In this study, Schüle et al. report an unexpected role of GADD45 proteins in regulation of the cycling of ESCs in the 2C state. Using methylome analysis of Gadd45 triple-mutant ESCs, they found a role for GADD45 in demethylation of specific TET targets and partial deregulation of ZGA genes at the two-cell stage.
Keywords: demethylation, GADD45, 5-hydroxymethylcytosine, TET, two-cell embryo, ZGA, ZSCAN4
Abstract
Mouse embryonic stem cell (ESC) cultures contain a rare cell population of “2C-like” cells resembling two-cell embryos, the key stage of zygotic genome activation (ZGA). Little is known about positive regulators of the 2C-like state and two-cell stage embryos. Here we show that GADD45 (growth arrest and DNA damage 45) proteins, regulators of TET (TET methylcytosine dioxygenase)-mediated DNA demethylation, promote both states. Methylome analysis of Gadd45a,b,g triple-knockout (TKO) ESCs reveal locus-specific DNA hypermethylation of ∼7000 sites, which are enriched for enhancers and loci undergoing TET–TDG (thymine DNA glycosylase)-mediated demethylation. Gene expression is misregulated in TKOs, notably upon differentiation, and displays signatures of DNMT (DNA methyltransferase) and TET targets. TKOs manifest impaired transition into the 2C-like state and exhibit DNA hypermethylation and down-regulation of 2C-like state-specific genes. Gadd45a,b double-mutant mouse embryos display embryonic sublethality, deregulated ZGA gene expression, and developmental arrest. Our study reveals an unexpected role of GADD45 proteins in embryonic two-cell stage regulation.
Mouse embryonic stem cells (ESCs) are a model for the inner cell mass around implantation stage. ESCs are heterogeneous and contain subpopulations with different properties (Hayashi et al. 2008; Toyooka et al. 2008; Zalzman et al. 2010; Macfarlan et al. 2012). One of these subpopulations (1%–5%) is transcriptionally and epigenetically similar to the two-cell stage embryo and hence is referred to as “2C-like” (Zalzman et al. 2010; Macfarlan et al. 2012). The embryonic two-cell stage is a key phase of mouse development during which the major wave of zygotic genome activation (ZGA) occurs (for review, see Jukam et al. 2017; Eckersley-Maslin et al. 2018; Svoboda 2018). During this period, the bulk of the genome becomes transcriptionally active, which is accompanied by extensive chromatin modification. 2C-like ESCs exhibit unique molecular features of totipotent cleavage stage cells and, in chimeras, can contribute to both embryonic and extraembryonic derivatives, including trophoblast (Macfarlan et al. 2012; Ishiuchi et al. 2015; Choi et al. 2017; De Iaco et al. 2017; Whiddon et al. 2017; Rodriguez-Terrones et al. 2018). ESCs cycle in and out of this transient 2C-like state at least once within nine passages (Zalzman et al. 2010). Characteristic markers for the 2C-like state are murine endogenous retrovirus with leucine tRNA primer (MERVL) retrotransposon and zinc finger and SCAN domain-containing protein 4 (Zscan4) (Zalzman et al. 2010; Macfarlan et al. 2012). Thus, 2C-like ESCs model essential aspects of the two-cell stage embryo and ZGA (for review, see Ishiuchi and Torres-Padilla 2013; Eckersley-Maslin et al. 2018). Few positive regulators of the 2C-like state or ZGA are known, including the transcriptional regulators ZSCAN4 (Falco et al. 2007; Zalzman et al. 2010; Hirata et al. 2012; Amano et al. 2013), DUX (De Iaco et al. 2017; Hendrickson et al. 2017), STELLA (Huang et al. 2017), and TBX3 (Dan et al. 2013) as well as DPPA2 and DPPA4 (Eckersley-Maslin et al. 2019).
Here we report a role for the small gene family Gadd45a (growth arrest and DNA damage protein 45a), Gadd45b, and Gadd45g in regulation of the 2C-like state. GADD45α is a stress response protein, which interacts with the key enzymes of the DNA demethylation machinery: TET1 (TET methylcytosine dioxygenase 1) and TDG (thymine DNA glycosylase) (Barreto et al. 2007; Cortellino et al. 2011; Kienhöfer et al. 2015; Li et al. 2015). TET enzymes convert 5-methylcytosine (5mC) sequentially to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) (Kriaucionis and Heintz 2009; Tahiliani et al. 2009; Guo et al. 2011; He et al. 2011; Ito et al. 2011). DNA repair via TDG removes 5fC and 5caC to restore unmethylated cytosine (Cortázar et al. 2011; Cortellino et al. 2011; Shen et al. 2013). GADD45α is an adapter protein that tethers TET/TDG to sites of DNA demethylation, which functions in locus-specific DNA demethylation (Barreto et al. 2007; Li et al. 2010; Cortellino et al. 2011; Zhang et al. 2011a; Arab et al. 2014; Sabag et al. 2014). GADD45α recruits TET/TDG to specific sites in the genome via additional cofactors (Schäfer et al. 2013; Arab et al. 2014, 2019; Schäfer et al. 2018).
Since not only Gadd45a but also Gadd45b and Gadd45g promote DNA demethylation (Rai et al. 2008; Ma et al. 2009; Sen et al. 2010; Gavin et al. 2015; Jarome et al. 2015) and since single mouse mutants are viable (Hollander et al. 1999; Lu et al. 2001, 2004), this raises the question of whether the genes have overlapping roles in development and differentiation. To address this question, we generated and characterized Gadd45a,b,g triple-knockout (TKO) mouse ESCs. We found that GADD45 proteins are dispensable for maintaining pluripotency and self-renewal. However, methylome analysis indicates that GADD45 proteins are required for DNA demethylation of specific loci and normal gene expression. Moreover, GADD45 proteins promote the 2C-like state, and Gadd45a,b double-mutant mouse embryos show partial deregulation of ZGA genes at the two-cell stage and developmental arrest. Collectively, the results indicate that GADD45 proteins act redundantly to promote locus-specific demethylation as well as embryonic two-cell stage.
Results
Gadd45 TKO ESCs are pluripotent and self-renew
We generated homozygous deletions in Gadd45a, Gadd45b, and Gadd45g in ESCs using the CRISPR/Cas9 system (Jinek et al. 2012; Cong et al. 2013; Mali et al. 2013). Six gRNAs were cotransfected, two for each Gadd45 gene, to create 300- to 700-bp deletions between the 5′ untranslated region and the second intron, covering the start codon (Fig. 1A). Out of 276 colonies obtained after selection, three independent Gadd45 TKO ESC clones were obtained (Supplemental Fig. S1A). Sequencing confirmed deletion of the respective genomic regions in the TKO ESCs (Supplemental Fig. S1B), and Western blot and mass-spectrometry analysis showed that both GADD45α and GADD45β were undetectable in TKO ESCs (Supplemental Fig. S1C,D). GADD45γ was undetectable in both wild-type and mutant ESCs (Supplemental Fig. S1E), and even if truncated GADD45γ protein was expressed, it would be nonfunctional, since deletion of exons 1 and 2 includes the dimerization domains (amino acids 43–86) required for GADD45γ function (Zhang et al. 2011b). To generate three independent wild-type ESC control clones, ESCs were transfected with Cas9 and the selection marker but without specific gRNAs.
Figure 1.

Gadd45 TKO ESCs are pluripotent and show normal global levels of DNA modifications. (A) Scheme of the CRISPR/Cas9-mediated Gadd45 knockout strategy. Numbers indicate exons, and red bars indicate the location of deletion. (B) Relative expression of representative pluripotency markers in control (Co) and Gadd45 TKO ESCs measured by quantitative PCR (qPCR). Expression values are relative to the average expression in control ESCs. (C) Hematoxylin/eosin staining of paraffin control and Gadd45 TKO teratoma sections. Representative examples for ectoderm, endoderm, and mesoderm derivatives are shown. (D–G) 5mC, 5hmC, 5fC, and 5caC levels in individual control ESC and Gadd45 TKO ESC clones determined by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Values are given as the percentage of total cytosine (C).
The Gadd45 TKO ESCs showed no apparent loss of pluripotency; Oct4, Nanog, and Sox2 expression was not reduced (Fig. 1B); and their morphology as well as growth rate were normal (Supplemental Fig. S2A–C). In teratoma assays, TKO ESCs gave rise to derivatives of all three germ layers (Fig. 1C).
Given the previously described functions of GADD45 proteins in active DNA demethylation, we analyzed global 5mC, 5hmC, 5fC, and 5caC levels by quantitative mass spectrometry (MS). Global levels of 5mC and its oxidative derivatives were only mildly affected in Gadd45 TKO ESCs (Fig. 1D–G). There was a slight increase of 5mC with a concomitant slight decrease of oxidized cytosine derivatives in TKO ESCs compared with control ESCs. This result is consistent with GADD45 proteins acting not in global but in locus-specific demethylation.
Mouse ESCs are a model for the inner cell mass (ICM) around implantation stage with relatively high methylation at promoters, enhancers, and bivalent loci (Habibi et al. 2013). However, ESCs can be reverted to a hypomethylated ground state more similar to preimplantation embryos using small molecule MEK and GSK3β inhibitors (“2i”) (Ficz et al. 2013; Leitch et al. 2013) as well by vitamin C (Blaschke et al. 2013). Hence, we induced global DNA demethylation via vitamin C or 2i treatment; however, global levels of cytosine modifications in Gadd45 TKO ESCs were changing similarly to control ESCs (Supplemental Fig. S2D,E).
We conclude that Gadd45 genes are dispensable for ESC maintenance, as is the case for Tet and Dnmt (DNA methyltransferase) genes (Lei et al. 1996; Okano et al. 1999; Tsumura et al. 2006; Dawlaty et al. 2011, 2014).
Loci undergoing TET-dependent oxidation are hypermethylated in TKO ESCs
To unravel DNA methylation changes, we performed whole-genome bisulfite sequencing (WGBS) of control and TKO ESCs to obtain base-pair-resolution methylomes. Control ESCs showed the characteristic bimodal distribution of CpG methylation, but the distribution in TKO ESCs was skewed toward higher methylation (Fig. 2A). To call differentially methylated CpG regions (DMRs) in TKO ESCs, we used a stringent cutoff of 5% false discovery rate (FDR) and >30% methylation difference on at least two CpGs. WGBS does not discriminate 5hmC from 5mC (Booth et al. 2012); hence, we may have underestimated the number of GADD45-dependent TET target sites. DMRs were broadly distributed on all 19 autosomes (data not shown). The DMR analysis identified 6904 hypermethylated, but only 34 hypomethylated, regions in TKO ESCs (Supplemental Fig. S3A). Thus, although global 5mC levels were only slightly increased as measured by liquid chromatography-tandem MS (LC-MS/MS), the skewed bimodal methylation pattern and the ∼200-fold bias toward hypermethylated DMRs indicate a locus-specific DNA hypermethylation in TKO ESCs.
Figure 2.
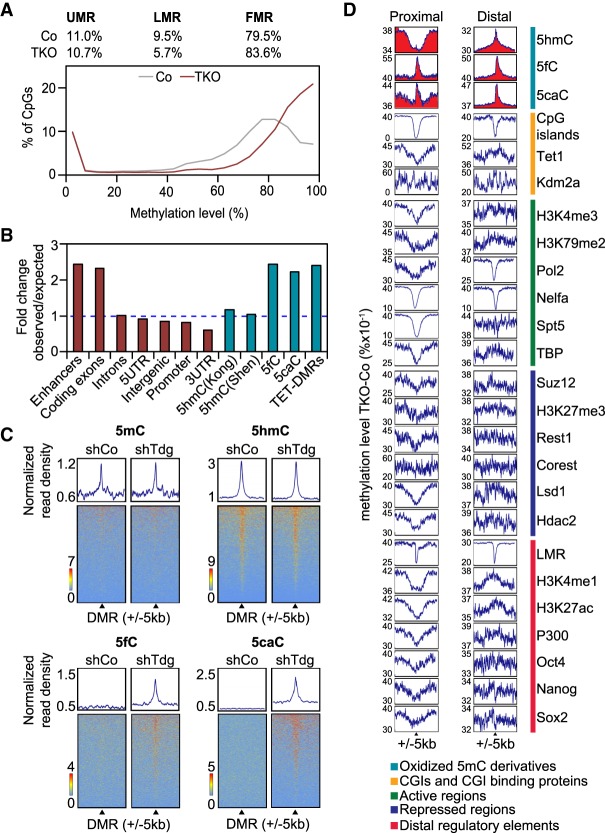
Loci undergoing TET-dependent oxidation are hypermethylated in Gadd45 TKO ESCs. (A) The bimodal methylation pattern of individual CpG sites in ESCs is skewed to a higher methylation level in Gadd45 TKO ESCs. Methylation levels are shown as the average of two biological replicates. (UMR) Unmethylated regions; (LMR) lowly methylated regions; (FMR) fully methylated regions as defined by Stadler et al. (2011). (B) Relative enrichment of Gadd45 TKO hypermethylated DMRs (hyper-DMRs) at various genomic elements (red), oxidative 5mC derivatives, and Tet-TKO hyper-DMRs (blue). (C) Heat maps of depicted DNA modifications (Shen et al. 2013) centered (±5 kb) on Gadd45 TKO hyper-DMRs (black triangles) in untreated and shTdg-treated ESCs. (D) Average methylation differences between Gadd45 TKO and control ESCs around centers (±5 kb) of annotated genomic features. Methylation differences are shown for proximal (within 1 kb of a gene transcription start site) and distal features. Red areas highlight oxidized 5mC derivatives.
We therefore focused on the hypermethylated DMRs (hyper-DMRs). The majority overlapped with intronic and intergenic regions (Supplemental Fig. S3B). There was ∼2.5-fold enrichment for enhancers and coding exons (Fig. 2B). Moreover, hyper-DMRs were enriched at sites marked by 5fC or 5caC after Tdg knockdown (Shen et al. 2013) and, to a lesser extent, at sites marked by 5hmC (Shen et al. 2013; Kong et al. 2016). Hyper-DMRs were also enriched for TET-dependent hyper-DMRs (Lu et al. 2014). Positional correlation analysis centered on hyper-DMRs revealed prominent overlap with sites marked by 5hmC (Fig. 2C). Hyper-DMRs also overlapped with 5fC/5caC peaks accumulating in Tdg knockdown ESCs (Shen et al. 2013), indicating that hyper-DMRs are targets of TET/TDG-mediated cytosine oxidation and excision in ESCs. In contrast, there was little overlap with 5mC sites, indicating that the association of hyper-DMRs occurred specifically with oxidized cytosines.
In another positional correlation analysis, we plotted the average levels of methylation change between TKO and control ESCs against the center of genomic features derived from a wide panel of published genome-wide mapping data sets in ESCs, including 5mC oxidative derivatives, DNA-binding factors, and major histone modifications. Moreover, we divided the analysis between proximal and distal elements with regard to gene transcription start sites. This analysis corroborated that the main co-occurrence of hypermethylation in TKO ESCs was with sites marked by 5fC and 5caC in both proximal and distal sites (Fig. 2D, top three rows). Hypermethylation accumulated also at the center of 5hmC peaks but to a lower level and restricted to distal loci.
Conversely, CpG islands (CGIs), which typically occur at proximal sites, showed the lowest levels of methylation difference, consistent with the fact that they are constitutively unmethylated (Deaton and Bird 2011). Other genomic features correlated with promoter CGIs, such as Pol2, TBP, and transcription elongation factors (Nelfa and Spt5), followed this trend. In general, this methylation signature parallels the signature in Tet1,2,3 TKO ESCs (Lu et al. 2014), whereby TET-mediated DNA demethylation (1) occurs mainly at distal regulatory elements and (2) affects sites marked in control cells by 5fC and 5caC more pronounced than those marked by 5hmC. We conclude that the hypermethylation signature in TKO ESCs closely correlates with loci processed by TET/TDG, consistent with GADD45 proteins acting in locus-specific DNA demethylation.
We segmented hyper-DMRs and carried out transcription factor (TF)-binding motif analysis using HOMER (Fig. 3A). The most prominent hit in all DMRs was Klf5, a possible reader of methylated DNA (Spruijt et al. 2013; Liu et al. 2014), which promotes the pluripotent ESC state and is required for trophectoderm development (Ema et al. 2008; Parisi et al. 2008). Other prominent hits were Ets-like TF-binding elements (Ehf, Etv1, Fli1, and Elk4/1). Interestingly, hyper-DMRs overlapping with enhancers harboring 5hmC were enriched for motifs of Zscan4, a key regulator of the 2C-like state.
Figure 3.
Methylation-regulated genes are down-regulated in Gadd45 TKO ESCs. (A) Motif analysis of hyper-DMRs in Gadd45 TKO ESCs using HOMER (Heinz et al. 2010). (B) Differentially expressed (DE) genes (FDR 10%) identified by RNA sequencing (RNA-seq) in Gadd45 TKO ESCs versus control (Co) ESCs, which were untreated, 72-h vitamin C-treated (VitC), or 72-h 2i-treated. (C) Overlap of genes down-regulated and up-regulated in untreated Gadd45 TKO ESCs and genes up-regulated in Dnmt1,2,3 TKO ESCs (Karimi et al. 2011). (D) Scatter plot of the common deregulated genes (gray) in Gadd45 TKO and Tet1 knockdown ESCs (log2FC [log2 of fold change] vs. control ESCs) (Huang et al. 2014). (E) Differentially expressed (DE) genes (FDR 10%) identified by RNA-seq in Gadd45 TKO cells versus control cells upon monolayer differentiation (monolayer), embryoid body (EB) differentiation, or retinoic acid (RA) stimulation during EB differentiation. (F) Expression of selected GADD45-dependent genes in control and Gadd45 TKO ESCs measured by qPCR. Expression is relative to control ESCs. Data are presented as mean ± SD from n = 3 independent clones and n = 3 independent experiments. (G) GADD45-regulated genes are DNA methylation-sensitive. Relative expression levels of selected GADD45-dependent genes in Gadd45 TKO ESCs upon 48 h of DMSO (mock) or 5′-azadeoxycytidine (5′-Aza) treatment measured by qPCR. Expression is relative to DMSO-treated control ESCs. (H) Hypermethylation of GADD45-dependent genes. DNA methylation of the indicated CpGs in the Rhox2a promoter in control and Gadd45 TKO ESCs monitored by site-specific bisulfite sequencing.
Methylation-regulated genes are down-regulated in Gadd45 TKO ESCs
To identify genes differentially expressed upon Gadd45 deficiency, RNA sequencing (RNA-seq) analysis was carried out under three culture conditions: normal serum culture and two conditions inducing global demethylation: vitamin C and 2i treatment (Blaschke et al. 2013; Ficz et al. 2013; Leitch et al. 2013). A total of 135 genes was differentially expressed in Gadd45 TKO ESCs versus control ESCs during normal serum culture (FDR 10%). This number decreased sharply upon vitamin C or 2i treatment (Fig. 3B), supporting that gene deregulation in TKO ESCs is due directly or indirectly to DNA hypermethylation. Around 25% of deregulated genes overlapped with hyper-DMR-associated genes (Supplemental Fig. S3C), further indicating that only a moderate fraction of GADD45-dependent genes are methylation-regulated. This is in line with generally modest correlation between gene expression and DNA methylation in ESCs (Karimi et al. 2011; Lu et al. 2014). Genes down-regulated in Gadd45 TKO ESCs overlapped significantly with genes up-regulated in Dnmt1−/−/Dnmt3a−/−/Dnmt3b−/− TKO ESCs (Fig. 3C; Karimi et al. 2011). Consequently, localization of genes down-regulated in Gadd45 TKO ESCs is enriched on the X chromosome (Benjamini P = 2.2 × 10−6), as has been observed for up-regulated genes in Dnmt TKO ESCs (Fouse et al. 2008). These genes tend to be involved in germ cell regulation (Wang et al. 2001). Moreover, genes deregulated in Gadd45 TKO ESCs correlated with genes deregulated upon Tet1 knockdown in ESCs (Fig. 3D; Huang et al. 2014).
Previous studies showed that DNA methylation and demethylation play a more important role during differentiation than during pluripotency (Lei et al. 1996; Okano et al. 1999; Jackson et al. 2004; Sakaue et al. 2010; Dawlaty et al. 2014). We therefore subjected TKO ESCs to three differentiation protocols and analyzed transcriptome changes by RNA-seq. First, ESCs were differentiated for 8 d as embryoid bodies (EBs). Second, we differentiated ESCs for 6 d in serum-free monolayer culture. Third, ESCs were differentiated for 4 d as EBs and then treated for four more days with retinoic acid (RA). While the latter two protocols favor neuronal differentiation, the unguided EB culture allows differentiation into all three germ layers (Ying et al. 2003; Bibel et al. 2007). Gadd45a, Gadd45b, and Gadd45g were all expressed at varying levels under these differentiation regimes (Supplemental Fig. S3D).
The number of differentially expressed genes in TKO cells versus control cells was 907 upon monolayer differentiation and 659 in EBs (FDR 10%) (Fig. 3E), and thus gene deregulation in Gadd45 TKO cells is indeed increased upon differentiation (Fig. 3, cf. B and E). Only 22 genes were differentially expressed in RA-treated EBs, suggesting that the particular neural lineages induced by RA are less sensitive to Gadd45 deficiency. Only 48 genes were commonly deregulated in TKO cells between EB and monolayer differentiation (data not shown), indicating that the function of the Gadd45 genes is highly context-dependent. Interestingly, genes differentially expressed in Gadd45 TKO EBs were more than twice as likely to be marked by 5fC in their promoter regions compared with unaffected genes (Supplemental Fig. S3E), supporting that GADD45-dependent genes are prone to undergo active DNA demethylation during EB differentiation.
Gene ontology (GO) term analysis in TKO EBs showed that down-regulated genes were highly enriched for developmental terms such as system development, cell fate determination, cell migration, and axon guidance (Supplemental Fig. S4A). Less pronounced GO term enrichment was found for genes up-regulated in TKOs (Supplemental Fig. S4B). For genes down-regulated in monolayer differentiated TKO cells, GO term enrichment was also found for cell motility related terms (Supplemental Fig. S4C), whereas genes up-regulated were enriched for developmental and neuronal functions (Supplemental Fig. S4D). Despite sharing similar gene ontologies, the genes affected in Gadd45 TKO EBs were largely distinct from those affected in Gadd45 TKO monolayer cells (data not shown). We conclude that in differentiating ESCs, GADD45 proteins regulate genes related to developmental, neuronal, and cell motility function, consistent with the involvement of GADD45 proteins in the regulation of neural development (Ma et al. 2009; Huang et al. 2010; Kaufmann and Niehrs 2011).
We validated selected genes commonly deregulated in Gadd45 TKO, Dnmt TKO, and Tet1 knockdown ESCs by quantitative PCR (qPCR). Commonly deregulated genes did not significantly cluster in terms of GO enrichment (data not shown) but included various germline-specific (e.g., Rhox2a and Asz1) and pluripotency-related (e.g., Pramel6 and Pramel7) genes (Fig. 3F; Supplemental Fig. S5A).
To analyze the Gadd45 gene redundancy in differential gene expression, we conducted rescue experiments. Transient combined overexpression of Gadd45a, Gadd45b, and Gadd45g rescued down-regulation of a panel of misregulated genes in Gadd45 TKO ESCs and further increased their expression levels in control ESCs (Supplemental Fig. S5A). Not only combined but also individual Gadd45a, Gadd45b, or Gadd45g overexpression was effective in these rescue experiments, indicating that Gadd45 genes in ESCs function redundantly (Supplemental Fig. S5B,C). Genes down-regulated in Gadd45 TKO ESCs were also induced by 5′-deoxyazacytidine treatment (Fig. 3G), further supporting that gene down-regulation in TKO ESCs involved DNA hypermethylation.
To test directly for DNA hypermethylation in TKO ESCs, we analyzed the methylation status of regulatory elements in the vicinity of selected GADD45-dependent genes, which are shared with DNMT- and TET1-dependent genes. The majority of CpG dinucleotides in the Rhox2a promoter (Fig. 3H), the Pramel6 promoter (Supplemental Fig. S6A), and the Gm364 promoter (Supplemental Fig. S6B) were hypermethylated in Gadd45 TKO ESCs. In contrast, in two control gene promoters displaying high and low methylation levels, respectively (Sry and Rerg), no methylation changes were observed in TKO ESCs (Supplemental Fig. S6C,D).
We conclude that GADD45 proteins act redundantly to maintain normal expression and methylation levels of selected genes in ESCs and that this list of genes overlaps with TET1 and DNMT target genes.
Gadd45 TKO ESCs show impaired 2C-like state
We used the ESCAPE (Embryonic Stem Cell Atlas of Pluripotency Evidence) database (Xu et al. 2014), which integrates high-content data from ESCs, to conduct enrichment analysis of genes down-regulated in undifferentiated Gadd45 TKO ESCs (Fig. 4A). The top hit returned was a set of genes reported up-regulated upon Gadd45a overexpression (Nishiyama et al. 2009), corroborating the validity of the ESCAPE analysis. Among the other hits with similar significance were genes up-regulated upon misexpression of Zscan4 (Nishiyama et al. 2009), a TF whose recognition motif was enriched in hyper-DMRs (Fig. 3A). Zscan4 is a marker and regulator of the two-cell embryo (Falco et al. 2007). Consistent with the overlap between GADD45- and ZSCAN4-regulated genes, we found that genes up-regulated in 2C-like cells tend to be down-regulated in Gadd45 TKO ESCs, suggesting a requirement of Gadd45 genes in regulating the 2C-like state (Fig. 4B). Indeed, although only 97 genes in Gadd45 TKO ESCs were down-regulated (10% FDR), these were enriched in genes specifically expressed in 2C-like cells (Supplemental Fig. S7A; Macfarlan et al. 2012). In contrast, not a single gene up-regulated in Gadd45 TKO ESCs overlapped with the 2C gene set. 2C-specific genes were only modestly enriched in the vicinity of hyper-DMRs (Fig. 4C), which is expectable, since the 2C-like cells represent only a small fraction of the ESC population. Of these 178 hyper-DMR- and 2C-like-associated genes, six were also down-regulated in the Gadd45 TKO ESCs (Igfbp2, Inpp4b, Pramel6, Pramel7, Snhg11, and Tmem92). Other hypermethylated 2C genes may be down-regulated only upon 2C cycling and hence escape detection.
Figure 4.

Gadd45 TKO ESCs show impairment of the 2C-like state. (A) Enrichment analysis of genes down-regulated in Gadd45 TKO ESCs using the ESCAPE database (Xu et al. 2014). Enrichment analysis shows a significant overlap with genes deregulated (up or down) upon overexpression of, for example, Gadd45a or Zscan4c. (B) Scatter plot of the common deregulated genes (gray) in Gadd45 TKO ESCs and up-regulated in the 2C-like state (log2FC [log2 of fold change versus control ESCs]) (Macfarlan et al. 2012). (C) Overlap between 2C-associated (Macfarlan et al. 2012) and hyper-DMR-associated genes. (D) qPCR expression analysis of selected 2C-associated genes and retroviral elements in control (Co) and Gadd45 TKO ESCs 48 h after transfection with the indicated genes. Expression is relative to GFP transfected control ESCs. Data are presented as mean ± SD from n = 3 independent clones and n = 2 independent experiments. Statistical significance was tested with two-tailed paired Student's t-test. (E) Scatter plot showing Zscan4c::eGFP-positive cells in bulk control ESCs measured by flow cytometry analysis. Representative gates used for bulk analysis are boxed. (F) Flow cytometry analysis of Zscan4c::eGFP-positive cells in control and Gadd45 TKO ESCs. (G) Flow cytometry analysis of Zscan4c::eGFP-positive cells in control or Gadd45-TKO ESCs 48 h after transfection with the indicated genes. Data are presented as means ± SD from n = 3 independent clones and n = 2 independent experiments. Statistical significance was tested with two-tailed unpaired (TKO vs. control [Co]) or paired (BFP vs. Gadd45 overexpression) Student's t-test. (H) Flow cytometry analysis of Zscan4::eGFP-positive cells in control ESCs 48 h after transfection with the indicated genes. Statistical significance was tested with two-tailed paired Student's t-test. (I,J) 5mC and 5hmC levels in unsorted or Zscan4− or Zscan4+ sorted control ESCs and Gadd45 TKO ESC clones determined by LC-MS/MS. Values are the percentage of total cytosine (C).
To confirm these results, we monitored the expression of prominent 2C-associated genes in control ESCs and TKO ESCs after combined overexpression of Gadd45a, Gadd45b, and Gadd45g or GFP (Fig. 4D). Overexpression of Gadd45 genes not only rescued down-regulation of the majority of tested 2C-associated genes in TKO ESCs but also increased expression levels even in control ESCs. In contrast, expression of retroviral elements unrelated to the 2C status (intracisternal A particle) was unchanged.
To investigate the role of GADD45 in regulating the 2C-like state, we stably introduced a 2C reporter, Zscan4c::eGFP (Zalzman et al. 2010), in control and TKO ESCs (Fig. 4E). Flow cytometry revealed a significant reduction of Zscan4+ cells in TKO ESCs (Fig. 4F). Combined overexpression of the Gadd45 genes rescued the reduction of Zscan4+ cells in TKO ESCs to control levels (Fig. 4G). Likewise, individual overexpression of Gadd45a, Gadd45b, or Gadd45g increased the percentage of Zscan4+ cells as well as the percentage of cells harboring the mERVL 2C reporter (Macfarlan et al. 2012), suggesting redundant function of the GADD45 proteins in 2C regulation (Fig. 4H; Supplemental Fig. S7B). Interestingly, long-term 2i treatment (seven passages), which induces hypomethylation (Ficz et al. 2013; Leitch et al. 2013), abolished the observed difference in the 2C-like population (Supplemental Fig. S7C). In contrast, overexpression of Tet1, Tet2, or Tdg did not affect the frequency of Zscan4+ cells in TKO or control ESCs (Supplemental Fig. S7D–F).
Zscan4+ sorted cells show a global demethylation relative to unsorted ESCs (Eckersley-Maslin et al. 2016). We confirmed by MS that control Zscan4+ cells show reduced 5mC levels (Fig. 4I, cf. control “unsorted” and “Zscan4+”), while Zscan4+ TKO cells showed hypermethylation. No difference was found for 5hmC (Fig. 4J).
Gene expression analysis of TKO ESCs (Fig. 5A) revealed reduced RNA levels of 2C-specific genes in not only unsorted but also Zscan4+ TKO ESCs (e.g., Zscan4, Sp110, and Tcstv1). This reduction was not due to deficient maintenance of the 2C-like state but reflected deficient entry, since cycling out of the Zscan4+ state occurred with the same kinetics in control and TKO ESCs (Fig. 5B).
Figure 5.

Gadd45 TKO ESCs show reduced 2C-like gene expression and transdifferentiation potential. (A) qPCR expression analysis of selected 2C-associated genes in control (Co) and Gadd45 TKO ESCs in unsorted or Zscan4− or Zscan4+ fluorescence-activated cell sorting (FACS)-sorted cells. Expression is relative to the average expression in unsorted control ESCs. (IAP) Intracisternal A particle. (B) Reanalysis of Zcan4+ sorted control and TKO ESCs at the indicated time points using flow cytometry. (C) Dux overexpression restores the reduced number of 2C-like cells in Gadd45 TKO ESCs. Flow cytometry analysis of Zscan4c::eGFP-positive cells in control and Gadd45 TKO ESCs 48 h after transfection with the indicated genes and 24 h after doxycycline addition. Statistical significance was tested with two-tailed unpaired (TKO vs. control) or paired (luciferase vs. Dux overexpression) Student's t-test. (D) qPCR expression analysis of selected 2C-associated genes in untreated or BMP4-treated (8 d) control and Gadd45 TKO ESCs. Expression is relative to untreated control ESCs. Data are presented as mean ± SD from n = 3 independent clones and n = 3 independent experiments. (E,F) Impairment of trophectoderm transdifferentiation in Gadd45 TKO ESCs. Gadd45 TKO and control ESCs were treated for 8 d with BMP4. Induction of early (E) and late (F) trophectoderm (TE) marker genes is relative to BMP4-treated control ESCs. (UT) Untreated.
Among the down-regulated 2C genes was Dux, a key regulator of ESCs cycling into the 2C-like state (De Iaco et al. 2017; Hendrickson et al. 2017). This suggested that GADD45 may act upstream of DUX to promote the 2C-like state. Concordantly, overexpression of Dux using a doxycycline-inducible plasmid restored the reduced number of 2C-like cells in TKO ESCs (Fig. 5C). The expression level of repressors of the 2C-like state (Trim28, Lsd1, G9a, and Chaf1a) was not altered in unsorted or Zscan4+ or Zscan4− TKO ESCs (Supplemental Fig. S7G).
Finally, we explored the consequences of 2C misregulation in ESC transdifferentiation. While ESCs normally do not give rise to trophoblasts, they do sporadically transdifferentiate into this lineage, which can be further enhanced by BMP4 treatment (Beddington and Robertson 1989; Hayashi et al. 2010). Trophoblast transdifferentiation involves 2C cycling, since 2C-like cells are not lineage-restricted (Macfarlan et al. 2012), and cycling through the 2C-state is critical to restore the full developmental capacity in ESCs (Amano et al. 2013). We treated ESCs with BMP4, which induced 2C-associated genes (Fig. 5D) as well as the trophoblast stem cell markers Cdx2 and Elf5 (Fig. 5E), supporting that ESCs use the 2C-like state during transdifferentiation. Induction of both 2C and trophoblast markers was greatly reduced in TKO ESCs. Moreover, expression of placental markers induced by BMP4 (e.g., Serpin, Psg, and Prl gene families) was almost abolished in TKOs (Fig. 5F). The reduced transdifferentiation potential of Gadd45 TKO ESCs is consistent with impaired 2C-like cycling. Interestingly, Dnmt1 deficiency yields the opposite phenotype to Gadd45 deficiency; i.e., activation of trophoblast lineage markers (Cambuli et al. 2014). Our data are therefore in line with the notion that DNA methylation is a barrier to ESC-to-trophoblast transdifferentiation (Ng et al. 2008).
(Gadd45a/Gadd45b)−/− mice are sublethal and show partially impaired ZGA gene expression
Interrogating a database of early mouse transcriptome (Park et al. 2015) revealed that Gadd45a and Gadd45b belong to a “two-cell transient” cluster, showing a peak of expression specifically in two-cell embryos (Fig. 6A). Gadd45g belongs to the “major ZGA cluster” but shows low expression during cleavage stages. This raised the possibility that the impairment of 2C cycling in ESCs in fact reflects a role for GADD45 in the embryonic two-cell stage, coinciding with the major phase of ZGA (Eckersley-Maslin et al. 2018). Single Gadd45a, Gadd45b, or Gadd45g mutants are viable and fertile (Hollander et al. 1999; Lu et al. 2001, 2004), but our results in ESCs indicate that they may compensate for each other in 2C state regulation. As generation of triple mutants is challenging, we generated Gadd45a,b double-knockout (DKO) mice by intercrossing double-heterozygous animals. All nine possible genotypes were obtained at expected Mendelian ratios, except for the homozygous Gadd45a,b double mutants, whose frequency was 50% reduced (Fig. 6B, arrow). Surviving DKO mice showed normal body size but displayed phenotypic abnormalities characteristic of neural tube closure defects (NTDs) such as curly tail and spina bifida. Litter size of DKO intercrossing was 50% reduced compared with double-heterozygous crosses (Fig. 6C). At embryonic stage 13.5 (E13.5), 50% (n = 24) of DKO embryos resulting from double-heterozygous breeding and 80% (n = 37) of embryos from homozygous DKO breeding also showed, beyond curly tail and spina bifida, defects such as exencephaly and cranial hemorrhage (Fig. 6D,E; Supplemental Fig. S7H). Sublethality, exencephaly, and cranial hemorrhaging are also observed in Tet1,2 DKO mice (Dawlaty et al. 2013). The increased phenotypical abnormalities of DKO embryos resulting from breeding homozygous DKOs compared with double-heterozygous mice hints at a requirement of GADD45 in the germline, as is the case for TET1 (Yamaguchi et al. 2013).
Figure 6.
(Gadd45a/Gadd45b)−/− mice show embryonic sublethality. (A) Expression analysis of Gadd45a, Gadd45b, and Gadd45g in preimplantation embryos and ESCs from the Database of Transcriptome in Mouse Early Embryos (DBTMEE) (Park et al. 2015). (FPKM) Fragments per kilobase of exon model per million reads mapped. (B) Genotypic analysis of progenies from Gadd45a,b double-heterozygous (DHet) mice showing expected and observed Mendelian ratios. (C) (Gadd45a/Gadd45b)−/− (DKO) intercrossing show reduced litter size compared with intercrossed double-heterozygous animals. Data points indicate the number of pups per litter. Red lines indicate average litter size. Statistical significance was tested with two-tailed paired Student's t-test. (D) Images of E13.5 heterozygous and homozygous Gadd45a/Gadd45b mouse embryos. Scale bar, 2 mm. (E) Developmental abnormalities in DKO embryos showing curly tail (ct), exencephaly (ex), cranial hemorrhage (he), and spina bifida (sb). (F,G) RNA-seq differential expression analysis of repeats (F) and genes (G) in two-cell stage DKO embryos. Significantly deregulated (FDR 10%) repeats and genes are highlighted in blue and red, respectively. (H–J) Overlap of genes up-regulated and down-regulated in Gadd45a,b DKO two-cell stage embryos and genes normally up-regulated in two-cell stage embryos (H) or oocytes (I) or during ZGA (J) (Macfarlan et al. 2012). (K) In vitro development of wild-type (WT) and Gadd45a,b DKO preimplantation embryos isolated from three independent breedings. Development was scored 24 h after isolation. Representative microscopic images of the scored embryonic stages are shown.
To investigate the impact of Gadd45a,b deficiency on gene expression, we performed transcriptome analysis of two-cell stage embryos from Gadd45a,b DKO crosses. Transposable elements (TEs) were hardly affected in DKO embryos. There were no down-regulated and few up-regulated TE transcripts, including LINE1 elements (L1MCc and Lx2B) (Fig. 6F). Also, the number of deregulated nonrepetitive genes in DKOs was limited (n = 104, 10% FDR) (Fig. 6G), in accord with the subviability of DKO mice. However, misregulated genes showed a clear signature of impaired two-cell stage entry: First, genes down-regulated in DKOs correlated with genes up-regulated in two-cell stage embryos compared with oocytes (Macfarlan et al. 2012), while genes up-regulated in DKOs correlated with genes up-regulated in oocytes (Fig. 6H,I). Second, ZGA genes associated specifically with the 2C-like state (Macfarlan et al. 2012) overlapped with none of the up-regulated but seven down-regulated genes in DKOs (Fig. 6J). The enrichment of maternal and depletion of zygotic transcripts in DKO embryos indicates that GADD45α,β promote the embryonic two-cell stage. Hence, we monitored the development of wild-type and DKO preimplantation embryos in vitro. No differences were observed for development until the two-cell stage between wild-type and DKO embryos (data not shown). However, only ∼40% of the DKO embryos reached eight-cell stage versus ∼80% of the wild-type embryos (Fig. 6K). The affected DKO embryos remained at two- or four-cell stage or died. Incomplete penetrance of preimplantation defects are also observed in other mutant mice (Narducci et al. 2002). However, the impaired in vitro preimplantation development supports our findings in mouse ESCs showing that GADD45 proteins are involved in two-cell stage regulation.
Discussion
GADD45α,β,γ are adaptors for TET/TDG-mediated DNA demethylation, but in which physiological processes and at which genomic loci they mediate demethylation remain poorly understood. In addition, they show overlapping expression and hence may act functionally redundant, complicating their analysis. Here we present analyses of TKO ESCs and Gadd45a,b DKO mice showing that (1) GADD45α,β,γ are not required to maintain pluripotency and self-renewal in ESCs; however, (2) GADD45 proteins are required for locus-specific demethylation of ∼7000 sites, notably on enhancers and at sites harboring oxidized 5mC; (3) Gadd45 mutant ESCs display methylation-related gene misexpression; and (4) GADD45 proteins promote the 2C-like ESC state and two-cell embryo stage, regulating a subset of ZGA-specific genes.
GADD45 proteins are not required for pluripotency but are required for differentiation of ESCs
We found that Gadd45 TKO ESCs remain pluripotent and self-renew, as has been found for other DNA demethylation-deficient ESCs such as Tet1,2,3, and Tdg knockout ESCs (Cortázar et al. 2011; Dawlaty et al. 2014). A previous study (Li et al. 2015) reported that also Gadd45a,b DKO ESCs remain pluripotent. In agreement with this study, overall oxidized cytosine levels were mostly unchanged in Gadd45 TKO ESCs. This is expected because GADD45α functions in locus-specific rather than global demethylation (Schmitz et al. 2009; Schäfer et al. 2013; Arab et al. 2014,2019). Our rescue experiments indicate that all three GADD45 proteins can compensate for the loss of all three genes, supporting their functional redundancy.
While GADD45 proteins were dispensable for overall ESC maintenance, a subset of genes was down-regulated and hypermethylated in TKO ESCs. Moreover, DNA hypomethylating 2i, vitamin C, and 5-azadeoxycytidine treatment rescued this down-regulation, suggesting that it was the direct or indirect consequence of DNA hypermethylation. Similarly, previous reduced representation bisulfite sequencing identified 68 hypermethylated but no hypomethylated loci in Gadd45a,b DKO ESCs (Li et al. 2015). Most of these sites overlapped with 5hmC- and 5fC-enriched regions, corroborating a role in DNA demethylation. Taken together with the overlap between the genes misregulated in Gadd45 and Tet mutants, this supports the conclusion that these two protein families cooperate in enzymatic DNA demethylation (Arab et al. 2014, 2019; Kienhöfer et al. 2015; Li et al. 2015).
Even though GADD45 proteins act locally and DNMTs act globally, there was a significant overlap between genes misregulated in Gadd45 TKO ESCs and Dnmt TKO ESCs. However, despite millions of genomic sites that become unmethylated in Dnmt TKO ESCs, including at least 6100 promoters, only a few hundred genes are actually derepressed (Karimi et al. 2011). These “hot spot” genes misregulated in Dnmt TKO ESCs are enriched for a role in germ cell development and localize on the X chromosome. Likewise, genes in the MageA and Rhox clusters that are prominently reactivated in the DNMT TKO ESCs are down-regulated in Gadd45 TKO ESCs. Thus, even though DNMTs act globally, respective gene expression changes in ESCs occur only on hot spots, which are prone to react to DNA methylation changes, such as germ cell-specific genes. Indeed, TET1 is a prominent regulator of gene expression in germ cells (Yamaguchi et al. 2012; Hill et al. 2018), suggesting that ESCs recapitulate aspects of germ cell gene regulation via DNMT–TET-mediated methylation–demethylation.
In contrast to a limited role of GADD45 proteins during ESC pluripotency, gene deregulation in TKOs was greatly increased upon EB and monolayer differentiation. Neuronal genes were particularly affected, corroborating a role for Gadd45 genes during neural differentiation (Ma et al. 2009; Huang et al. 2010; Kaufmann and Niehrs 2011). Indeed, while Gadd45a,b DKO mice were viable, they were sublethal, with embryos showing gross abnormalities, including defects in neural tube closure and brain hemorrhage. This phenocopies Tet1,2 DKOs, which are also sublethal, with affected embryos showing exencephaly and cranial hemorrhaging and where the defects were attributed to reduced 5hmC, increased 5mC, and aberrant imprinting (Dawlaty et al. 2013).
TET/TDG processed sites are main targets of GADD45-mediated DNA demethylation
Our methylome analysis of TKO ESCs revealed ∼7000 hyper-DMRs, the greatest number of locus-specific methylation changes so far reported to be associated with GADD45 function. Since only 34 hypo-DMRs were detected (arguing against generalized methylation misregulation) and since the most significant association of the hyper-DMRs was with oxidized 5mC, we conclude that the hyper-DMRs arose as the result of impaired enzymatic DNA demethylation. This is consistent with direct interaction of GADD45α with both TET1 and TDG (Barreto et al. 2007; Cortellino et al. 2011; Kienhöfer et al. 2015; Li et al. 2015). Moreover, GADD45-dependent hyper-DMRs are enriched at TET-dependent hyper-DMRs (Lu et al. 2014) with the caveat that the number of called DMRs is not directly comparable due to differences in ESC lines and in DMR-calling algorithms used. Consistently, we reached conclusions for Gadd45 TKO ESCs similar to those that have been reported for Tet1,2,3 TKO ESCs (Lu et al. 2014). First, the most important concordance of hyper-DMRs was with sites marked by oxidized 5mC derivatives. Enrichment of hyper-DMRs at TDG 5fC and 5caC target sites concurs with the conclusion of a dual role of GADD45α to stimulate TET1 chemical processivity for iterative 5mC oxidation and enhance TDG processing of 5fC/5caC (Kienhöfer et al. 2015; Li et al. 2015). Second, enhancers are the most enriched genomic target of both GADD45- and TET-mediated DNA demethylation. Enhancers in ESCs tend to be hypomethylated (Stadler et al. 2011; Kieffer-Kwon et al. 2013; Ziller et al. 2013), and thus GADD45 plays a significant role in this phenomenon. Enhancers are also the main target of GADD45α-mediated DNA demethylation in mouse embryonic fibroblasts (MEFs) (Schäfer et al. 2018).
In both Tet and Gadd45 TKO ESCs, enhancer hypermethylation affected expression of relatively few genes, consistent with the minor role of 5mC in gene repression of early embryos and ESCs (Fouse et al. 2008; Bogdanović et al. 2011). The regulatory role of DNA demethylation for Gadd45 may manifest more upon later cell differentiation, as, e.g., mutation of Dnmt and Tet leads to differentiation defects in ESCs (Jackson et al. 2004; Dawlaty et al. 2014) and lethality in whole embryos (Okano et al. 1999; Dai et al. 2016). Hence, the dynamics of DNA methylation in ESCs may reflect a role as molecular memory for subsequent enhancer regulation in differentiating cells rather than in regulation of acute gene expression (Kim et al. 2018).
GADD45 proteins promote a 2C-like state and ZGA-specific gene expression
A main finding of this study is that Gadd45 TKO ESCs displayed impaired cycling into the 2C-like state, and Gadd45a,b DKOs showed partial gene misregulation at the two-cell stage as well as embryonic sublethality with developmental stage arrest. This suggests that 2C cycling defects in TKO ESCs mirror a role of GADD45 during two-cell stage development and ZGA. The 2C-like state of ESCs recapitulates key aspects of the two-cell stage mouse embryo both phenotypically and molecularly (for review, see Ishiuchi and Torres-Padilla 2013; Eckersley-Maslin et al. 2018). Concordantly, Gadd45 expression peaks in two-cell stage mouse embryos, and, while Gadd45a,b DKOs display only moderate gene misregulation, our results may underestimate their role, as Gadd45g could partially compensate for Gadd45a,b deficiency in DKOs (Bogdanović et al. 2011).
Entry into the 2C-like state is accompanied by genome-wide DNA demethylation (Eckersley-Maslin et al. 2016; Dan et al. 2017). However, ESCs cultured in serum are in a distinct, potentially artifactual epigenetic state: The de novo DNA methylation machinery, only transiently active in embryos during implantation, is instead continuously operating in ESCs (Brandeis et al. 1994; Lienert et al. 2011). Hence, down-regulation of 2C genes in TKO ESCs might rather reflect a role of GADD45 proteins during early two-cell stage in protecting against de novo methylation. This is because the genome of two-cell embryos is already hypomethylated due to global demethylation occurring in zygotes (for review, see Lee et al. 2014; Messerschmidt et al. 2014; Eckersley-Maslin et al. 2018) and requires TET3 to protect against de novo DNA methylation (Amouroux et al. 2016). It is this protection against de novo DNA methylation that may require GADD45.
In apparent contradiction to these findings, transition to the 2C-like state in Tet TKO ESCs is actually enhanced, as TET proteins repress type III ERVs and 2C-specific genes (Lu et al. 2014). Moreover, Tet TKO mouse embryos develop past implantation stage to gastrulae, albeit with altered expression of a few hundred genes at the blastocyst stage (Dai et al. 2016). If GADD45 acts via TETs to affect 2C-like state and two-cell embryo development, how can this discrepancy be explained? First, Tet TKO mouse embryos were generated by crossing mice with TET-deficient germ cells (Dai et al. 2016) and hence with zygotes that had adapted to a state completely devoid of TET enzymatic activity. Furthermore, close inspection of Tet1,3 DKO mice showed unexpected variable expression of ∼150 genes in eight-cell stage embryos, among them the Zscan4 cluster (Kang et al. 2015). In fact, acute Tet1,2,3 knockdown in oocytes leads to developmental arrest at two-cell stage with severe ZGA gene misregulation (M Wossidlo, pers. comm.). Second, TETs have a dual function involving both their catalytic and noncatalytic gene-repressive function, the latter being the dominant gene regulatory mode of TET1 in ESCs (Williams et al. 2011). The repressive role involves recruitment of KAP1/TRIM28 by TETs, a negative 2C-like state regulator (Lu et al. 2014). Consistent with a dual role, Tet-deficient ESCs and epiblast-derived stem cells can display either up-regulation or down-regulation of Zscan4 cluster expression, depending on culture conditions (Yang et al. 2016; Khoueiry et al. 2017). Similarly, for LINE1 regulation, TETs play a dual role in ESCs, activating and silencing expression via LINE1 promoter demethylation and via recruiting the SIN3A repressor, respectively (de la Rica et al. 2016).
In analogy, TET enzymes may play a dual role in the regulation of the key developmental transition at the two-cell stage: First, a repressive one, whereby possibly TET represses LINE1 elements whose transcripts silence Dux expression and thereby the two-cell state (Percharde et al. 2018). Second, a promoting one, as supported by our study, whereby GADD45 targets TETs to promote the two-cell stage via demethylation and protection against de novo methylation.
Materials and methods
Statistics
Unless indicated otherwise, statistical significance was tested with unpaired two-tailed Student's t-test with three biological replicates. Unless indicated otherwise, data are presented as mean ± standard deviation from three independent clones (P < 0.05 [*], P < 0.005 [**], and P < 0.0005 [***]).
GO enrichment analysis for the ontology “biological process” was carried out using GOrilla (Eden et al. 2009) using the parameter “two unranked lists of genes” and the whole-mouse transcriptome as a background list.
Commonly deregulated genes were identified using publicly available data sets. Overlaps were identified using ENSEMBL or RefSeq IDs if available and gene symbols if not. Statistical significance of overlaps was determined using hypergeometric test unless indicated otherwise (P< 0.05 [∗], P < 0.005 [∗∗], and P < 0.0005 [∗∗∗]). Only genes detectable by both platforms used were considered. For scatter plots, only genes that were significantly deregulated in both respective studies are shown.
qPCR
Total RNA was isolated using a Qiagen RNeasy minikit with on-column DNase digest (Qiagen). First strand cDNA was generated using SuperScript II reverse transcriptase (Invitrogen). Real-time PCR was performed in technical duplicates using Roche LightCycler480 probe master and primers in combination with predesigned monocolor hydrolysis probes of the Roche universal probe library (UPL). For quantification, the Roche LC480 quantification software module was used. Expression levels first were normalized to Gapdh or Tbp expression, followed by normalization to control conditions specific to the individual experiment, as indicated.
Quantitative MS of DNA modifications
Genomic DNA sample preparation and quantification of 5mC and its oxidative derivatives was carried out as described before (Schomacher et al. 2016).
ESC culture, treatment, and differentiation
ESCs were cultured on gelatinized cell culture vessels in LIF-conditioned DMEM supplemented with 15% ESC-grade FBS, 2 mM L-glutamine, 50 U/mL penicillin/streptomycin, 1× NEAA, 1 mM sodium pyruvate, and 100 µM β-mercaptoethanol at 37°C, 5% CO2, and 21% O2, with daily medium changes. Cells were passaged every 2 d once reaching ∼60% confluency with a ratio of 1:8. For ESC culture on MEF feeder, ESCs were preplated in a 12-well plate with CF-1 MEF feeder (American Type Culture Collection). For growth curve analysis, cells were counted in duplicates using a TC10 automated cell counter (Bio-Rad).
For EB differentiation, 3.5 × 106 mouse ESCs were plated on nonadherent 10-cm bacterial dishes (Greiner) in 15 mL of CA medium (Bibel et al. 2007). CA medium was changed every other day. For RA-induced EB differentiation, CA medium was supplemented with RA (final concentration 5 µM) at days 4 and 6 of differentiation. EBs were harvested after 8 d of differentiation.
For monolayer differentiation, 5000 ESCs per square centimeter were plated on gelatinized cell culture plates in regular ESC medium on the evening before differentiation. On the next morning, cells were washed twice with PBS, and medium was changed to N2B27 medium (50% advanced DMEM/F12, 50% neurobasal, 2 mM L-glutamine, 50 U/mL penicillin/streptomycin, 50 µg/mL BSA, 0.5× N2 supplement [Invitrogen], 0.5× B27-supplement [Invitrogen]). Medium was changed on days 3 and 5 of differentiation. Cells were harvested after 6 d of differentiation.
For 2i treatment, 230,000 ESCs were plated per six-well plate in regular ESC medium. On the next day, cells were washed twice with PBS and cultivated up to 72 h in 2i medium (N2B27 medium, 1 µM PD0325901, 3 µM CHIR99021, 4% LIF supernatant).
For vitamin C treatment, 230.000 ESCs per well were plated in six-well plates in regular ESC medium. The next day, vitamin C (L-ascorbic acid 2-phosphate sesquimagnesium salt) was added to a final concentration of 100 µg/mL. Cells were incubated in vitamin C-containing medium for up to 72 h and passaged once during that time.
For 5-azacytidine treatment, ESCs were incubated in 10 µM 5-deoxyazacytidine in ESC medium for 48 h.
For transdifferentiation, ESCs were resuspended in DMEM supplemented with 15% knockout serum replacement (Invitrogen), 2 mM L-glutamine, 50 U/mL penicillin/streptomycin, 1× NEAA, 1 mM Na-pyruvate, 100 µM β-mercaptethanol medium, and 10 ng/mL recombinant hBMP-4 (R&D Systems) after passaging and plated at a density of 104 cells per square centimeter on gelatinized cell culture vessels. Medium was changed on days 2, 4, 6, and 7 of transdifferentiation. Cells were harvested after 8 d.
For transient overexpression, preplated ESCs were transfected with Lipofectamine 2000 (Thermo Fisher) according to the manufacturer's recommendations. Medium was renewed on the next day, and cells were harvested 48 h after transfection.
CRISPR/Cas9-mediated knockout and introduction of stable-expressing Zscan4c::eGFP
ESCs were seeded on 10-cm dishes (1.45 × 106 each) and transfected on the next day with either (1) 13.5 µg of empty px330 vector and 1.5 µg of pPuro or (2) 2.25 µg of each of the six gRNA-containing plasmids (see the Supplemental Material) and 1.5 µg of pPuro using 45 µL of Lipofectamine 2000 (Thermo Fisher) in a volume of 1.5 mL of OptiMEM but otherwise as described above. ESCs were passaged from one 10-cm dish to two 15-cm dishes the following day. Cells were selected with 2 µg/mL puromycin from 48 h after transfection and kept under selection until the day of freezing. Forming ESC colonies were transferred to gelatinized 48-well plates 6 d after passaging. On the next day, colonies were washed once with 500 µL of PBS, dissociated with 100 µL of 0.25% trypsin for 90 sec at 37°C, quenched with 800 µL of ESC medium, and plated on a new 48-well plate. Three days later, cells were passaged and partially used for genotyping PCR (for primers, see the Supplemental Material). For transfection of Zscan4c::eGFP, 1 × 106 ESCs were seeded on gelatinized 10-cm dishes and transfected 4 h after seeding using Lipofectamine 2000 (Thermo Fisher). Cells were selected with 5 µg/mL blasticidin 48 h after transfection for 11 d, expanded, and frozen for further analysis. For transfection of 2C::tdTomato reporter (Addgene, 40281) 1 × 106 ESCs were seeded on gelatinized 10-cm dishes and transfected 4 h after seeding using Lipofectamine 2000 (Thermo Fisher). Cells were selected with 150 µg/mL hygromycin 48 h after transfection for 7 d, expanded, and frozen for further analysis.
Plasmids
pZscan4-Emerald was a kind gift from M. Ku (Zalzman et al. 2010). pCW57.1_Luciferase and pCW57.1_mDuxCA-3xHA were a kind gift from B. Cairns (Hendrickson et al. 2017). The expression constructs in this study were pCS2 + Flag_hTDG (Schomacher et al. 2016), pCS2 + GFP (Barreto et al. 2007), pCS2 + myc-MmGadd45a (M Gierl, unpubl.), pCS2 + myc-MmGadd45b (M Gierl, unpubl.), pCS2 + myc-MmGadd45g (Gierl et al. 2012), and pRKW2-mTet2 (Ko et al. 2010). The catalytic domain of mouse Tet1 was inserted into pCS2+ as an N-terminal HA tag expression construct (A Ernst, unpubl.).
Teratoma assays
ESCs were sent cryoconserved to EPO Berlin GmbH, where they were thawed and passaged twice before transplantation. Cells were resuspended in PBS, mixed with Matrigel, and transplanted into the flanks of three NSG mice per mouse ESC clone. Tumor weight and size were measured twice per week. Animals injected with the same ESC clone were sacrificed once the average tumor size reached ∼1.0 cm3. Tumors were excised, weighed, and cut. One-third was shock-frozen, and two-thirds were fixed in formalin. Formalin-fixed tumors were paraffin-embedded, sectioned, and stained with hematoxylin and eosin.
Flow cytometry analysis and fluorescence-activated cell sorting (FACS)
Cells were detached using 0.25% trypsin and resuspended in PBS containing 2.5% ESC-grade and 5 mM EDTA (pH 8). Suspended cells were then analyzed by the BD LSRFortessaSORP flow cytometry system using DiVa software. For downstream analysis on sorted ESCs, cells were sorted according to the fluorescence intensity of eGFP into PBS using BD FACSAria III SORP with a 85-µm nozzle. Data analysis was performed with FlowJo software (version 10.5.3).
WGBS
Genomic DNA from control and Gadd45 TKO ESCs was purified using a QIAamp DNA minikit (Qiagen) according to the manufacturer's recommendations. An additional RNase A treatment (10 mg/mL; Qiagen) was done after cell lysis. WGBS library preparation was carried out using the TruSeq PCR-free library preparation kit (LT) and the Epitect kit (Qiagen) for bisulfite conversion. Sequencing was performed on Illumina HiSeq X in paired-end mode (PE150).
RNA-seq
ESC samples
Total RNA was isolated using the RNeasy minikit (Qiagen) with on-column DNase digest. Next-generation sequencing (NGS) library preparation was performed using Illumina's TruSeq stranded mRNA HT sample preparation kit with dual indexing following the standard protocol (Illumina, 15031047 revision D). Libraries were profiled with a DNA 1000 chip on an Agilent 2100 Bioanalyzer and quantified using the Qubit dsDNA HS assay kit on a Qubit 2.0 fluorometer (Life Technologies). All 36 samples were pooled in equimolar ratio and sequenced on eight HiSeq 2000 lanes for 35 cycles plus an additional 16 cycles for the i7 and i5 index reads.
Isolation and culture of preimplantation embryos
For RNA-seq, two-cell stage embryos were collected from 3-wk-old wild-type (n = 4) and (Gadd45a/Gadd45b)−/− (n = 4) superovulated females 20 h after the appearance of the vaginal plug. Two-cell stage embryos coming from the same litter were pooled and considered as one biological sample. Embryos were collected in M2 medium (Sigma) supplemented with 0.3 mg/mL hyaluronidase (Sigma), washed twice with PBS, and directly transferred into lysis buffer (Smart Seq version 4 ultralow input RNA kit for sequencing, Takara). Sample preparation was done according to the manufacturer's recommendations. cDNA was amplified using 11 PCR cycles. NGS library preparation was performed using NuGEN's Ovation ultralow system V2 1-96 (2014). Libraries were prepared with a starting amount of 2.36 ng of fragmented cDNA and were amplified in 11 PCR cycles. Libraries were profiled in a high-sensitivity DNA on a 2100 Bioanalyzer (Agilent Technologies) and quantified using the Qubit dsDNA HS assay kit in a Qubit 2.0 fluorometer (Life Technologies). All eight samples were pooled in equimolar ratio and sequenced on one NextSeq 500 high-output FC SR for 85 cycles plus seven cycles for the index read.
For the in vitro development assay, mouse preimplantation embryos were collected as described above and then cultured in EmbryoMax human tubal fluid medium (Millipore) for 48 h at 37°C and 5% CO2. The experiment was performed in biological triplicates using three independent breedings for wild-type and (Gadd45a/Gadd45b)−/− animals.
Animal experiments
Gadd45a and Gadd45b knockout mice were kindly provided by M.C. Hollander (Hollander et al. 1999; Gupta et al. 2005). Both strains were backcrossed several generations into the C57BL/6N background and interbred to obtain Gadd45a+/−/Gadd45b+/− (DHet) mice, which were further intercrossed to generate wild-type, Gadd45a+/−/Gadd45b+/−, and Gadd45a−/−/Gadd45b−/− (DKO) animals from a homogeneous genetic background. Mice were housed under 12:12 light/dark cycles and provided with ad libitum food and water in accordance with national and European guidelines. For embryo isolation at stage E13.5, timed matings were set up between DHet mice and DKO animals. All procedures were performed with the approval of the ethical committees on animal care and use of the federal states of Rheinland-Pfalz, Germany.
Data availability
All NGS data have been deposited in the NCBI's Gene Expression Omnibus (GEO) under superseries accession number GSE127720.
Supplementary Material
Acknowledgments
We thank C. Christopoulou for assistance with the rescue experiments. We thank M.C. Hollander for Gadd45a and Gadd45b mutant mice. B. Cairns, M. Ku, A. Rao, and H. Richly kindly provided reagents. We thank C. Scholz for technical support. Contributions by the Institute of Molecular Biology Flow Cytometry, Genomics, Proteomics, and Microscopy Core Facilities and the German Cancer Research Center (DKFZ) High-Throughput Sequencing Unit are gratefully acknowledged. M.U.M. was supported by a Natural Sciences and Engineering Research Council of Canada Postdoctoral Fellowship (NSERC-PDF). This work was supported by a European Research Council advanced grant (“Demethylase”).
Author contributions: K.M.S. conceived and conducted most of the 2C-related experiments. M.L. generated and characterized the TKO ESCs. V.V. carried out all knockout mouse analyses. T.A., M.U.M., and E.K. carried out bioinformatics analyses. M.U.M. performed LC-MS/MS measurements. All authors analyzed and discussed the data. C.N. conceived and coordinated the study and wrote the paper with contribution from K.M.S. and M.L.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.325696.119.
References
- Amano T, Hirata T, Falco G, Monti M, Sharova LV, Amano M, Sheer S, Hoang HG, Piao Y, Stagg CA, et al. 2013. Zscan4 restores the developmental potency of embryonic stem cells. Nat Commun 4: 1966 10.1038/ncomms2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amouroux R, Nashun B, Shirane K, Nakagawa S, Hill PW, D'Souza Z, Nakayama M, Matsuda M, Turp A, Ndjetehe E, et al. 2016. De novo DNA methylation drives 5hmC accumulation in mouse zygotes. Nat Cell Biol 18: 225–233. 10.1038/ncb3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arab K, Park YJ, Lindroth AM, Schäfer A, Oakes C, Weichenhan D, Lukanova A, Lundin E, Risch A, Meister M, et al. 2014. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell 55: 604–614. 10.1016/j.molcel.2014.06.031 [DOI] [PubMed] [Google Scholar]
- Arab K, Karaulanov E, Musheev M, Trnka P, Schäfer A, Grummt I, Niehrs C. 2019. GADD45A binds R-loops and recruits TET1 to CpG island promoters. Nat Genet 51: 217–223. 10.1038/s41588-018-0306-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto G, Schäfer A, Marhold J, Stach D, Swaminathan SK, Handa V, Döderlein G, Maltry N, Wu W, Lyko F, et al. 2007. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature 445: 671–675. 10.1038/nature05515 [DOI] [PubMed] [Google Scholar]
- Beddington RS, Robertson EJ. 1989. An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development 105: 733–737. [DOI] [PubMed] [Google Scholar]
- Bibel M, Richter J, Lacroix E, Barde Y-A. 2007. Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nat Protoc 2: 1034–1043. 10.1038/nprot.2007.147 [DOI] [PubMed] [Google Scholar]
- Blaschke K, Ebata KT, Karimi MM, Zepeda-Martínez JA, Goyal P, Mahapatra S, Tam A, Laird DJ, Hirst M, Rao A, et al. 2013. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 500: 222–226. 10.1038/nature12362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanović O, Long SW, van Heeringen SJ, Brinkman AB, Gómez-Skarmeta JL, Stunnenberg HG, Jones PL, Veenstra GJC. 2011. Temporal uncoupling of the DNA methylome and transcriptional repression during embryogenesis. Genome Res 21: 1313–1327. 10.1101/gr.114843.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. 2012. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 336: 934–937. 10.1126/science.1220671 [DOI] [PubMed] [Google Scholar]
- Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. 1994. Sp1 elements protect a CpG island from de novo methylation. Nature 371: 435–438. 10.1038/371435a0 [DOI] [PubMed] [Google Scholar]
- Cambuli F, Murray A, Dean W, Dudzinska D, Krueger F, Andrews S, Senner CE, Cook SJ, Hemberger M. 2014. Epigenetic memory of the first cell fate decision prevents complete ES cell reprogramming into trophoblast. Nat Commun 5: 5538 10.1038/ncomms6538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YJ, Lin C-P, Risso D, Chen S, Kim TA, Tan MH, Li JB, Wu Y, Chen C, Xuan Z, et al. 2017. Deficiency of microRNA miR-34a expands cell fate potential in pluripotent stem cells. Science 355: eaag1927 10.1126/science.aag1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortázar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, et al. 2011. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature 470: 419–423. 10.1038/nature09672 [DOI] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. 2011. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146: 67–79. 10.1016/j.cell.2011.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H-Q, Wang B-A, Yang L, Chen J-J, Zhu G-C, Sun M-L, Ge H, Wang R, Chapman DL, Tang F, et al. 2016. TET-mediated DNA demethylation controls gastrulation by regulating Lefty-Nodal signalling. Nature 538: 528–532. 10.1038/nature20095 [DOI] [PubMed] [Google Scholar]
- Dan J, Li M, Yang J, Li J, Okuka M, Ye X, Liu L. 2013. Roles for Tbx3 in regulation of two-cell state and telomere elongation in mouse ES cells. Sci Rep 3: 3492 10.1038/srep03492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J, Rousseau P, Hardikar S, Veland N, Wong J, Autexier C, Chen T. 2017. Zscan4 inhibits maintenance DNA methylation to facilitate telomere elongation in mouse embryonic stem cells. Cell Rep 20: 1936–1949. 10.1016/j.celrep.2017.07.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Ganz K, Powell BE, Hu Y-C, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi S-W, Page DC, et al. 2011. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9: 166–175. 10.1016/j.stem.2011.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, Gao Q, Powell BE, Li Z, Xu M, et al. 2013. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell 24: 310–323. 10.1016/j.devcel.2012.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Breiling A, Le T, Barrasa MI, Raddatz G, Gao Q, Powell BE, Cheng AW, Faull KF, Lyko F, et al. 2014. Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev Cell 29: 102–111. 10.1016/j.devcel.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A. 2011. CpG islands and the regulation of transcription. Genes Dev 25: 1010–1022. 10.1101/gad.2037511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A, Planet E, Coluccio A, Verp S, Duc J, Trono D. 2017. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet 49: 941–945. 10.1038/ng.3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Rica L, Deniz Ö, Cheng KCL, Todd CD, Cruz C, Houseley J, Branco MR. 2016. TET-dependent regulation of retrotransposable elements in mouse embryonic stem cells. Genome Biol 17: 234 10.1186/s13059-016-1096-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckersley-Maslin MA, Svensson V, Krueger C, Stubbs TM, Giehr P, Krueger F, Miragaia RJ, Kyriakopoulos C, Berrens RV, Milagre I, et al. 2016. MERVL/Zscan4 network activation results in transient genome-wide DNA demethylation of mESCs. Cell Rep 17: 179–192. 10.1016/j.celrep.2016.08.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckersley-Maslin MA, Alda-Catalinas C, Reik W. 2018. Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat Rev Mol Cell Biol 19: 436–450. 10.1038/s41580-018-0008-z [DOI] [PubMed] [Google Scholar]
- Eckersley-Maslin M, Alda-Catalinas C, Blotenburg M, Kreibich E, Krueger C, Reik W. 2019. Dppa2 and Dppa4 directly regulate the Dux-driven zygotic transcriptional program. Genes Dev 33: 194–208. 10.1101/gad.321174.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema M, Mori D, Niwa H, Hasegawa Y, Yamanaka Y, Hitoshi S, Mimura J, Kawabe Y-i, Hosoya T, Morita M, et al. 2008. Krüppel-like factor 5 is essential for blastocyst development and the normal self-renewal of mouse ESCs. Cell Stem Cell 3: 555–567. 10.1016/j.stem.2008.09.003 [DOI] [PubMed] [Google Scholar]
- Falco G, Lee S-L, Stanghellini I, Bassey UC, Hamatani T, Ko MSH. 2007. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev Biol 307: 539–550. 10.1016/j.ydbio.2007.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Hore TA, Santos F, Lee HJ, Dean W, Arand J, Krueger F, Oxley D, Paul Y-L, Walter J, et al. 2013. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell 13: 351–359. 10.1016/j.stem.2013.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, van Neste L, Jaenisch R, Fan G. 2008. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell 2: 160–169. 10.1016/j.stem.2007.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin DP, Kusumo H, Sharma RP, Guizzetti M, Guidotti A, Pandey SC. 2015. Gadd45b and N-methyl-D-aspartate induced DNA demethylation in postmitotic neurons. Epigenomics 7: 567–579. 10.2217/epi.15.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierl MS, Gruhn WH, von Seggern A, Maltry N, Niehrs C. 2012. GADD45G functions in male sex determination by promoting p38 signaling and Sry expression. Dev Cell 23: 1032–1042. 10.1016/j.devcel.2012.09.014 [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming G-l, Song H. 2011. Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle 10: 2662–2668. 10.4161/cc.10.16.17093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M, Gupta SK, Balliet AG, Hollander MC, Fornace AJ, Hoffman B, Liebermann DA. 2005. Hematopoietic cells from Gadd45a- and Gadd45b-deficient mice are sensitized to genotoxic-stress-induced apoptosis. Oncogene 24: 7170–7179. 10.1038/sj.onc.1208847 [DOI] [PubMed] [Google Scholar]
- Habibi E, Brinkman AB, Arand J, Kroeze LI, Kerstens HHD, Matarese F, Lepikhov K, Gut M, Brun-Heath I, Hubner NC, et al. 2013. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell 13: 360–369. 10.1016/j.stem.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Sousa Lopes Sd, Tang F, Lao K, Surani MA. 2008. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell 3: 391–401. 10.1016/j.stem.2008.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Furue MK, Tanaka S, Hirose M, Wakisaka N, Danno H, Ohnuma K, Oeda S, Aihara Y, Shiota K, et al. 2010. BMP4 induction of trophoblast from mouse embryonic stem cells in defined culture conditions on laminin. In Vitro Cell Dev Biol Anim 46: 416–430. 10.1007/s11626-009-9266-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y-F, Li B-Z, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333: 1303–1307. 10.1126/science.1210944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson PG, Doráis JA, Grow EJ, Whiddon JL, Lim J-W, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, et al. 2017. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet 49: 925–934. 10.1038/ng.3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill PWS, Leitch HG, Requena CE, Sun Z, Amouroux R, Roman-Trufero M, Borkowska M, Terragni J, Vaisvila R, Linnett S, et al. 2018. Epigenetic reprogramming enables the transition from primordial germ cell to gonocyte. Nature 555: 392–396. 10.1038/nature25964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata T, Amano T, Nakatake Y, Amano M, Piao Y, Hoang HG, Ko MSH. 2012. Zscan4 transiently reactivates early embryonic genes during the generation of induced pluripotent stem cells. Sci Rep 2: 208 10.1038/srep00208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander MC, Sheikh MS, Bulavin DV, Lundgren K, Augeri-Henmueller L, Shehee R, Molinaro TA, Kim KE, Tolosa E, Ashwell JD, et al. 1999. Genomic instability in Gadd45a-deficient mice. Nat Genet 23: 176–184. 10.1038/13802 [DOI] [PubMed] [Google Scholar]
- Huang HS, Kubish GM, Redmond TM, Turner DL, Thompson RC, Murphy GG, Uhler MD. 2010. Direct transcriptional induction of Gadd45γ by Ascl1 during neuronal differentiation. Mol Cell Neurosci 44: 282–296. 10.1016/j.mcn.2010.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Chavez L, Chang X, Wang X, Pastor WA, Kang J, Zepeda-Martínez JA, Pape UJ, Jacobsen SE, Peters B, et al. 2014. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc Natl Acad Sci 111: 1361–1366. 10.1073/pnas.1322921111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Kim JK, Do DV, Lee C, Penfold CA, Zylicz JJ, Marioni JC, Hackett JA, Surani MA. 2017. Stella modulates transcriptional and endogenous retrovirus programs during maternal-to-zygotic transition. Elife 6: e22345 10.7554/eLife.22345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiuchi T, Torres-Padilla M-E. 2013. Towards an understanding of the regulatory mechanisms of totipotency. Curr Opin Genet Dev 23: 512–518. 10.1016/j.gde.2013.06.006 [DOI] [PubMed] [Google Scholar]
- Ishiuchi T, Enriquez-Gasca R, Mizutani E, Bošković A, Ziegler-Birling C, Rodriguez-Terrones D, Wakayama T, Vaquerizas JM, Torres-Padilla M-E. 2015. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat Struct Mol Biol 22: 662–671. 10.1038/nsmb.3066 [DOI] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. 2011. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333: 1300–1303. 10.1126/science.1210597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M, Krassowska A, Gilbert N, Chevassut T, Forrester L, Ansell J, Ramsahoye B. 2004. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol Cell Biol 24: 8862–8871. 10.1128/MCB.24.20.8862-8871.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarome TJ, Butler AA, Nichols JN, Pacheco NL, Lubin FD. 2015. NF-κB mediates Gadd45β expression and DNA demethylation in the hippocampus during fear memory formation. Front Mol Neurosci 8: 539 10.3389/fnmol.2015.00054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukam D, Shariati SAM, Skotheim JM. 2017. Zygotic genome activation in vertebrates. Dev Cell 42: 316–332. 10.1016/j.devcel.2017.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Lienhard M, Pastor WA, Chawla A, Novotny M, Tsagaratou A, Lasken RS, Thompson EC, Surani MA, Koralov SB, et al. 2015. Simultaneous deletion of the methylcytosine oxidases Tet1 and Tet3 increases transcriptome variability in early embryogenesis. Proc Natl Acad Sci 112: E4236–E4245. 10.1073/pnas.1510510112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi MM, Goyal P, Maksakova IA, Bilenky M, Leung D, Tang JX, Shinkai Y, Mager DL, Jones S, Hirst M, et al. 2011. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8: 676–687. 10.1016/j.stem.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann LT, Niehrs C. 2011. Gadd45a and Gadd45g regulate neural development and exit from pluripotency in Xenopus. Mech Dev 128: 401–411. 10.1016/j.mod.2011.08.002 [DOI] [PubMed] [Google Scholar]
- Khoueiry R, Sohni A, Thienpont B, Luo X, Velde JV, Bartoccetti M, Boeckx B, Zwijsen A, Rao A, Lambrechts D, et al. 2017. Lineage-specific functions of TET1 in the postimplantation mouse embryo. Nat Genet 49: 1061–1072. 10.1038/ng.3868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer-Kwon K-R, Tang Z, Mathe E, Qian J, Sung M-H, Li G, Resch W, Baek S, Pruett N, Grøntved L, et al. 2013. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell 155: 1507–1520. 10.1016/j.cell.2013.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienhöfer S, Musheev MU, Stapf U, Helm M, Schomacher L, Niehrs C, Schäfer A. 2015. GADD45a physically and functionally interacts with TET1. Differentiation 90: 59–68. 10.1016/j.diff.2015.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Tan Y, Ma W, Merkurjev D, Destici E, Ma Q, Suter T, Ohgi K, Friedman M, Skowronska-Krawczyk D, et al. 2018. Pluripotency factors functionally premark cell-type-restricted enhancers in ES cells. Nature 556: 510–514. 10.1038/s41586-018-0048-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. 2010. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468: 839–843. 10.1038/nature09586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Tan L, Lv R, Shi Z, Xiong L, Wu F, Rabidou K, Smith M, He C, Zhang L, et al. 2016. A primary role of TET proteins in establishment and maintenance of De Novo bivalency at CpG islands. Nucleic Acids Res 44: 8682–8692. 10.1093/nar/gkw529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. 2009. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324: 929–930. 10.1126/science.1169786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Hore TA, Reik W. 2014. Reprogramming the methylome: erasing memory and creating diversity. Cell Stem Cell 14: 710–719. 10.1016/j.stem.2014.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H, Oh SP, Okano M, Jüttermann R, Goss KA, Jaenisch R, Li E. 1996. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122: 3195–3205. [DOI] [PubMed] [Google Scholar]
- Leitch HG, McEwen KR, Turp A, Encheva V, Carroll T, Grabole N, Mansfield W, Nashun B, Knezovich JG, Smith A, et al. 2013. Naive pluripotency is associated with global DNA hypomethylation. Nat Struct Mol Biol 20: 311–316. 10.1038/nsmb.2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhao M, Yin H, Gao F, Wu X, Luo Y, Zhao S, Zhang X, Su Y, Hu N, et al. 2010. Overexpression of the growth arrest and DNA damage-induced 45α gene contributes to autoimmunity by promoting DNA demethylation in lupus T cells. Arthritis Rheum 62: 1438–1447. 10.1002/art.27363 [DOI] [PubMed] [Google Scholar]
- Li Z, Gu T-P, Weber AR, Shen J-Z, Li B-Z, Xie Z-G, Yin R, Guo F, Liu X, Tang F, et al. 2015. Gadd45a promotes DNA demethylation through TDG. Nucleic Acids Res 43: 3986–3997. 10.1093/nar/gkv283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienert F, Wirbelauer C, Som I, Dean A, Mohn F, Schübeler D. 2011. Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet 43: 1091–1097. 10.1038/ng.946 [DOI] [PubMed] [Google Scholar]
- Liu Y, Olanrewaju YO, Zheng Y, Hashimoto H, Blumenthal RM, Zhang X, Cheng X. 2014. Structural basis for Klf4 recognition of methylated DNA. Nucleic Acids Res 42: 4859–4867. 10.1093/nar/gku134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Yu H, Chow C, Li B, Zheng W, Davis RJ, Flavell RA. 2001. GADD45γ mediates the activation of the p38 and JNK MAP kinase pathways and cytokine production in effector TH1 cells. Immunity 14: 583–590. 10.1016/S1074-7613(01)00141-8 [DOI] [PubMed] [Google Scholar]
- Lu B, Ferrandino AF, Flavell RA. 2004. Gadd45β is important for perpetuating cognate and inflammatory signals in T cells. Nat Immunol 5: 38–44. 10.1038/ni1020 [DOI] [PubMed] [Google Scholar]
- Lu F, Liu Y, Jiang L, Yamaguchi S, Zhang Y. 2014. Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev 28: 2103–2119. 10.1101/gad.248005.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang M-H, Guo JU, Kitabatake Y, Chang M-l, Pow-anpongkul N, Flavell RA, Lu B, Ming G-l, Song H. 2009. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 323: 1074–1077. 10.1126/science.1166859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. 2012. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63. 10.1038/nature11244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt DM, Knowles BB, Solter D. 2014. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev 28: 812–828. 10.1101/gad.234294.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narducci MG, Fiorenza MT, Kang S-M, Bevilacqua A, Di Giacomo M, Remotti D, Picchio MC, Fidanza V, Cooper MD, Croce CM, et al. 2002. TCL1 participates in early embryonic development and is overexpressed in human seminomas. Proc Natl Acad Sci 99: 11712–11717. 10.1073/pnas.182412399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng RK, Dean W, Dawson C, Lucifero D, Madeja Z, Reik W, Hemberger M. 2008. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat Cell Biol 10: 1280–1290. 10.1038/ncb1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Xin L, Sharov AA, Thomas M, Mowrer G, Meyers E, Piao Y, Mehta S, Yee S, Nakatake Y, et al. 2009. Uncovering early response of gene regulatory networks in ESCs by systematic induction of transcription factors. Cell Stem Cell 5: 420–433. 10.1016/j.stem.2009.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257. 10.1016/S0092-8674(00)81656-6 [DOI] [PubMed] [Google Scholar]
- Parisi S, Passaro F, Aloia L, Manabe I, Nagai R, Pastore L, Russo T. 2008. Klf5 is involved in self-renewal of mouse embryonic stem cells. J Cell Sci 121: 2629–2634. 10.1242/jcs.027599 [DOI] [PubMed] [Google Scholar]
- Park S-J, Shirahige K, Ohsugi M, Nakai K. 2015. DBTMEE: a database of transcriptome in mouse early embryos. Nucleic Acids Res 43: D771–D776. 10.1093/nar/gku1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percharde M, Lin C-J, Yin Y, Guan J, Peixoto GA, Bulut-Karslioglu A, Biechele S, Huang B, Shen X, Ramalho-Santos M. 2018. A LINE1-nucleolin partnership regulates early development and ESC identity. Cell 174: 391–405.e19. 10.1016/j.cell.2018.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. 2008. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell 135: 1201–1212. 10.1016/j.cell.2008.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Terrones D, Gaume X, Ishiuchi T, Weiss A, Kopp A, Kruse K, Penning A, Vaquerizas JM, Brino L, Torres-Padilla M-E. 2018. A molecular roadmap for the emergence of early-embryonic-like cells in culture. Nat Genet 50: 106–119. 10.1038/s41588-017-0016-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabag O, Zamir A, Keshet I, Hecht M, Ludwig G, Tabib A, Moss J, Cedar H. 2014. Establishment of methylation patterns in ES cells. Nat Struct Mol Biol 21: 110–112. 10.1038/nsmb.2734 [DOI] [PubMed] [Google Scholar]
- Sakaue M, Ohta H, Kumaki Y, Oda M, Sakaide Y, Matsuoka C, Yamagiwa A, Niwa H, Wakayama T, Okano M. 2010. DNA methylation is dispensable for the growth and survival of the extraembryonic lineages. Curr Biol 20: 1452–1457. 10.1016/j.cub.2010.06.050 [DOI] [PubMed] [Google Scholar]
- Schäfer A, Karaulanov E, Stapf U, Döderlein G, Niehrs C. 2013. Ing1 functions in DNA demethylation by directing Gadd45a to H3K4me3. Genes Dev 27: 261–273. 10.1101/gad.186916.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer A, Mekker B, Mallick M, Vastolo V, Karaulanov E, Sebastian D, Lippen Cvd, Epe B, Downes DJ, Scholz C, et al. 2018. Impaired DNA demethylation of C/EBP sites causes premature aging. Genes Dev 32: 742–762. 10.1101/gad.311969.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz K-M, Schmitt N, Hoffmann-Rohrer U, Schäfer A, Grummt I, Mayer C. 2009. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol Cell 33: 344–353. 10.1016/j.molcel.2009.01.015 [DOI] [PubMed] [Google Scholar]
- Schomacher L, Han D, Musheev MU, Arab K, Kienhöfer S, von Seggern A, Niehrs C. 2016. Neil DNA glycosylases promote substrate turnover by Tdg during DNA demethylation. Nat Struct Mol Biol 23: 116–124. 10.1038/nsmb.3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA. 2010. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature 463: 563–567. 10.1038/nature08683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Wu H, Diep D, Yamaguchi S, D'Alessio AC, Fung H-L, Zhang K, Zhang Y. 2013. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 153: 692–706. 10.1016/j.cell.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PWTC, Bauer C, Münzel M, Wagner M, Müller M, Khan F, et al. 2013. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152: 1146–1159. 10.1016/j.cell.2013.02.004 [DOI] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. 2011. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480: 490–495. 10.1038/nature10716 [DOI] [PubMed] [Google Scholar]
- Svoboda P. 2018. Mammalian zygotic genome activation. Semin Cell Dev Biol 84: 118–126. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935. 10.1126/science.1170116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H. 2008. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development 135: 909–918. 10.1242/dev.017400 [DOI] [PubMed] [Google Scholar]
- Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S-i, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda HR, et al. 2006. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 11: 805–814. 10.1111/j.1365-2443.2006.00984.x [DOI] [PubMed] [Google Scholar]
- Wang PJ, McCarrey JR, Yang F, Page DC. 2001. An abundance of X-linked genes expressed in spermatogonia. Nat Genet 27: 422–426. 10.1038/86927 [DOI] [PubMed] [Google Scholar]
- Whiddon JL, Langford AT, Wong C-J, Zhong JW, Tapscott SJ. 2017. Conservation and innovation in the DUX4-family gene network. Nat Genet 49: 935–940. 10.1038/ng.3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PAC, Rappsilber J, Helin K. 2011. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473: 343–348. 10.1038/nature10066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Ang Y-S, Sevilla A, Lemischka IR, Ma'ayan A. 2014. Construction and validation of a regulatory network for pluripotency and self-renewal of mouse embryonic stem cells. PLoS Comput Biol 10: e1003777 10.1371/journal.pcbi.1003777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Hong K, Liu R, Shen L, Inoue A, Diep D, Zhang K, Zhang Y. 2012. Tet1 controls meiosis by regulating meiotic gene expression. Nature 492: 443–447. 10.1038/nature11709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Shen L, Liu Y, Sendler D, Zhang Y. 2013. Role of Tet1 in erasure of genomic imprinting. Nature 504: 460–464. 10.1038/nature12805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Guo R, Wang H, Ye X, Zhou Z, Dan J, Wang H, Gong P, Deng W, Yin Y, et al. 2016. Tet enzymes regulate telomere maintenance and chromosomal stability of mouse ESCs. Cell Rep 15: 1809–1821. 10.1016/j.celrep.2016.04.058 [DOI] [PubMed] [Google Scholar]
- Ying Q-L, Stavridis M, Griffiths D, Li M, Smith A. 2003. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol 21: 183–186. 10.1038/nbt780 [DOI] [PubMed] [Google Scholar]
- Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee S-L, Stagg CA, Hoang HG, Yang H-T, Indig FE, et al. 2010. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature 464: 858–863. 10.1038/nature08882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R-p, Shao J-z, Xiang L-x. 2011a. GADD45A protein plays an essential role in active DNA demethylation during terminal osteogenic differentiation of adipose-derived mesenchymal stem cells. J Biol Chem 286: 41083–41094. 10.1074/jbc.M111.258715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Fu S, Liu X, Zhao X, Zhang W, Peng W, Wu C, Li Y, Li X, Bartlam M, et al. 2011b. Crystal structure of human Gadd45γ corrected reveals an active dimer. Protein Cell 2: 814–826. 10.1007/s13238-011-1090-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Müller F, Donaghey J, Tsai LT-Y, Kohlbacher O, Jager Pd, Rosen ED, Bennett DA, Bernstein BE, et al. 2013. Charting a dynamic DNA methylation landscape of the human genome. Nature 500: 477–481. 10.1038/nature12433 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All NGS data have been deposited in the NCBI's Gene Expression Omnibus (GEO) under superseries accession number GSE127720.