Abstract
The integrated stress response (ISR) tunes the rate of protein synthesis. Control is exerted by phosphorylation of the general translation initiation factor eIF2. eIF2 is a GTPase, that becomes activated by eIF2B, a two-fold symmetric and heterodecameric complex that functions as eIF2’s dedicated nucleotide exchange factor. Phosphorylation converts eIF2 from a substrate into an inhibitor of eIF2B. We report cryoEM structures of eIF2 bound to eIF2B in the dephosphorylated state. The structures reveal that the eIF2B decamer is a static platform upon which one or two flexible eIF2 trimers bind and align with eIF2B’s bipartite catalytic centers to catalyze nucleotide exchange. Phosphorylation refolds eIF2α, allowing it to contact eIF2B at a different interface and, we surmise, thereby sequesters it into a non-productive complex.
One Sentence Summary:
Structures of translation factors eIF2 and eIF2B reveal the mechanism of nucleotide exchange and its phosphoregulation during stress.
Numerous factors regulate translation of the genetic code into proteins, including eukaryotic translation initiation factor 2 (eIF2), a GTPase composed of α, β, and γ subunits. During initiation, eIF2 binds tRNAMet and GTP to form a ternary complex that scans mRNAs for start codons. Following start codon detection, eIF2γ hydrolyzes its GTP and translation initiates. For eIF2 reactivation, GDP is replaced by GTP upon catalysis by a dedicated guanine nucleotide exchange factor (GEF), eIF2B.
eIF2 and eIF2B control translation initiation. Stress-responsive kinases phosphorylate eIF2α at conserved Ser51, transforming eIF2 from substrate into a competitive GEF inhibitor. Phosphoregulation of eIF2 is known as the integrated stress response (ISR)(1). Once activated, the ISR reduces overall protein synthesis, while enhancing translation of a small subset of mRNAs in response to cellular threats, including protein misfolding, infection, inflammation, and starvation(1–3).
eIF2B comprises two copies each of an α, β, γ, δ, and ɛ subunit that assemble into a two-fold symmetric heterodecamer(4,5). The eIF2Bε subunit contains the enzyme’s catalytic center and associates closely with eIF2Bγ. Two copies each of the eIF2Bβ and δ subunits form the complex’s core, bridged by two eIF2Bα subunits across the symmetry interface(4,6). Genetic and biochemical studies identified residues responsible for eIF2B’s catalytic activity and suggested how eIF2 binding to eIF2B may differ following eIF2α-S51 phosphorylation(4,7–10). Yet, how eIF2 recognizes eIF2B and how eIF2B catalyzes nucleotide exchange remained unknown, as did the transformation of eIF2 from a substrate to high-affinity inhibitor of eIF2B following its phosphorylation.
A potent small-molecule, drug-like inhibitor of the integrated stress response, ISRIB, allays the effects of eIF2α phosphorylation by activating eIF2B(11–13). Upon adding ISRIB, cells undergoing the ISR resume translation(12,13). When administered to rodents, ISRIB enhances cognition and ameliorates cognitive deficits caused by traumatic brain injury(14) and prion-induced neurodegeneration(15). Furthermore, eIF2B activation rescues cognitive and motor function in mouse models of leukoencephalopathy with vanishing white matter disease (VWMD), a fatal familial disorder associated with mutations spread over all eIF2B subunits(16).
ISRIB bridges the symmetric interface of two eIF2B subcomplexes to enhance the formation of the decameric eIF2B holoenzyme(17,18), enhancing available GEF activity by promoting higher-order assembly of the eIF2B decamer. However, it has remained enigmatic why decameric eIF2B would be more active than its unassembled subcomplexes. To explore this question, we determined structures of eIF2B bound with both its substrate, eIF2α,β,γ, and inhibitor, eIF2α•P.
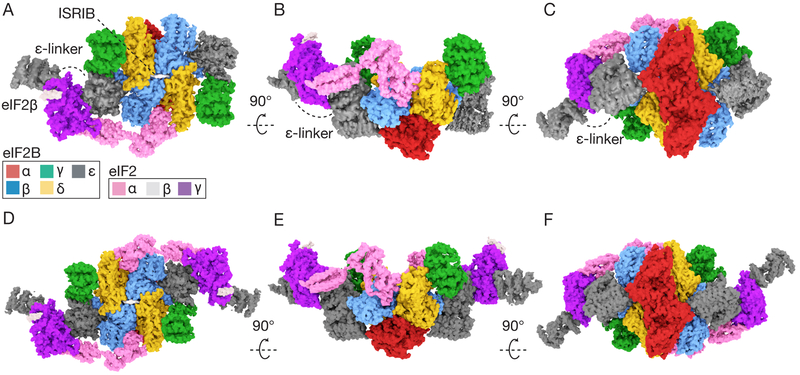
We co-expressed all five subunits of human eIF2B in E. coli and all three subunits of human eIF2 in S. cerevisiae (Fig.S1A–B). The yeast expression strain lacked GCN2, the eIF2 kinase, to ensure expression of homogenously non-phosphorylated eIF2(19). We incubated ISRIB and purified eIF2 at concentrations near the Michaelis constant of the nucleotide exchange reaction (Km = ~1.5μM,(17)) and added an inter-amine crosslinker to stabilize complexes before sample vitrification and cryoEM analysis (Fig.S2A–C). We resolved two structures: eIF2B bound asymmetrically to a single eIF2 trimer and eIF2B bound symmetrically to two eIF2 trimers (Figs.1A–F,S3A,S4–5,TableS1–S3).
Fig. 1. eIF2B heterodecamer bound to one or two eIF2 heterotrimers.
(A-C) Orthogonal views of a single versus (D-E) a pair of elongated eIF2 heterotrimers bound to ISRIB-stabilized eIF2B decamers. ISRIB density is rendered in white.
Snaking across the surface of eIF2B, we observed density consistent in size and shape with eIF2 subunits and the previously unresolved eIF2Bε HEAT domain. Comparison with homologous structures of eIF2α and eIF2γ revealed that the assembled eIF2•eIF2B complex retained similarity to the structures of these individually analyzed domains (20, 21)(Fig. 2,Fig.S6). We only resolved a single helix of eIF2β (Fig. 1A,D,Fig. 2A), consistent with other studies(20,21). In both reconstructions, all five subunits of eIF2B can be superimposed on previously determined structures lacking eIF2 (RMSD of ~0.6Å)(17). Thus, eIF2B retained its overall arrangement when bound to one or two eIF2s (Fig. 1A–F), indicating that eIF2 binds via equivalent modes to both sides of a static eIF2B scaffold with no allostery in eIF2B upon eIF2 engagement. This is consistent with non-cooperative kinetics reported for nucleotide exchange by eIF2B decamers(17).
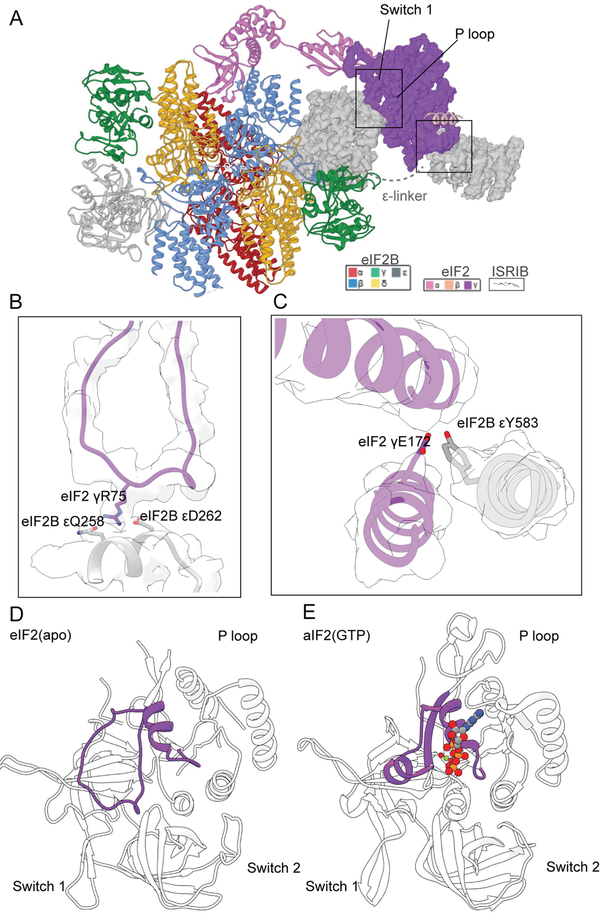
Fig. 2. The bipartite basis of guanine nucleotide exchange by eIF2B.
(A) Structural model of a single eIF2 heterotrimer bound to eIF2B decamer, emphasizing the cryoEM density for eIF2γ and its interactions with eIF2Bε. (B) Comparison of an aIF2 structure bound to GTP (PDB: 4RCY) and (C) GDP (PDB: 4RD6) with the open, nucleotide-free state of eIF2 (D) reported here.
Bound to eIF2B, eIF2 adopted an extended 150Å conformation (Figs.1–2) with eIF2’s central nucleotide-binding γ-subunit flanked by its α- and β-subunits at its opposing ends. eIF2γ contains classical GTP-binding motifs, including the nucleobase-binding G4 motif, the phosphate-binding P-loop, and switch helices 1 and 2. eIF2B recognizes eIF2 via coincident binding of both eIF2α and eIF2γ. Binding to both eIF2 subunits involves bipartite elements of eIF2B(Figs. 1,2A–C).
First, bipartite recognition of eIF2γ involves two domains of eIF2Bε that function together to splay open the nucleotide-binding site. Our nucleotide-free cryoEM model is similar to the γ-subunit of GTP-bound aIF2 from S.solfataricus(24) (Fig.2D–E, average RMSD ~2.3Å). Yet, surrounding the GTP binding pocket, the structures diverged considerably with the P-loop in eIF2B•eIF2 partially occluding the nucleotide-binding site (RMSD of ~12Å). Prior work implicated the HEAT domain in catalysis(23,24). In agreement, eIF2γ interacts with the HEAT domain, including a partially hydrophobic surface that includes eIF2Bε Y583(Fig. 2C). On the opposing side of the nucleotide-binding pocket, the central core of eIF2Bε engaged with an open loop conformation of Switch 1. This change appears due to electrostatic interactions between eIF2γ R75 in Switch 1 and Q258 and D262 in eIF2Bε. Thus, both eIF2Bε’s HEAT domain and core collaborate to open the nucleotide-binding site (Fig. 2B–D).
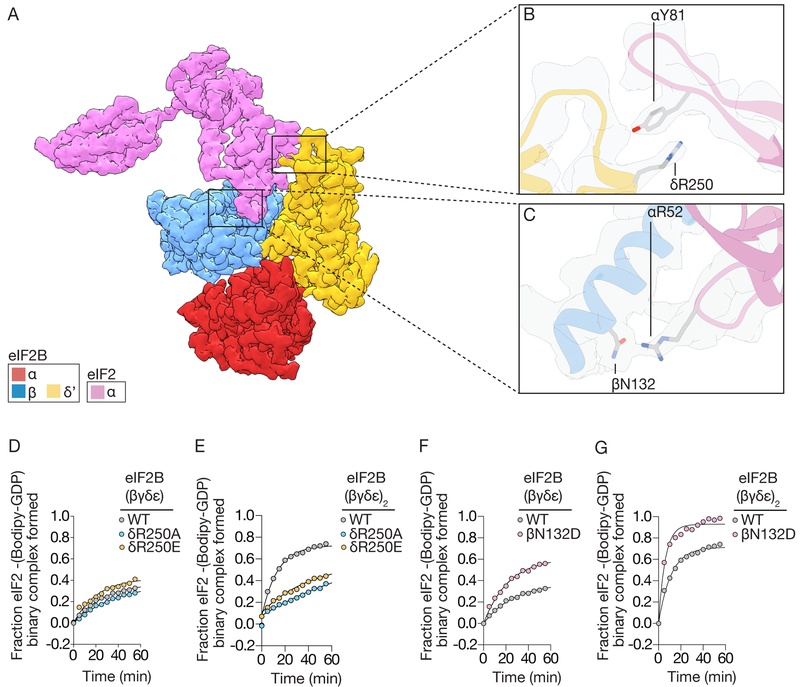
The second example of bipartite recognition concerns eIF2α binding in the cleft between eIF2Bβ and eIF2Bδ’ (δ’ indicates the δ-subunit from the opposing tetramer, Figs.1–3). Notably, this binding site only exists when two tetramers of eIF2B(βγδε) associate to form the symmetry interface in octameric eIF2B(βγδε)2. eIF2α contains two structured domains separated by a flexible linker(Fig. 1–2,S6). The N-terminus consists of an OB-fold, common in tRNA-binding proteins(20). The OB-fold is further elaborated with a positively-charged loop (the S-loop), while the C-terminal αβ-fold connects eIF2α to eIF2γ. The S-loop harbors S51 and is responsible for all of the resolvable contacts between eIF2α and eIF2B’s β subunit (Fig. 3A). Prior work implicated a conserved ‘K79GYID83’ motif in eIF2α as being important for eIF2B binding(10). Of note, an interaction between Y81 was well-resolved adjacent to the equally prominent R250 on eIF2Bδ’ (Fig. 3B). When we mutated R250 to either alanine or glutamate, neither mutation affected the residual GEF activity displayed by dissociated tetramers (Fig. 3D;R250A kobs=0.013min−1,R250E kobs=0.023min−1,wild-type kobs=0.016min−1), while both mutants diminished the GEF activity of the ISRIB-stabilized eIF2B octamer when compared to wild-type (Fig. 3E;R250A kobs=0.012min−1,R250E kobs=0.017min−1,wild-type kobs=0.063min−1). This is consistent with the notion that unphosphorylated eIF2α interacts with the trans-tetramer only upon assembly of octameric or decameric eIF2B across its symmetry interface.
Fig. 3. The bipartite basis of eIF2α recognition and assembly-stimulated activity.
(A) cryoEM density for eIF2α bound to the regulatory subcomplex (α,β,δ or RSC) of eIF2B. (B) Density and zoom-in detail of a cation-π interaction between eIF2Bδ and eIF2α. (C) polar interactions between eIF2Bβ and the S-loop of eIF2α. (D,F) GEF activity of wildtype versus mutated eIF2B(βγδε) tetramers, and (E,G) ISRIB-stabilized eIF2B(βγδε)2 octamers measured by loading of fluorescent GDP onto eIF2.
On the cis-tetramer, eIF2α’s positively-charged S-loop binds negatively charged and polar residues along the exposed surface of eIF2Bβ. This binding site is consistent with yeast studies suggesting that mutations in this site compromise eIF2 binding(7). Examination of the structure identified a potential hydrogen bond between eIF2Bβ N132 and eIF2α R52 (Fig. 3C). We substituted N132 with aspartate, anticipating that the introduced charge complementarity would enhance binding to eIF2α R52. When compared to wild-type eIF2B tetramers, eIF2B-βN132D tetramers and ISRIB-stabilized octamers indeed proved to be gain-of-function mutations, exhibiting ~two-fold enhanced GEF activity (Figs.3F–G,S1D, eIF2B(βγδε) βN132D kobs=0.044 min−1, eIF2B(βγδε)2 βN132D kobs=0.169 min−1). This is consistent with eIF2B tetramers possessing reduced activity when compared to assembled octameric or decameric holo-eIF2B. eIF2α binding in the cleft between tetramers further supports the notion that ISRIB enhances eIF2B’s GEF activity by promoting higher-order assembly.
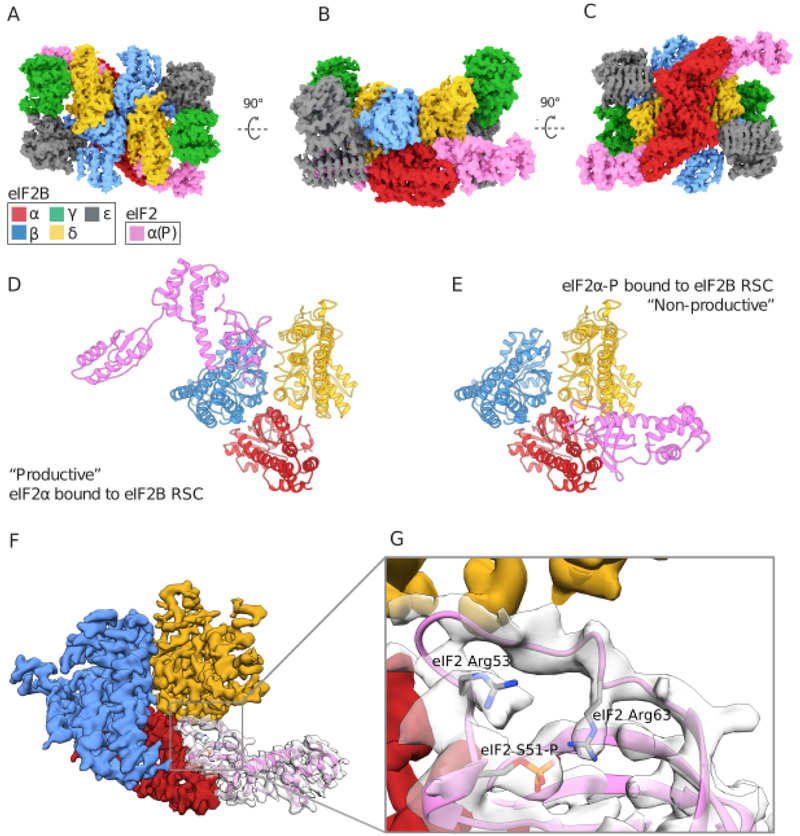
To understand how eIF2α phosphorylation on S51 transforms eIF2 from substrate to inhibitor, we co-expressed the isolated eIF2α subunit in E.coli with the kinase domain of PERK (Fig.S1C). We incubated pre-assembled eIF2B decamers with an excess of eIF2α-P, followed by crosslinking and vitrification. Reconstruction of the eIF2B decamer adorned with a two copies of eIF2α-P (Figs. 4A,S7–S8,Tables S1–S3), revealed eIF2α-P bridging the interface between eIF2Bδ and eIF2Bα(Fig. 4A). Intriguingly, we observed no overlap between the binding sites of non-phosphorylated eIF2α described above and eIF2α-P (Fig. 4B–C).
Fig. 4. The structural basis of phosphoregulation by the ISR.
(A-C) Orthogonal views of a pair of S51-phosphorylated eIF2α subunits bound to the eIF2B decamer. (D) Comparison of the productive binding mode of non-phosphorylated eIF2α, versus (E) the non-productive and non-overlapping binding mode of phosphorylated eIF2α. (F-G) CryoEM density and interpretation of the phosphorylated eIF2α binding mode and refolded conformation of the S-loop, placing the S51 phosphate moiety near eIF2 α R53 and R63.
Density for both eIF2α S51-P and two arginines positioned ~4Å away, eIF2α R53 and R63, were well-resolved and suggestive of an electrostatic coordination responsible for phosphorylation-induced refolding of the S-loop (MovieS1,Fig. 4F–G,S8), as initially observed by Kashiwagi, et al (this issue). The phosphorylation-induced rearrangement also positions hydrophobic residues on eIF2α for potential interactions with hydrophobic residues on eIF2B (including eIF2α I55, I58, and L61 and eIF2Bδ L314, A315, A318, and F322).
This structural model agrees with analyses in yeast and mammalian systems. First, eIF2Bα is dispensable for viability in yeast, yet eIF2Bα deletion impairs phospho-inhibition of eIF2B, consistent with the subunit’s role in binding eIF2α-P(27). Point mutations with identical phenotypes cluster at the interface between eIF2Bα and eIF2Bδ, e.g., eIF2Bα F239 and eIF2Bδ M506 and P508(28, 29). Importantly, eIF2Bδ L314 complements the hydrophobic surface of the eIF2α S-loop that is exposed upon refolding, and mutation of the equivalent position in S. cerevisiae, L381Q, impairs the ISR in yeast(28). These data validate the phosphorylation-induced refolding and relocation of eIF2α-P observed here.
Our analyses reveal the mechanistic basis of eIF2B’s nucleotide exchange activity and suggest how phosphorylation converts eIF2 from substrate to inhibitor. The non-phosphorylated form of eIF2 binds to a composite surface created only in the assembled decamer, allowing both the core and the flexibly attached HEAT domain of eIF2Bε to engage its target in concert for enhanced GEF activity.
By contrast, eIF2α-P adopts a new conformation and suggests how the S-loop may become incompatible for binding to the site where nonphosphorylated eIF2α binds as a substrate (Movie S1). Phosphorylation thus enables a distinct binding mode on the opposite side of eIF2B where eIF2α-P lies exiled at the interface of eIF2Bα and eIF2Bδ. In eIF2•P, the rearrangement of the S-loop derives from an intramolecular electrostatic interaction between R63 and R53 and the phosphate, which also exposes a hydrophobic surface upon phosphorylation-induced refolding. We surmise that this new binding mode is nonproductive for nucleotide exchange on eIF2-P and sequesters the catalytic domains into an inhibited state that prevents the catalytic moieties of eIF2Bε from properly engaging in productive nucleotide exchange.
Supplementary Material
Acknowledgments:
We thank E. Pavolcak, J. Peschek, E. Karagöz, M. Boone, V. Belyy, G. Narlikar, R. Vale, , members of the Walter and Frost labs, Asarnow, D. for creating PyEM, Green, E for help with cisTEM, Thomas, P. for computational support, Melo, A. for help with Blender. ; The UCSF Center for Advanced CryoEM, which are supported by NIH grants S10OD020054 and 1S10OD021741 and the Howard Hughes Medical Institute (HHMI); Z. Yu and H. Chou of the CryoEM Facility at the HHMI Janelia Research Campus NIH grant 1S10OD021596–01 for computational support; and G. Pavitt for the GP6452 yeast strain used in the purification of eIF2. A Titan X Pascal used for this research was donated by the NVIDIA Corporation.
Funding: This work was supported by an HHMI Faculty Scholar grant (A.F.) and by Calico Life Sciences LLC, the Rogers Family Foundation, the Weill Foundation, and HHMI (P.W.). L.R.K. was supported by a graduate research fellowship from the N.S.F. A.F. is a Chan Zuckerberg Biohub Investigator, and P.W. is an Investigator of HHMI.
Footnotes
Competing interests: P.W. is an inventor on U.S. Patent 9708247 held by the Regents of the University of California that describes ISRIB and its analogs. Rights to the invention have been licensed by UCSF to Calico.
Accession numbers: Accession numbers for the human eIF2•eIF2B structures are as follows: EMD-0649, EMD-0651, EMD-0664 (density maps; Electron Microscopy Data Bank) and PDB-6O81, PDB-6O85, PDB-6O9Z (coordinates of atomic models; Protein Data Bank).
Data availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the supplementary materials, and the structural data is available in public databases. All of the raw cryo-EM data is also available upon request. The GP6452 yeast strain is available under a material transfer agreement with the University of Manchester.
Supplementary Materials:
Materials and Methods
Figures S1-S7
Tables S1-S3
Movie S1
References (38–46)
References and Notes:
- 1.Harding HP et al. , Mol. Cell 11, 619–33 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Palam LR, Baird TD, Wek RC, J. Biol. Chem 286, 10939–10949 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vattem KM, Wek RC, Proc. Natl. Acad. Sci 101, 11269–11274 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kashiwagi K et al. , Nature. 531, 122–125 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Gordiyenko Y et al. , Nat. Commun 5, 3902 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuhle B, Eulig NK, Ficner R, Nucleic Acids Res. 43, gkv930 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dev K et al. , Mol. Cell. Biol 30, 5218–33 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pavitt GD, V Ramaiah K, Kimball SR, Hinnebusch AG, Genes Dev. 12, 514–26 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimball SR, Fabian JR, Pavitt GD, Hinnebusch AG, Jefferson LS, J. Biol. Chem 273, 12841–5 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Krishnamoorthy T, Pavitt GD, Zhang F, Dever TE, Hinnebusch AG, Mol. Cell. Biol 21, 5018–5030 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sidrauski C et al. , Elife. 2 e00498 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sidrauski C et al. , Elife. 4 e07314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sekine Y et al. , Science. 348, 1027–30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou A et al. , Proc. Natl. Acad. Sci. U. S. A 114, E6420–E6426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halliday M et al. , Cell Death Dis. 6, e1672 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong YL et al. , bioRxiv, 462820 (2018). [Google Scholar]
- 17.Tsai JC et al. , Science. 359, eaaq0939 (2018).29599213 [Google Scholar]
- 18.Zyryanova AF et al. , Science. 359, 1533–1536 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Almeida RA et al. , PLoS One. 8, e53958 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito T, Marintchev A, Wagner G, Structure. 12, 1693–1704 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Dhaliwal S, Hoffman DW, J. Mol. Biol 334, 187–195 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Eliseev B et al. , Nucleic Acids Res. 46, 2678–2689 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beilsten-Edmands V et al. , Cell Discov., 1 15020 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubiez E, Aleksandrov A, Lazennec-Schurdevin C, Mechulam Y, Schmitt E, Nucleic Acids Res. 43, 2946–2957 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boesen T, Mohammad SS, Pavitt GD, Andersen GR, J. Biol. Chem 279, 10584–10592 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Wei J et al. , Protein Cell. 1, 595–603 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elsby R et al. , J. Virol 85, 9716–25 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pavitt GD, Yang W, Hinnebusch AG, Mol. Cell. Biol 17, 1298–313 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vazquez de Aldana CR, Hinnebusch AG, Mol. Cell. Biol 14, 3208–22 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crespillo-Casado A et al. , J. Biol. Chem 293, 7766–7776 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng SQ et al. , Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang K, J. Struct. Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Grant T, Rohou A, Grigorieff N, Elife. 7 (2018), doi:10.7554/eLife.35383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersen EF et al. , J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Simonetti A et al. , Mol. Cell 63, 206–217 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Lohkamp B, Scott WG, Cowtan K, Acta Crystallogr. Sect. D Biol. Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD et al. , Acta Crystallogr. Sect. D Biol. Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen VB et al. , Acta Crystallogr. Sect. D Biol. Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ludtke SJ, Methods Enzymol. 579, 159–89 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.