Abstract
The principal immune mechanism against biotrophic pathogens in plants is the resistance (R)-gene-mediated defence1. It was proposed to share components with the broad-spectrum basal defence machinery2. However, the underlying molecular mechanism is largely unknown. Here we report the identification of novel genes involved in R-gene-mediated resistance against downy mildew in Arabidopsis and their regulatory control by the circadian regulator, CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1). Numerical clustering based on phenotypes of these gene mutants revealed that programmed cell death (PCD) is the major contributor to resistance. Mutants compromised in the R-gene-mediated PCD were also defective in basal resistance, establishing an interconnection between these two distinct defence mechanisms. Surprisingly, we found that these new defence genes are under circadian control by CCA1, allowing plants to ‘anticipate’ infection at dawn when the pathogen normally disperses the spores and time immune responses according to the perception of different pathogenic signals upon infection. Temporal control of the defence genes by CCA1 differentiates their involvement in basal and R-gene-mediated defence. Our study has revealed a key functional link between the circadian clock and plant immunity.
The life cycles of biotrophic pathogens of plants are intimately linked with host metabolism controlled by the cycle of day and night. Hence, their interactions with the host may be dictated by the circadian clock. This is especially likely as plants do not have specialized immune cells and their immune responses have to be finely balanced with other cellular functions. However, a link between the circadian clock and plant defence has never been firmly established3.
The major plant defence strategy against biotrophic pathogens is resistance (R)-gene-mediated immunity. Detection of a pathogen-encoded virulence effector by the R protein triggers programmed cell death (PCD) and several other physiological responses collectively known as the hypersensitive response4. The effector-specific R-gene-mediated resistance may share components with the broad-spectrum basal defence machinery2. But how these components are differentially regulated is still unclear.
We chose to study resistance against Hyaloperonospora arabidopsidis (Hpa) because this obligate biotrophic oomycete pathogen causes downy mildew disease on Arabidopsis leaves through clearly defined infection steps5, allowing better dissection of the corresponding resistance mechanisms blocking these steps. The Arabidopsis Columbia (Col-0) accession is resistant to the Hpa Emwa1 isolate due to the presence of the R gene, RPP4 (ref. 6). We performed a time-course expression profiling of wild type and rpp4 (Supplementary Fig. 1) in response to Hpa Emwa1 infection. Based on the phenotype development and defence marker gene expression (Supplementary Fig. 2a, b), we identified 106 genes differentially expressed in wild type and rpp4 at 2 days post inoculation (dpi) (Supplementary Fig. 2c). These candidate genes were induced earlier than the previously reported immune regulators including EDS5, PAD4, PBS3, ICS1, NDR1 and EDS1 (ref. 7), which were known to function downstream of R gene activation.
We inoculated the T-DNA insertion mutants (from the ABRC and NASC Stock Centres) of these 106 candidate genes with Hpa Emwa1 and identified 22 mutants that displayed enhanced susceptibility compared to wild type based on sporangiophore growth and other disease symptoms (for example, chlorosis) by microscopic inspection. For most of the 22 genes, at least two homozygous mutant T-DNA alleles were tested (Supplementary Fig. 3 and Supplementary Tables 1 and 2).
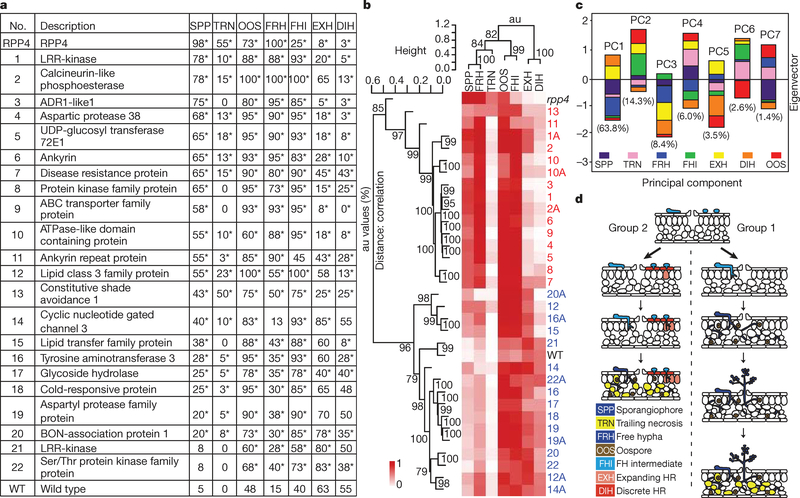
To identify specific resistance defects in the mutants, we stained the infected plants with lactophenol trypan blue (LTB) 7 dpi and scored for the occurrence of the seven phenotypes represented in Supplementary Fig. 4a. As shown in Supplementary Fig. 4b, the rpp4 mutant had the highest percentage of leaves with sporangiophores (SPP), confirming that its resistance to Hpa Emwa1 is completely compromised as SPP indicates completion of the infection cycle. Wild type had the highest score of discrete hypersensitive response (DIH), which was defined by the small cluster of infected host cells that underwent PCD, a phenotype associated with R-gene-mediated resistance. The phenotype scores are also presented numerically in Fig. 1a and the mutants are ranked on the basis of their SPP scores.
Figure 1 |. Phenotypic analyses discovered two distinct RPP4-mediated resistance responses against Hpa Emwa1.
a, Phenotype scores (percentage in 40 leaves per genotype). SPP, sporangiophore; TRN, trailing necrosis; OOS, oospore; FRH, free hypha; FHI, free hyphal intermediate; EXH, expanding hypersensitive response; DIH, discrete hypersensitive response. *P<0.05. b, Mutants were clustered on the basis of their phenotype scores in Fig. 1a. Second allele, ‘A’. Group 1, red; Group 2, blue. au, Approximately unbiased P-values (0–100%, the higher the number the more significant). c, Eigenvectors derived from PCA. The percentage of phenotypic variations captured by each PC is shown. d, A diagram showing that the Group 1 mutants are defective in RPP4-mediated PCD, whereas the Group 2 mutants are compromised in formation of physical/chemical barriers with intact PCD.
Hierarchical clustering of the mutants using their phenotype scores (Fig. 1a) put these 22 gene mutants into two groups (Fig. 1b). Similar groupings are also obtained with data from three biological replicates (Supplementary Fig. 5). Eigenvectors derived from principal component analysis indicate that 63.8% of the phenotype variations could be accounted for by PC1 with indicators of resistance, DIH and expanding hypersensitive response (EXH), as positive contributors and disease phenotypes, SPP and free hypha (FRH), as negative contributors (Fig. 1c). If PC2 was also considered, six out of the seven phenotypes had significant contributions.
The Group 1 mutants (red numbers in Fig. 1b and Supplementary Fig. 5) seem to be defective in R-gene-mediated PCD (low DIH and EXH scores) with high disease symptoms (FRH and SPP). In contrast, the Group 2 mutants (blue numbers) appeared to be intact in PCD with high EXH and DIH scores and milder symptoms (low FRH and SPP scores). To determine the resistance defects in Group 2 mutants, we examined them for other R-gene-mediated physiological responses, such as accumulation of phenolic compounds involved in cell wall strengthening against pathogen penetration and deposition of callose after Hpa Emwa1 inoculation. We found that Mutant 16 (tyrosine aminotransferase 3) was defective in phenolic compound accumulation at the site of pathogen penetration (Supplementary Fig. 6a) and Mutants 12 (lipase class 3 family protein) and 14 (cyclic nucleotide gated channel 3) showed a deficiency in callose deposition in response to Hpa Emwa1 similar to that observed in rpp4 (Supplementary Fig. 6b).
Collectively, these observations suggest that RPP4 regulates at least two separate responses (Fig. 1d): Group 1 genes are required for R-mediated PCD, as mutations in these genes led to low DIH and EXH scores and formation of FRH, TRN, and SPP. The Group 2 genes are probably involved in defence responses other than PCD, such as callose deposition and phenolic compound accumulation. Loss of these latter functions resulted in pathogen penetration even in the presence of PCD. One important conclusion from these data is that PCD is the predominant resistance response against Hpa Emwa1 because the Group 1 mutants were more susceptible, on the basis of the SPP scores (except Mutant 12), than the Group 2 mutants (Fig. 1a, b). This is supported by the eigenvector composition where PC1 (DIH and EXH) was the major contributor to the phenotypic variations (Fig. 1c). This finding is consistent with the fact that Hpa Emwa1 is an obligate biotrophic pathogen. Suicidal death of the host cell means the end of the pathogen life cycle. The functional diversity of the Group 1 genes indicates that RPP4-mediated PCD is orchestrated by changes in multiple biological processes, rather than a single triggering event.
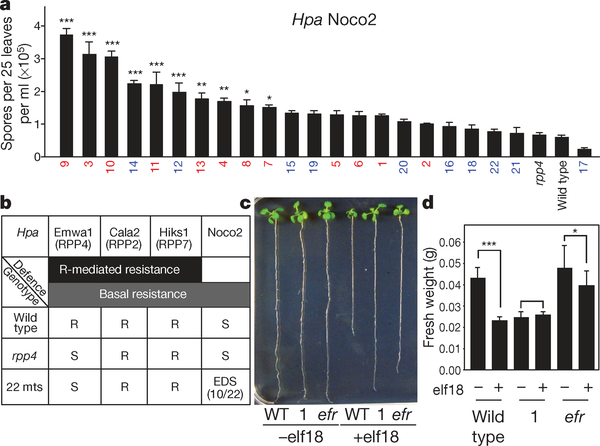
We also subjected the 22 defence gene mutants to infection by the virulent isolate, Hpa Noco2, to which a cognate R gene is absent in Col-0. We found that 10 of the mutants displayed significantly enhanced disease susceptibility (Fig. 2a) demonstrating that these defence genes are involved in both R gene-specific PCD and general basal resistance. To determine whether the observed defect in Hpa Emwa1 resistance is RPP4-specific or due to compromised basal defence, we infected the mutants with Hpa isolates, Cala2 and Hiks1, which are known to have cognate R genes, RPP2 and RPP7, respectively, in the Col-0 background7,8. None of the mutants showed compromised resistance (Fig. 2b and Supplementary Table 1) indicating that the deficiency in resistance against Hpa Emwa1 is RPP4 gene-specific. However, a few of the mutants did show defects in RPS2-mediated resistance to a bacterial pathogen Pseudomonas syringae pv. maculicola ES4326 carrying AvrRpt2 (Psm ES 4326/AvrRpt2) (Supplementary Fig. 7a). Three of them were also hypersusceptible to Psm ES4326 in the absence of the AvrRpt2 signal (Supplementary Fig. 7b).
Figure 2 |. Some of the RPP4-mediated resistance mutants are also compromised in basal defence.
a, Enhanced disease susceptibility to Hpa Noco2 based on sporangiospore count 7 dpi (n=3). b, Summary of the infection tests on the 22 defence gene mutants (22 mts) using different Hpa isolates. S, susceptible; R, resistant; EDS, enhanced disease susceptibility. c, Root length measurements 9 days after elf18 treatment (n=3). efr, efl18 receptor mutant. d, Fresh weight measurements 6 days after elf18 treatment (n=3). *P<0.05, **P<0.01, ***P<0.001.
We next subjected all of the 22 mutants to microbial-associated molecular pattern (MAMP) treatments including EF-Tu (elf18) and flagellin (flg22) to examine the interconnection between R-gene-mediated resistance and MAMP-triggered basal immunity. We found that Mutant 1, mutated in the leucine-rich repeat receptor-like kinase (LRR-RLK; AT1G35710), was insensitive to elf18 (Fig. 2c, d). Because Mutant 1 also showed the highest level of susceptibility to Hpa Emwa1 (Fig. 1a), we propose that this LRR-RLK is a link between MAMP-signalling and RPP4-mediated PCD and resistance. Although MAMP-triggered immunity is not typically associated with PCD, MAMP signalling components have been implicated previously in PCD. Mutation of the Arabidopsis BRI1-associated receptor kinase 1 (BAK1), which is a MAMP-coreceptor required for responses to elf18 and flg22 (ref. 9), was shown to cause spreading necrosis upon pathogen challenge, indicating that BAK1 is an inhibitor of PCD10. However, silencing BAK1 in Nicotiana benthamiana blocked the cell death induced by the oomycete elicitor INF1 (ref. 11), consistent with our finding that a MAMP signalling component is involved in PCD resistance against oomycete infection.
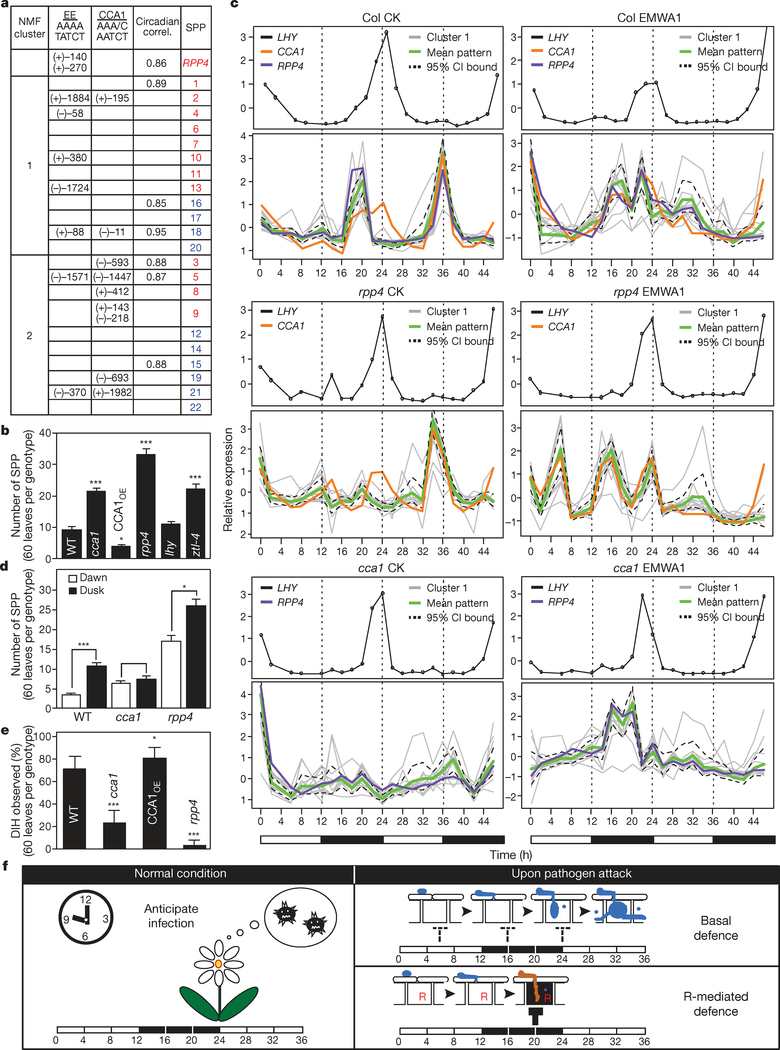
Our genetic data showed that R-mediated resistance and basal defence share common components. This raises the question of how activation of similar sets of genes causes PCD in RPP4-specific resistance against Hpa Emwa1 and non-specific basal resistance against Hpa Noco2. To understand the differential regulation of these immune mechanisms, we analysed the promoter regions of these 22 genes. Using the Athena program (http://www.bioinformatics2.wsu.edu/Athena/), we found significant enrichment of the ‘evening element’, which is regulated both positively and negatively by the circadian regulator, CCA1 (refs 12, 13). Further examination showed that 14 of the 22 genes contain either evening element and/or the CCA1-binding site and/or have rhythmic expression patterns (Fig. 3a)14. Interestingly, the promoter region of RPP4 also contains two evening elements and its expression shows a circadian rhythm.
Figure 3 |. The circadian regulator, CCA1, controls the defence gene expression and the timing of immune responses.
a, Enrichment of evening element (EE) (P<10−5). NMF, non-negative matrix factorization; CCA1, CCA1-binding sites; Circadian correl., circadian correlations14. +, sense; −, antisense. b, SPP count 7 dpiby Hpa Emwa1 (n=3). c, Time-course expression of NMF Cluster 1 genes. CI, confidence interval; CK, control; EMWA1, Hpa Emwa1 inoculated. White bars, day; black bars, night. d, SPP count after Hpa Emwa1 infection at dawn or dusk (n=3). e, Occurrence of DIH 7 dpi by Hpa Emwa. f, A model showing circadian regulation of the defence genes in anticipation of infection under normal conditions, in basal and R-gene-mediated resistance. The blocked arrows represent defence against infection.
To confirm the involvement of the circadian clock in defence, we first examined the responses of clock mutants to Hpa Emwa1. The infection was carried out at dawn, the time when Hpa spores are normally disseminated in nature15. The cca1 mutant (Salk_067780) and ztl-4 (a mutant of ZEITLUPE)16 showed compromised resistance whereas a CCA1-overexpression line (CCA1OE)17 showed enhanced resistance (Fig. 3b). Surprisingly, lhy, the mutant of the CCA1 homologue, LATE AND ELONGATED HYPOCOTYL (LHY)18 responded as wild type.
We next examined the expression patterns of all 22 defence genes in wild type, rpp4 and cca1 every 2 h in a 46-h time-course, with and without infection by Hpa Emwa1. Because of the large number of samples involved, we used the innovative high throughout RNA annealing selection ligation-sequencing (RASL-seq) technology19 for expression analysis (Supplementary Table 3 and Methods).
As shown in Fig. 3c, consistent with the genetic data, the rhythmic expression of LHY was not significantly perturbed by infection in either wild type or rpp4. This indicates that RPP4-mediated defence does not disrupt the overall running of the clock, but rather engages CCA1. This specific sensitivity of CCA1 to infection conditions was confirmed using a transgenic line expressing CCA1:LUC (ref. 16, Supplementary Fig. 8a). To eliminate the effects of light changes on CCA1 expression, we also performed infection in transgenic plants carrying the CCA1:LUC and LHY:LUC reporters16 under the free-running light cycles (Supplementary Fig. 8b). Similar to the RASL-seq results, LHY:LUC expression remained unchanged, whereas CCA1:LUC expression was significantly induced and became arrhythmic upon Hpa Emwa1 challenge.
Conveniently, the stable expression pattern of LHY served as an internal control for the quality of RNA preparations and RASL-seq. Based on the non-negative matrix factorization (NMF) algorithm20, the 22 RPP4-regulated genes fit best into two clusters (Fig. 3a and Supplementary Figs 9 and 10). The membership distance of each gene to its cluster is illustrated by the circle radius in Supplementary Fig. 11a. These two clusters corresponded roughly to the two phenotypic groups determined through genetic analysis (Figs 1 and 3a). Most of the Cluster 1 genes containing evening element in their promoters are involved in R-gene-mediated PCD and were therefore the focus of further concern (Fig. 3c). The expression patterns of the Cluster 2 genes are shown in Supplementary Fig. 11b.
Consistent with the fact that evening element is enriched in the Cluster 1 gene promoters (Fig. 3a), the weighted mean expression of these genes largely overlaps with the expression patterns of CCA1 (Fig. 3c). In wild-type control (Col CK), Cluster 1 genes showed a rhythmic expression pattern with a single sharp peak every evening. In cca1 (cca1 CK), the expression peaks were greatly diminished, confirming that CCA1 is an activator of these defence genes.
The rhythmic expression of the defence genes in the absence of pathogen indicates that plants are programmed to ‘anticipate’ infection according to a circadian schedule. The CCA1-mediated pulse expression of the defence genes coincides with the time of Hpa sporulation which mainly occurs at night and the time of spore dissemination which takes place at dawn15. To test this, we performed Hpa Emwa1 infection not only at the normal ‘dawn’ infection time but also at ‘dusk’. We found that if the plants were inoculated at dusk, when infection was unexpected, significantly higher levels of susceptibility were observed in both wild type and rpp4 (Fig. 3d). CCA1 clearly has a role in conferring resistance at dawn because in cca1, more Hpa Emwa1 growth was observed compared to wild type. However, no further increase in susceptibility was observed in cca1 if inoculation was carried at dusk because CCA1 and the CCA1-regulated defence genes are not expressed at this time.
In response to Hpa Emwa1 infection, Cluster 1 genes showed drastically different expression patterns in wild type (Col EMWA1) and rpp4 (rpp4 EMWA1) (Fig. 3c). Without RPP4, the expression of the defence genes peaked at the 6-, 16- and 24-h time points which coincided with the expected time of Hpa spore germination, formation of penetration hyphae and establishment of primary haustoria in mesophyll cells, respectively5. This pattern of expression may explain how these defence genes contribute to the basal resistance against Hpa infection. Consistent with CCA1 having a role in basal resistance, CCA1oe was more resistant to Hpa Noco2 than wild type (Supplementary Fig. 12). However, understanding the signalling events between the pathogen and CCA1 leading to this specific timing of defence gene expression will require future research.
In the presence of RPP4, the 6-h expression peak was diminished (Col EMWA1, Fig. 3c). The subsequent perception of the pathogen effector by RPP4 led to the gradual and sustained expression of defence genes. We propose that the prolonged expression of these defence genes, which are normally pulse-expressed at dawn, results in PCD of the infected host cells and pathogen resistance. Consistent with it being a key positive regulator of Cluster 1 defence genes involved in RPP4-mediated PCD, knocking out CCA1 function significantly lowered the average DIH score (a measure of host cell death) in cca1 after Hpa Emwa1 infection (Fig. 3e).
How RPP4 interacts with CCA1 to control the defence gene expression requires further investigation. The spatial resolution of the expression data from the Hpa Emwa1-infected samples, homogenized from both infected and uninfected cells, was not enough to allow detailed dissection of the contribution of RPP4, CCA1 and unknown pathogenic signals. Nevertheless, RPP4 clearly is not only a target gene of CCA1, but also a partner of CCA1 in regulating the defence genes as their patterns of expression were disturbed in rpp4 (Fig. 3c and Supplementary Fig. 2c).
Establishment of a molecular link between the plant circadian clock and R-mediated defence reveals a new interface between the plant host and biotrophic pathogens. Although the interactions between R genes and the circadian clock have yet to be studied genetically and at the molecular level, this study indicates a central role of the circadian clock in balancing growth and defence. As summarized in Fig. 3f, we hypothesize that the Cluster 1 genes are pulse-expressed to minimize adverse effects to the host in anticipation of infection under normal conditions and during basal defence. In contrast, detection of a pathogenic effector by the R protein may disrupt this control, leading to PCD of the infected cell and R-mediated resistance, which is a much stronger and signal-specific immune response. There is also an increasing body of evidence indicating that animal immune response is influenced by the circadian clock21. Understanding the molecular link between the circadian clock and immunity therefore has broad implications in biology.
METHODS SUMMARY
Hyaloperonospora arabidopsidis (Hpa) propagation and inoculation were performed as described6,22. Ten-day-old plants were inoculated with the asexual spores suspension (5 × 105 spores per ml) of Hpa. Unless specified, the Hpa infection was always performed at dawn of the growth chamber’s photoperiod. Hpa Emwa1-inoculated samples were collected at 0, 0.5, 2 and 4 days post inoculation (dpi). ATH1 GeneChip (Affymetrix) was used for microarray. The arrays were normalized and analysed as described previously23. Disease phenotypes were scored after trypan blue staining at 7 dpi24. Significance of the phenotypic scores was determined based on binomial distribution. Disease phenotypic analysis was performed using hierarchical clustering with distance measured by the standard correlation (average linkage; scale 0–1). The significance of the clustering (bootstrap 100,000 times) was measured by the approximately unbiased P-values (0–100%, the higher the number the more significant25). Callose deposition was detected after aniline blue staining26. Accumulation of phenolic compounds was examined under ultraviolet illumination (Leica). Root length and fresh weight assays for elf18 sensitivity were performed as described previously9. The evening element enrichment was determined based on hypergeometric distribution. Samples for RASL-seq were prepared according to ref. 19. Non-negative matrix factorization algorithm was used to cluster the genes20. RNA extraction was performed as described previously27. cDNA synthesis (Superscript III, Invitrogen) and quantitative PCR (SYBR Green, Qiagen) were performed according to the manufacturer’s protocols. For Pseudomonas infection, 4-week-old plants were inoculated with 10 mM MgCl2 or Pseudomonas syringae maculicola ES4326 with or without the effector AvrRpt2 (OD600=0.001). The in planta bacterial growth was measured at 3 dpi. For diurnal luciferase measurement, protein was extracted and bioluminescence intensity was measured using the Luciferase Assay System (Promega) according to manufacturer’s protocol. Ten-day-old plate-grown plants were used for free-running test (details in Methods).
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
METHODS
Arabidopsis and Hyaloperonospora arabidopsidis (Hpa) growth conditions
Arabidopsis seedlings were grown for 10 days at 16–18 °C, 12-h day length, 80–100% relative humidity before Hpa infection through spray of a spore suspension (5 × 105 spores per ml in distilled H2O) at dawn according to the photoperiod of the plant growth chamber. Hpa Emwa1 and Hpa Noco2 were subcultured and inocula prepared using methods modified from previous reports6,22.
RNA extraction and quantitative PCR analysis
RNA extraction was performed as described previously27. cDNA synthesis (Superscript III, Invitrogen) and quantitative PCR (SYBR Green PCR kit, Qiagen) were performed according to the manufacturer’s protocols.
Microarray
Ten-day-old wild-type and rpp4 seedlings were inoculated with Hpa Emwa1 and samples were collected at 0, 0.5, 2 and 4 days after inoculation. Total RNA was isolated from the frozen material using the Qiagen RNeasy kit. RNA probes were labelled using the GeneChip Eukaryotic Small Sample Target Labelling Assay Version II and hybridized on the Affymetrix ATH1 GeneChip (Santa Clara). Two biological replicates were performed. The data presented in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE22274 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE22274).
Normalization and mixed-model analysis
The mixed-model software used to normalize globally all arrays and to identify differentially expressed probe sets was as described previously23. Expression indices were used to calculate P- and q-values for pairwise comparisons of all probe sets across all treatments. R2 values for the CEL files are as follows (Col_0d: 1 vs 2(0.98); Col_0.5d: 1 vs 2(0.97); Col_2d: 1 vs 2(0.98); Col_4d: 1 vs 2(0.96); rpp4_0d, 1 vs 2 (0.98); rpp4_0.5d, 1 vs 2 (0.97); rpp4_2d, 1 vs 2 (0.98); rpp4_4d, 1 vs 2 (0.97)).
Phenotyping mutants in response to Hpa infection
Seven days after inoculation with Hpa Emwa1 infection, phenotypes were scored following lactophenol trypan blue staining24. Leaves were vacuum-infiltrated twice in a solution of phenol, lactic acid, glycerol and water (1:1:1:1) plus 2.5 mg ml−1 trypan blue. The tubes containing the samples were placed in a boiling water bath for 2 min and allowed to cool for overnight. The leaves were destained in the chloral hydrate solution and then treated with 70% glycerol. Whole leaves were analysed and photographed with a MZ8 stereo microscope (Leica) and a PM-C35 camera (Olympus). Detailed examination of Hpa structures was conducted with an Olympus BX60F compound microscope and differential interference contrast (DIC) optics. Leaves were stained for callose as described with modifications26. To visualize callose, leaves were cleared in a solution of ethanol and acetic acid (3:1), stained with 0.02% aniline blue in 100 mM sodium phosphate buffer (pH 9) for 1h, and examined with an Axio imager wide field fluorescence microscope (Zeiss). To detect phenolic compounds, leaves were examined under ultraviolet fluorescent illumination (Leica DMRB). To measure Hpa Noco2 infection, infected leaves were collected in 1 ml water, and sporangiospores were counted.
elf18 and flg22 treatment
Wild-type Arabidopsis (Col-0), efr and candidate mutant plants were grown on MS11% sucrose medium (pH 5.7) plate with 1 μM elf18 or MS alone as a control under continuous light for 9 days for root length assay9. For fresh weight assay, plants were grown on MS11% sucrose medium (pH 5.7) plate for 4 days (16/8 light/dark cycle), transferred into water containing 50 nM elf18 or water as a control, and fresh weight was measured 6 days after elf18 treatment. flg22 (10 nM) was infiltrated into 3-week-old plants for the induction of callose deposition.
Promoter element analysis
The statistical significance of over-represented transcription factor (TF) binding elements was calculated using a hypergeometric probability model. The following equation was used to provide the P-values:
N is the total number of promoters in the genome, n is the number of promoters in the genome containing the specified TF-binding element, m is the size of the selected set of promoters, and x is the number of promoters with the specified element in the selected set. Because multiple hypotheses were tested in the analysis, the Bonferroni correction was used. The genome-wide occurrences of these elements in the promoters are used as controls.
RASL-seq
The growth conditions (12/12 light/dark cycle, 16–18 °C, 80–100% humidity), which were optimized for Hpa infection, were different from those used in traditional circadian studies12,13. Samples were collected every 2 h after inoculation and the remaining plants were kept to ensure successful pathogen inoculation. Total RNA for each sample (1 μg) was used for RASL-seq. Primer (gene-specific with flanking 5′ or 3′ universal sequences) annealing to mRNA and ligation were carried out according to ref. 19. Bar-coded primers were then added to each sample to convert the ligated products to individual libraries, which were pooled from all samples and subjected to multiplex sequencing using Solexa GAII (Illumina).
RASL-seq data analysis
The readings from RASL-seq were assumed Poisson distribution. Only those samples with mean readings significantly above zero (Pr (mean=0)<0.01) were considered for further analysis. The reading for each sample was first divided by the corresponding reading of control, ubiquitin 5 (UBQ5; AT3G62250), and then standardized. The resulting matrix was used for clustering analysis.
Non-negative matrix factorization (NMF) algorithm20 was used to cluster the genes. The number of the clusters was determined by comparing the cophenetic correlation coefficient for a range of cluster numbers (from 2 to 22). The cophenetic correlation coefficient is a measurement of how faithfully the result of NMF clustering preserves the pairwise distances between the original data points. As shown in Supplementary Fig. 9, two clusters generated the highest cophenetic correlation coefficient, which means two clusters can reflect the original data more faithfully than more clusters. Divergence was used as the update rule and cost measurement. Minimum of the data was subtracted from the data matrix to ensure that there were no negative numbers in the matrix. Because the NMF algorithm iteratively updates the decomposition of the data matrix, 300 runs with 10,000 iterations/run were performed to reach the convergence (Supplementary Fig. 10). The membership indicators from NMF clustering were used as weights to calculate the weighted mean expression pattern shown in Fig. 3c. The weights were also used to determine the radii of circles in Supplementary Fig. 11a. Smaller radius indicates a higher membership of the gene to the corresponding cluster.
Bioluminescence detection
Protein was extracted and bioluminescence intensity measured using the Luciferase Assay System (Promega) according to the manufacturer’s manual. A Victor3 (PerkinElmer) multilabel reader was used to detect the bioluminescence. Substrate (100 μl) was added using an automatic injector. After 3 s shaking, 2 s delay, the signal was captured for 20 s. Log10 transformation was performed to the raw signals to ensure the normal distribution of the data. After subtraction of the blank, the data were normalized according to the total protein concentrations determined by the Bradford method (Bio-Rad). The resulting data were then standardized.
Free-running test
Seeds were sterilized in 2% Plant Preservative Mixture (PPM, Plant Cell Technology) in the dark at 4 °C for 4 days before plating on MS plate (3% sucrose, 1.5% agar) and grown in a 12/12 h light/dark growth chamber for 9 days. At the dawn and the dusk of the ninth day, 2.5 mM luciferin in 0.05% Triton-X 100 was sprayed onto the seedlings. At the dawn of the tenth day, the seedlings were treated by distilled H2O or Hpa Emwa1 before being placed in a constant light chemiluminescence box. The bioluminescence signals were captured by CCD.
Supplementary Material
Acknowledgements
We thank S. Brady for performing mixed model ANOVA of the data and advice on data analyses; J. Li for sharing the protocol for elf18 treatment, E. Tobin for providing the CCA1OE transgenic line, R. McClung for CCA1:LUC, LHY:LuC, ztl-4 lines. H. Lu for helpful discussion of the work, F. Ausubel, P. Benfey, S. Brady, J. Siedow and R. Mohan for critiquing the manuscript This work was supported by a grant from NSF (MCB-0519898) to X.D. and a grant (HG004659) to X.-D.F.
Footnotes
The authors declare no competing financial interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Jones JD & Dangl JL The plant immune system. Nature 444, 323–329(2006). [DOI] [PubMed] [Google Scholar]
- 2.Tao Y et al. Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15, 317–330 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roden LC & Ingle RA Lights, rhythms, infection: the role of light and the circadian clock in determining the outcome of plant-pathogen interactions. Plant Cell 21, 2546–2552 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lam E, Kato N & Lawton M Programmed cell death, mitochondria and the plant hypersensitive response. Nature 411, 848–853 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Donofrio NM & Delaney TP Abnormal callose response phenotype and hypersusceptibility to Peronospoara parasitica in defence-compromised Arabidopsis nim1–1 and salicylate hydroxylase-expressing plants. Mol. Plant Microbe Interact 14, 439–450 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Holub EB, Beynon JL & Crute IR Phenotypic and genotypic characterization of interactions between isolates of Peronospora parasitica and accessions of Arabidopsis thaliana. Mol. Plant Microbe Interact 7, 223–239 (1994). [Google Scholar]
- 7.van der Biezen EA, Freddie CT, Kahn K, Parker JE & Jones JD Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J. 29, 439–451 (2002). [DOI] [PubMed] [Google Scholar]
- 8.McDowell JM et al. Downy mildew (Peronospora parasitica) resistance genes in Arabidopsis vary in functional requirements for NDR1, EDS1, NPR1 and salicylic acid accumulation. Plant J. 22, 523–529 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Chinchilla D et al. Aflagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Kemmerling B et al. The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr. Biol 17, 1116–1122 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Heese A et al. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl Acad. Sci. USA 104, 12217–12222 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harmer SL et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Harmer SL & Kay SA Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell 17, 1926–1940 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michael TP et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet 4, e14 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slusarenko A & Schlaich NL Downy Mildew of Arabidopsis thaliana caused by Hyaloperonospora parasitica (formerly Peronospora parasitica). Mol. Plant Pathol 4, 159–170 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Salome PA & McClung CR PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17, 791–803 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang ZY & Tobin EM Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Schaffer R et al. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93, 1219–1229 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Yeakley JM et al. Profiling alternative splicing on fiber-optic arrays. Nature Biotechnol. 20, 353–358 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Tamayo P et al. Metagene projection for cross-platform, cross-species characterization of global transcriptional states. Proc. Natl Acad. Sci. USA 104, 5959–5964 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bryant PA, Trinder J & Curtis, N. Sick and tired: does sleep have a vital role in the immune system? Nature Rev. Immunol 4, 457–467 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Tor M et al. Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew resistance genes. Plant Cell 14, 993–1003 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brady SM et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Bowling SA, Clarke JD, Liu Y, Klessig DF & Dong X The cpr5 mutant of Arabidopsis expresses both NPR1-dependentand NPR1-independent resistance. Plant Cell 9, 1573–1584(1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki R &Shimodaira, H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Adam L & Somerville SC Genetic characterization of five powdery mildew resistance loci in Arabidopsis thaliana. Plant J. 9, 341–356 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Cao H, Bowling SA, Gordon S & Dong X Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.