Abstract
To develop a deep learning system based on 3D convolutional neural networks (CNNs), and to automatically predict EGFR‐mutant pulmonary adenocarcinoma in CT images. A dataset of 579 nodules with EGFR mutation status labels of mutant (Mut) or wild‐type (WT) was retrospectively analyzed. A deep learning system, namely 3D DenseNets, was developed to process 3D patches of nodules from CT data, and learn strong representations with supervised end‐to‐end training. The 3D DenseNets were trained with a training subset of 348 nodules and tuned with a development subset of 116 nodules. A strong data augmentation technique, mixup, was used for better generalization. We evaluated our model on a holdout subset of 115 nodules. An independent public dataset of 37 nodules from the cancer imaging archive (TCIA) was also used to test the generalization of our method. Conventional radiomics analysis was also performed for comparison. Our method achieved promising performance on predicting EGFR mutation status, with AUCs of 75.8% and 75.0% for our holdout test set and public test set, respectively. Moreover, strong relations were found between deep learning feature and conventional radiomics, while deep learning worked through an enhanced radiomics manner, that is, deep learned radiomics (DLR), in terms of robustness, compactness and expressiveness. The proposed deep learning system predicts EGFR‐mutant of lung adenocarcinomas in CT images noninvasively and automatically, indicating its potential to help clinical decision‐making by identifying eligible patients of pulmonary adenocarcinoma for EGFR‐targeted therapy.
Keywords: convolutional neural networks, deep learning, EGFR, mixup training technique, radiomics
1. INTRODUCTION
Lung cancer is one of the leading causes of cancer‐related death.1 Non‐small cell lung cancer (NSCLC) accounts for more than 80% lung cancer cases, where adenocarcinoma is the most common histological subtype.2 With the advancements of genomics, several genomic alternations, such as oncogenic ALK rearrangements,3 ROS1 rearrangement,4 KRAS mutations,5 and sensitizing EGFR mutations,6 have been identified as predictive and prognostic markers for NSCLC. Targeted therapy with these molecular markers has become an essential part of precision medicine for lung cancer.
Small molecule tyrosine kinase inhibitors (TKIs) that target specific EGFR mutations have resulted in improved progression‐free survival (PFS) and higher objective radiographic response rate in patients with EGFR mutations than standard chemotherapy.7, 8, 9 Moreover, approximately 80% patients with EGFR‐mutant lung cancer respond to EGFR TKIs therapy (at initial treatment).10 However, the administration of EGFR TKIs (for example, gefitinib) on the patients without EGFR mutations, not just showed no effect but even resulted in worse PFS and unnecessary costs compared to platinum‐based chemotherapy,11 which highlights the importance of identifying eligible patients for the first‐line TKI therapy.
Mutation profiling after biopsies or surgical resections has become a standard and informative medical procedure. However, the high cost and invasiveness of the approaches and repeating tumor sampling strongly limit the applicability of molecular testing. Besides, the poor DNA quality, intratumoral heterogeneity and long turnaround time raise much concern on the balance of costs and benefits.12, 13 Notwithstanding re‐biopsy is feasible in clinical practice, it is an invasive operation and faces the same challenges voiced by previous studies.14, 15 These defects enormously limit the practicability of precision medicine at scale.
Alternatively, noninvasive markers are promising for predicting EGFR mutation.16, 17 Cancers with different genotypes drive specific biological processes involved in the development and progression of tumors, thus ultimately leading to different phenotypes. In other words, it is potentially feasible to predict the genotypes by identifying specific phenotypes. There have been studies investigating the phenotype‐genotype associations to identify associated genomic changes,18, 19 based on tumor morphology on computed tomography (CT)20, 21 and magnetic resonance imaging (MRI).22 Radiomics,20 that encodes tumor phenotypes with innumerable quantitative features using predefined image analysis algorithms. In particular, previous studies have demonstrated that certain radiomic features are associated with EGFR mutations status, suggesting that those identified features may be driven by somatic mutations.16, 23 Although these studies have achieved impressive performances, especially when combined with clinical information,17, 23, 24 conventional radiomics‐based methods are born with three main challenges. Firstly, conventional radiomics methods require strict procedures, including detection, segmentation, feature extraction, selection etc,25 which is tedious and time‐consuming. Secondly, radiomic features are susceptible to the manual segmentation as well as CT scanning parameters, thus interobserver reproducibility analysis is necessary. Finally, hand‐craft radiomic features are expressive but may be not enough for high‐level tasks.
On the other hand, deep neural networks, or Deep Learning, have achieved remarkable success in several important problems of artificial intelligence (AI), for example, natural image classification26 and human language translation.27 Deep convolutional neural networks (CNNs), a family of neural networks, have shown incredible effectiveness in several tasks of natural image computer vision and medical image computing.28, 29 As powerful algorithms of representation learning, CNNs largely reduce the necessities of hand‐craft feature engineering. Our previous study has proven the effectiveness and efficacy of deep learning in predicting the invasiveness of lung adenocarcinomas from CT images.30 In this regard, we addressed the problem of CT‐based EGFR mutation prediction by deep neural networks, to make our system automatic, robust and accurate.
In this study, we aimed to develop a deep learning system to predict the EGFR mutation status of lung adenocarcinoma based on CT images by integrating recent advances in deep supervised learning, such as dense connections31 and mixup training,32 to significantly reduce the empirical risks of overfitting. Our method is a labor‐saving strategy without the requirement of precise nodule segmentation, and also expected to obtain more stable performance due to the enhanced nature of the employed learning algorithms.
2. MATERIALS AND METHODS
We developed a novel analytic framework based on deep learning of the CT imaging phenotypes of lung adenocarcinoma to predict EGFR mutation status (see Figure 1). This retrospective study was approved by the Institutional Review Board of our institution (NO.20170103), which waived the requirement for patients' informed consent referring to the CIOMS guideline.
Figure 1.
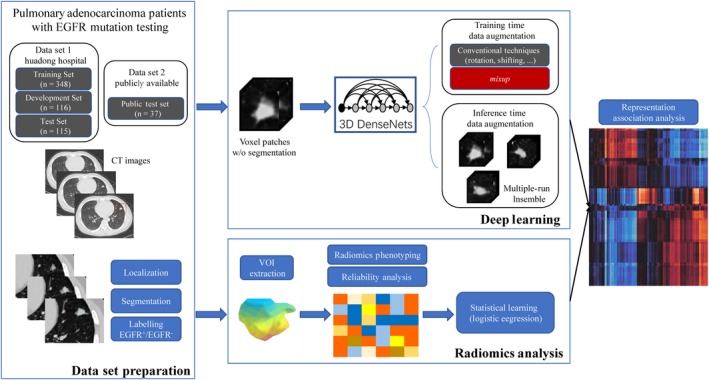
Overall pipeline for this study. A local CT dataset (HdH Dataset) and a public dataset selected from TCIA database (TCIA Dataset) of lung adenocarcinoma patients with EGFR mutation testing were used. Nodules were manually localized, segmented, and labelled as EGFR mutant (Mut)/wild‐type (WT). For deep learning, 3D DenseNets were trained using the training subset. A strong data augmentation technique, mixup, was used for better regularization. Expressive representations, that is, deep learned radiomics (DLR), for the nodules were end‐to‐end learned during the training procedure. Meanwhile, conventional radiomics analysis following the common practice was carried out for performance comparison, and association study between the 3D deep leaning and conventional radiomics was performed by calculating the pairwise correlation coefficients
2.1. Data collection
This study used local data collected in Huadong Hospital (HdH Dataset) and publicly available data selected from TCIA (TCIA Dataset).33, 34 In HdH Dataset, the inclusion criteria were (a) patient receiving thin‐slice chest CT (0.75‐1.5 mm) scan prior to biopsies or surgical treatment, (b) with pathology reports for the diagnosis of pulmonary adenocarcinoma, and (c) with detailed EGFR mutation testing reports. Only one malignant nodule was studied for each patient due to the availability of EGFR testing report. Among the 579 lung adenocarcinoma patients, 308 were EGFR‐mutant (Mut) and 271 were EGFR wild‐type (WT). The detailed characteristics of patients and lesions in HdH Dataset are presented in Table 1. CT acquisition parameters are described in Appendix S1. In the TCIA Dataset, 37 nodules were selected to validate the stability and generalization of the analytic framework. The inclusion criteria were (a) CT with slice thickness ≤1.5 mm (to avoid data inconsistency), (b) with EGFR mutations testing reports, (c) with pathology reports for the diagnosis of lung adenocarcinoma, (d) and lesions that could be certainly identified as the resected or biopsied lesions. The detailed information of the TCIA Dataset is described in Table S1 .
Table 1.
Characteristics of patients and lesions in HdH dataset
| Characteristics | Number | Percentage |
|---|---|---|
| Gender | ||
| Male | 245 | 42.3 |
| Female | 334 | 57.6 |
| Mean age (range) (y) | ||
| Male | 61.8 ± 11.6 (29‐85) | ‐ |
| Female | 58.4 ± 11.9 (22‐85) | ‐ |
| Total | 59.8 ± 11.9 (22‐85) | ‐ |
| Mean size (range) (cm) | 1.8 (0.3‐8.6) | ‐ |
| Location | ||
| Right lobe | 342 | 59.1 |
| Left lobe | 237 | 40.9 |
| Pathology | ||
| Adenocarcinoma in situ | 31 | 5.4 |
| Minimally invasive adenocarcinoma | 157 | 27.1 |
| Invasive adenocarcinoma | 391 | 67.5 |
| TMN classification (eighth edition) | ||
| 0 | 31 | 5.4 |
| I A‐B | 356 | 61.5 |
| II A‐B | 7 | 1.2 |
| III A‐C | 10 | 1.7 |
| IV A‐B | 175 | 30.2 |
| EGFR Mut | 308 | 53.2 |
2.2. EGFR mutation profiling
Drug target‐associated mutations on EGFR exons 18, 19, 20, and 21 were examined using a PCR‐based amplification‐refractory mutation system (ARMS) with Human EGFR Gene Mutations Fluorescence Polymerase Chain Reaction Diagnostic Kit (ADx‐EG01, AmoyDx). Wild‐type EGFR in this study referred to no mutation detected among those loci.
2.3. Data annotation and pretreatment
Volumes of interest (VOIs) of the enrolled nodules in both datasets were manually delineated at voxel level by a radiologist with 4‐year experience in chest CT interpretation using medical image processing and navigation software 3D Slicer (version 4.8.0, Brigham and Women's Hospital), and subsequently confirmed (modified or re‐delineated) by another radiologist with 12‐year experience in chest CT interpretation. DICOM‐format images were imported into the software, subsequently the images with VOI information were exported into NII format for further analysis. Each segmented nodule was assigned a specific EGFR mutation label (Mut or WT) according to the corresponding EGFR mutation testing report.
Deep learning is known to require heavy tuning. Thus, we randomized the HdH Dataset into training, development, and test subsets, containing 60%, 20%, and 20% (only once) of each EGFR categories (Mut or WT), respectively. The training set was used for training the neural networks. The development set was used for tuning the hyperparameters, such as determining early stopping, discovering the most suitable neural architectures, and choosing the best model snapshots. Once we chose the best model using the development set, the holdout test set was used for fairly evaluating the performance of our method. To further validate the stability and generalization of our method, we evaluate our model on the independent TCIA Dataset, without any fine tuning.
2.4. EGFR mutation status prediction with conventional radiomics
We investigated the performance of the conventional radiomics method using the two datasets. Following the common practice of radiomics method,23 the radiomic features were extracted with numbers of image analysis algorithms. Specifically, 475 radiomics features, including 50 histogram features, 325 co‐occurrence matrix features, and 100 run lengths matrix features, were automatically extracted using MATLAB 2016b for all delineated nodules in the two datasets. The detailed radiomics extraction methodology is described in Appendix S1.
Radiomic features require refined manual segmentation of nodules. Even so, many radiomic features are not stable due to different segmentation manners. To ensure the reproducibility of radiomics, 50 nodules in the HdH Dataset were randomly selected for independent segmentation by two radiologists. The 475 radiomics features of the 50 nodules were evaluated with interclass correlation coefficient (ICC) analysis using the “irr” package in R software (version 3.4.3). Features with an ICC > 0.8 were considered reliable. Finally, a total of 401 features were selected.
Logistic regression was used to model the relation between the radiomic features and EGFR status with the widely used scikit‐learn library in Python.17, 35 Due to the simple hyperparameter setting of logistic regression, the training and development sets were merged into a “train‐dev” set. Ten‐fold cross validation search was performed on the train‐dev set with 1000 randomly sampled values of regularization term C. The trained model was used for scoring the HdH test Dataset and TCIA Dataset. We also tried heavy search for models and hypermeters with an automated machine learning tool (AutoML),36 built upon widely used scikit‐learn Python package37; however it did not yield better performance. Note that the feature selection procedure to reduce the redundancy between the features had been included in the AutoML search procedure. Although there may exist a better model theoretically, the heavy search process implied conventional radiomic features are not strong enough representations.
2.5. EGFR status prediction with 3D DenseNets
To take the advantage of deep representation learning, we designed a deep learning‐based framework for analyzing the EGFR mutation status of nodules. Taking into account the characteristics of the used data, we have adopted the following principle to design the models:
3D: CT images were presented in 3D, and the 3D views provided critical visual information from a practical point of radiology;
Parameter‐efficient: the model should be compact and easy to train with limited data;
Data‐efficient: our training method should lead to less overfitting;
Automatic: at inference stage, our model should be easy to use, given approximate locations of nodules, rather than the manual segmentation.
Following these principles, we adopted 3D DenseNets, which were derived from the powerful deep convolutional neural networks 2D DenseNets.31 DenseNets have achieved great success in 2D natural images and medical images,38 which elegantly reuse the features from lower layers, thus exploring high‐level representations in an efficient way. Our 3D DenseNets used same notations but 3D convolutions, with a growth rate k = 16, a compression rate θ = 2, and a bottleneck B = 4. Batch Normalization39 and Leaky ReLU40 (α = 0.1) were used together as activation functions. The resulting neural networks contained only 0.55 M parameters, making the training compatible with limited data. We implemented the neural networks with Keras 2.1.541 and TensorFlow 1.4.0.42 The detailed structure of the 3D DenseNets is depicted in Table 2.
Table 2.
3D DenseNet architectures for EGFR mutation classification
| Layer | Tensor size | Building blocks | |
|---|---|---|---|
| Input | 48 × 48 × 48 × 1 | ||
| Convolution | 48 × 48 × 48 × 32 | 3 × 3 × 3 conv | |
| Pooling | 24 × 24 × 24 × 32 | 2 × 2 × 2 average pool | |
| Dense Block (1) | 24 × 24 × 24 × 80 |
|
|
| Compression and Pooling (1) | 12 × 12 × 12 × 40 |
|
|
| Dense Block (2) | 12 × 12 × 12 × 136 |
|
|
| Compression and Pooling (2) | 6 × 6 × 6 × 68 |
|
|
| Dense Block (3) | 6 × 6 × 6 × 132 |
|
|
| Compression and Pooling (3) | 3 × 3 × 3 × 66 |
|
|
| Dense Block (4) | 3 × 3 × 3 × 114 |
|
|
| Global Pooling (DLR) | 114 | 3 × 3 × 3 average pool | |
| Output | 1 | sigmoid |
The inputs of the proposed 3D DenseNets were cubic patches of 48 × 48 × 48 mm, generated by (pre‐processed) chest CT scans and the coordinates of the approximate mass centers of nodule. In practice, the coordinates can be marked by radiologists manually, or by automatic nodule detection systems.43 Our method did not require the manually refined segmentation, which were usually done by the users (radiologists) manually. The preprocessing followed a “standard” procedure: the input patches were converted into Hounsfield units, followed by resizing the volumetric data into spacing of 1 × 1 × 1 mm by trilinear interpolation, clipping the voxel intensity into , quantifying the density into grayscale, and transforming the values to by the mapping . We further applied multiple data augmentation techniques to represent nodules in different views:
Moving the centers by small amounts in [−m, m] pixels in the three axes
Reordering the axes and rotation by 90° increments
Left‐right flipping.
2.6. Training for the deep learning models
During training, we applied the above data augmentation techniques with m = 8. These conventional techniques could effectively increase the training data size, yet not enough for a good training convergence. We further used a novel data augmentation technique: mixup 32 to stabilize the training. mixup showed remarkable improvements in state‐of‐the‐art natural image classification neural networks. To our knowledge, this is the first comprehensive study introducing mixup into medical image computing.
The mixup produces extra informative training samples by the following easy‐to‐implement data augmentation routine. Given , α is a predefined hyperparameter, and (xi, yi) and (xj, yj) are two input‐label pairs from the training distribution, then extra training samples are obtained by the following equations:
α defines the strength of mixup by controlling the beta distribution; when α = 0, mixup ceases to be effective. In our experiments, we set α = 0.9 for modest usage of mixup. As shown later in the Results section, mixup was critical for enabling the training of deep neural networks with limited data on this task.
We trained the 3D DenseNets with binary cross‐entropy loss:
where y is the ground truth of the input, and is the prediction given by the models. “Kaiming uniform” method44 was used for initializing the neural networks. During training, we sampled 20 positive and 20 negative samples (with a batch size of 40) for balanced optimization. A Nesterov‐momentum SGD45 with momentum = 0.9 was used as neural optimizer. The initial learning rate was 3 × 10−2, and decayed by a factor of 1/3 at the end of epoch = 30, 60, 100, and 150. No weight decay nor dropout46 was used. All hyperparameters were manually tuned on the HdH development set. A model snapshot was saved at the end of each epoch. Finally, the best model of a single run was selected by the following heuristics:
initially, select five candidates with the highest AUCs on the development set;
then, choose one of the candidates with the lowest absolute difference of training and development AUCs.
Two hundred epochs were usually enough for a good convergence. In our experiments, the best model snapshot was generated at the end of epoch 131.
2.7. Inference for the deep learning models
Representing nodules in multiple views not only increases the training data size, but also effectively reduces empirical variance of inference by the ensemble trick. In practice, given an input, we forwarded the well‐trained neural network using the mentioned conventional data augmentation (m = 3) with 100 runs. Then, probability scores of EGFR Mut were obtained by averaging the multiple runs:
We used the last‐layer (the DLR layer in Table 1) 114‐d outputs of the trained 3D DenseNet as the learned representations given nodules, that is, the deep learned radiomics (DLR). More robust DLR was obtained by averaging the feature outputs of multiple runs. Note the DLR and the probability scores could be obtained within a single neural network forward pass.
3. RESULTS
3.1. Performance of deep learning on EGFR mutation status prediction
We compared the performance between deep learning and conventional radiomics on predicting EGFR mutation status of lung adenocarcinoma, which implied the associations of EGFR genotype and radiomic phenotype. The AUC, which is insensitive of class skews, was used for evaluation. As depicted in Table 3 and Figure 2C, the 3D DenseNets (with mixup, ensemble) outperformed our conventional radiomics‐based method (P = 0.021, DeLong test47). Several prior studies were also listed in Table 3. Since they used nonshared datasets independently, their results were only for reference only. On our holdout test set (HdH test Dataset) containing 115 patients and the TCIA Dataset of 37 patients, 3D DenseNets achieved AUCs of 75.8% and 75.0%, respectively. Considering the class‐imbalance and threshold bias, we believed the threshold‐free AUROC was the most appropriate metric. For reference, we applied a threshold that maximizes the accuracy to obtain the sensitivity, specificity, accuracy, and precision (or positive predictive value). These results were depicted in Table S2. Notably, the data distribution of the TCIA Dataset differed much from that of the HdH Dataset in terms of patients and imaging; the comparable performances on the two datasets indicated the robustness of our method. More importantly, unlike conventional radiomics methods that require labor‐intensive manual segmentation masks, our deep learning method was compatible with approximate locations of the nodules instead.
Table 3.
Presentation of our method and several previous studies in terms of methods, datasets, and resulting classification AUCs
| Method | Training #Patients | Test #Patients | #EGFR muta | AUC (%) |
|---|---|---|---|---|
| Radiomics23 | 353 | 352 | 183 (24.0%)b | 69 |
| + clinical information | 75 | |||
| Radiomics17 | 298 | NAc | 137 (46.0%) | 64.7 |
| + clinical information | 70.9 | |||
| Radiomics18 | 47 | NAc | 19 (40.4%) | 67.0 |
| Radiomics (This study) | 464 (HdH train‐dev Dataset) | 115 (HdH test Dataset) | 62 (53.9%) | 64.5 |
| 3D DenseNets w/mixup, ensemble (This study) | 348 (HdH training Dataset) | 115 (HdH test Dataset) | 62 (53.9%) | 75.8 |
| Radiomics (this study) | 464 (HdH train‐dev Dataset) | 37 (TCIA Dataset) | 9 (24.3%) | 68.7 |
| 3D DenseNets w/mixup, ensemble (this study) | 348 (HdH training Dataset) | 37 (TCIA Dataset) | 9 (24.3%) | 75.0 |
Shown as the number of cases (percentage).
Estimated using the proportion of EGFR Mut on the entire data set, rather than the test set.
The evaluation results are based on multivariate statistical analysis, rather than the practice of training – validation (development) – test in machine learning. Since the prior studies listed in the above table used nonshared datasets independently, the results are for reference only.
Figure 2.
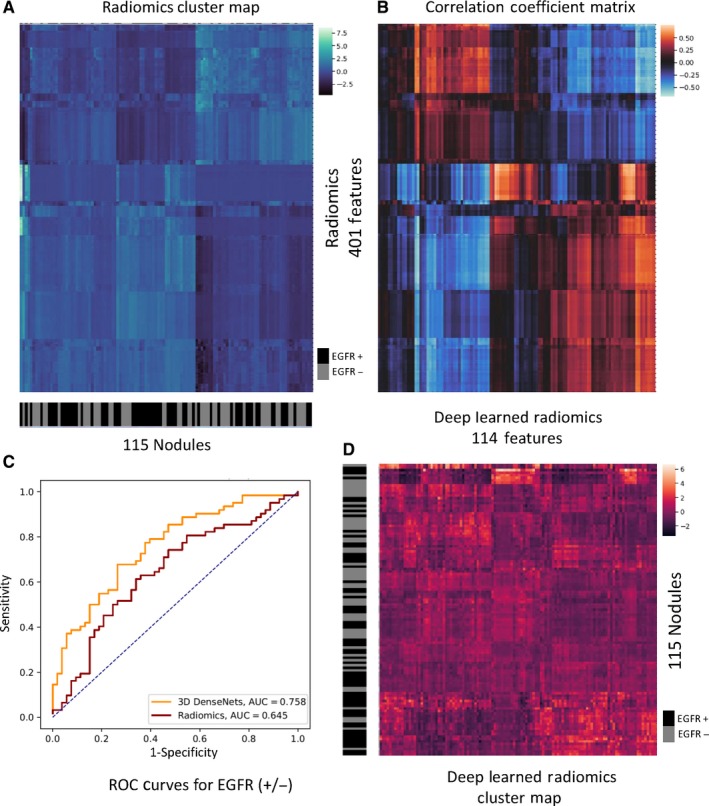
Visualization of conventional radiomics, deep learned radiomics (DLR) and their associations. A, Cluster map of conventional radiomics, with 115 nodules on the x‐axis and 401 radiomic features on the y‐axis. Each feature was normalized into zero mean and unit standard variance. The nodules of a same cluster (adjacent columns) shared similar radiomic features in Euclidean space. The semantic label EGFR Mut/WT of each nodule was shown on the black‐grey bar below the x‐axis. B, Correlation coefficient matrix for conventional radiomics and DLR. Note the radiomic features (y‐axis in A) and the DLR features (x‐axis in D) were both aligned with the correlation coefficient matrix. C, The classification ROC curves of our radiomics‐based and DLR‐based methods. The brighter (red or blue) blocks show the higher correlation. The black denotes no correlation. D, Cluster map of DRL with 115 nodules on the y‐axis and 114 radiomic features on the x‐axis. Each feature was normalized into zero mean and unit standard variance. The nodules of a same cluster (adjacent rows) shared similar DLR in Euclidean space. The semantic label EGFR Mut/WT of each nodule was shown on the black‐grey bar on the left of the y‐axis
3.2. Potential association between the conventional radiomics and deep learned radiomics
Radiomics analysis provides important medical insights. However, conventional radiomics requires manual segmentation, which is a tedious process in practice. Instead, our method based on 3D deep learning predicts the labels automatically with even better performance. To our knowledge, few prior studies compare these two methods together. Assumed that deep learning networks particularly learn the representation of radiomics with end‐to‐end training, we investigated the relation between the both, and discuss the reason why deep learning provided better discriminative performance on this problem.
To this end, we analyzed 401 conventional radiomic features (ICC > 0.8), against 114 deep learned radiomics (DLR) features extracted from 3D DenseNets (see the “Inference for the deep learning models” section). As illustrated in Figure 2A,D, the unsupervised clusters of both the conventional radiomics and DLR matched much with the semantic labels (Mut/WT); in other words, the continuous regions on the black‐grey bars shared numbers of similar features respectively. However, the cluster map of DLR had obviously shaper contrast than that of conventional radiomics, which indicated the DLR features are more representative and compact than the hand‐craft features. It well explained the higher classification performance of DLR than conventional radiomics.
Moreover, we associated DLR and conventional radiomics, by visualizing the pairwise Pearson correlation coefficients on the HdH test Dataset with a cluster map in Figure 2B. Though the radiomic features and DLR features do not highly correlate (correlation coefficients range from −0.5 to +0.5), the cluster map indicated relatively high correlation (bright red or blue) between DLR and radiomics. It is worth noting that, most of the radiomic features were potentially correlated with at least one DLR feature, while certain DLR features showed low correlation with all the radiomic features, which potentially corresponded to the extra high‐level information with more hints on modeling the EGFR status. Considering that our DLR‐based method showed better performance, it is reasonable to assume that the DLR features encoded automatically not only almost all information of the (highly correlated) conventional radiomics with even more compact representation (more informative with lower feature dimension), but also critical discriminative information not in the conventional radiomics.
To verify our assumption further, we used the 114 DLR features together with the 401 radiomics features to fit a cross validation searched logistic regression model as in our conventional radiomics analysis. An automatic machine learning tool (AutoML)36 was also applied for searching a best‐performing model. The resulting model achieved AUCs of 71.2% and 74.2% on the HdH test Dataset and TCIA Dataset, respectively, which performs poorer than using DLR alone. It implied that, combining the conventional radiomics and DLR naïvely could not boost the performance, due to the high correlation between the both. Note that the feature selection procedure to reduce the redundancy between the features had been included in the AutoML search procedure.
3.3. Ensemble vs vanilla (no ensemble)
The inference‐stage data augmentation by representing a nodule in multiple views, was not only reasonable from a medical perspective, but also made sense in our empirical results. As depicted in Table 4, this simple ensemble technique was beneficial to reducing the inference variance. The vanilla term referred to inference with a single forward. The ensemble inference produced a more stable performance, on the HdH Dataset (training, development, test) and TCIA Dataset, though the vanilla inference might produce even better results in some cases.
Table 4.
Dataset summary and prediction performance of deep learning systems on HdH Dataset (training, development and test) and TCIA Dataset
| Dataset | #Patients | EGFR+ a | AUC (w/mixup, ensemble) (%) | AUC (w/mixup, vanilla) (%) | AUC (w/o mixup, ensemble) (%) | AUC (w/o mixup, vanilla) (%) |
|---|---|---|---|---|---|---|
| HdH training dataset | 348 | 185 (53.2%) | 76.7 | 76.0 | 71.0 | 70.1 |
| HdH development dataset | 116 | 61 (52.6%) | 74.1 | 74.6 | 69.2 | 70.4 |
| HdH test dataset | 115 | 62 (53.9%) | 75.8 | 76.8 | 67.9 | 67.9 |
| TCIA dataset | 37 | 9 (24.3%) | 75.0 | 68.3 | 70.6 | 71.4 |
Shown as the number of cases (percentage).
3.4. Mixup vs no mixup
The mixup technique was critical for training our 3D DenseNets. As shown in Table 4, the models with mixup training significantly outperformed those without in all of our experiments. The models without mixup training follows the same model selection procedure.
We explained the reason for these remarkable differences with the surprisingly strong regularization effects of mixup training. As demonstrated in Figure 3, the learning curves with mixup training were much smoother, and no severe overfitting was observed. On the contrary, the training without mixup was not stable; besides, there existed typical overfitting, making the model selection impractical. Considering the unreasonable effectiveness of mixup training, it could potentially be a standard technique in medical image computing in the future.
Figure 3.
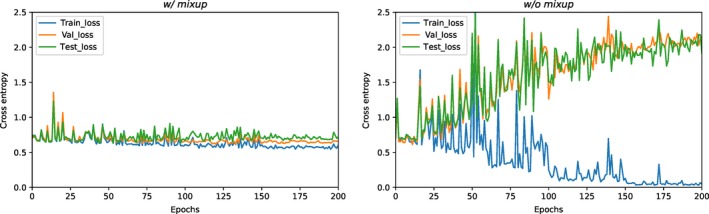
The learning curves of the best models with mixup training and those without, in terms of binary cross‐entropy loss. The losses on the HdH training, development (val) set and test Dataset were shown on the figures. “epochs” on the x‐axis means the training consumes once the entire training set
3.5. The t‐SNE visualization of the deep learned radiomics
To explore the manifold structure of the DLR intuitively, we visualized the DLR features on the HdH test Dataset using t‐Distributed Stochastic Neighbor Embedding (t‐SNE)48 as illustrated in Figure 4. No distinct pattern was shown on the t‐SNE visualization, which was reasonable considering the difficulty of using the phenotype information to predict the genotype expression. Even so, two clusters of EGFR Mut and EGFR WT could be found, see Figure 4. Most of the DLR features outside of the contours of the two clusters were assigned a score with high uncertainty by the 3D DenseNets. Nevertheless, the DLR features were shown to be meaningful and representative despite the difficulty of the task.
Figure 4.
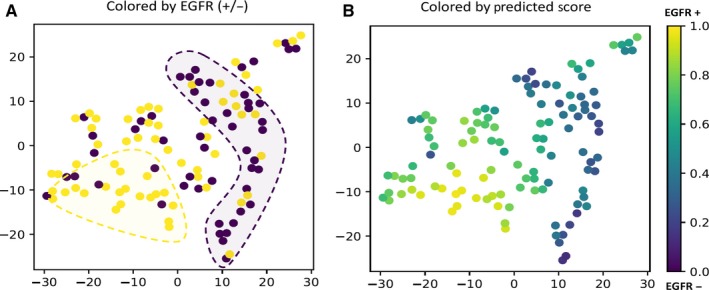
t‐SNE visualization of the deep learned radiomics in a 2D space on the HdH test Dataset. A, The t‐SNE visualization scatter plot colored by the EGFR labels. Clusters can be found labeled by the ground truth. B, The t‐SNE visualization scatter plot colored by the EGFR probability score predicted by the 3D DenseNets. The prediction scores are consistent with ground truth
4. DISCUSSION
This study developed a deep supervised learning approach to predict EGFR‐mutant lung adenocarcinoma in CT images. Compared with conventional radiomics, the models are less prone to overfitting on the limited training data and show better prediction performance. The deep learning method is also labor‐saving since it does not require precise segmentation of nodules. More importantly, we empirically find that the deep learning models learn more representative features than conventional radiomics, named deep learned radiomics (DLR), which are the keys to better analytic performances.
Our method outperformed previous studies and established a new methodology on this task. Using imaging information only, our deep 3D DenseNets achieved AUCs of 75.8% and 75.0% on the HdH test Dataset and independent public TCIA Dataset, respectively, which were better than the best of previous radiomics method (69%), and on par with their model combining additional clinical information (75%). We analyzed the associations between DLR and conventional radiomics, and empirically showed that DLR features were more representative and learned extra imaging information compared to conventional radiomics. Furthermore, we introduced the mixup training to our deep learning method, which suggested “unreasonable effectiveness” in regularizing the training of the deep neural networks with limited data.
Decoding image phenotypes using radiomics to predict tumor genotypes, called radio‐genomics as well, shows promising performances and outcomes in precision medicine research. A few recent studies have investigated radiomics analysis to noninvasively predict EGFR mutation status.16, 23 For example, Rios et al23 discriminate EGFR Mut and WT cases with an AUC of 69%, with an improvement on AUC (75%) when combining clinical information. Despite these impressive results, radiomics‐based methods are limited with expressiveness of the hand‐craft features; besides, the requirement of manual segmentation makes it difficult to apply in practical clinical contexts.
In contrast, our deep learning method is automatic in predicting the EGFR mutation status, which only requires an approximate location of nodule instead of labor‐intensive voxel‐wise segmentation. More importantly, the resulting DLR features are learned automatically with end‐to‐end training. In our association analysis, we found that some DLR features were not correlated with any radiomic features in our experiments, implying extra information extracted by our 3D DenseNets. Powered by dense connections31 and mixup training,32 our method achieved a state‐of‐the‐art performance. Considering that sensitizing EGFR mutations are more likely found in Asian patients,2 a selected subset from TCIA database33 was used for testing the generality of our system. Our deep learning system showed robust performance on the public data set.
Radiomic features, defined by image analysis algorithms on numbers of visual characteristics, such as shape, volume, and intensity, are low‐level features without high‐level semantic information. However, these features are naturally “grey‐box” interpretable. On the other hand, DLR features are semantically high‐level thanks to the end‐to‐end learning, yet deep learning models are known to be black‐box artificial intelligence lack of the desirable interpretability, especially in the medical contexts. Our association analysis not only provides insight on understanding the DLR features, but also approaches opening the black‐box of deep learning in medical image computing by a grey‐box model. However, it is hardly possible to identify the specific correlation between radiomics and DLR, we leave this for further exploration.
There were limitations in our study. Due to resource limitation, EGFR mutations were detected with ARMS‐PCR covering specific loci, thus EGFR mutations were narrowly defined in this study. Further studies with mutations detected by second‐generation sequencing are needed. Besides, information within the CT images is relative limited; integrating more available information of patients, such as clinicopathological facets, blood testing result, proteomics, and even the lifestyles, into the models can be beneficial to inferring the genotypes in multiple views. Recently, liquid biopsy, using blood as opposed to tumor samples for molecular analysis, was developed to identify EGFR mutations with promising results.49, 50 Taking advantage of these valuable information or integrating different promising approaches may improve the performance of predicting the EGFR mutation status. We consider it as a future direction. Moreover, the sample size of the independent validation was still small, and our deep leaning approach should be tested in larger cohorts. Also, this was a single‐center study, our deep learning systems desire larger datasets with more diversity. Further improvement can be made to practically help scalable precision medicine; however, the methodology established by this study, could serve as a paradigm for future studies. Lastly, only thin‐slice CT images were included in the current study to mitigate the radiomic feature variabilities. However, whether the deep learning method could perform better with different slice thickness CT images is worth further investigating. Actually, this is one of our ongoing research.
In conclusion, the proposed deep learning system predicts EGFR‐mutant lung adenocarcinoma in CT images automatically and noninvasively with promising performance, indicating the potential to help clinical decision‐making by identifying eligible patients of pulmonary adenocarcinoma for EGFR‐targeted therapy. The association study between conventional radiomics and deep learned radiomics discusses the relation between the both, and approaches toward a grey‐box explanation methodology of black‐box model.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
AUTHORS CONTRIBUTION
Conception and design: W. Zhao, J. Yang, Y. Hua, M. Li. Development of methodology: W. Zhao, J. Yang, D. Bi, B.Ni, M.Xu, Y. Hua, M. Li. Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc): W. Zhao, J. Yang, X. Zhu, Y. Sun, L. Jin, C. li, P. Gao, Y.Hua, M. Li. Analysis and interpretation of data (eg, statistical analysis, biostatistics, computational analysis):W. Zhao, J. Yang, D.Bi, Y. Hua, M. Li. Writing, review, and/or revision of the manuscript: All authors Administrative, technical, or material support (ie, reporting or organizing data, constructing databases): W. Zhao J. Yang, P. Wang, Y. Hua, M. Li. Study supervision: W. Zhao, B. Ni, P. Wang, Y. Hua, M. Li. Other (algorithm and software development): J. Yang.
ETHICAL APPROVAL
This retrospective study was approved by the Institutional Review Board of our institution (NO.20170103), which waived the requirement for patients’ informed consent referring to the CIOMS guideline.
Supporting information
ACKNOWLEDGMENTS
The authors would like to thank Dongfang Tang and Feng Gao for assisting in manuscript writing. We thank Yuxiang Ye and Liang Ge in Diannei Technology for generous help in data and insightful discussion.
Zhao W, Yang J, Ni B, et al. Toward automatic prediction of EGFR mutation status in pulmonary adenocarcinoma with 3D deep learning. Cancer Med. 2019;8:3532–3543. 10.1002/cam4.2233
Zhao and Yang contributed equally to this work.
Funding information
This study was supported by the Research Program of Shanghai Hospital Development Center SHDC22015025 (M. Li), the National Key Research and Development Program of China 2017YFC0112800 (P. Wang), 2017YFC0112905 (M. Li), the National Science Foundation of China 61502301 (B. Ni) and the Medical Imaging Key Program of Wise Information Technology of 120, Health Commission of Shanghai 2018ZHYL0103 (M. Li). This study was supported by SJTU‐UCLA Joint Center for Machine Perception and Inference (B. Ni and J. Yang). The study was also partially supported by China's Thousand Youth Talents Plan, STCSM 17511105401, 18DZ2270700 (B. Ni).
Contributor Information
Yanqing Hua, Email: 362186805@qq.com.
Ming Li, Email: minli77@163.com.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. [DOI] [PubMed] [Google Scholar]
- 2. Ettinger DS, Wood DE, Aisner DL, et al. Cell lung cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2017;15(4):504‐535. [DOI] [PubMed] [Google Scholar]
- 3. Kwak EL, Bang Y‐J, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non‐small‐cell lung cancer. N Engl J Med. 2010;363(18):1693‐1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1‐rearranged non‐small‐cell lung cancer. N Engl J Med. 2014;371(21):1963‐1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meng D, Yuan M, Li X, et al. Prognostic value of K‐RAS mutations in patients with non‐small cell lung cancer: a systematic review with meta‐analysis. Lung Cancer. 2013;81(1):1‐10. [DOI] [PubMed] [Google Scholar]
- 6. Pao W, Girard N. New driver mutations in non‐small‐cell lung cancer. Lancet Oncol. 2011;12(2):175‐180. [DOI] [PubMed] [Google Scholar]
- 7. Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31(27):3327‐3334. [DOI] [PubMed] [Google Scholar]
- 8. Li T, Kung HJ, Mack PC, Gandara DR. Genotyping and genomic profiling of non‐small‐cell lung cancer: implications for current and future therapies. J Clin Oncol. 2013;31(8):1039‐1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non‐small‐cell lung cancer with mutated EGFR. N Engl J Med. 2010;362(25):2380‐2388. [DOI] [PubMed] [Google Scholar]
- 10. Miller VA, Riely GJ, Zakowski MF, et al. Molecular characteristics of bronchioloalveolar carcinoma and adenocarcinoma, bronchioloalveolar carcinoma subtype, predict response to erlotinib. J Clin Oncol. 2008;26(9):1472‐1478. [DOI] [PubMed] [Google Scholar]
- 11. Mok TS, Wu Y‐L, Thongprasert S, et al. Gefitinib or carboplatin‐paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947‐957. [DOI] [PubMed] [Google Scholar]
- 12. Bedard PL, Hansen AR, Ratain MJ, Siu LL. Tumour heterogeneity in the clinic. Nature. 2013;501(7467):355‐364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Mol Diagn. 2013;15(4):415‐453. [DOI] [PubMed] [Google Scholar]
- 14. Kim TO, Oh IJ, Kho BG, et al. Feasibility of re‐biopsy and EGFR mutation analysis in patients with non‐small cell lung cancer. Thorac Cancer. 2018;9(7):856‐864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kobayashi K, Naoki K, Manabe T, et al. Comparison of detection methods of EGFR T790M mutations using plasma, serum, and tumor tissue in EGFR‐TKI‐resistant non‐small cell lung cancer. Onco Targets Ther. 2018;11:3335‐3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yip S, Kim J, Coroller TP, et al. Associations between somatic mutations and metabolic imaging phenotypes in non‐small cell lung cancer. J Nucl Med. 2017;58(4):569‐576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu Y, Kim J, Balagurunathan Y, et al. Radiomic features are associated with EGFR mutation status in lung adenocarcinomas. Clin Lung Cancer. 2016;17(5):441‐448.e446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aerts H, Grossmann P, Tan Y, et al. Defining a radiomic response phenotype: a pilot study using targeted therapy in NSCLC. Sci Rep. 2016;6:33860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Valentini MC, Mellai M, Annovazzi L, et al. Comparison among conventional and advanced MRI, (18)F‐FDG PET/CT, phenotype and genotype in glioblastoma. Oncotarget. 2017;8(53):91636‐91653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563‐577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aerts H, Velazquez ER, Leijenaar R, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li Y, Liu X, Xu K, et al. MRI features can predict EGFR expression in lower grade gliomas: a voxel‐based radiomic analysis. Eur Radiol. 2018;28(1):356‐362. [DOI] [PubMed] [Google Scholar]
- 23. Rios Velazquez E, Parmar C, Liu Y, et al. Somatic mutations drive distinct imaging phenotypes in lung cancer. Can Res. 2017;77(14):3922‐3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tu W, Sun G, Fan LI, et al. Radiomics signature: a potential and incremental predictor for EGFR mutation status in NSCLC patients, comparison with CT morphology. Lung Cancer. 2019;132:28‐35. [DOI] [PubMed] [Google Scholar]
- 25. Aerts HJ. The potential of radiomic‐based phenotyping in precision medicine: a review. JAMA Oncol. 2016;2:1636‐1642. [DOI] [PubMed] [Google Scholar]
- 26. Krizhevsky A, Sutskever I, Hinton G.ImageNet classification with deep convolutional neural networks. International Conference on Neural Information Processing Systems; 2012.
- 27. Vaswani A, Shazeer N, Parmar N, et al. Attention is all you need. 2017.
- 28. Esteva A, Kuprel B, Novoa RA, et al. Dermatologist‐level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115‐118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316(22):2402‐2410. [DOI] [PubMed] [Google Scholar]
- 30. Zhao W, Yang J, Sun Y, et al. 3D deep learning from ct scans predicts tumor invasiveness of subcentimeter pulmonary adenocarcinomas. Cancer Res. 2018;78(24):6881‐6889. [DOI] [PubMed] [Google Scholar]
- 31. Huang G, Liu Z, Maaten L, Weinberger K. Densely connected convolutional networks. 2016.
- 32. Zhang H, Cisse M, Dauphin Y, Lopez‐Paz D. Mixup: beyond empirical risk minimization. 2017.
- 33. Gevaert O, Xu J, Hoang CD, et al. Non‐small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data–methods and preliminary results. Radiology. 2012;264(2):387‐396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26(6):1045‐1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pedregosa F, Gramfort A, Michel V, et al. Scikit‐learn: machine learning in python. J Mach Learn Res. 2012;12(10):2825‐2830. [Google Scholar]
- 36. Feurer M, Klein A, Eggensperger K, Springenberg J, Blum M, Hutter F. Efficient and robust automated machine learning. Ecol Inform. 2015;30:49‐59. [Google Scholar]
- 37. Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit‐learn: machine learning in Python. J Mac Learn Res. 2011;12:2825‐2830. [Google Scholar]
- 38. Rajpurkar P, Irvin J, Zhu K, et al. Radiologist‐level pneumonia detection on chest x‐rays with deep learning. 2017.
- 39. Ioffe S, Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift. 2015:448‐456.
- 40. Xu B, Wang N, Chen T, Li M. Empirical evaluation of rectified activations in convolutional network. Computer Science. 2015.
- 41. Keras CF. GitHub repository. 2015.
- 42. Abadi M, Barham P, Chen J, et al. TensorFlow: a system for large‐scale machine learning. 2016.
- 43. Dou Q, Chen H, Jin Y, Lin H, Qin J, Heng P. Automated pulmonary nodule detection via 3D ConvNets with online sample filtering and hybrid‐loss residual learning. International Conference on Medical Image Computing and Computer‐Assisted Intervention; 2017.
- 44. He K, Zhang X, Ren S, Sun J. Delving deep into rectifiers: surpassing human‐level performance on imagenet classification. 2015;1026‐1034.
- 45. Sutskever I, Martens J, Dahl G, Hinton G. On the importance of initialization and momentum in deep learning. Presented at: International Conference on International Conference on Machine Learning; 2013.
- 46. Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15:1929‐1958. [Google Scholar]
- 47. DeLong ER, DeLong DM, Clarke‐Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837‐845. [PubMed] [Google Scholar]
- 48. Pérezgonzález A, Vergara M, Sanchobru JL, et al. Visualizing data using t‐SNE. J Mach Learn Res. 2015;9(2605):2579‐2605. [Google Scholar]
- 49. Rolfo C, Mack PC, Scagliotti GV, et al. Liquid Biopsy for Advanced Non‐Small Cell Lung Cancer (NSCLC): a statement paper from the IASLC. J Thorac Oncol. 2018;13(9):1248‐1268. [DOI] [PubMed] [Google Scholar]
- 50. Buder A, Hochmair MJ, Schwab S, et al. Cell‐free plasma DNA‐guided treatment with osimertinib in patients with advanced EGFR‐mutated NSCLC. J Thorac Oncol. 2018;13(6):821‐830. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


