In this study, Pearson et al. investigated the transcriptional mechanisms that promote remodeling in adipose tissue during the cold. Their findings demonstrate that transcriptional coregulator TLE3 regulates thermogenic gene expression in beige adipocytes through inhibition of EBF2 transcriptional activity.
Keywords: adipocytes, beige adipocytes, development, diabetes, metabolism, TLE3, thermogenesis
Abstract
Prolonged cold exposure stimulates the recruitment of beige adipocytes within white adipose tissue. Beige adipocytes depend on mitochondrial oxidative phosphorylation to drive thermogenesis. The transcriptional mechanisms that promote remodeling in adipose tissue during the cold are not well understood. Here we demonstrate that the transcriptional coregulator transducin-like enhancer of split 3 (TLE3) inhibits mitochondrial gene expression in beige adipocytes. Conditional deletion of TLE3 in adipocytes promotes mitochondrial oxidative metabolism and increases energy expenditure, thereby improving glucose control. Using chromatin immunoprecipitation and deep sequencing, we found that TLE3 occupies distal enhancers in proximity to nuclear-encoded mitochondrial genes and that many of these binding sites are also enriched for early B-cell factor (EBF) transcription factors. TLE3 interacts with EBF2 and blocks its ability to promote the thermogenic transcriptional program. Collectively, these studies demonstrate that TLE3 regulates thermogenic gene expression in beige adipocytes through inhibition of EBF2 transcriptional activity. Inhibition of TLE3 may provide a novel therapeutic approach for obesity and diabetes.
Subcutaneous white adipose tissue (WAT) undergoes substantial remodeling in response to nutrient availability and changes in ambient temperature. Cold exposure promotes the recruitment of thermogenic adipocytes—a long-term adaptive mechanism that facilitates thermogenesis (Young et al. 1984). Two distinct subtypes of thermogenic adipocytes have been found in mammals: brown and beige (Pisani et al. 2011; Wu et al. 2012). Many similarities exist between the two subtypes, including the presence of small multilocular lipid droplets, increased mitochondrial number, and the characteristic expression of the mitochondrial uncoupling protein 1 (UCP1) (Cannon et al. 1982; Bouillaud et al. 1985). UCP1 transports protons across the inner mitochondrial membrane without using this potential energy for ATP synthesis (Fedorenko et al. 2012), causing the chemical energy of substrates consumed by the cell in the TCA cycle to be released as heat (Orava et al. 2011). Beige adipocytes are also capable of generating heat through a futile creatine kinase cycle (Kazak et al. 2015). Interest in brown adipose tissue (BAT) was catalyzed in the early 2000s, when advancements in positron emission tomography (PET) scanning demonstrated the presence of BAT in adult humans (Hany et al. 2002; Cypess et al. 2009; Saito et al. 2009; Virtanen et al. 2009).
Despite their functional similarities, beige adipocytes arise from a lineage separate from brown adipocytes and are distinctively located as clusters of cells within WAT depots (Seale et al. 2008; Wu et al. 2012; Ikeda et al. 2018). Not all white adipose tissue is prone to acquire beige adipocytes: Subcutaneous depots such as the inguinal WAT (iWAT) recruit beige adipocytes in response to long-term cold exposure or after stimulation by β3 adrenergic receptor agonists (de Jesus et al. 2001), while much of the visceral adipose tissue, such as the epididymal WAT (eWAT), is resistant to acquiring beige adipocytes.
There has been considerable dispute over the identity of thermogenic tissue in adult humans, but mounting evidence points to the existence of BAT (Cypess et al. 2013) as well as populations of cells bearing markers of beige adipocytes (Sharp et al. 2012; Wu et al. 2012). Pathological conditions such as pheochromocytoma that lead to excess catecholamine production are associated with a greater abundance of beige adipocytes that is thought to promote a hypermetabolic state (Vergnes et al. 2016; Scheele and Nielsen 2017). Since the presence of these thermogenic tissues is inversely related to metabolic disorders and obesity (Orava et al. 2011; Sidossis and Kajimura 2015), understanding the regulatory mechanisms that govern the development and activity of thermogenic adipocytes has become an area of intense therapeutic interest.
All adipocyte subtypes require peroxisome proliferator-activated receptor γ (PPARγ), which is necessary and sufficient to drive adipocyte differentiation (Tontonoz et al. 1994). A prior genome-wide survey of PPARγ-binding profiles from primary cells isolated from three separate tissues (iWAT, eWAT, and BAT) differentiated in vitro showed an overlap in binding and similar intensities at the majority of PPARγ-binding sites (Siersbaek et al. 2012). However, this study also revealed a subset of depot-specific binding sites that correlated well with depot-specific gene expression (Siersbaek et al. 2012). These findings suggested that PPARγ likely works in concert with other transcription factors and transcriptional coregulators to regulate cell type-specific gene expression, although the identities of these auxiliary factors remain to be fully elucidated.
Among the transcription factors known to work in concert with PPARγ to determine adipocyte identity are those of the early B-cell factor (EBF) family. Both EBF1 and EBF2 participate in early adipogenesis (Akerblad et al. 2005; Jimenez et al. 2007), and it has been demonstrated recently that they also play important roles in differentiated adipocytes. EBF1 positively regulates insulin-stimulated glucose uptake and lipogenesis (Griffin et al. 2013; Gao et al. 2014), while EBF2 is selectively expressed in brown adipocytes and facilitates the binding of PPARγ to brown fat-specific genes to promote thermogenesis (Rajakumari et al. 2013; Lee and Ge 2014). Notably, in WAT, Ebf2 overexpression enhances beige adipocyte recruitment (Stine et al. 2016). EBF2 was shown recently to affect chromatin access via recruiting the chromatin remodeling complex BAF to brown fat genes (Shapira et al. 2017). The EBF DNA-binding motif associates with BAT-specific PPARγ-binding sites (Rajakumari et al. 2013) and enhances expression of genes important for mitochondrial function (Shapira et al. 2017). There are a number of known regulators of EBF2. For example, Blnc1 is a long noncoding RNA (lncRNA) that forms a ribonucleoprotein complex with EBF2 to promote thermogenic adipocyte differentiation and function (Zhao et al. 2014). ZFP423, which is preferentially expressed in white adipocytes, has been shown to inhibit EBF2 activity, causing inhibition of beige adipocyte differentiation (Shao et al. 2016).
The transducin-like enhancer of split (TLE) protein family is a highly conserved group of transcriptional coregulators (Fisher and Caudy 1998; Chen and Courey 2000). Groucho, the TLE ortholog in Drosophila melanogaster, was first identified as a key regulator of neuronal development (Chen and Courey 2000). A high-throughput screen identified TLE3 as a PPARγ cofactor that promotes white-specific gene expression (Villanueva et al. 2011, 2013). TLE3 has been shown to disrupt the interaction between PRDM16 and PPARγ, causing the BAT of Tle3 transgenic mice to appear more like WAT, with larger lipid droplets and higher levels of WAT-selective transcripts (Villanueva et al. 2013). During preadipocyte differentiation, TLE3 activates PPARγ to promote differentiation of all adipocyte subtypes and to sustain the lipid storage transcriptional program after differentiation. Tle3 is expressed in all adipocytes, but its expression is highest in WAT when compared with BAT.
Here we report that conditional deletion of Tle3 in adipocytes promotes the recruitment of beige adipocytes, increases energy expenditure, and leads to improved glucose metabolism and insulin sensitivity. Analysis of genome-wide occupancy of TLE3 in adipocytes suggests a direct role for this cofactor in regulating the nuclear-encoded mitochondrial gene expression program. Moreover, TLE3 binds to genomic regions that also associate with EBF transcription factors. We define a functional interaction between TLE3 and EBF2 in which TLE3 attenuates the EBF2-driven transcriptional program. Our findings outline a transcriptional mechanism that governs mitochondrial gene expression and the programming of beige adipocytes.
Results
Conditional deletion of Tle3 in adipocytes promotes a beige adipocyte transcriptional program
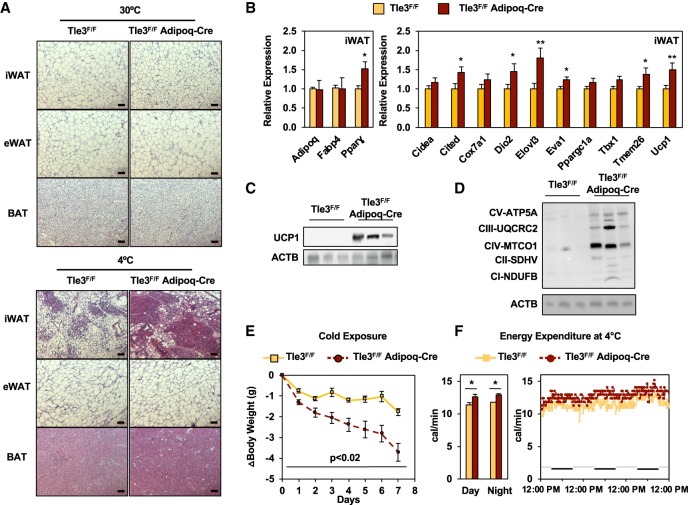
To test the impact of TLE3 on whole-body energy metabolism, we generated mice with a conditional deletion of Tle3 in adipocytes. We acclimated the Tle3F/F Adiponectin-Cre (Tle3F/F Adipoq-Cre) mice and their littermate controls (aged 3–4 mo) to one of two temperature conditions—thermoneutrality (30°C) or cold exposure (4°C)—for 7 d. At the conclusion of the study, the iWAT, eWAT, and BAT depots were fixed, sectioned, and stained with hematoxylin and eosin (H&E). Under thermoneutral conditions, we observed no differences in adipose histology between control and Tle3F/F Adipoq-Cre mice (Fig. 1A, top panel). However, when mice were subjected to cold exposure, both groups had reduced lipid droplet size in iWAT, but, strikingly, there were more multilocular cells, consistent with beige adipocytes in the Tle3F/F Adipoq-Cre mice (Fig. 1A, bottom panel). In contrast, both eWAT and BAT showed similar histological features between control and Tle3F/F Adipoq-Cre mice in the setting of cold exposure (Fig. 1A). No alterations were observed in liver morphology at thermoneutrality or at 4°C (Supplemental Fig. S1A). We measured gene expression within iWAT following 4 d of cold exposure and found increased expression of thermogenic-associated genes, such as Cited, Dio2, Elovl3, Eva1, Tmem26, and Ucp1, while the genes encoding general adipocyte markers such as Adipoq and Fabp4 remained unchanged (Fig. 1B). In these circumstances, the changes were specific to iWAT, as no changes in gene expression were observed between control and Tle3F/F Adipoq-Cre mice in the eWAT or BAT depots during cold exposure (Supplemental Fig. S1B). After 1 wk at thermoneutrality, there were no significant differences identified in the thermogenic gene expression program in iWAT; although several thermogenesis-associated genes showed trends of increased expression in the Tle3F/F Adipoq-Cre mice, the overall levels were low after 1 wk of thermoneutrality (Supplemental Fig. S1C).
Figure 1.
Conditional deletion of Tle3 in adipocytes promotes beige adipocyte programming and whole-body energy expenditure. (A) Histological analysis of iWAT, eWAT, and BAT at thermoneutrality (30°C) and cold (4°C) for control (Tle3F/F) and knockout (Tle3F/F Adipoq-Cre) mice. Scale bar, 500 µm. (B) Expression of adipogenic- and thermogenic-associated genes from the iWAT, measured by real-time PCR after 4 d of cold exposure and normalized to 36B4. (C) UCP1 protein expression in iWAT after 4 d of cold exposure. (D) Protein expression for oxidative phosphorylation proteins in iWAT after 4 d of cold exposure. (E) Time-dependent changes in body weight for Tle3F/F and Tle3F/F Adipoq-Cre mice housed at 4°C. n = 4 per group. (F) Energy expenditure as measured by indirect calorimetry in the comprehensive laboratory animal-monitoring system (CLAMS) metabolic cages for Tle3F/F and Tle3F/F Adipoq-Cre mice. n = 4 per group. Data are presented as mean ± SEM. Significance was analyzed by two-tailed Student's t-test. (*) P < 0.05; (**) P < 0.01.
The greater recruitment of beige adipocytes was further verified by the finding of increased UCP1 protein in iWAT of Tle3F/F Adipoq-Cre mice (Fig. 1C). As beige adipocytes are rich in mitochondria, we also measured mitochondrial content by blotting for proteins from each of the electron transport chain complexes. After 4 d of cold exposure, the iWAT from Tle3F/F Adipoq-Cre mice expressed more protein from each subunit than the littermate controls, with marked increases in the cytochrome oxidase complex IV (Fig. 1D).
Loss of Tle3 in adipocytes increases whole-body energy expenditure
We next proceeded to monitor the animal during cold exposure and observed greater weight loss over time in the Tle3F/F Adipoq-Cre mice as compared with Tle3F/F controls (Fig. 1E). To assess whether these changes in body weight were due to decreased food consumption or increased energy expenditure, we placed age-matched mice in metabolic cages at 4°C for a 3-d period. We observed no differences in body weight or body composition between the two groups prior to cold exposure (Supplemental Fig. S1D). Tle3F/F Adipoq-Cre mice also showed an increase in energy expenditure at 4°C in both day and night when compared with the control group (Fig. 1F). Measurements of O2 consumption and CO2 production (Supplemental Fig. S1E) revealed a modest but consistent increase in the cold. During this period, we did not find measurable differences in food consumption or activity (Supplemental Fig. S1F).
TLE3 regulates glucose utilization and insulin sensitivity
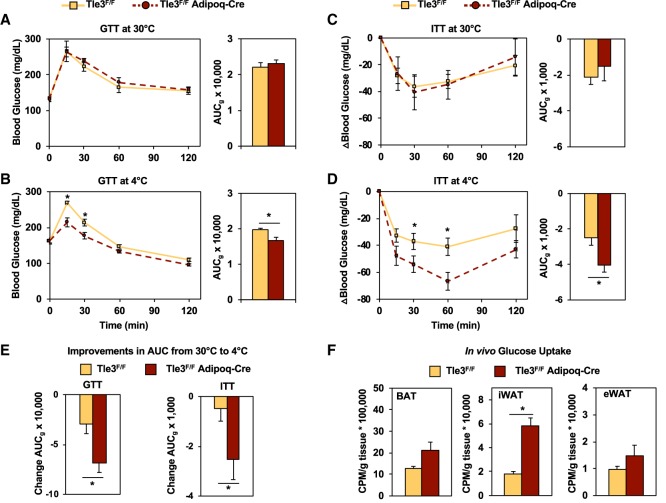
We next assessed whether having more beige adipocytes in the setting of Tle3 deletion promoted glucose utilization. As increased beige adipose tissue and BAT have been linked previously to improvements in glucose handling (Lee et al. 2010; Stanford et al. 2013), we asked whether the Tle3F/F Adipoq-Cre mice had improved glucose clearance. We performed glucose tolerance tests (GTTs) on Tle3F/F Adipoq-Cre and control mice that had been acclimated to thermoneutrality for 1 wk, followed by 4 d of cold exposure and a second GTT. No differences were observed in the glucose excursion curves at 30°C (Fig. 2A). After cold acclimation, the Tle3F/F Adipoq-Cre mice had improved glucose handling and significantly lower glucose area under the curve (AUCg) (Fig. 2B). We performed an insulin tolerance test (ITT) following the same procedure, and while no differences were seen between the two groups at 30°C, cold acclimation improved insulin sensitivity to a significant degree in the Tle3F/F Adipoq-Cre group (Fig. 2C,D). We did note that both groups of mice demonstrated improved glucose handling in both GTTs and ITTs after cold acclimation, but, in both cases, the change was greatest in the Tle3F/F Adipoq-Cre group (Fig. 2E). No significant differences were observed in plasma insulin levels between the two groups after cold exposure (Supplemental Fig. S2A).
Figure 2.
Deletion of Tle3 in adipocytes improves glucose handling with cold exposure. (A) GTT and AUGg on control (Tle3F/F) and knockout (Tle3F/F Adipoq-Cre) mice after 1 wk at thermoneutrality (30°C). n = 6–7 per group. (B) GTT and AUCg following 4 d in cold (4°C). n = 6–7 per group. (C) ITT and AUCg on Tle3F/F and Tle3F/F Adipoq-Cre mice after 1 wk at thermoneutrality and cold. n = 6–7 per group. (D) ITT and AUCg following 4 d in the cold (4°C). n = 6–7 per group. (E) Change in measured AUCg for GTT and ITT from thermoneutrality to after cold exposure. (F) Uptake of 2-[U-14C]-deoxyglucose in BAT, iWAT, and eWAT for Tle3F/F and Tle3F/F Adipoq-Cre mice following 4 d of cold exposure. n = 4 per group. Data are presented as mean ± SEM. Significance was analyzed by two-tailed Student's t-test. (*) P < 0.05.
Under these conditions, beige adipocytes represent a small fraction of the total adipocyte content of the body. This led us to ask whether beige adipocytes were abundant enough to account for these changes in glucose handling. With cold exposure, we observed increases in the expression of Glut1 and Glut4 transporters in the iWAT depot of Tle3F/F Adipoq-Cre mice but not in the BAT or eWAT (Supplemental Fig. S2B), but this only indicates that the tissue is poised to augment glucose clearance. To directly test the contribution of beige adipocytes to the phenotype of Tle3F/F Adipoq-Cre mice, we performed in vivo glucose uptake assays with uniformly labeled 14C-2-deoxyglucose in mice housed for 4 d at 4°C. Thirty minutes following radiolabeled glucose administration, the animals were euthanized, tissues were excised, and the radioactive content of each tissue was measured. Glucose uptake in Tle3-deficient iWAT was increased threefold compared with controls, while BAT and eWAT had minimal changes (Fig. 2F). We note that the skeletal muscle in the Tle3F/F Adipoq-Cre mice trended toward increased glucose uptake, suggesting that the glucose improvements may be affected by the interplay between fat and muscle, although these data did not reach statistical significance (Supplemental Fig. S2C). These data suggest that the changes in whole-body glucose handling observed in the Tle3F/F Adipoq-Cre mice may be driven by uptake into beige adipose tissue and that increased beige adipose tissue is capable of regulating systemic glucose metabolism.
TLE3 intrinsically regulates the thermogenesis and glucose clearance in beige adipocytes
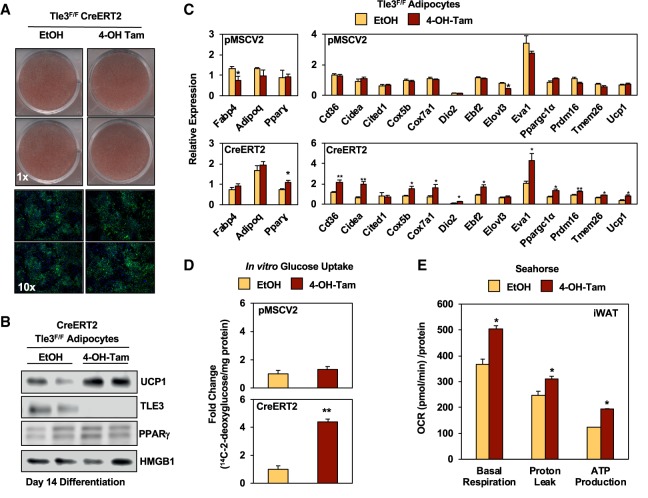
Beige adipocytes rely on both intrinsic regulation and systemic signaling to control their recruitment. To ascertain whether the phenotypic changes that we observed in vivo were due to alterations in TLE3-dependent gene transcription, we used immortalized Tle3F/F iWAT preadipocytes with a tamoxifen-inducible Cre (pMSCV2-CreERT2). Because TLE3 is known to be involved in early adipocyte differentiation (Villanueva et al. 2011), we chose to knock out Tle3 4 d after the differentiation cocktail was administered and harvest cells on the 10th day. To ensure that the differentiation potential was not affected by knocking out Tle3 at this time point, we stained for lipids using both Oil Red O and Bodipy and found similar lipid accumulation in Tle3-knockout cells when compared with controls (Fig. 3A). In this in vitro system, the loss of TLE3 led to increased UCP1 protein but no change in PPARγ protein levels (Fig. 3B). Furthermore, these protein changes were accompanied by increases in other thermogenesis-associated transcripts, including Cd36, Cidea, Cox5b, Cox7a1, Dio2, Ebf2, Eva1, Ppargc1a, Prdm16, Tmem26, and Ucp1, while no changes were observed in transcript levels of the adipocyte differentiation markers Fabp4 and Adipoq. In contrast, no changes were observed in the thermogenesis gene program in the empty vector (pMSCV2) cells when treated with tamoxifen (Fig. 3C). Similar to our in vivo findings, we found increased expression of the Glut4 glucose transporter in the tamoxifen-treated CreERT2 cells that was not observed in the empty vector cells (Supplemental Fig. S3A). To further ensure that tamoxifen treatment itself was not responsible for the changes in gene expression, Tle3F/F cells were infected with Cre-expressing adenovirus, where we observed increases in Ucp1 expression (Supplemental Fig. S3B).
Figure 3.
Deletion of Tle3 promotes the programming of beige adipocytes and glucose clearance in vitro. (A) Oil Red O (top) and DAPI/Bodipy (bottom) staining of Tle3F/F iWAT adipocytes expressing CreERT2 and treated with either EtOH (control) or 4-hydroxytamoxifen on day 4 after administration of differentiation cocktail and stained on day 10. (B) Protein expression of UCP1 and TLE3 by immunoblot in Tle3F/F CreERT2 iWAT adipocytes differentiated in culture. (C) Gene expression measured by real-time PCR in Tle3F/F pMSCV2 (empty vector) and CreERT2 iWAT adipocytes differentiated in culture. Data were normalized to 36B4. n = 3 per group. (D) In vitro glucose uptake of 2-[U-14C]-deoxyglucose in Tle3F/F iWAT adipocytes expressing either pMSCV2 or CreERT2. n = 4 per group. (E) Oxygen consumption rate (OCR) of Tle3F/F CreERT2 iWAT adipocytes, as measured by Seahorse XF analyzer. n = 14 per group. Data are presented as mean ± SEM. Significance was analyzed by two-tailed Student's t-test. (*) P < 0.05; (**) P < 0.01.
Having established an in vitro model for the beige adipocyte recruitment that we observed in vivo, we next performed in vitro glucose uptake with uniformly labeled 14C-2-deoxyglucose to determine whether the glucose clearance observed previously was due to intrinsic changes or whether Tle3-deficient iWAT was responding to systemic signaling. Upon the administration of tamoxifen to differentiated CreERT2-expressing cells, the loss of TLE3 causes approximately a fourfold increase in glucose uptake, while no change was observed pMSCV2 cells treated with tamoxifen (Fig. 3D). We also observed that the loss of TLE3 led to increases in oxygen consumption during basal respiratory conditions, isolated proton leak, and ATP production (Fig. 3E), demonstrating a rise in energy utilization in beige adipocytes lacking TLE3.
TLE3 promotes diet-induced weight gain and glucose intolerance
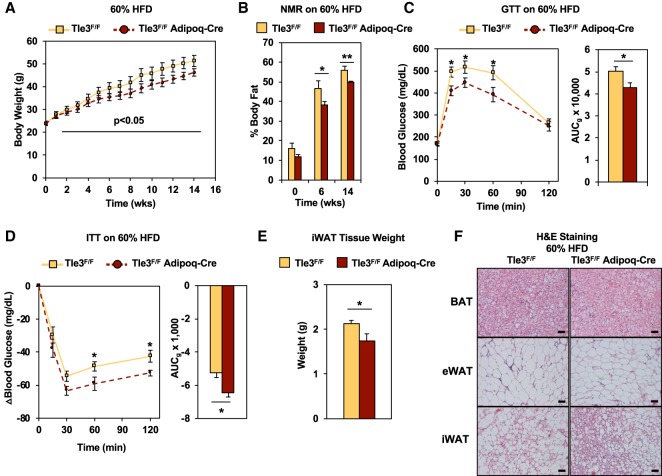
We appreciate that 4°C cold exposure represents a severe thermogenic stimulus and wanted to understand whether the loss of adipose TLE3 could affect systemic metabolism in the absence of a cold stimulus. Therefore, the Tle3F/F Adipoq-Cre mice and their littermate controls were placed on a high-fat diet (HFD; 60% fat; 5.2 kcal/g) for a period of 14 wk and housed at ambient temperature (22°C–24°C). We monitored body weight weekly and found that the Tle3F/F Adipoq-Cre mice were protected against weight gain on a HFD (Fig. 4A). Analysis of body composition revealed that the control group had higher body fat beginning after 6 wk on a HFD, and the difference was maintained until the end of the study (Fig. 4B). To test whether loss of TLE3 in this setting was also associated with improved glucose clearance, we performed a GTT. We observed elevated fasting glucose levels in both groups, but the Tle3F/F Adipoq-Cre mice showed enhanced glucose excursion (Fig. 4C). When compared with littermate controls, Tle3F/F Adipoq-Cre mice also showed enhanced insulin response on an ITT (Fig. 4D). Notably, the iWAT fat pads of the Tle3F/F Adipoq-Cre mice weighed less than those of the control group (Fig. 4E), while tissue weight of BAT, eWAT, and the liver were similar between control and Tle3F/F Adipoq-Cre mice (Supplemental Fig. S4A). The gross morphology demonstrates that iWAT fat depots appeared smaller in the Tle3F/F Adipoq-Cre mice, but no other differences in tissues between the two groups were visible (Supplemental Fig. S4B). Histology indicated that the Tle3F/F Adipoq-Cre mice also had more beige adipocytes than the control group on a HFD (Fig. 4F). Together, these findings suggest that TLE3 contributes to the impairment of diet-associated weight gain and glucose intolerance.
Figure 4.
Conditional deletion of TLE3 in adipocytes protects against diet-induced weight gain and glucose intolerance. (A) Time course of body weight for control (Tle3F/F) and knockout (Tle3F/F Adipoq-Cre) mice housed at ambient temperature on a 60% HFD for 14 wk. n = 6–8 per group. (B) Body fat composition by NMR in Tle3F/F and Tle3F/F Adipoq-Cre groups on a 60% HFD over time. n = 6–8 per group. (C) GTT and AUGg on Tle3F/F and Tle3F/F Adipoq-Cre mice after 14 wk on a 60% HFD. n = 6–7 per group. (D) ITT and AUCg on Tle3F/F and Tle3F/F Adipoq-Cre mice after 10 wk on a 60% HFD. n = 3 per group. (E) Weight of individual iWAT tissue depots. n = 10 per group. (F) Histology of adipose depots after 14 wk on a 60% HFD. Data are presented as mean ± SEM. Significance was analyzed by two-tailed Student's t-test. (*) P < 0.05; (**) P < 0.01.
Genome-wide analysis reveals preferential association of TLE3 with WAT genes
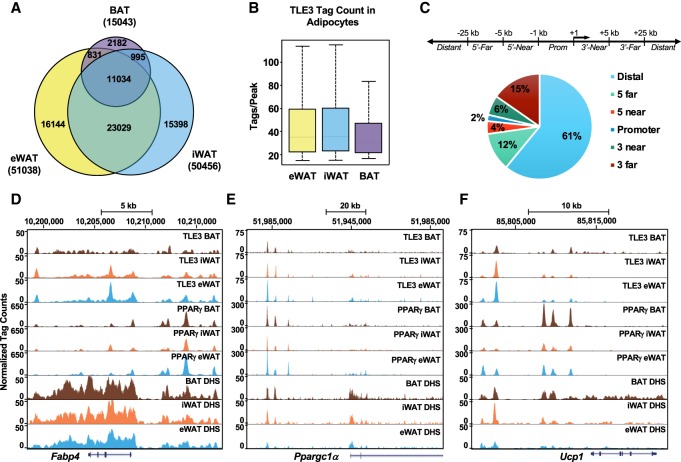
TLE3 was initially identified as a transcriptional coactivator of PPARγ during early adipocyte differentiation (Villanueva et al. 2011). To develop a broader understanding of TLE3's role in regulating gene expression, we performed genome-wide chromatin immunoprecipitation (ChIP) followed by deep sequencing (ChIP-seq). The specificity of the antibody was verified using differentiated preadipocytes immortalized from iWAT stromal vascular fractions of Tle3 floxed mice. Cells were treated with adenovirus expressing Cre-recombinase or LacZ, and ChIP with the TLE3 antibody was performed. Quantitative ChIP-PCR was performed on known TLE3-binding sites near Pparγ, Fabp4, Adipoq, Aqp7, and Angptl4. The limited recovery after TLE3 depletion indicates limited cross-reactivity of the TLE3 antibody (Supplemental Fig. S5A). Our ChIP-seq analysis was performed in eWAT-, iWAT-, and BAT-derived adipocytes using cultures with at least 50% differentiation and showing similar expression levels of general adipocyte markers (Supplemental Fig. S5B). We found that TLE3 had a greater number of binding sites in both eWAT and iWAT as compared with BAT in terms of both number of binding sites (Fig. 5A) and binding intensity (Fig. 5B). These findings are consistent with the previous report showing lower TLE3 expression in BAT (Villanueva et al. 2013). Although TLE3 mapped to many common sites within the three adipocyte subtypes, we also identified unique binding sites for each adipocyte subtype (Fig. 5A).
Figure 5.
Genome-wide profiling of TLE3 in adipocytes reveals increased binding in WAT. (A) Venn diagram demonstrating the number of TLE3-binding sites in eWAT, iWAT, and BAT, filtered for >15 tags and >10-fold above input. (B) Binding intensity at sites occupied by TLE3 in differentiated adipocytes from eWAT, iWAT, and BAT. (C) Genomic distribution of TLE3 relative to the distance from transcriptional start sites (TSSs). (D) Genome browser tracks of Fabp4 for TLE3 and PPARγ in eWAT, iWAT, and BAT. (E) Genome browser tracks of Ppargc1α for TLE3 and PPARγ in eWAT, iWAT, and BAT. (F) Genome browser tracks of Ucp1 for TLE3 and PPARγ in eWAT, iWAT, and BAT.
The genomic distribution of TLE3 mapped principally to distal regions of the mouse genome (Fig. 5C), an observation that is consistent with the TLE/Groucho family being located primarily at enhancers (Chambers et al. 2017; Dali et al. 2018). There was nearly equal distribution of binding upstream of and downstream from the transcriptional start site (TSS) (Supplemental Fig. S5C). Despite unique elements of TLE3 binding in each tissue, the distribution of TLE3 was largely similar between iWAT, eWAT, and BAT (Supplemental Fig. S5D). Through our genomic analysis, we found that 61% of TLE3-binding sites are >25 kb distal from the TSS. TLE3 was shown previously by ChIP-qPCR to occupy the Fabp4 enhancer/promoter. A closer examination of our ChIP-seq data set revealed several TLE3-binding sites near the Fabp4 gene. Using data from a PPARγ ChIP-seq of differentiated progenitors of eWAT, iWAT, and BAT (Siersbaek et al. 2012), we observed that TLE3 occupancy overlapped with multiple prominent PPARγ-binding sites (Fig. 5D). TLE3 was reported previously to interact with PPARγ at the promoters of key adipocyte target genes (Villanueva et al. 2011). To determine the overlap in binding sites between TLE3 and PPARγ, we aligned TLE3 and PPARγ ChIP-seq data sets and found that 23% of TLE3 WAT-binding sites overlap with PPARγ, while 41% of PPARγ WAT-binding sites overlap with TLE3 (Supplemental Fig. S5E). PPARγ WAT-binding sites were defined as PPARγ-binding sites shared between eWAT and iWAT. Interestingly, genes annotated to overlapping TLE3- and PPARγ-binding sites included genes involved in several metabolic pathways (Supplemental Fig. S5F), and motif analysis in these overlapping binding sites suggests a possible interaction between TLE3, PPARγ, and other known adipocyte transcription factors (Supplemental Fig. S5G). Examples of the distribution of TLE3-binding sites relative to PPARγ can be seen at Fabp4, Ppargc1a, and Ucp1 (Fig. 5D–F). When we examined other genes associated with thermogenesis, such as Ppargc1a and Ucp1, we noted multiple nearby TLE3-binding sites overlapping with putative regulatory regions as detected by DNase I hypersensitivity sites (DHS) (Fig. 5E,F). Together, the observations demonstrate that both Ppargc1a and Ucp1 are elevated with the loss of TLE3 (Fig. 3C), and, since these genes are also bound by TLE3, it raises the possibility that TLE3 might be acting to repress their expression.
Loss of TLE3 stimulates the thermogenic gene expression program in iWAT
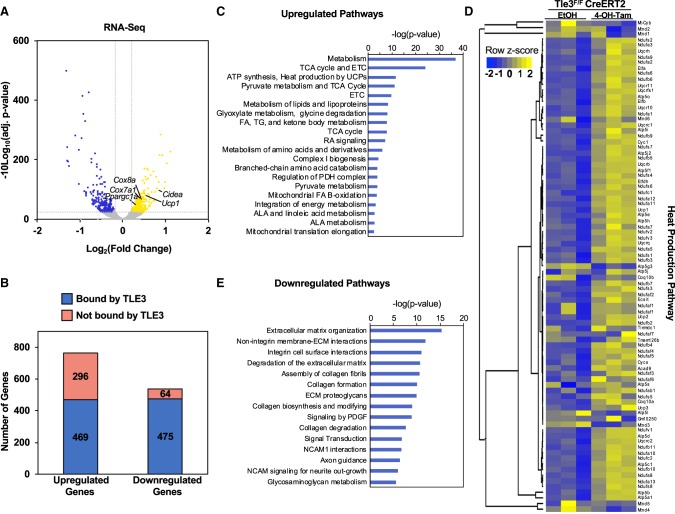
Thus far, we had observed changes in thermogenic genes by real-time PCR in vitro and in vivo and, through ChIP-seq, found that TLE3 binds in proximity to these genes but had only limited insights into how the overall transcriptional landscape of the cells was changing in the absence of TLE3. To define the global changes in gene expression provoked by alteration in TLE3 expression, we used the tamoxifen-inducible Tle3F/F preadipocytes expressing CreERT2. After 4 d of differentiation, we knocked out Tle3 by tamoxifen administration, harvested the cells on the 10th day of differentiation, and then assessed changes in gene expression by RNA sequencing (RNA-seq). Consistent with our previous findings, Tle3 deletion led to the up-regulation of genes associated with the thermogenic gene expression program, including Cidea, Ucp1, and Ppargc1a (Fig. 6A). Of the 765 genes up-regulated in the TLE3 knockout cells, 469 may be direct targets of TLE3, as their expression increased in the absence of TLE3 and they have a TLE3-binding site nearby. We note that, among these genes, there is an overrepresentation of genes that have a role in mitochondrial function, including those that are associated with the heat production pathway, including Aldh4a1, Cox7a1, Ndufc2, and Cox5a (Supplemental Fig. S6A). Of the 539 genes that were down-regulated in the Tle3 knockout, 475 have a TLE3-binding site nearby (Fig. 6B). The distribution of TLE3-binding sites associated with TLE3-regulated genes shows a greater number of binding sites downstream from the TSS when compared with the number of upstream binding sites (Supplemental Fig. S6B). Our finding that up-regulated and down-regulated genes have proximal TLE3-binding sites is consistent with a dual role of TLE3 in activating or inhibiting gene expression (Villanueva et al. 2011).
Figure 6.
TLE3 represses TCA cycle and thermogenic gene program in iWAT. (A) Volcano plot showing differentially expressed genes in beige adipocytes isolated from Tle3F/F mice expressing CreERT2. Cells were treated with EtOH (control) or 4-hydroxytamoxifen on day 4 after differentiation and harvested on day 10. Adjusted P-value of <0.01; log2 ratio of differential expression >|0.2|. (B) Stacked bar graph demonstrating genes that are differentially regulated with the loss of TLE3 with or without a TLE3-binding site. RNA-seq genes with adjusted P-value of <0.05, no distance from TSS cutoff applied. (C) Gene ontology analysis of 475 genes whose expression is increased with loss of TLE3 and which have at least one TLE3 occupancy site identified by ChIP-seq. Reactome pathways (P < 0.0005) are listed in order of decreasing P-value. (D) Gene ontology analysis of 469 genes whose expression is decreased with loss of TLE3 and which have at least one TLE3 occupancy site identified by ChIP-seq. Reactome pathways (P < 0.0005) are listed in order of decreasing P-value. (E) Heat map of genes associated with Reactome pathway R-MMU-163200 (respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins) in Tle3F/F iWAT adipocytes expressing CreERT2 and treated with either EtOH (control) or 4-hydroxytamoxifen. Z-score reflects the log2 ratio. n = 3 per group.
To characterize the gene program regulated by TLE3, we performed gene ontology analysis using the subset of genes in iWAT that were repressed by TLE3 and had an associated TLE3 site. We found overrepresentation of genes that encode proteins in metabolic pathways and are involved in energy utilization (Fig. 6C). Notably, the pathway encompassing heat production by the uncoupling proteins was ranked third highest in significance among those pathways, supporting the notion that TLE3 acts physiologically to counteract this program. A heat map of the changes for all genes included in this pathway is shown (Fig. 6D; Supplemental Table S1).
We also performed gene ontology analysis on genes that were decreased with the loss of TLE3 and had an associated TLE3 site. This analysis showed an enrichment for pathways associated with the extracellular matrix, although the statistical significance of this association was lower in comparison with pathways that were up-regulated by Tle3 deletion (Fig. 6E). Using Tle3F/F adipocytes expressing CreERT2, we verified that the loss of TLE3 does repress both Col6a1 and Col6a2 and, using iWAT from HFD-fed mice, saw a decrease in expression of Mmp19 (Supplemental Fig. S6C) and noted that these genes have TLE3-binding sites nearby (Supplemental Fig. S6D). Together, these data suggest that TLE3 has a role in promoting adipose tissue fibrosis and that the glucose improvements seen in Tle3 knockout mice might partially be due to decreases in the fibrotic gene program.
TLE3 motif analysis predicts functional interaction with EBFs
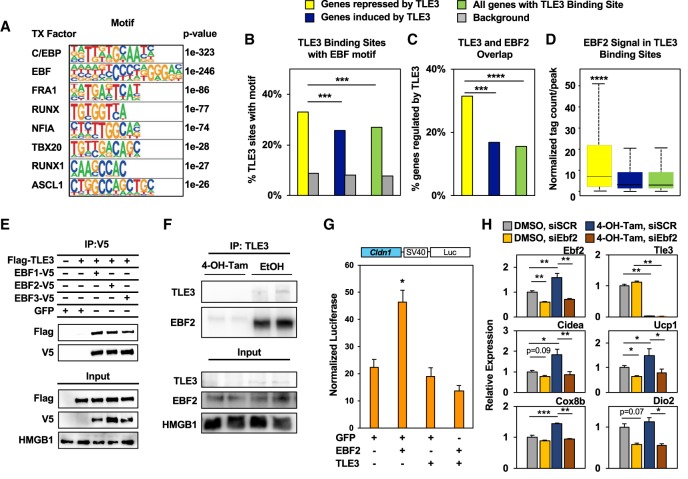
Because TLE3 lacks a DNA-binding domain, its ability to modulate gene expression depends on interactions with DNA-binding transcription factors. To identify putative transcription factors that recruit TLE3, we completed de novo motif analysis on the top 1000 genomic regions identified by TLE3 ChIP-seq and found sequences that were associated with C/EBP, EBF, FRA1, RUNX, NFIA, TBX20, RUNX1, and ASCL1 motifs (Fig. 7A). Analysis of individual adipocyte subtypes showed slight variations in the list of transcription factors. Of particular interest was the finding that C/EBP and EBF motifs were consistently highly enriched in sites where TLE3 was bound (Supplemental Fig. S7A).
Figure 7.
Analysis of TLE3 binding demonstrates repressive interaction with EBF2. (A) De novo motif analysis of the top 1000 TLE3 occupancy sites in WAT and their assigned closest matching transcription factor, listed in order of decreasing P-value. (B) Known motif analysis of the EBF motif in TLE3-binding sites and matching background sequences within 15 kb of the indicated gene groups. (C) Overlap between TLE3- and EBF2-binding sites within 15 kb of the indicated gene groups with randomly selected GC content-matching background sequences. (D) EBF2 ChIP-seq signal in TLE3-binding sites within 15 kb of the indicated gene groups. (E) Coimmunoprecipitation of TLE3 and EBF proteins from 293 cells transfected with Flag-TLE3 and one of the following: EBF1-V5, EBF2-V5, or EBF3-V5. Lysates were immunoprecipitated with anti-V5 antibody. (F) Coimmunoprecipitation of TLE3 and EBF2 from Tle3F/F iWAT adipocytes expressing CreERT2 and treated with either EtOH (control) or 4-hydroxytamoxifen. Lysates were immunoprecipitated with anti-TLE3 antibody. (G) Analysis of Cldn1 enhancer activation of luciferase reporter by coexpression of EBF2, TLE3, or both in beige adipocytes differentiated in vitro. n = 3 per group. (H) Gene expression of Tle3F/F iWAT adipocytes expressing CreERT2. Cells treated with DMSO (control) or 4-hydroxytamoxifen on day 4 of differentiation and either siSCR or siEBF2 on day 12 of differentiation and harvested on day 14 of differentiation. n = 3 per group. Data are presented as mean ± SEM. Significance was analyzed by Fisher's exact test (B,C), Wilcoxon rank sum test (D), or Student's t-test (G,H). (*) P < 0.05; (**) P < 0.01; (***) P < 2.2 × 10−8; (****) P < 2.2 × 10−16.
To understand the mechanisms by which TLE3 modulates the recruitment of beige adipocytes in inguinal white adipocytes, we analyzed TLE3-binding sites that overlap with PPARγ. We found that the C/EBP and the EBF motif were enriched at PPARγ sites that are associated with TLE3 (Supplemental Fig. S7B). Given the potential cross-talk of these transcription factors, we next analyzed putative target genes of TLE3 in iWAT adipocytes (i.e., genes that were repressed or induced by TLE3 knockout and display nearby TLE3 binding) (Supplemental Fig. S7C). Interestingly, the EBF motif was present in a higher fraction of TLE3-binding sites near genes that were repressed by TLE3 compared with genes that were enhanced by TLE3 (Fig. 7B). This was not observed for the C/EBP motif, where we found a similar number of C/EBP motifs in genes stimulated or repressed by TLE3 (Supplemental Fig. S7D). Also, some of the transcripts regulated by TLE3 have been reported to be direct targets of EBF2 (Rajakumari et al. 2013). We therefore analyzed the coincidence of TLE3 and EBF2 binding near genes regulated by TLE3 on a genome-wide level. We compared ChIP-seq data sets from EBF2 in BAT (Shapira et al. 2017) with the TLE3-binding profile in iWAT and found a greater overlap between TLE3- and EBF2-binding sites near genes that were repressed by TLE3 compared with genes enhanced by TLE3 (Fig. 7C). The binding intensity of EBF2 was higher in TLE3-binding sites near repressed genes (Fig. 7D), suggesting that cross-talk between TLE3 and EBF2 occurs primarily near genes directly repressed by TLE3.
To test the functional connection between TLE3 and EBFs, we explored interactions between TLE3 and EBF1, EBF2, and EBF3. Cells were transfected with Flag-TLE3 in the presence or absence of V5-tagged EBF proteins, immunoprecipitated with anti-V5 antibody, and immunoblotted with anti-Flag antibody. We found that TLE3 coimmunoprecipitated with EBF1, EBF2, and EBF3 (Fig. 7E). Seeking whether the endogenous proteins would interact, we also used Tle3F/F iWAT preadipocytes treated with either ethanol or tamoxifen and immunoprecipitated the cell lysates with an antibody that detects TLE3. Blotting for EBF2 demonstrated a specific interaction between TLE3 and EBF2 (Fig. 7F). Because we observed that TLE3 and EBF2 colocalized near several putative target genes, we sought to investigate how TLE3 influenced the transcriptional activity of EBF2. We used the ChIP-seq data of TLE3 and EBF2 (Lai et al. 2017) to identify a site where both TLE3 and EBF2 colocalized in proximity to BAT-selective markers and identified Cldn1 (Supplemental Fig. S7E). Day 2 differentiated beige adipocytes were transfected with a luciferase reporter driven by the 0.5-kb enhancer element upstream of the Cldn1 gene. This enhancer element was defined as a DHS site that has overlapping TLE3 and EBF2 occupancy (Supplemental Fig. S7E). The cells were also transfected with an Ebf2 expression vector, Tle3 expression vector, or both. We found that EBF2 activated the Cldn1 reporter and that reporter expression was inhibited to baseline levels when TLE3 was also expressed (Fig. 7G). Finally, we performed Ebf2 knockdown experiments in Tle3F/F iWAT adipocytes expressing CreERT2. Cells were treated with either DMSO or tamoxifen on day 4 of differentiation and subsequently transfected with either a control scramble siRNA (siSCR) or an siRNA targeting Ebf2 (siEbf2) on day 12. Cells were harvested on day 14 of differentiation to measure expression of genes under the control of EBF2 (Fig. 7H). Ebf2 mRNA levels were reduced, and we observed a corresponding decrease in expression levels of several beige fat-associated genes, including Cidea, Cox8b, Dio2, and Ucp1. In the control siSCR group, the loss of TLE3 caused an increase in mRNA levels of those same genes. However, the loss of TLE3 failed to induce expression in the case of Ebf2 knockdown. Together, these findings suggest that the effects on thermogenic gene expression seen with the loss of TLE3 are mediated through EBF2.
Discussion
Subcutaneous WAT has a tremendous capacity to change its morphology in response to the cold. Mice that are acclimated to the cold acquire thermogenic cells that are rich in mitochondria and are believed to play a long-term adaptive role in thermogenesis. The mechanisms that regulate the mitochondrial gene expression program in beige adipocytes are incompletely understood. Here we outlined a role for the transcriptional cofactor TLE3 in regulating the mitochondrial beige adipocyte program. The loss of Tle3 in adipocytes promotes the recruitment of beige adipocytes in the iWAT tissue depot in response to long-term cold exposure, as shown by gross histological analysis, transcriptional changes, and increased expression of UCP1. Our findings further support the concept that increasing beige adipocyte number can lead to improvements in glucose metabolism and insulin sensitivity. We demonstrated that TLE3 regulates the mitochondrial gene expression program in beige adipocytes and provide evidence indicating that this occurs through countering the action of EBF2, a transcription factor that promotes the recruitment of thermogenic cells (Rajakumari et al. 2013; Stine et al. 2016). These studies highlight that mechanisms that promote the recruitment of beige adipocytes can prevent weight gain associated with a HFD.
Our findings strongly support the contention that promoting an increase in beige adipocyte activity is sufficient to have beneficial effects on systemic glucose metabolism. We found that loss of Tle3 improved glucose metabolism in the setting of cold exposure but not at thermoneutrality and that this effect was correlated with the abundance of beige adipocytes. We found differences in histology and gene expression consistent with beige adipocytes between control and Tle3-deficient mice during cold exposure but not thermoneutrality, strongly suggesting that increasing the amount of beige fat alone can lead to improvements in glucose metabolism. When testing to see which adipose tissue depots were primarily involved in clearing glucose, we found that the iWAT depots had the greatest increase in glucose uptake. Thus, despite the fact that they represent a small fraction of the total adipocyte population, beige adipocytes can exert marked effects on systemic energy balance.
Our data suggest that the increase in glucose utilization provoked by Tle3 deletion is likely due to increased expression of glucose transporters and genes involved in mitochondrial oxidative phosphorylation. Gene ontology analysis pointed to broad changes in expression in metabolic pathways in the absence of TLE3, including components of the electron transport chain, heat production, fatty acid oxidation, and the TCA cycle. Loss of Tle3 also led to enhanced energy expenditure under conditions where mice had more beige adipocytes. In the cold, mice lacking Tle3 in adipocytes lost more weight, an outcome that was largely attributed to increased energy expenditure. This was also seen in vitro, where beige adipocytes lacking Tle3 had increased basal respiration and uncoupling. When mice were challenged with a HFD, they were protected against diet-induced weight gain at ambient temperature, a condition that leads to mild activation of thermogenesis. Improvements in glucose metabolism were also associated with reduced expression of a subset of adipose tissue fibrosis genes, which has been implicated recently in improving systemic glucose metabolism (Hasegawa et al. 2018). Although we suspect that beige adipocytes contribute to protection against weight gain, it is possible that loss of Tle3 in BAT is also contributing to changes in body weight (Villanueva et al. 2013). These findings parallel those that are observed with the loss of ZFP423, where there is increased abundance of beige adipocytes in subcutaneous WAT (Shao et al. 2016). Like ZFP423, TLE3 expression in adipose tissue reprograms BAT to WAT (Villanueva et al. 2013). Future studies exploring the relationship between TLE3 and ZFP423 may provide new insights into the transcriptional control of white adipocytes.
Although TLE3 has been studied broadly during development, there has heretofore been little known about its genome-wide distribution in adipocytes. By integrating RNA-seq and ChIP-seq analysis, we identified putative genomic targets of TLE3. Most of these were found in distal genomic regions that were >25 kb from the TSS. Given that TLE3 lacks a DNA-binding domain, we performed motif analysis on genomic regions associated with TLE3 and found that C/EBP and EBF motifs were highly associated with TLE3 binding. Our analysis also supports an overlap in genome-wide occupancy of TLE3 and PPARγ, where 23% of TLE3-binding sites were associated with PPARγ, while 41% of PPARγ-binding sites overlap with TLE3. After analyzing TLE3-binding sites near genes regulated by TLE3 in beige adipocytes, we found that EBF2 and TLE3 colocalized near genes activated with the loss of TLE3. This led us to investigate the cross-talk between TLE3 and EBFs. We found that TLE3 interacted with EBF2, repressed EBF2 target genes, and repressed EBF2 transcriptional activity in a reporter assay and an Ebf2 knockdown experiment. Furthermore, we found that genes that are repressed by TLE3 were more likely to contain an EBF motif compared with those that were enhanced by TLE3. There was a high degree of overlap between TLE3- and EBF2-binding sites near genes that were repressed by TLE3, and the TLE3-binding intensity was consistently higher at sites near TLE3-repressed genes. Previously, we found that TLE3 disrupts the interaction between PPARγ and PRDM16. One would suspect that a similar mechanism may be involved, in which TLE3 disrupts the interaction between EBF2 and PRDM16. Together, our findings suggest a new mechanism by which TLE3 attenuates the beige adipocyte gene expression program by serving as a molecular brake on EBF2.
In conclusion, we identified TLE3 as a negative regulator of EBF2 and the beige adipocyte program, whose actions block energy expenditure, glucose utilization, and mitochondrial energetics. Developing therapeutics that target TLE3 may provide opportunities to prevent weight gain associated with diet.
Materials and methods
Animals
A conditional knockout allele for Tle3 was generated on a 129/OlaHsd and C57BL/6J background, as has been described (Villanueva et al. 2013). Tle3flox/flox mice were bred for nine generations to a C57BL/6J background. To obtain age-matched littermates, Tle3flox/flox mice were bred to Tle3flox/flox Adipoq-Cre BAC transgenic mice bred for nine generations by the Jackson Laboratories to a C57BL/6J background. Male mice age 3–4 mo that expressed the Adipoq-Cre transgene were compared with male littermate controls lacking the Cre transgene.
Mice were housed at 22°C–24°C under a 12-h light/dark cycle with ad libitum access to food and water, except when food was withdrawn for an experiment. Animals were maintained on a Teklad global soy protein-free diet (2920x) except where noted otherwise.
Cell culture
Preadipocytes isolated from the stromal vascular fraction of inguinal white fat depots were isolated as described previously (Rodríguez-Cuenca et al. 2007); cells were immortalized by retroviral expression of the SV40 large T antigen (Addgene, 13970) using a hygromycin selection marker and expressing MSCV CreERT2 (Addgene, 22776) using a puromycin selection marker. Adenovirus was amplified using Phoenix-ECO cells. Adenovirus vectors were purified by CsCl gradient.
Preadipocytes were plated in a 12-well plate in DMEM, 10% FBS, 20 nM insulin, and 1 nM T3 with a seeding density of 0.1 × 106 cells per well. Upon confluence, cells were differentiated using DMEM medium containing 10% fetal bovine serum (FBS) (RBMI, FBS-BBT-5XM), 20 nM insulin, 1 nM T3, 0.5 mM isobutylmethylxanthine, 0.5 µM dexamethasone, 0.125 mM indomethacin, and 1 µM rosiglitazone. After 2 d, the medium was changed to DMEM, 10% FBS, 20 nM insulin, 1 nM T3, and 1 µM rosiglitazone. On the fourth day, cells were given either 500 nM 4-hydroxytamoxifen to induce knockout of TLE3 or an equivalent volume of ethanol or DMSO as a control.
Cell staining
After 10 d of differentiation, cells were washed three times in 1× PBS, fixed with freshly prepared PBS + 4% formaldehyde for 1 h, and then incubated with DAPI (Thermo Fisher, D3571) and 1 µg/mL Bodipy (Thermo Fisher, D3922) for 20 min. Cells were subsequently washed three times in 1× PBS and visualized by GFP and DAPI channels. Following DAPI and Bodipy visualization, cells were stained using stock solution Oil Red O (0.5 g in 100 mL of isopropanol; Sigma-Aldrich, O0625) diluted to a freshly prepared working solution (6 mL of stock solution and 4 mL of ddH2O) and incubated for 1 h.
Western blot analysis
Cells were homogenized using radioimmunoprecipitation assay (RIPA) buffer (Boston Bioproducts, Inc.) plus proteinase inhibitor cocktail (Sigma Aldrich, 04693124001) and lysed using a 25-gauge needle. Protein was extracted from flash-frozen tissues in RIPA buffer with protease inhibitor cocktail using a glass dounce. Cell or tissue homogenate was spun at 10,000g for 10 min at 4°C, and the concentration of the supernatant was measured using the Pierce BCA protein assay kit (Thermo Fisher, 23225). Protein samples (10–20 µg per lane) were denatured in Laemmli loading buffer, heated for 20 min at 70°C, resolved by SDS-PAGE gels (typically 10%), and transferred to nitrocellulose membranes (GE Healthcare). Membranes were probed for TLE3 (Proteintech, 11372-1-AP; Abcam, ab213596), UCP1 (Abcam, ab10983), PPARγ (Proteintech, 22061-1-AP), β-actin (Cell Signaling, 4970 and 3700), HMGB1 (Abcam, ab18256), V5 (Thermo Fisher, 46-0705), EBF2 (R&D Systems, AB213596), or Flag M2 (Millipore-Sigma, F3165).
For analysis of mitochondrial oxidative phosphorylation proteins, protein samples were denatured in Laemmli loading buffer, heated for 20 min at 50°C, resolved by SDS-PAGE gels (5%–20% gradient; Bio-Rad), and transferred to a PVDF membrane (Bio-Rad) in CAPS buffer (10 mM CAPS, 10% methanol at pH 11.0). Membrane was probed using the Total OXPHOS rodent antibody cocktail (Abcam, ab110413).
Gene expression
Total RNA was isolated using Trizol reagents (Thermo Fisher, 15596018) and reverse-transcribed using SS VILO Mastermix (Thermo Fisher, 11755500). Gene expression of TLE3 was quantified using TaqMan Universal Master Mix II (Applied Biosystems, 4440038) and Quant Studio 6 instrument (Applied Biosystems by Invitrogen). All other transcripts were quantified by real-time qPCR using KAPA SYBR Fast Rox Low (Kapa Biosystems, KK4621). Primer sequences are shown in Supplemental Table S2.
In vitro glucose uptake
Tle3F/F iWAT preadipocytes were plated in a 12-well plate, allowed to reach confluence, and then stimulated to differentiate. After 2 d, the medium was changed to DMEM, 10% FBS, 20 nM insulin, 1 nM T3, and 1 µM rosiglitazone. On the fourth day, cells were given either 500 nM 4-hydroxytamoxifen to induce knockout of Tle3 or an equivalent volume of ethanol as a control. On the 10th day, the cells were incubated in DMEM, 10% FBS, 20 nM insulin, and 1 nM T3 (no rosiglitazone) for 3 h at 37°C. Cells were washed twice in warm 1× PBS solution and incubated with 0.1 uCi of 2-[14C(U)]-deoxy-D-glucose (Perkin Elmer, NEC720A050UC) per well with unlabeled 2-deoxy-D-glucose up to 6.5 mM total glucose concentration. Negative control wells were also given 20 µM cytochalasin B. After 5 min of incubation, cells were washed three times in 1× PBS solution at 4°C, and then 1 mL of PBS with 1% SDS was added to each well to lyse the cells. After 10 min of incubation, the cell mixture was mixed by pipette. A sample was taken for measuring protein concentration, and another sample was taken to measure radioactivity by liquid scintillation counter in Ultima Gold MV (Perkin Elmer, 6013159). Scintillation counts were normalized to total protein concentration.
Dual-luciferase reporter assay
iWAT preadipocytes were plated and differentiated with the differentiation cocktail described above. Four days after differentiation, the cells were trypsinized and plated in a 48-well plate with a seeding density of 20,000 cells per well. After 24 h, the cells were cotransfected with 50 ng of pGL3-firefly luciferase reporter vector (Promega, E1960) driven by the Cldn1 enhancer element, 1 ng of Renilla luciferase vector (Promega, E1960), and separate expression vectors containing 50 ng of GFP, 50 ng of Ebf2, and/or 100 ng of TLE3. Cldn1 enhancer element was cloned out using the following primers: Fwd-GTGGCGATGACAAGGTGATA and Rev-ATAGCTACTCAGTTCCACATTCC. Cells were transfected using Lipofectamine 3000. The next day, cells were washed and lysed. Twenty microliters of cell lysate was incubated with the luciferase assay substrate, and both firefly and Renilla luciferase activities were measured. Renilla luciferase activity was used as an assay control.
Knockdown of Ebf2
siRNA to Ebf2 was purchased (IDT) in the TriFECTa RNAi kit (mm.Ri.Ebf2.13.1, mm.Ri.Ebf2.13.2, and mm.Ri.Ebf2.13.3). Scrambled siRNA (NC1) or a mixture of siEbf2 oligos was transfected into mature beige adipocytes using Lipofectamine RNAiMAX (Invitrogen) for 4 h on day 12 of differentiation.
Prolonged cold exposure and thermoneutrality
For cold experiments, male mice age 3–4 mo were put for 4 d at 2°C–8°C. For thermoneutrality experiments, male mice age 3–4 mo were put for 1 wk at 30°C.
Energy expenditure
Energy expenditure, food and water intake, and ambulatory activity were determined by using comprehensive laboratory animal-monitoring system (CLAMS; Columbus Instruments). Animals were housed individually within the metabolic cages on a 12-h light/dark cycle with ad libitum access to food (chow diet) and water. System temperature was set at 4°C and remained at 4°C–5°C throughout the run. Energy expenditure was calculated as a function of oxygen consumption and carbon dioxide production in the CLAMS cages. Energy expenditure = [3.815 + 1.232 × (VCO2/VO2)] × VO2.
Histology
Adipose tissue was fixed in 4% buffered formaldehyde and embedded in paraffin. Sections were stained with H&E using standard protocols.
GTT and ITT
A GTT was administered to fasting mice for 6 h with free access to water during fasting, and fasting blood glucose levels were measured using a glucometer by tail bleeds. Mice were then given an intraperitoneal injection of glucose (1 mg/g). Blood glucose levels were measured in intervals (0, 15, 30, 60, 120 min after injection) to determine how quickly the glucose was cleared from the blood.
An ITT was administered in a similar manner but using intraperitoneally injected insulin (Lilly, NDC 0002-8215-17 HI-213) at 0.75 U/kg or 1.0 U/kg for mice on a HFD.
In vivo glucose uptake
Male TLE3F/F Adipoq-Cre mice and their littermate controls were placed for 4 d at 2°C–8°C with free access to food and water. On the fourth day, mice were fasted for 6 h and given a tail vein injection of 4 uCi of 2-[14C(U)]-dexoxy-D-glucose (Perkin Elmer, NEC720A050UC) in 1× PBS. Blood samples were taken at 0, 5, 10, 20, and 30 min to monitor clearance of radioactive glucose from the blood. After 30 min, animals were sacrificed, and tissues were weighed and dissolved in 1 mL of NaOH for 1 h at 60°C. Samples were neutralized. One part of homogenate was used to determine the content of unphosphorylated glucose. This sample was deproteinated by adding equivalent volumes of 0.3 M Ba(OH)2 and 0.3 M ZnSO4 and centrifuged, and the 14C content of the supernatant was measured by liquid scintillation counting. Another part of the homogenate was used to determine the content of total glucose. Insoluble proteins in this sample were removed by adding 6% HClO4 and centrifuging, and the 14C content of the supernatant was measured by liquid scintillation counting. The amount of phosphorylated 2-[14C(U)]-deoxy-D-glucose was then determined by subtracting the radioactivity counts of the unphosphorylated sample from radioactivity counts of the total sample and normalizing to tissue weight.
HFD
Where noted, male Tle3F/F Adipoq-Cre and littermate controls were put on a 60% HFD (Research Diets, D12492) starting at 2 mo of age and maintained for 14 wk. Weight was measured weekly.
Body composition/MRI measurements
Composition of lean tissue, fat mass, and fluid was analyzed using the Bruker Minispec LF50 body composition analyzer.
TLE3 ChIP-seq
Cells from the stromal vascular fraction of BAT (pooled interscapular, cervical, and axillary depots) as well as the eWAT and iWAT of male NMRI mice were obtained and differentiated with rosiglitazone as described previously (Siersbaek et al. 2012). Mature adipocytes were analyzed at day 7, and only cultures with at least a 50% degree of differentiation were used.
For preparation of material for TLE3 ChIP-seq, eWAT-, iWAT-, and BAT-derived adipocytes were treated with 2 mM disuccinimidyl glutarate (Proteochem) for 15 min followed by cross-linking with 1% formaldehyde for 30 min. Cross-linking was stopped by addition of glycine. Nucleus isolation, chromatin preparation, and ChIP were performed as described previously (Siersbaek et al. 2012). The ChIP was performed using anti-TLE3 (Proteintech, 11372-1-AP). ChIP-seq libraries were constructed according to the manufacturer's instructions (Illumina) as described in Nielsen and Mandrup (2014).
TLE3 ChIP-seq data analysis
ChIP-seq data sets were mapped to the mm9 genome with STAR (Dobin et al. 2013) set to not map across splice junctions, as described previously (Brier et al. 2017), and tag directory generation, peak calling/annotations, and motif analyses were performed with HOMER (Heinz et al. 2010). TLE3-binding sites were called using “-style factor” and “-F 10” settings with all other parameters set at default in eWAT-, iWAT-, and BAT-derived adipocytes, and overlapping peaks were merged into one master peak file. Subsequently, TLE3 peaks in each condition were identified as having >15 normalized tags per peak in a 230-bp window and 10-fold more tags relative to the input control. For known and de novo motif analyses, we used HOMER's findMotifsGenome.pl with default settings in the indicated groups of TLE3-binding sites. The background comparison used randomly selected regions that had similar GC content. For comparison with PPARγ- and EBF2-binding profiles, data sets were obtained from the Gene Expression Omnibus (GEO) under accession numbers GSE41481 and GSE97116, respectively. Mapping and peak calling were performed with a strategy similar to that used for TLE3 ChIP-seq data. PPARγ profiles were divided into binding sites (>25 tags per peak and 10-fold more tags than input) with high TLE3 binding (i.e., 25 tags per window and 10-fold more tags than input) and no TLE3 binding (<15 tags per window). To determine genomic co-occurence of EBF2 and TLE3 binding, we defined TLE3-binding sites near the indicated gene groups and used HOMER's mergePeaks program to count the literal overlaps of binding sites. Furthermore, EBF2 tag counts were summarized in a 230-bp window in TLE3-binding sites near the indicated gene groups.
The University of California at Santa Cruz (UCSC) Genome Browser (Kent et al. 2002) was used for visualization of genome tracks.
EBF2 ChIP-seq for genome tracks
EBF2 ChIP-seq data from immortalized BAT preadipocytes differentiated for 2 d in cultures were obtained from GSM1913014 (Lai et al. 2017).
DNase-seq for genome tracks
The DNase-seq data from in vitro differentiated eWAT, iWAT, and BAT primary adipocytes is available under GEO accession number GSE122417.
Gene expression profile—input data
Intact poly(A) RNA was purified from total RNA samples (100–500 ng) with oligo(dT) magnetic beads, and stranded mRNA sequencing libraries were prepared as described using the Illumina TruSeq stranded mRNA library preparation kit (RS-122-2101 and RS-122-2102). Purified libraries were qualified on an Agilent Technologies 2200 TapeStation using a D1000 ScreenTape assay (catalog nos. 5067-5582 and 5067-5583). The molarity of adapter-modified molecules was defined by qPCR using the Kapa Biosystems Kapa library quantification kit (KK4824). Individual libraries were normalized to 10 nM, and equal volumes were pooled in preparation for Illumina sequence analysis.
Sequencing libraries (25 pM) were chemically denatured and applied to an Illumina HiSeq v4 single-read flow cell using an Illumina cBot. Hybridized molecules were clonally amplified and annealed to sequencing primers with reagents from an Illumina HiSeq SR cluster kit v4-cBot (GD-401-4001). Following transfer of the flow cell to an Illumina HiSeq 2500 instrument (HCS version 2.2.38 and RTA version 1.18.61), a 50-cycle single-read sequence run was performed using HiSeq SBS kit version 4 sequencing reagents (FC-401-4002).
Gene expression profile—analysis
Fastq sequences were aligned with Novoalign (version 2.8) against the mouse genome (UCSC release mm9) with splice junctions. Alignments to splice junctions were converted to genomic coordinates with USeq SamTranscriptomeParser (version 8.8.8). RNA-seq quality was evaluated by running Picard CollectRnaSeqMetrics. Differential gene expression was identified using the USeq application DefinedRegionDifferentialSeq (version 8.8.8), which counts alignment tags over collapsed Ensembl gene annotation, executes the R DESeq2 application, filters the gene results at log2 ratio of 1.0 and −10log10(adjusted P) of 20, and reports the results.
The resulting gene lists were used for functional enrichment analysis (gene ontology) using the Panther classification system (Mi et al. 2017) and Reactome pathway (Fabregat et al. 2018) databases.
Heat map
The heat map was created at Shinyheatmap (http://www.shinyheatmap.com) using Euclidiean distance metrics and complete linkage algorithms (Khomtchouk et al. 2017).
Data availability
RNA-seq data are available from GEO under accession number GSE116894. ChIP-seq data are available from GEO under accession number GSE116767.
Statistical analysis
Data are presented as mean ± SEM unless stated otherwise. Student's t-test was used to determined significance unless stated otherwise.
Study approval
Animal experiments were conducted with the approval of the Institutional Animal Care and Use Committee (IACUC) of the University of Utah.
Supplementary Material
Acknowledgments
We thank members of the Diabetes and Metabolism Center and the Biochemistry Department at the University of Utah for useful discussion and feedback. We are grateful to the Metabolic Phenotyping Core, a part of the Health Sciences Cores at the University of Utah, for work performed and access to instrumentation, and the High-Throughput Genomics Core at the Huntsman Cancer Institute of the University of Utah for their assistance with performing RNA-seq and data analysis. This study was supported by 1R01DK103930, R03DK103089, K01DK097285, DRC, P30DK020579, 5T32DK091317, and P30CA042014. Work in the Mandrup laboratory was supported by grants from the Lundbeck Foundation, the Novo Nordisk Foundation, and the Danish Independent Research Council-Natural Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), the National Cancer Institute (NCI), or the National Institutes of Health (NIH).
Author contributions: S.P., A.L., P.R., J.S., S.L., and C.J.V. performed experiments. S.P., A.L, P.R., P.T., S.M., and C.J.V. conceived the experiments and analyzed data. S.P. and C.J.V. wrote the manuscript. P.T., S.M., and C.J.V. provided resources. C.J.V. secured funding.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.321059.118.
Freely available online through the Genes & Development Open Access option.
References
- Akerblad P, Mansson R, Lagergren A, Westerlund S, Basta B, Lind U, Thelin A, Gisler R, Liberg D, Nelander S, et al. 2005. Gene expression analysis suggests that EBF-1 and PPARγ2 induce adipogenesis of NIH-3T3 cells with similar efficiency and kinetics. Physiol Genomics 23: 206–216. 10.1152/physiolgenomics.00015.2005 [DOI] [PubMed] [Google Scholar]
- Bouillaud F, Ricquier D, Thibault J, Weissenbach J. 1985. Molecular approach to thermogenesis in brown adipose tissue: cDNA cloning of the mitochondrial uncoupling protein. Proc Natl Acad Sci 82: 445–448. 10.1073/pnas.82.2.445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brier AB, Loft A, Madsen JGS, Rosengren T, Nielsen R, Schmidt SF, Liu Z, Yan Q, Gronemeyer H, Mandrup S. 2017. The KDM5 family is required for activation of pro-proliferative cell cycle genes during adipocyte differentiation. Nucleic Acids Res 45: 1743–1759. 10.1093/nar/gkw1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon B, Hedin A, Nedergaard J. 1982. Exclusive occurrence of thermogenin antigen in brown adipose tissue. FEBS Lett 150: 129–132. 10.1016/0014-5793(82)81319-7 [DOI] [PubMed] [Google Scholar]
- Chambers M, Turki-Judeh W, Kim MW, Chen K, Gallaher SD, Courey AJ. 2017. Mechanisms of Groucho-mediated repression revealed by genome-wide analysis of Groucho binding and activity. BMC Genomics 18: 215 10.1186/s12864-017-3589-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Courey AJ. 2000. Groucho/TLE family proteins and transcriptional repression. Gene 249: 1–16. 10.1016/S0378-1119(00)00161-X [DOI] [PubMed] [Google Scholar]
- Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, et al. 2009. Identification and importance of brown adipose tissue in adult humans. N Engl J Med 360: 1509–1517. 10.1056/NEJMoa0810780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cypess AM, White AP, Vernochet C, Schulz TJ, Xue R, Sass CA, Huang TL, Roberts-Toler C, Weiner LS, Sze C, et al. 2013. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat Med 19: 635–639. 10.1038/nm.3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dali R, Verginelli F, Pramatarova A, Sladek R, Stifani S. 2018. Characterization of a FOXG1:TLE1 transcriptional network in glioblastoma-initiating cells. Mol Oncol 12: 775–787. 10.1002/1878-0261.12168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jesus LA, Carvalho SD, Ribeiro MO, Schneider M, Kim SW, Harney JW, Larsen PR, Bianco AC. 2001. The type 2 iodothyronine deiodinase is essential for adaptive thermogenesis in brown adipose tissue. J Clin Invest 108: 1379–1385. 10.1172/JCI200113803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, et al. 2018. The Reactome pathway knowledgebase. Nucleic Acids Res 46: D649–D655. 10.1093/nar/gkx1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorenko A, Lishko PV, Kirichok Y. 2012. Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat mitochondria. Cell 151: 400–413. 10.1016/j.cell.2012.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher AL, Caudy M. 1998. Groucho proteins: transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes Dev 12: 1931–1940. 10.1101/gad.12.13.1931 [DOI] [PubMed] [Google Scholar]
- Gao H, Mejhert N, Fretz JA, Arner E, Lorente-Cebrian S, Ehrlund A, Dahlman-Wright K, Gong X, Stromblad S, Douagi I, et al. 2014. Early B cell factor 1 regulates adipocyte morphology and lipolysis in white adipose tissue. Cell Metab 19: 981–992. 10.1016/j.cmet.2014.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin MJ, Zhou Y, Kang S, Zhang X, Mikkelsen TS, Rosen ED. 2013. Early B-cell factor-1 (EBF1) is a key regulator of metabolic and inflammatory signaling pathways in mature adipocytes. J Biol Chem 288: 35925–35939. 10.1074/jbc.M113.491936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hany TF, Gharehpapagh E, Kamel EM, Buck A, Himms-Hagen J, von Schulthess GK. 2002. Brown adipose tissue: a factor to consider in symmetrical tracer uptake in the neck and upper chest region. Eur J Nucl Med Mol Imaging 29: 1393–1398. 10.1007/s00259-002-0902-6 [DOI] [PubMed] [Google Scholar]
- Hasegawa Y, Ikeda K, Chen Y, Alba DL, Stifler D, Shinoda K, Hosono T, Maretich P, Yang Y, Ishigaki Y, et al. 2018. Repression of adipose tissue fibrosis through a PRDM16–GTF2IRD1 complex improves systemic glucose homeostasis. Cell Metab 27: 180–194.e6. 10.1016/j.cmet.2017.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Maretich P, Kajimura S. 2018. The common and distinct features of brown and beige adipocytes. Trends Endocrinol Metab 29: 191–200. 10.1016/j.tem.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez MA, Akerblad P, Sigvardsson M, Rosen ED. 2007. Critical role for Ebf1 and Ebf2 in the adipogenic transcriptional cascade. Mol Cell Biol 27: 743–757. 10.1128/MCB.01557-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazak L, Chouchani ET, Jedrychowski MP, Erickson BK, Shinoda K, Cohen P, Vetrivelan R, Lu GZ, Laznik-Bogoslavski D, Hasenfuss SC, et al. 2015. A creatine-driven substrate cycle enhances energy expenditure and thermogenesis in beige fat. Cell 163: 643–655. 10.1016/j.cell.2015.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res 12: 996–1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khomtchouk BB, Hennessy JR, Wahlestedt C. 2017. shinyheatmap: ultra fast low memory heatmap web interface for big data genomics. PLoS ONE 12: e0176334 10.1371/journal.pone.0176334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai B, Lee JE, Jang Y, Wang L, Peng W, Ge K. 2017. MLL3/MLL4 are required for CBP/p300 binding on enhancers and super-enhancer formation in brown adipogenesis. Nucleic Acids Res 45: 6388–6403. 10.1093/nar/gkx234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Ge K. 2014. Transcriptional and epigenetic regulation of PPARγ expression during adipogenesis. Cell Biosci 4: 29 10.1186/2045-3701-4-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P, Greenfield JR, Ho KK, Fulham MJ. 2010. A critical appraisal of the prevalence and metabolic significance of brown adipose tissue in adult humans. Am J Physiol Endocrinol Metab 299: E601–E606. 10.1152/ajpendo.00298.2010 [DOI] [PubMed] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD. 2017. PANTHER version 11: expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45: D183–D189. 10.1093/nar/gkw1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Mandrup S. 2014. Genome-wide profiling of transcription factor binding and epigenetic marks in adipocytes by ChIP-seq. Methods Enzymol 537: 261–279. 10.1016/B978-0-12-411619-1.00014-8 [DOI] [PubMed] [Google Scholar]
- Orava J, Nuutila P, Lidell ME, Oikonen V, Noponen T, Viljanen T, Scheinin M, Taittonen M, Niemi T, Enerback S, et al. 2011. Different metabolic responses of human brown adipose tissue to activation by cold and insulin. Cell Metab 14: 272–279. 10.1016/j.cmet.2011.06.012 [DOI] [PubMed] [Google Scholar]
- Pisani DF, Djedaini M, Beranger GE, Elabd C, Scheideler M, Ailhaud G, Amri EZ. 2011. Differentiation of human adipose-derived stem cells into ‘brite’ (brown-in-white) adipocytes. Front Endocrinol (Lausanne) 2: 87 10.3389/fendo.2011.00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S, Wu J, Ishibashi J, Lim HW, Giang AH, Won KJ, Reed RR, Seale P. 2013. EBF2 determines and maintains brown adipocyte identity. Cell Metab 17: 562–574. 10.1016/j.cmet.2013.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Cuenca S, Monjo M, Gianotti M, Proenza AM, Roca P. 2007. Expression of mitochondrial biogenesis-signaling factors in brown adipocytes is influenced specifically by 17β-estradiol, testosterone, and progesterone. Am J Physiol Endocrinol Metab 292: E340–E346. 10.1152/ajpendo.00175.2006 [DOI] [PubMed] [Google Scholar]
- Saito M, Okamatsu-Ogura Y, Matsushita M, Watanabe K, Yoneshiro T, Nio-Kobayashi J, Iwanaga T, Miyagawa M, Kameya T, Nakada K, et al. 2009. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes 58: 1526–1531. 10.2337/db09-0530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheele C, Nielsen S. 2017. Metabolic regulation and the anti-obesity perspectives of human brown fat. Redox Biol 12: 770–775. 10.1016/j.redox.2017.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. 2008. PRDM16 controls a brown fat/skeletal muscle switch. Nature 454: 961–967. 10.1038/nature07182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao M, Ishibashi J, Kusminski CM, Wang QA, Hepler C, Vishvanath L, MacPherson KA, Spurgin SB, Sun K, Holland WL, et al. 2016. Zfp423 maintains white adipocyte identity through suppression of the beige cell thermogenic gene program. Cell Metab 23: 1167–1184. 10.1016/j.cmet.2016.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira SN, Lim HW, Rajakumari S, Sakers AP, Ishibashi J, Harms MJ, Won KJ, Seale P. 2017. EBF2 transcriptionally regulates brown adipogenesis via the histone reader DPF3 and the BAF chromatin remodeling complex. Genes Dev 31: 660–673. 10.1101/gad.294405.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp LZ, Shinoda K, Ohno H, Scheel DW, Tomoda E, Ruiz L, Hu H, Wang L, Pavlova Z, Gilsanz V, et al. 2012. Human BAT possesses molecular signatures that resemble beige/brite cells. PLoS One 7: e49452 10.1371/journal.pone.0049452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidossis L, Kajimura S. 2015. Brown and beige fat in humans: thermogenic adipocytes that control energy and glucose homeostasis. J Clin Invest 125: 478–486. 10.1172/JCI78362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbaek MS, Loft A, Aagaard MM, Nielsen R, Schmidt SF, Petrovic N, Nedergaard J, Mandrup S. 2012. Genome-wide profiling of peroxisome proliferator-activated receptor γ in primary epididymal, inguinal, and brown adipocytes reveals depot-selective binding correlated with gene expression. Mol Cell Biol 32: 3452–3463. 10.1128/MCB.00526-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford KI, Middelbeek RJ, Townsend KL, An D, Nygaard EB, Hitchcox KM, Markan KR, Nakano K, Hirshman MF, Tseng YH, et al. 2013. Brown adipose tissue regulates glucose homeostasis and insulin sensitivity. J Clin Invest 123: 215–223. 10.1172/JCI62308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stine RR, Shapira SN, Lim HW, Ishibashi J, Harms M, Won KJ, Seale P. 2016. EBF2 promotes the recruitment of beige adipocytes in white adipose tissue. Mol Metab 5: 57–65. 10.1016/j.molmet.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Spiegelman BM. 1994. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell 79: 1147–1156. 10.1016/0092-8674(94)90006-X [DOI] [PubMed] [Google Scholar]
- Vergnes L, Davies GR, Lin JY, Yeh MW, Livhits MJ, Harari A, Symonds ME, Sacks HS, Reue K. 2016. Adipocyte browning and higher mitochondrial function in periadrenal but not SC fat in pheochromocytoma. J Clin Endocrinol Metab 101: 4440–4448. 10.1210/jc.2016-2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva CJ, Waki H, Godio C, Nielsen R, Chou WL, Vargas L, Wroblewski K, Schmedt C, Chao LC, Boyadjian R, et al. 2011. TLE3 is a dual-function transcriptional coregulator of adipogenesis. Cell Metab 13: 413–427. 10.1016/j.cmet.2011.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva CJ, Vergnes L, Wang J, Drew BG, Hong C, Tu Y, Hu Y, Peng X, Xu F, Saez E, et al. 2013. Adipose subtype-selective recruitment of TLE3 or Prdm16 by PPARγ specifies lipid storage versus thermogenic gene programs. Cell Metab 17: 423–435. 10.1016/j.cmet.2013.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto NJ, Enerback S, et al. 2009. Functional brown adipose tissue in healthy adults. N Engl J Med 360: 1518–1525. 10.1056/NEJMoa0808949 [DOI] [PubMed] [Google Scholar]
- Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, Khandekar M, Virtanen KA, Nuutila P, Schaart G, et al. 2012. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 150: 366–376. 10.1016/j.cell.2012.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P, Arch JR, Ashwell M. 1984. Brown adipose tissue in the parametrial fat pad of the mouse. FEBS Lett 167: 10–14. 10.1016/0014-5793(84)80822-4 [DOI] [PubMed] [Google Scholar]
- Zhao XY, Li S, Wang GX, Yu Q, Lin JD. 2014. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol Cell 55: 372–382. 10.1016/j.molcel.2014.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data are available from GEO under accession number GSE116894. ChIP-seq data are available from GEO under accession number GSE116767.