Abstract
Pancreatic cancer is one of the deadliest malignancies with few early detection tests or effective therapies. PI3K‐AKT signaling is recognized to modulate cancer progression. We previously identified that a genetic variant in PKN1 increased pancreatic cancer risk through the PKN1/FAK/PI3K/AKT pathway. In order to investigate the associations between genetic variations in that pathway and pancreatic cancer prognosis, we conducted a two‐stage survival analysis in a total of 547 Chinese pancreatic cancer patients. Consequently, a variant, rs13167294 A>C in PIK3R1, was significantly associated with poor survival in both stages and with hazard ratio being 1.32 (95% CI = 1.13‐1.56, P = 0.0007) in the combined analysis. Function annotation and prediction suggested that genetic variants in this locus might affect overall survival of pancreatic cancer patients by regulating PIK3R1 expression.
Keywords: genetic variant, genome‐wide association study, pancreatic cancer survival, PI3K
1. INTRODUCTION
Pancreatic ductal adenocarcinoma (hereafter referred to as pancreatic cancer, PC) is a highly lethal human cancer. In 2018, an estimated 432 243 patients died of the disease, with the fatality rate reaching up to 94.2%.1 Due to the lack of early detection methods and effective treatments, the prognosis has been dismal over the past decades. In fact, the survival times vary with each patient and are partly explained by traditional clinical and pathological features.2 Moreover, germline variants, ie, single nucleotide polymorphisms (SNP) have been demonstrated associated with overall survival with increasing evidence.3, 4, 5, 6, 7
PI3K‐AKT signaling is a front and center pathway that is often overactivated in a wide range of tumor types. It triggers a cascade of response, from cell growth to survival and motility, that drive tumor initiation to progression.8 Recently, we discovered that a germline variant in PKN1 gene might perturb the PKN1/FAK/PI3K/AKT pathway and lead to pancreatic tumorigenesis.9 Our study bridged the core elements of PI3K‐AKT signaling with PKN1/FAK, and highlighted the role of the germline variant on this pathway played in PC development. PKN1 and FAK had been reported to play a part in cancer progression and metastasis.10, 11 But the association between genetic variants in this extended pathway and patient survival remained elusive.
Therefore, we hypothesized that genetic variants in the PKN1/FAK/PI3K/AKT pathway were associated with the survival of PC patients, and we tested this hypothesis with a two‐stage survival analysis in a total of 547 subjects.
2. MATERIALS AND METHODS
2.1. Study subjects
We performed a two‐stage study to search for genetic variants that correlated with PC survival. In the discovery stage, we included 341 patients with complete follow‐up information and genotyped in both of our previous genome‐wide association studies (GWAS) (ChinaPC) 12 and exome‐wide association studies (EWAS).9 These subjects were recruited from the Cancer Hospital, Chinese Academy of Medical Sciences, Beijing, between 2000 and 2011. As for the replication stage, 206 PC patients, from Tongji Hospital of Huazhong University of Science and Technology, Wuhan, were enrolled in our study. Characteristics and clinical information including age, gender, smoking and drinking status and tumor stage were obtained from patients’ medical records. Smoking status dichotomized subjects into two categories as we did in former studies 13, 14, 15: (a) Patients who had smoked ≥ 1 cigarette per day and for ≥ 1 year before cancer diagnosis were regarded as smokers (current or former); (b) The rest were nonsmokers. A similar classification went for drinking status: (a) Subjects who drank more than twice a week and for ≥ 1 year were considered as drinkers (current or former); (b) Others were nondrinkers. Tumor stages were harmonized into three categories: (a) local disease amenable to surgical resection; (b) locally advanced disease with extra‐pancreatic extension rendering it unresectable, but without distant metastases; and (c) distant metastatic disease. Survival time was measured from the date of surgery to last follow‐up or death, and the follow‐up information was obtained through telephone calls. Informed consents were obtained from all individuals.
2.2. Selection of genes and SNPs
Through literature research, we found a total of 16 genes as follows that are closely related to the PKN1/FAK/PI3K/AKT pathway. Among which, RAC1, RHOA, RHOB and CASP3 are upstream genes encoding small G proteins or caspase‐3 that activate PKN1.16, 17, 18, 19, 20 The other 12 genes (PDK1, PKN1, FAK, PIK3CA, PIK3CB, PIK3CD, PIK3R1, PIK3R2, PIK3R3, AKT1, AKT2, AKT3) are members of the PKN1/FAK/PI3K/AKT phosphorylation pathway.21, 22, 23, 24 The regulatory relationships are illustrated in Figure S1. We retrieved all of the experimentally genotyped SNPs (minor allele frequency ≥ 0.05) from the GWAS and EWAS dataset within the above genes or their ±10 kilobase flanking regions. All selected SNPs passed the quality control.
2.3. Genotyping
Genomic DNA was isolated from peripheral‐blood cells. In the discovery stage, samples were genotyped with the Affymetrix GeneChip Human Mapping 6.0 set and the Illumina HumanExome Beadchip.9, 12 For the replication stage, four variants were subsequently genotyped using a TaqMan assay on the ABI PRISM 7900 HT platform (Applied Biosystems, Inc).
2.4. Function annotation and prediction
All SNPs in linkage disequilibrium (LD) (r 2 > 0.2, 1000G Phase 1 Asian population) with the tag SNP in a ±200 kb flanking region were selected for function annotation and prediction. RegulomeDB (http://www.regulomedb.org/) and HaploReg v4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php), two online function annotation softwares especially for noncoding genetic variants, were used. Those SNPs with RegulomeDB score ≤ 2 (the threshold was set with reference to literature 25, 26), and located in DNase hypersensitive sites or histone modification marker peaks were predicted to be of regulatory function. GTEx eQTL Calculator (https://gtexportal.org/home/testyourown) was used for expression quantitative trait loci analysis. SurvExpress (http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp) was used to assess the association between gene expression and survival time.
2.5. Statistical analysis
Cox proportional hazards regression was used to measure the effects of candidate SNPs on PC survival, in different genetic models (additive, dominant and recessive) with adjustment for age, gender, smoking and drinking status and stage of disease. Kaplan‐Meier survival estimates were plotted and P values were assessed using the log‐rank test. Survival analyses using “coxph” function in “Survival package” were performed with R (3.5.2). For all analyses, P < 0.05 was regarded as statistical significance and all tests were two‐sided.
3. RESULTS
3.1. Characteristics of study subjects
The current study included a discovery dataset of 341 PC patients from our previous GWAS and EWAS studies,9, 12 and a replication stage of 206 PC patients recruited form Wuhan. The characteristics are shown in Table 1. Demographic characteristics including age, gender, smoking and drinking status showed no significant influence on survival time in two stages and combined analysis. Clinical stage of disease, consistent with general knowledge, was strongly correlated with survival time.
Table 1.
Selected characteristics of study subjects
| Discovery stage (N = 341) | Replication stage (N = 206) | Combined stage (N = 547) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of Patients (%) | No. of Death (%) | MST (months) | P a | No. of Patients (%) | No. of Death (%) | MST (months) | P a | No. of Patients (%) | No.of Death (%) | MST (months) | P a | |
| Age at diagnosis | 0.4360 | 0.3489 | 0.3718 | |||||||||
| ≤60 | 153 (44.87) | 136 (88.89) | 7.4 | 93 (45.1) | 67 (72.0) | 9.2 | 335 (61.2) | 279 (83.3) | 8.2 | |||
| >60 | 188 (55.13) | 176 (93.62) | 7.2 | 113 (54.9) | 88 (77.9) | 8.3 | 212 (38.8) | 188 (88.7) | 7.6 | |||
| Gender | 0.3208 | 0.9921 | 0.1731 | |||||||||
| Male | 202 (59.24) | 180 (89.11) | 7.4 | 133 (64.6) | 99 (74.4) | 8.6 | 246 (45.0) | 203 (82.5) | 7.9 | |||
| Female | 139 (40.76) | 132 (94.96) | 7.2 | 73 (35.4) | 56 (76.7) | 9.8 | 301 (55.0) | 264 (87.7) | 7.8 | |||
| Smoking status | 0.5021 | 0.4905 | 0.3473 | |||||||||
| Yes | 74 (21.70) | 68 (91.89) | 6.9 | 52 (25.2) | 43 (82.7) | 8.6 | 421 (77.0) | 356 (84.6) | 7.3 | |||
| No | 267 (78.30) | 244 (91.39) | 7.6 | 154 (74.8) | 112 (72.7) | 8.9 | 126 (23.0) | 111 (88.1) | 8.0 | |||
| Drinking status | 0.9305 | 0.9296 | 0.9963 | |||||||||
| Yes | 62 (18.18) | 56 (90.32) | 6.8 | 38 (18.4) | 30 (78.9) | 10.3 | 447 (81.7) | 381 (85.2) | 8.1 | |||
| No | 279 (81.82) | 256 (91.76) | 7.4 | 168 (81.6) | 125 (74.4) | 8.5 | 100 (18.3) | 86 (86.0) | 7.8 | |||
| Clinical stage | 1.98E‐08 | 3.37E‐15 | 9.56E‐21 | |||||||||
| Local | 72 (21.11) | 61 (84.72) | 9.3 | 54 (26.2) | 29 (53.7) | 18.8 | 126 (23.0) | 90 (71.4) | 13.9 | |||
| Locally advanced | 151 (44.28) | 133 (88.08) | 8.9 | 87 (42.2) | 70 (80.5) | 9.2 | 238 (43.5) | 203 (85.3) | 9.1 | |||
| Metastatic | 118 (34.60) | 118 (100.00) | 5.1 | 65 (31.6) | 56 (86.2) | 4.2 | 183 (33.5) | 174 (95.1) | 4.8 | |||
Abbreviation: MST, median survival time.
Calculated using the log‐rank test.
3.2. Multivariate analyses of associations between SNPs and patient survival
The design flowchart of the present study was illustrated in Figure 1. At first, a total of 203 genotyped common SNPs (minor allele frequency > 0.05) in GWAS (ChinaPC) and EWAS were selected for which were located in the 16 PKN1/FAK/PI3K/AKT pathway genes or their 10 kb flanking regions. Next, the associations between the 203 SNPs with PC survival were assessed with Cox proportional hazards regression under an additive model adjusting for age, gender, smoking and drinking status and clinical stage. Among these SNPs, four were significantly associated with patient survival and passed for validation in an independent population (Table 2). As a result, only one SNP, rs13167294 in PIK3R1, was successfully replicated and remained significantly associated with survival time in the combined analysis. The minor C‐allele conferred poorer prognosis for patients, and the per‐allele hazard ratio for which was 1.32 (95% CI = 1.13‐1.56, P = 0.0007) in the combined analysis. rs13167294 A>C was also correlated with decreased survival under a dominant model in discovery, replication and combined stages (Table 3). Further stratification analyses showed that the significant correlation was particularly in patients with metastatic stage of disease. As for patients in locally advanced stage, the minor C‐allele also indicated worse survival than the reference, but the association of SNP and survival was not statistically significant possibly due to the relatively small sample size. Further research with larger sample size is still needed to draw firm conclusion about the effects of rs13147249 in different stages (Table S1). Kaplan‐Meier curves illustrated significant associations between this variation and survival especially in additive and dominant models (Figure 2).
Figure 1.
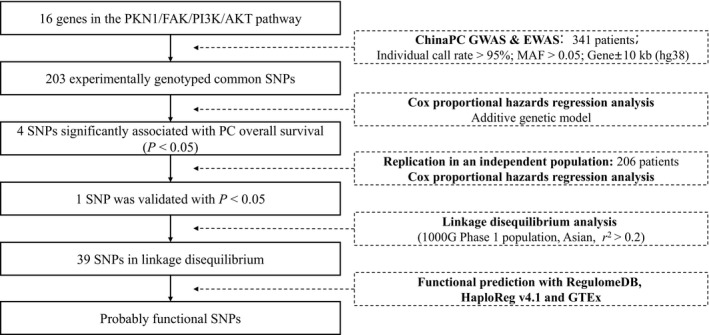
Research workflow for SNPs in the PKN1/FAK/PI3K/AKT pathway genes. EWAS, exome‐wide association study; GWAS, genome‐wide association study; MAF, minor allele frequency; SNP, single nucleotide polymorphism
Table 2.
Analysis of the four SNPs that passed the screen of discovery stage
| SNP | Allelea | Gene | Discovery stage | Replication stage | Combined stage | |||
|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | P b | HR (95% CI) | P b | HR (95% CI) | P b | |||
| rs13167294 | A>C | PIK3R1 | 1.31 (1.07‐1.61) | 0.0104 | 1.45 (1.10‐1.91) | 0.0076 | 1.32 (1.13‐1.56) | 0.0007 |
| rs7839119 | C>A | FAK | 0.76 (0.62‐0.95) | 0.0138 | 1.02 (0.76‐1.36) | 0.8905 | 0.83 (0.70‐0.99) | 0.0369 |
| rs4983387 | G>A | ZBTB42 c | 1.21 (1.02‐1.42) | 0.0252 | 1.13 (0.87‐1.46) | 0.3525 | 1.18 (1.03‐1.35) | 0.0192 |
| rs342105 | T>C | RHOB | 1.57 (1.01‐2.44) | 0.0471 | 0.94 (0.57‐1.55) | 0.8068 | 1.13 (0.82‐1.56) | 0.4703 |
Abbreviations: HR, hazard ratio; SNP, single nucleotide polymorphism.
Reference allele/effect allele.
Calculated using Cox regression in an additive genetic model adjusting for age, gender, smoking, drinking status and clinical stage.
This SNP locates in the exon of ZBTB42, and 6.1kb upstream of AKT1.
Table 3.
Association between rs13167294 and overall survival in pancreatic cancer patients
| Discovery stage | Replication stage | Combined stage | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. (%) | MST | HR (95% CI) | P a | No. (%) | MST | HR (95% CI) | P a | No. (%) | MST | HR (95% CI) | P a | |
| PIK3R1 rs13167294 | ||||||||||||
| AA | 229 (67.2) | 8.32 | 1.00 (Reference) | 125 (60.7) | 10.62 | 1.00 (Reference) | 354 (64.7) | 8.95 | 1.00 (Reference) | |||
| AC | 103 (30.2) | 6.27 | 1.34 (1.05‐1.72) | 0.0192 | 72 (35.0) | 7.60 | 1.44 (1.02‐2.04) | 0.0373 | 175 (32.0) | 6.69 | 1.36 (1.11‐1.65) | 0.0026 |
| CC | 9 (2.6) | 4.83 | 1.57 (0.80‐3.07) | 0.1913 | 9 (4.4) | 3.75 | 2.13 (1.02‐4.43) | 0.0434 | 18 (3.3) | 4.67 | 1.63 (1.00‐2.67) | 0.0512 |
| Additive model | 1.31 (1.07‐1.61) | 0.0104 | 1.45 (1.10‐1.91) | 0.0076 | 1.32 (1.13‐1.56) | 0.0007 | ||||||
| Dominant model | 1.36 (1.07‐1.73) | 0.0117 | 1.51 (1.08‐2.10) | 0.0151 | 1.38 (1.14‐1.67) | 0.0010 | ||||||
| Recessive model | 1.45 (0.74‐2.82) | 0.2812 | 1.88 (0.92‐3.87) | 0.0859 | 1.48 (0.91‐2.42) | 0.1119 | ||||||
Abbreviation: HR, hazard ratio; MST, median survival time.
Calculated using Cox regression adjusting for age, gender, smoking, drinking status and clinical stage.
Figure 2.
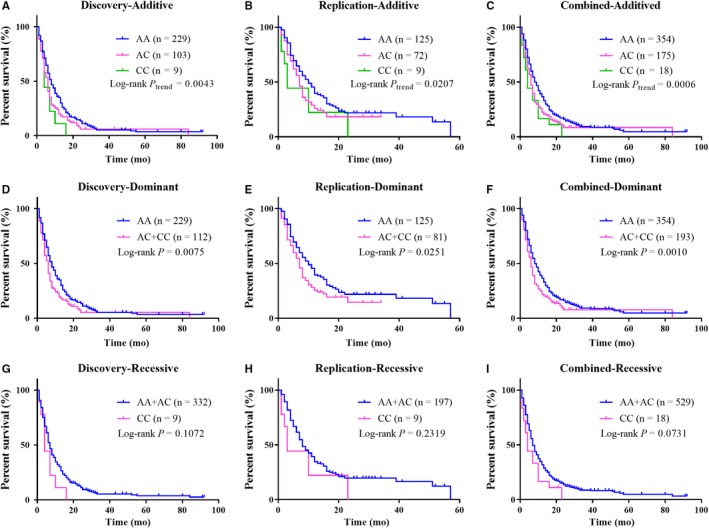
Kaplan‐Meier curves of survival in pancreatic cancer patients by rs13167294 genotypes, in different stages, assuming (a‐c) additive, (d‐f) dominant and (g‐i) recessive models
3.3. Functional variants potentially affect PC prognosis by regulating PIK3R1 expression
In order to further seek functional variants related to the identified locus, we retrieved 39 SNPs in LD (r 2 > 0.2, 1000G Phase 1 Asian population) with rs13167294. These SNPs were all noncoding variants (38 intronic and one in the 5′UTR) of PIK3R1. So we utilized RegulomeDB and HaploReg v4.1 to assess whether each one of the 39 SNPs was functional. Those SNPs with RegulomeDB score ≤ 2, and located in DNase hypersensitive sites or histone modification marker peaks (ie rs1819986 and rs6876003) were predicted to be of regulatory function probably by regulating the expression of PIK3R1 (Table S2). Utilizing the GTEx eQTL Calculator, we found that the associations between genotypes and PIK3R1 expressions in pancreas tissue were significant for six SNPs (ie rs6876003, rs6890202, rs1819987, rs6894871, rs6861401 and rs1010793). Taking account of results from the above estimates, the variant rs6876003 was most likely functional by affecting PIK3R1 expression (Table S2). Next, we took advantage of SurvExpress to assess the association between PIK3R1 expressions and overall survival in PC patients. The higher expression of PIK3R1 suggested worse survival in two public datasets: Stratford Yeh Pancreatic GSE21501 and Pancreatic Cancer‐AU (PACA‐AU‐ICGC‐June 2016, Australian Pancreatic Cancer Genome Initiative) (Figure S2). Taken together, genetic variants such as rs6876003 might influence PC prognosis by regulating PIK3R1 mRNA expression levels.
4. DISCUSSION
In the present study, we performed a two‐stage survival analysis to specifically estimate the association between overall survival in PC patients and genetic variants of the 16 genes belonging to the PKN1/FAK/PI3K/AKT pathway. In the discovery stage, we screened 203 corresponding variants and found four of prognostic significance. Among which was only one variant, rs13167294 in PIK3R1, successfully validated in an independent population in the replication stage. For the combined analysis of 547 patients in total, rs13167294 remained significantly correlated with PC survival under both additive and dominant genetic model. Subsequent in silico functional prediction suggested variants in the locus tagged by rs13167294 might affect survival by regulation of PIK3R1 expression.
In the discovery stage, we found four potentially associated SNPs with the lowest P value of 0.0104. Partly due to the relatively small sample size, the P values would be no longer significant after multiple comparisons correction. In fact, the aim of the discovery stage was to screen out the candidate SNPs as completely as possible. Therefore, we did not adopt corrected P values for multiple comparisons, in case the actual associated SNPs were excluded by the strict threshold. As for the replication and combined stages, we took Bonferroni correction for the four candidate SNPs, and found that rs13167249 was significantly associated with survival even at the corrected threshold of P < 0.0125.
It is well recognized that the PI3K‐AKT signaling plays critical roles in metastasis of cancers including PC.27, 28, 29, 30 It promotes cellular events such as invasion, migration and epithelial‐mesenchymal transition via regulation of different transcription factors.31, 32, 33 PIK3R1, which encodes the 85 kD regulatory subunit of PI3K, functions primarily as a regulator of the p110α catalytic product. Its mutations and abnormal expression were reported to impair PI3K function, and further cause disease.34, 35, 36, 37 An integrative survival‐based molecular profiling of human pancreatic cancer had identified PIK3R1 expression as a putative clinical biomarker for the disease outcome.38 But little was known about the cis‐regulatory element of this gene. Our current study identified that several genetic variants in PIK3R1 might regulate the expression of this gene. Take the most likely functional variant rs6876003 for instance, this intronic variant locates in a DNase I hypersensitive site, where histone modification predicts an active enhancer function. It potentially affects TF binding since it resides in some motifs, thus supporting its regulatory function. In particular, the genotype‐phenotype correlation demonstrated the modulation of PIK3R1 expression levels by rs6876003, suggesting that the findings were biologically plausible that this variant played a role in PI3K signaling and even PC prognosis. It should be admitted that the association between PC survival and rs6876003 were not completely represented by the result of the tagSNP rs13167294 since the LD relationship is not strong enough (r 2 = 0.32). However, the reason for choosing a weak LD threshold is to find, as much as possible, potentially functional SNP for further research. As for the eQTL analysis, we only found available data from pancreatic normal tissues in the public databases. Genotype‐phenotype data from cancer tissues was lacking, which was a limitation of the present study. It is also worth noting that other variants in the LD list might also be functional such as those with RegulomeDB score > 2. In fact, the bioinformatics tools just offered us some clues, and biological experiment is needed to prove them in the future.
In summary, we identified a variant in PIK3R1 associated with PC outcome. Further studies of larger sample size are required to validate our findings.
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
Supporting information
ACKNOWLEDGEMENTS
This work is supported by the National Natural Science Foundation of China NSFC‐81673256, the National High‐Tech Research and Development Program of China 2014AA020609, Program for HUST Academic Frontier Youth Team and the National Program for Support of Top‐notch Young Professionals for Xiaoping Miao.
Zhu Y, Ke J, Gong Y, et al. A genetic variant in PIK3R1 is associated with pancreatic cancer survival in the Chinese population. Cancer Med. 2019;8:3575–3582. 10.1002/cam4.2228
REFERENCES
- 1. Ferlay J, Colombet M, Soerjomataram I, et al. GLOBOCAN sources and methods. Int J Cancer. 2018;144:1941-1953. 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 2. Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605‐1617. 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 3. Couch FJ, Wang X, Bamlet WR, de Andrade M, Petersen GM, McWilliams RR. Association of mitotic regulation pathway polymorphisms with pancreatic cancer risk and outcome. Cancer Epidemiol Biomarkers Prev. 2010;19:251‐257. 10.1158/1055-9965.EPI-09-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Willis JA, Olson SH, Orlow I, et al. A replication study and genome‐wide scan of single‐nucleotide polymorphisms associated with pancreatic cancer risk and overall survival. Clin Cancer Res. 2012;18:3942‐3951. 10.1158/1078-0432.CCR-11-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dong X, Javle M, Hess KR, Shroff R, Abbruzzese JL, Li D. Insulin‐like growth factor axis gene polymorphisms and clinical outcomes in pancreatic cancer. Gastroenterology. 2010;139:464‐473. e461‐463. 10.1053/j.gastro.2010.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu C, Kraft P, Stolzenberg‐Solomon R, et al. Genome‐wide association study of survival in patients with pancreatic adenocarcinoma. Gut. 2014;63:152‐160. 10.1136/gutjnl-2012-303477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhu Y, Zhai K, Ke J, et al. BRCA1 missense polymorphisms are associated with poor prognosis of pancreatic cancer patients in a Chinese population. Oncotarget. 2017;8:36033‐36039. 10.18632/oncotarget.16422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vivanco I, Sawyers CL. The phosphatidylinositol 3‐Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489‐501. 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 9. Chang J, Tian J, Zhu Y, et al. Exome‐wide analysis identifies three low‐frequency missense variants associated with pancreatic cancer risk in Chinese populations. Nat Commun. 2018;9:3688 10.1038/s41467-018-06136-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jiang H, Hegde S, Knolhoff BL, et al. Targeting focal adhesion kinase renders pancreatic cancers responsive to checkpoint immunotherapy. Nat Med. 2016;22:851‐860. 10.1038/nm.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang C‐S, Melhuish TA, Spencer A, et al. The protein kinase C super‐family member PKN is regulated by mTOR and influences differentiation during prostate cancer progression. Prostate. 2017;77:1452‐1467. 10.1002/pros.23400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu C, Miao X, Huang L, et al. Genome‐wide association study identifies five loci associated with susceptibility to pancreatic cancer in Chinese populations. Nat Genet. 2012;44:62‐66. 10.1038/ng.1020. [DOI] [PubMed] [Google Scholar]
- 13. Li J, Chang J, Tian J, et al. A rare variant P507L in TPP1 interrupts TPP1‐TIN2 interaction, influences telomere length, and confers colorectal cancer risk in chinese population. Cancer Epidemiol Biomarkers Prev. 2018;27:1029‐1035. 10.1158/1055-9965.EPI-18-0099. [DOI] [PubMed] [Google Scholar]
- 14. Chang J, Tian J, Yang Y, et al. A rare missense variant in TCF7L2 associates with colorectal cancer risk by interacting with a GWAS‐identified regulatory variant in the MYC enhancer. Cancer Res. 2018;78:5164‐5172. 10.1158/0008-5472.CAN-18-0910. [DOI] [PubMed] [Google Scholar]
- 15. Chang J, Zhong R, Tian J, et al. Exome‐wide analyses identify low‐frequency variant in CYP26B1 and additional coding variants associated with esophageal squamous cell carcinoma. Nat Genet. 2018;50:338‐343. 10.1038/s41588-018-0045-8. [DOI] [PubMed] [Google Scholar]
- 16. Takahashi M, Mukai H, Toshimori M, Miyamoto M, Ono Y. Proteolytic activation of PKN by caspase‐3 or related protease during apoptosis. Proc Natl Acad Sci U S A. 1998;95:11566‐11571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Flynn P, Mellor H, Casamassima A, Parker PJ. Rho GTPase control of protein kinase C‐related protein kinase activation by 3‐phosphoinositide‐dependent protein kinase. J Biol Chem. 2000;275:11064‐11070. [DOI] [PubMed] [Google Scholar]
- 18. Owen D, Lowe PN, Nietlispach D, et al. Molecular dissection of the interaction between the small G proteins Rac1 and RhoA and protein kinase C‐related kinase 1 (PRK1). J Biol Chem. 2003;278:50578‐50587. 10.1074/jbc.M304313200. [DOI] [PubMed] [Google Scholar]
- 19. Park YH, Wood G, Kastner DL, Chae JJ. Pyrin inflammasome activation and RhoA signaling in the autoinflammatory diseases FMF and HIDS. Nat Immunol. 2016;17:914‐921. 10.1038/ni.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hutchinson CL, Lowe PN, McLaughlin SH, Mott HR, Owen D. Differential binding of RhoA, RhoB, and RhoC to protein kinase C‐related kinase (PRK) isoforms PRK1, PRK2, and PRK3: PRKs have the highest affinity for RhoB. Biochemistry. 2013;52:7999‐8011. 10.1021/bi401216w. [DOI] [PubMed] [Google Scholar]
- 21. Gagliardi PA, Puliafito A, Primo L. PDK1: At the crossroad of cancer signaling pathways. Semin Cancer Biol. 2018;48:27‐35. 10.1016/j.semcancer.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 22. Sulzmaier FJ, Jean C, Schlaepfer DD. FAK in cancer: mechanistic findings and clinical applications. Nat Rev Cancer. 2014;14:598‐610. 10.1038/nrc3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. The PI3K pathway in human disease. Cell. 2017;170:605‐635. 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Collazos A, Michael N, Whelan RD, et al. Site recognition and substrate screens for PKN family proteins. Biochem J. 2011;438:535‐543. 10.1042/BJ20110521. [DOI] [PubMed] [Google Scholar]
- 25. Harismendy O, Notani D, Song X, et al. 9p21 DNA variants associated with coronary artery disease impair interferon‐gamma signalling response. Nature. 2011;470:264‐268. 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Emison ES, Garcia‐Barcelo M, Grice EA, et al. Differential contributions of rare and common, coding and noncoding Ret mutations to multifactorial Hirschsprung disease liability. Am J Hum Genet. 2010;87:60‐74. 10.1016/j.ajhg.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stoll V, Calleja V, Vassaux G, Downward J, Lemoine NR. Dominant negative inhibitors of signalling through the phosphoinositol 3‐kinase pathway for gene therapy of pancreatic cancer. Gut. 2005;54:109‐116. 10.1136/gut.2004.046706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang L, Yin J, Wang X, et al. C‐Type lectin‐like receptor 2 suppresses AKT signaling and invasive activities of gastric cancer cells by blocking expression of phosphoinositide 3‐kinase subunits. Gastroenterology. 2016;150: 1183‐1195. e1116. 10.1053/j.gastro.2016.01.034. [DOI] [PubMed] [Google Scholar]
- 29. Dey N, De P, Leyland‐Jones B. PI3K‐AKT‐mTOR inhibitors in breast cancers: from tumor cell signaling to clinical trials. Pharmacol Ther. 2017;175:91‐106. 10.1016/j.pharmthera.2017.02.037. [DOI] [PubMed] [Google Scholar]
- 30. Steelman LS, Abrams SL, Whelan J, et al. Contributions of the Raf/MEK/ERK, PI3K/PTEN/Akt/mTOR and Jak/STAT pathways to leukemia. Leukemia. 2008;22:686‐707. 10.1038/leu.2008.26. [DOI] [PubMed] [Google Scholar]
- 31. Xue G, Restuccia DF, Lan Q, et al. Akt/PKB‐mediated phosphorylation of Twist1 promotes tumor metastasis via mediating cross‐talk between PI3K/Akt and TGF‐beta signaling axes. Cancer Discov. 2012;2:248‐259. 10.1158/2159-8290.CD-11-0270. [DOI] [PubMed] [Google Scholar]
- 32. Yoshikawa Y, Takano O, Kato I, Takahashi Y, Shima F, Kataoka T. Ras inhibitors display an anti‐metastatic effect by downregulation of lysyl oxidase through inhibition of the Ras‐PI3K‐Akt‐HIF‐1alpha pathway. Cancer Lett. 2017;410:82‐91. 10.1016/j.canlet.2017.09.017. [DOI] [PubMed] [Google Scholar]
- 33. Wu CY, Carpenter ES, Takeuchi KK, et al. PI3K regulation of RAC1 is required for KRAS‐induced pancreatic tumorigenesis in mice. Gastroenterology. 2014;147:1405‐1416. e1407. 10.1053/j.gastro.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cheung LW, Yu S, Zhang D, et al. Naturally occurring neomorphic PIK3R1 mutations activate the MAPK pathway, dictating therapeutic response to MAPK pathway inhibitors. Cancer Cell. 2014;26:479‐494. 10.1016/j.ccell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cheung LW, Hennessy BT, Li J, et al. High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer elucidates a novel mechanism for regulation of PTEN protein stability. Cancer Discov. 2011;1:170‐185. 10.1158/2159-8290.CD-11-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. He S, Zhang J, Zhang W, Chen F, Luo R. FOXA1 inhibits hepatocellular carcinoma progression by suppressing PIK3R1 expression in male patients. J Exp Clin Cancer Res. 2017;36:175 10.1186/s13046-017-0646-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lu Y, Lemon W, Liu PY, et al. A gene expression signature predicts survival of patients with stage I non‐small cell lung cancer. PLoS Medicine. 2006;3:e467 10.1371/journal.pmed.0030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Donahue TR, Tran LM, Hill R, et al. Integrative survival‐based molecular profiling of human pancreatic cancer. Clin Cancer Res. 2012;18:1352‐1363. 10.1158/1078-0432.CCR-11-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


