufficient to make a transcript resistant to high doses of CHX.
We instead propose that ribosome queuing is responsible for the increased efficiency of non-AUG translation that is observed upon treatment with high doses of CHX (Fig. 5A). Consistent with this model, perturbation of PIC formation, loading, or scanning rendered non-AUG translation sensitive to CHX (Figs. 5, 6), likely because the queue was no longer able to efficiently form. Moreover, we found that non-AUG translation was resistant to other elongation inhibitors that can stall ribosomes within coding sequences but not those that stall elongation immediately following initiation (Fig. 7). It thus appears that a number of protein synthesis inhibitors can initiate formation of queues and that the location of the queue is a critical determinant of whether it can drive increased initiation from start codons that are normally poorly used. Notably, a natural form of ribosome queuing has been proposed recently to regulate polyamine synthesis via stimulating translation of an inhibitory uORF (Ivanov et al. 2018). As with that prior study, we found that PIC queuing can increase translation from non-AUG codons, but it remains possible that queuing can also increase translation from certain AUG codons that are poorly used. We further found that the strength of the elongation block is a key determinant of whether the queue can lead to increased translational output. When the elongation block is too strong, the observed increase in initiation is overshadowed and protein levels decrease. For example, we found that non-AUG translation is inhibited (albeit not as robustly as canonical translation) when elongation is very strongly blocked by the irreversible inhibitor emetine (EME) or high concentrations of ASN or BVD (Supplemental Fig. S11C).
Beyond regulating polyamine synthesis (Ivanov et al. 2018), we suggest that ribosome queuing may play a broader role in controlling cellular non-AUG translation events. For example, in Huntington's disease, Gln-tRNACAG can become depleted due to translation of the expanded CAG repeats within the huntingtin mRNA (Girstmair et al. 2013). It is tempting to speculate that this may cause a ribosome queue to form on the huntingtin mRNA that drives further initiation of RAN translation (Bañez-Coronel et al. 2015). Further studies should reveal critical insights into the mechanisms that control natural ribosome queues, especially the cellular signaling pathways (e.g., the inhibitory phosphorylation of eEF2) and molecular trigger events (e.g., depletion of a specific charged tRNA or the presence of ribosome pause sites). It is nevertheless important to keep in mind that such ribosome/PIC queues formed due to elongation blocks may be naturally limited by simultaneous down-regulation of initiation. For example, starvation is known to induce elongation inhibition via phosphorylation of eEF2, but initiation is also limited via down-regulation of mTOR activity (for review, see Sabatini 2017). Such coregulation likely prevents the formation of ribosome queues transcriptome-wide while still allowing the potential for transcript-specific regulation. Future studies characterizing these processes may ultimately allow the development of therapeutics that modulate queues and ultimately inhibit pathogenic non-AUG translation.
Materials and methods
Cell culture
HeLa and HEK293T cells were obtained from American Type Culture Collection and grown at 37°C and 5% CO2. HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing high glucose (Thermo Fisher Scientific) supplemented with 10% (v/v) fetal bovine serum (FBS), 1% (v/v) nonessential amino acids, and 1% (v/v) penicillin–streptomycin on standard tissue culture-treated dishes. HEK293T cells were maintained in DMEM containing high glucose supplemented with 10% (v/v) FBS and 1% (v/v) penicillin–streptomycin on CellBIND-treated dishes (Corning). IMR90 cells were a kind gift from David L. Spector (Cold Spring Harbor Laboratory) and were maintained in DMEM containing high glucose supplemented with 10% (v/v) FBS and 1% (v/v) penicillin–streptomycin on standard tissue culture-treated dishes.
Inhibitors
The following compounds were obtained from Sigma: CHX (100 mg/mL stock in DMSO; C1988), PURO (10 mg/mL stock in water; P9620), ANS (10 mg/mL stock in DMSO; A5862), EME (100 mg/mL stock in water; E2375), and HHT (10 mg/mL stock in DMSO; SML1091). LTM (5 mM stock in DMSO; 506291) was obtained from Millipore. BVD (10 mM stock in DMSO) and DIDB (10 mM in DMSO) were kind gifts from Tin Tin Su (University of Colorado at Boulder) and Malia Potts (St. Jude Children's Research Hospital), respectively. NSC119893 (50 mM stock in DMSO) was obtained from the National Cancer Institute Development Therapeutics Program.
Reporter plasmids
pcDNA3.1(+)-based plasmids expressing nLuc with a C-terminal 3XFlag tag were described previously (Kearse et al. 2016). The FFLuc expression plasmid was obtained from Promega (pGL4.13). Full sequences of the inserts for all plasmids are in the Supplemental Material.
Reporter assays
HeLa and HEK293T cells were seeded in 96-well plates with 100 µL of medium. After 24 h, cells (∼50% confluency) were transfected with 100 ng of plasmid DNA (50 ng of nLuc plasmid, 50 ng of pGL4.13 [FFLuc]) using Viafect (Promega) at a 2:1 (reagent:DNA) ratio following the manufacturer's protocol. IMR90 cells were seeded in 96-well plates with 100 µL of medium, grown until ∼50% confluent (often 4–5 d), and then transfected using Viafect at a 4:1 ratio. Twenty-four hours after transfection, cells were either lysed in well with 100 µL of Glo lysis buffer (Promega) for 10 min and frozen at −80°C (for the control) or treated with 100 µg/mL CHX (or other inhibitors as described in the figure legends) for the indicated amounts of time before lysis/sample freezing. Luciferase assays were then performed on thawed samples by mixing equal volumes of cell lysate with either ONE-Glo (Promega) for FFLuc or Nano-Glo (Promega) for nLuc in black 96-well plates for 5 min. Luminescence was measured on a GloMax Explorer multimode plate reader (Promega). The cotransfected FFLuc was used as an internal control to confirm protein synthesis inhibition but was not used for normalization except where indicated. In assays using NSC119893, fresh medium with inhibitors was replaced 1.5 h after the initial treatment, as described previously (Robert et al. 2006).
Western blot analysis
HeLa cells were seeded in 12-well plates with 1.5 mL of medium. After 24 h, cells (∼50% confluency) were transfected with 20 or 450 ng of plasmid (20 ng of AUG-encoded reporters, 450 ng of non-AUG-encoded reporters) using Viafect (Promega) at a 2:1 (reagent:DNA) ratio. Different amounts of AUG- and non-AUG-encoding plasmids were transfected to prevent overexposure of the higher-expressing AUG-encoded reporters. Twenty-four hours after transfection, cells were either lysed in well in 250 µL of RIPA buffer (for the control) or treated with 100 µg/mL CHX for the indicated amount of time before lysis. SDS-PAGE and transfer onto PVDF were performed using NuPAGE reagents according to the manufacturer's protocol. Membranes were blocked with 5% nonfat dry milk for 30 min before incubation in primary antibody (in TBST, 0.02% NaN3) overnight. The following antibodies were used: mouse anti-Flag M2 (1:1000; Sigma, F8104), mouse anti-PURO (1:2000; Millipore, MABE343), mouse antivinculin (1:1000; Sigma, V9131), mouse anti-α-Tubulin (1:1000; Sigma, T6074), and rabbit anti-DAP5 (1:1000; Cell Signaling, 5169). Sheep antimouse IgG/HRP (GE Healthcare, NA931) and donkey antirabbit IgG/HRP (GE Healthcare, NA934) were used at 1:10,000 in TBST.
Northern blot analysis
HeLa cells were seeded in 12-well plates with 1.5 mL of medium. After 24 h, cells (∼50% confluency) were transfected with 500 ng of plasmid (250 ng of nLuc reporter, 250 ng of pGL4.13, which served as a transfection control) using Viafect (Promega) at 2:1 (reagent:DNA). Twenty-four hours after transfection, total RNA was harvested with TRIzol (Thermo Fisher Scientific). Ten micrograms of total RNA was separated on a 1.2% denaturing formaldehyde agarose gel using NorthernMax reagents (Thermo Fisher Scientific). Northern blots were then performed as described previously (Tatomer et al. 2017).
PURO labeling
Cells were seeded in 12-well dishes as described above. After 24 h, the medium was replaced with 1.5 mL of fresh medium containing 1 or 100 µg/mL CHX, and cells were incubated for 1 or 24 h before 1.5 µL of 10 mg/mL PURO was added (final concentration of 10 µg/mL). Plates were mixed gently and placed back in the incubator for 10 min. Cells lysates were then prepared and analyzed by Western blot as described above.
Crystal violet cell counting and metabolism assays
HeLa cells were plated in 12-well dishes as described above and grown for 24 h before adding the indicated inhibitors. At the indicated time points, cells were then washed gently with ice-cold PBS, fixed with 100% methanol for 15 min, and subsequently stained with 0.25% (w/v) crystal violet in 70% methanol for 5 min as described previously (Feoktistova et al. 2016). For each treatment and time point, four images of three separate wells were collected. Cells were counted in batch using ImageJ.
To measure cell metabolism, cells were grown in tissue culture-treated black 96-well plates and analyzed using the real-time MT assay (Promega) following the manufacturer's instructions. The MT assay reagents were added to cells for 3 h before adding inhibitors and recording the initial time point.
Ribosome profiling and RNA-seq
HeLa cells were seeded in duplicate for each time point in 10-cm plates with 10 mL of medium. After 24 h, cells (∼50% confluency) were transfected with 5 µg of GUG-nLuc plasmid DNA using Viafect (Promega) at a 2:1 (reagent:DNA) ratio following the manufacturer's protocol. Twenty-four hours later, medium was replaced with fresh medium containing 0.1% DMSO (vehicle) for 15 min or 100 µg/mL CHX for 15 min or 24 h. At each time point, cells were placed on ice, washed with 10 mL of ice-cold PBS with 100 µg/mL CHX, and lysed in plate in 400 µL of cold lysis buffer (20 mM Tris at pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 1% Triton X-100, 50 µg/mL EME, 25 U/mL Turbo DNase [Thermo Fisher Scientific]). Cells were then scraped off the plate, transferred to a 1.5-mL tube, and incubated for 5 min on ice. Cell debris was pelleted at 16,000g for 10 min at 4°C. The resulting supernatant was then snap-frozen in liquid nitrogen before storing at −80°C. Ribosome profiling and RNA-seq were performed and analyzed as described previously (Ingolia et al. 2012; Dobin et al. 2013; Jiang et al. 2014; Xiao et al. 2016) with minor modifications described in the Supplemental Material. All ribosome profiling and RNA-seq data sets generated in this study are available for download from Gene Expression Omnibus (GSE125086).
Polysome analysis and RT-qPCR
HeLa cells were seeded in 10-cm plates for 24 h and treated with 100 µg/mL CHX for 15 min or 24 h. Cells were pelleted, lysed, and then layered on top of a linear 10%–50% buffered sucrose gradient followed by centrifugation and fractionation. Full details are in the Supplemental Material.
Supplementary Material
Acknowledgments
We thank Gideon Dreyfuss, Ben Garcia, Zissimos Mourelatos, and members of the Wilusz laboratory for valuable discussions and advice. Tin Tin Su (University of Colorado at Boulder) and Malia Potts (St. Jude Children's Research Hospital) kindly provided BVD and DIDB, respectively. This work was supported by National Institutes of Health grants K99-GM126064 (to M.G.K.), R35-GM119735 (to J.E.W.), NCI-P30-CA016520 (to J.E.W.), T32-GM007315 (to K.M.G.), F31-NS100302 (to K.M.G.), R01-GM105707 (to A.C.G.), R01-NS099280 (to P.K.T.), R01-NS086810 (to P.K.T.), and R37-GM059425 (to R.G.). D.H.G. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2280-16). P.K.T. is supported by Veterans Affairs Biomedical Laboratory Research and Development grants 1I21BX001841 and 1I01BX003231 and the Michigan Discovery Fund. R.G. is a Howard Hughes Medical Institute Investigator. J.E.W. is a Rita Allen Foundation Scholar. Illumina sequencing was conducted at the Genetic Resources Core Facility at the Johns Hopkins Institute of Genetic Medicine.
Author contributions: M.G.K. and J.E.W. conceived and designed the experiments. M.G.K. performed all experiments with help from J.C. (RocA experiments), D.L. (Northern blots), C.N. (Western blots), and K.M.G. (RAN translation reporters). D.H.G. and R.G. performed and analyzed the ribosome profiling and RNA-seq experiments. A.C.G. and P.K.T. helped with initial experimental design and data interpretation. M.G.K. and J.E.W. wrote the manuscript with input from all coauthors.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.324715.119.
References
- Andreev DE, O'Connor PB, Loughran G, Dmitriev SE, Baranov PV, Shatsky IN. 2017. Insights into the mechanisms of eukaryotic translation gained with ribosome profiling. Nucleic Acids Res 45: 513–526. 10.1093/nar/gkw1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud E, Touriol C, Boutonnet C, Gensac MC, Vagner S, Prats H, Prats AC. 1999. A new 34-kilodalton isoform of human fibroblast growth factor 2 is cap dependently synthesized by using a non-AUG start codon and behaves as a survival factor. Mol Cell Biol 19: 505–514. 10.1128/MCB.19.1.505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bañez-Coronel M, Ayhan F, Tarabochia AD, Zu T, Perez BA, Tusi SK, Pletnikova O, Borchelt DR, Ross CA, Margolis RL, et al. 2015. RAN translation in Huntington disease. Neuron 88: 667–677. 10.1016/j.neuron.2015.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, Ahmad Y, Gierliński M, Charrière F, Lamont D, Scott M, Barton G, Lamond AI. 2012. A quantitative spatial proteomics analysis of proteome turnover in human cells. Mol Cell Proteomics 11: M111.011429 10.1074/mcp.M111.011429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugler B, Amalric F, Prats H. 1991. Alternative initiation of translation determines cytoplasmic or nuclear localization of basic fibroblast growth factor. Mol Cell Biol 11: 573–577. 10.1128/MCB.11.1.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Wang S, Mestre AA, Fu C, Makarem A, Xian F, Hayes LR, Lopez-Gonzalez R, Drenner K, Jiang J, et al. 2018. C9ORF72 GGGGCC repeat-associated non-AUG translation is upregulated by stress through eIF2α phosphorylation. Nat Commun 9: 51 10.1038/s41467-017-02495-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary JD, Ranum LP. 2017. New developments in RAN translation: insights from multiple diseases. Curr Opin Genet Dev 44: 125–134. 10.1016/j.gde.2017.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couderc B, Prats H, Bayard F, Amalric F. 1991. Potential oncogenic effects of basic fibroblast growth factor requires cooperation between CUG and AUG-initiated forms. Cell Regul 2: 709–718. 10.1091/mbc.2.9.709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev SE, Terenin IM, Andreev DE, Ivanov PA, Dunaevsky JE, Merrick WC, Shatsky IN. 2010. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J Biol Chem 285: 26779–26787. 10.1074/jbc.M110.119693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennis HL, Lubin M. 1964. Cycloheximide: aspects of inhibition of protein synthesis in mammalian cells. Science 146: 1474–1476. 10.1126/science.146.3650.1474 [DOI] [PubMed] [Google Scholar]
- Feoktistova M, Geserick P, Leverkus M. 2016. Crystal violet assay for determining viability of cultured cells. Cold Spring Harb Protoc 2016: pdb.prot087379 10.1101/pdb.prot087379 [DOI] [PubMed] [Google Scholar]
- Garreau de Loubresse N, Prokhorova I, Holtkamp W, Rodnina MV, Yusupova G, Yusupov M. 2014. Structural basis for the inhibition of the eukaryotic ribosome. Nature 513: 517–522. 10.1038/nature13737 [DOI] [PubMed] [Google Scholar]
- Gentilella A, Morón-Duran FD, Fuentes P, Zweig-Rocha G, Riaño-Canalias F, Pelletier J, Ruiz M, Turón G, Castaño J, Tauler A, et al. 2017. Autogenous control of 5′ TOP mRNA stability by 40S ribosomes. Mol Cell 67: 55–70. 10.1016/j.molcel.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko MV, Gladyshev VN. 2014. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res 42: e134 10.1093/nar/gku671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girstmair H, Saffert P, Rode S, Czech A, Holland G, Bannert N, Ignatova Z. 2013. Depletion of cognate charged transfer RNA causes translational frameshifting within the expanded CAG stretch in huntingtin. Cell Rep 3: 148–159. 10.1016/j.celrep.2012.12.019 [DOI] [PubMed] [Google Scholar]
- Gismondi A, Caldarola S, Lisi G, Juli G, Chellini L, Iadevaia V, Proud CG, Loreni F. 2014. Ribosomal stress activates eEF2K-eEF2 pathway causing translation elongation inhibition and recruitment of terminal oligopyrimidine (TOP) mRNAs on polysomes. Nucleic Acids Res 42: 12668–12680. 10.1093/nar/gku996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokal PK, Cavanaugh AH, Thompson EA. 1986. The effects of cycloheximide upon transcription of rRNA, 5 S RNA, and tRNA genes. J Biol Chem 261: 2536–2541. [PubMed] [Google Scholar]
- Green KM, Linsalata AE, Todd PK. 2016. RAN translation-What makes it run? Brain Res 1647: 30–42. 10.1016/j.brainres.2016.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KM, Glineburg MR, Kearse MG, Flores BN, Linsalata AE, Fedak SJ, Goldstrohm AC, Barmada SJ, Todd PK. 2017. RAN translation at C9orf72-associated repeat expansions is selectively enhanced by the integrated stress response. Nat Commun 8: 2005 10.1038/s41467-017-02200-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, et al. 2012. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem Biol 7: 1848–1857. 10.1021/cb3002478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann SR. 1994. Regulation and function of non-AUG-initiated proto-oncogenes. Biochimie 76: 880–886. 10.1016/0300-9084(94)90190-2 [DOI] [PubMed] [Google Scholar]
- Hann SR, King MW, Bentley DL, Anderson CW, Eisenman RN. 1988. A non-AUG translational initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt's lymphomas. Cell 52: 185–195. 10.1016/0092-8674(88)90507-7 [DOI] [PubMed] [Google Scholar]
- Hann SR, Sloan-Brown K, Spotts GD. 1992. Translational activation of the non-AUG-initiated c-myc 1 protein at high cell densities due to methionine deprivation. Genes Dev 6: 1229–1240. 10.1101/gad.6.7.1229 [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6: 1099–1108. 10.1016/S1097-2765(00)00108-8 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. 2017. Structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem Sci 42: 589–611. 10.1016/j.tibs.2017.03.004 [DOI] [PubMed] [Google Scholar]
- Hussmann JA, Patchett S, Johnson A, Sawyer S, Press WH. 2015. Understanding biases in ribosome profiling experiments reveals signatures of translation dynamics in yeast. PLoS Genet 11: e1005732 10.1371/journal.pgen.1005732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka H, Olsen HS, Sonenberg N. 1997. A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J 16: 817–825. 10.1093/emboj/16.4.817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802. 10.1016/j.cell.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. 2012. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc 7: 1534–1550. 10.1038/nprot.2012.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Firth AE, Michel AM, Atkins JF, Baranov PV. 2011. Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences. Nucleic Acids Res 39: 4220–4234. 10.1093/nar/gkr007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Shin BS, Loughran G, Tzani I, Young-Baird SK, Cao C, Atkins JF, Dever TE. 2018. Polyamine control of translation elongation regulates start site selection on antizyme inhibitor mRNA via ribosome queuing. Mol Cell 70: 254–264. 10.1016/j.molcel.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Floor SN, Ingolia NT. 2016. Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature 534: 558–561. 10.1038/nature17978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R, Standart N. 2015. The awesome power of ribosome profiling. RNA 21: 652–654. 10.1261/rna.049908.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Lei R, Ding SW, Zhu S. 2014. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15: 182 10.1186/1471-2105-15-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse MG, Wilusz JE. 2017. Non-AUG translation: a new start for protein synthesis in eukaryotes. Genes Dev 31: 1717–1731. 10.1101/gad.305250.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse MG, Green KM, Krans A, Rodriguez CM, Linsalata AE, Goldstrohm AC, Todd PK. 2016. CGG repeat-associated non-AUG translation utilizes a cap-dependent scanning mechanism of initiation to produce toxic proteins. Mol Cell 62: 314–322. 10.1016/j.molcel.2016.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Park SM, Park JH, Keum SJ, Jang SK. 2011. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J 30: 2454–2464. 10.1038/emboj.2011.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolitz SE, Lorsch JR. 2010. Eukaryotic initiator tRNA: finely tuned and ready for action. FEBS Lett 584: 396–404. 10.1016/j.febslet.2009.11.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar AA, Gross SR, Barth-Baus D, Strachan R, Hensold JO, Goss Kinzy T, Merrick WC. 2005. Novel characteristics of the biological properties of the yeast Saccharomyces cerevisiae eukaryotic initiation factor 2A. J Biol Chem 280: 15601–15611. 10.1074/jbc.M413728200 [DOI] [PubMed] [Google Scholar]
- Kozak M. 1989. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol Cell Biol 9: 5134–5142. 10.1128/MCB.9.11.5134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1990. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci 87: 8301–8305. 10.1073/pnas.87.21.8301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Strumpf N, Deiss LP, Berissi H, Kimchi A. 1997. DAP-5, a novel homolog of eukaryotic translation initiation factor 4G isolated as a putative modulator of γ interferon-induced programmed cell death. Mol Cell Biol 17: 1615–1625. 10.1128/MCB.17.3.1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Iadevaia V, Averous J, Taylor PM, Zhang Z, Proud CG. 2014. Impairing the production of ribosomal RNA activates mammalian target of rapamycin complex 1 signalling and downstream translation factors. Nucleic Acids Res 42: 5083–5096. 10.1093/nar/gku130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low WK, Dang Y, Schneider-Poetsch T, Shi Z, Choi NS, Merrick WC, Romo D, Liu JO. 2005. Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol Cell 20: 709–722. 10.1016/j.molcel.2005.10.008 [DOI] [PubMed] [Google Scholar]
- Packham G, Brimmell M, Cleveland JL. 1997. Mammalian cells express two differently localized Bag-1 isoforms generated by alternative translation initiation. Biochem J 328: 807–813. 10.1042/bj3280807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody DS. 1989. Translation initiation at non-AUG triplets in mammalian cells. J Biol Chem 264: 5031–5035. [PubMed] [Google Scholar]
- Pestova TV, Hellen CU. 2003. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev 17: 181–186. 10.1101/gad.1040803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarto N, Talarico D, Florkiewicz R, Rifkin DB. 1991. Selective expression of high molecular weight basic fibroblast growth factor confers a unique phenotype to NIH 3T3 cells. Cell Regul 2: 699–708. 10.1091/mbc.2.9.699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F, Kapp LD, Khan SN, Acker MG, Kolitz S, Kazemi S, Kaufman RJ, Merrick WC, Koromilas AE, Lorsch JR, et al. 2006. Initiation of protein synthesis by hepatitis C virus is refractory to reduced eIF2.GTP.Met-tRNAiMet ternary complex availability. Mol Biol Cell 17: 4632–4644. 10.1091/mbc.e06-06-0478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini DM. 2017. Twenty-five years of mTOR: uncovering the link from nutrients to growth. Proc Natl Acad Sci 114: 11818–11825. 10.1073/pnas.1716173114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, Liu JO. 2010. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol 6: 209–217. 10.1038/nchembio.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellier C, Buijsen RA, He F, Natla S, Jung L, Tropel P, Gaucherot A, Jacobs H, Meziane H, Vincent A, et al. 2017. Translation of expanded CGG repeats into FMRpolyG is pathogenic and may contribute to fragile X tremor ataxia syndrome. Neuron 93: 331–347. 10.1016/j.neuron.2016.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendoel A, Dunn JG, Rodriguez EH, Naik S, Gomez NC, Hurwitz B, Levorse J, Dill BD, Schramek D, Molina H, et al. 2017. Translation from unconventional 5′ start sites drives tumour initiation. Nature 541: 494–499. 10.1038/nature21036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirokikh NE, Preiss T. 2018. Translation initiation by cap-dependent ribosome recruitment: recent insights and open questions. Wiley Interdiscip Rev RNA 9: e1473 10.1002/wrna.1473 [DOI] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV. 2010. Activities of ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev 24: 1787–1801. 10.1101/gad.1957510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck SR, Jiang V, Pavon-Eternod M, Prasad S, McCarthy B, Pan T, Shastri N. 2012. Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science 336: 1719–1723. 10.1126/science.1220270 [DOI] [PubMed] [Google Scholar]
- Tabet R, Schaeffer L, Freyermuth F, Jambeau M, Workman M, Lee CZ, Lin CC, Jiang J, Jansen-West K, Abou-Hamdan H, et al. 2018. CUG initiation and frameshifting enable production of dipeptide repeat proteins from ALS/FTD C9ORF72 transcripts. Nat Commun 9: 152 10.1038/s41467-017-02643-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Maruyama M, Tokuzawa Y, Murakami M, Oda Y, Yoshikane N, Makabe KW, Ichisaka T, Yamanaka S. 2005. Evolutionarily conserved non-AUG translation initiation in NAT1/p97/DAP5 (EIF4G2). Genomics 85: 360–371. 10.1016/j.ygeno.2004.11.012 [DOI] [PubMed] [Google Scholar]
- Tatomer DC, Liang D, Wilusz JE. 2017. Inducible expression of eukaryotic circular RNAs from plasmids. Methods Mol Biol 1648: 143–154. 10.1007/978-1-4939-7204-3_11 [DOI] [PubMed] [Google Scholar]
- Touriol C, Bornes S, Bonnal S, Audigier S, Prats H, Prats AC, Vagner S. 2003. Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol Cell 95: 169–178. 10.1016/S0248-4900(03)00033-9 [DOI] [PubMed] [Google Scholar]
- Walden WE, Godefroy-Colburn T, Thach RE. 1981. The role of mRNA competition in regulating translation. I. Demonstration of competition in vivo. J Biol Chem 256: 11739–11746. [PubMed] [Google Scholar]
- Wang Y, Ringquist S, Cho AH, Rondeau G, Welsh J. 2004. High-throughput polyribosome fractionation. Nucleic Acids Res 32: e79 10.1093/nar/gnh077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR, Girard M, Latham H, Darnell JE. 1966. Ribosome formation in HeLa cells in the absence of protein synthesis. J Mol Biol 19: 373–382. 10.1016/S0022-2836(66)80011-6 [DOI] [PubMed] [Google Scholar]
- Wilson JE, Pestova TV, Hellen CU, Sarnow P. 2000. Initiation of protein synthesis from the A site of the ribosome. Cell 102: 511–520. 10.1016/S0092-8674(00)00055-6 [DOI] [PubMed] [Google Scholar]
- Xiao Z, Zou Q, Liu Y, Yang X. 2016. Genome-wide assessment of differential translations with ribosome profiling data. Nat Commun 7: 11194 10.1038/ncomms11194 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.