Abstract
Recent advances in neuro-technologies have revolutionized knowledge of brain structure and functions. Governments and private organizations worldwide have initiated several large-scale brain connectome projects, to further understand how the brain works at the systems levels. Most recent projects focus on only brain neurons, with the exception of an early effort to reconstruct the 302 neurons that comprise the whole body of the small worm, Caenorhabditis elegans. However, to fully elucidate the neural circuitry of complex behavior, it is crucial to understand brain interactions with the whole body, which can be achieved only by mapping the whole-body connectome. In this article, we discuss the current state of connectomics study, focusing on novel optical approaches and related imaging technologies. We also discuss the challenges encountered by scientists who endeavor to map these whole-body connectomes in large animals.
This article is based on a Presidential Lecture presented at the 2016 Annual Meeting.
Introduction
Understanding the functions of the brain, which supports our consciousness and mental processes, is the ultimate challenge of the biological sciences (Kandel et al., 2012). Efforts to develop this understanding resulted in modern neuroscience and began with the technology developments initiated by Camillo Golgi in 1873. Golgi's staining method led to the innovative work of Santiago Ramón y Cajal, who drew hundreds of exquisite illustrations of neuronal cells and their networks (De Carlos and Borrell, 2007). More than a century after Cajal's work, we have gained considerable knowledge regarding the molecular and cellular processes underlying neuronal functioning. However, the functions of a nervous system cannot be fully understood without knowing its connectome, which is the comprehensive map of neuronal connections (Sporns et al., 2005; Lichtman et al., 2008; Pastrana, 2013). To accomplish this goal, technological advances are required for mapping and studying neural circuits (Jorgenson et al., 2015). In recent decades, neuroscience has taken large steps toward this goal. Several technologies have been developed, enabling study of the structures and functions associated with large-scale brain circuits at an unprecedented speed, as well as analyses of the enormous datasets generated from these studies.
Despite these advances, there is still a discrepancy between current connectomics approaches and the ultimate goal of understanding brain functions. We propose that, to fully understand how the brain works, a detailed map between neural circuitry, neural activity, and complex behavior must be constructed. Therefore, connectomics studies must be constructed in situ and expanded beyond the brain (C. W. Lin et al., 2015), by investigating whole-body connectomes (Bidaye et al., 2014); the brain cannot function in isolation from interactions with the external environment and other internal body parts.
Whole-body connectomics involves all levels of imaging technologies, including macroscopic MRI, which reveals connectivity between brain regions; microscopic light microscopy, which images neuronal circuits at the cellular levels; and nanoscopic electron microscopy (EM), which identifies synapses and intracellular structures. This review focuses on the optical technologies that are designed for high throughput mapping with reasonable image acquisition speed (Osten and Margrie, 2013), rather than providing a comprehensive discussion of all related brain imaging technologies. Considering the compatibility with various genetic labeling and manipulation techniques, we believe that optical imaging will provide feasible solutions for mapping the whole-body connectomes of large animals in the near future (Lemon et al., 2015). Here, we review the current technologies available for mapping the brain connectome and discuss the challenges associated with mapping whole-body connectomes.
Key technologies
The first step to mapping a connectome is to image large tissue structures at the cellular level. After a connectome is constructed, its benefit is determined by how it can be used to study brain functions. Moreover, because of the scale of data, a connectomics project must be supported by advanced data analysis and management technologies. Therefore, we discuss the relevant technologies using the following three categories: structure, function, and informatics.
Structure
Connectomics studies begin with acquisition of high-resolution structural brain images, which enable the cellular-level connectome to be reconstructed. The images must also be acquired rapidly, so that the whole project can be accomplished within a meaningful timeframe. Therefore, neuroscientists must overcome challenges related to four domains: neuron labeling, optical tissue clearing, large volume tomography, and high-resolution microscopy. In this section of our review, we discuss the key technologies that are designed to overcome these challenges.
Neuron labeling
Neuronal structures must be labeled, either genetically or chemically, before they can be imaged using an optical microscope. Despite its broad application, Golgi staining is limited because cell labeling is random, only postmortem fixed tissues can be labeled, and it is difficult to accomplish 3D reconstruction for use in a modern connectome study, where a large volume of comprehensive fine neural circuitry construction is required. To overcome these limitations, several genetic labeling methods have been developed.
Mosaic analysis with a repressible cell marker (MARCM) and its modified versions remove the GAL80 transcriptional repressor during cell division and allow GAL4 to activate cell marker expression in a few individuals, and within a specific subset of neurons (Lee and Luo, 1999, 2001; Zong et al., 2005). To distinctly image many neurons in a sample, Brainbow has been developed for combinatorial multicolor stochastic labeling, which exclusively expresses only one of three or four spectrum-separated fluorescent proteins in each neuron (Livet et al., 2007; Hadjieconomou et al., 2011; Hampel et al., 2011; Richier et al., 2015). Recently, MultiColor FlipOut was used to label neurons using two different recombinases to achieve control of labeling density, using a controlled number of color combinations (Nern et al., 2015).
Labeling two or more connected neurons within a functional circuit is useful to information flow analyses. Such demand is fulfilled by trans-synaptic labeling techniques, which use viruses as the tracers (Callaway, 2008). This viral tracing can be anterograde, retrograde, or bidirectional and can even be designed to cross only one synapse (Wickersham et al., 2007). Although tissue degradation is a potential methodological limitation, viral tracing is widely used in studies of neural circuitry, including the mouse connectome project by the Allen Institute (Oh et al., 2014).
Optical tissue clearing
The next challenge in reconstructing the 3D brain structure is to acquire and process images from deep tissues, which are usually opaque. Although the challenge may be partially resolved by mechanical sectioning, it inevitably produces tissue loss or distortion, making image alignment between sections problematic. In contrast, optical tissue clearing techniques minimize light scattering, thereby providing an alternative method of acquiring fluorescent images from deep tissue. However, traditional organic-based clearing requires dehydration, which results in tissue deformation. Moreover, the whole clearing process takes several days, which reduces the emission of fluorescent proteins due to their instability in the organic solution (Dodt et al., 2007; Renier et al., 2014). The first breakthrough in the clearing process resulted from the invention of an aqueous clearing solution, FocusClear, which produces minimal tissue deformation and is compatible with most immunolabeling fluorescent dyes (Chiang et al., 2001; Liu and Chiang, 2003). This development led to initiation of the first large-scale Drosophila brain connectome project, which has acquired more than 20,000 high-resolution images of single brain neurons, which are hosted in the FlyCircuit database (Chiang et al., 2011) and several other neural image databases (Osumi-Sutherland et al., 2012; Parekh and Ascoli, 2013). In the past few years, several other clearing techniques have been developed, including additional aqueous solutions (Ke et al., 2013; Kuwajima et al., 2013; Costantini et al., 2015), urea-assisted hydration (Hama et al., 2011), and hydrogel embedding (Chung et al., 2013; Yang et al., 2014). The CLARITY technique (Chung et al., 2013) is considered to be suitable for high-resolution connectome reconstruction. Using CLARITY, the brain is embedded in hydrogel while the major light-scattering substances (tissue lipids) are removed by SDS via electrophoresis. The samples are subsequently cleared in FocusClear or 80% glycerol, allowing imaging of fluorescence up to a depth of 3–4 mm in the mouse brain. The rapid development of tissue clearing technologies has also enabled whole-body clearing, which is achieved by intracardiac perfusion of tissue-clearing agents (Tainaka et al., 2014; Yang et al., 2014; Hama et al., 2015; Pan et al., 2016). In summary, tissue clearing technologies enable 3D in situ imaging of large tissues and have become an important alternative to standard histological examinations using thin tissue slices.
Large-tissue tomography
Despite recent advances, the tissue clearing technologies have a fundamental limitation: the working distance of optical microscopy. Without physical sectioning, an objective lens with high numerical aperture (≥1.2) for resolving submicron-resolution details does not have enough working distance (≥0.28 mm) to visualize cellular structures that are more than several millimeters deep. This limitation can be partially addressed by shrinkage-mediated imaging, which reduces the tissue volume after clearing (Pan et al., 2016). However, serial sectioning is still required for 3D imaging and reconstruction of large tissues.
Serial sectioning has been used with transmission EM to reconstruct the whole-body connectome of Caenorhabditis elegans (White et al., 1986; Xu et al., 2013) and larval Drosophila (Ohyama et al., 2015) as well as the partial optic medulla connectome in Drosophila (Takemura et al., 2013). However, the use of transmission EM in connectome reconstruction is limited by the labor-intensive tissue sectioning process and the difficulty using it in large nervous systems. These issues are addressed by serial block-face scanning EM, which incorporates an ultramicrotome that automatically and repeatedly sections a thin slice from the top surface of the sample block after each image is acquired (Denk and Horstmann, 2004). Serial block-face scanning EM greatly improves the speed of image acquisition; however, the postacquisition data analyses are extremely labor intensive, as they include tracing, segmentation, synapse identification, and circuit reconstruction. Therefore, although EM imaging has an advantage over light microscopy in unambiguously identifying synapses and other structures at the nanometer level, the time-consuming imaging acquisition and postprocessing make it a challenge for EM to reconstruct the connectome of large animals, such as mammals. Another advantage of EM is to uniquely determine the identity of a neuron through detailed morphology and connectivity. However, it is still difficult for EM to detect neuronal properties (e.g., neurotransmitters) due to lack of specific markers producing distinguishable electron dense signals.
Optical microscopy combining automated serial sectioning allows 3D reconstruction of large tissues, such as the whole mouse or even the whole human brain, although resolution is compromised. It scans faster, enables imaging deeper in the tissue, overcomes the limitations related to the working distance of the high-resolution objective lens, and provides multichannel images with distinguishable labeling for easier image processing. Array tomography achieves a depth-independent immunofluorescent image by ultrathin sectioning to improve spatial resolution on the z-axis (Micheva and Smith, 2007; Tapia et al., 2012). Using automated serial sectioning, knife-edge scanning microscopy (Mayerich et al., 2008) and micro-optical sectioning tomography (Li et al., 2010) image fine structures on the knife edge to increase the speed of image acquisition in large-volume tissues. Using serial two-photon tomography, an automated system that removes the surface tissue after two-photon imaging (Ragan et al., 2012), the Allen Institute for Brain Science has constructed a mesoscale connectome atlas of the mouse brain with cellular level resolution (Oh et al., 2014). These optical imaging technologies have advanced the connectome frontiers from the brain to the whole body, and from small animals to large animals. Furthermore, we envision a continual trend of developing new technologies. For example, confocal microscopy combining tissue clearing and automated sectioning should further improve the speed of 3D image acquisition for large tissues, with improved image quality, as well as minimal tissue distortion and loss.
Advanced optical microscopy
With advances in neuron labeling, tissue clearing, and large-tissue tomography technologies, the next major challenge is determining how to acquire images with scanning speeds and spatial resolutions that are sufficient for detailed connectome reconstruction. Acquiring images from large tissue volumes is time-consuming using conventional confocal microscopy, due to the nature of point scanning. Moreover, low temporal resolution limits the ability of confocal microscopy to capture the progress of cellular processes that occur within millisecond timeframes. This issue is addressed by more advanced optical imaging technologies, including light field microscopy (Levoy et al., 2006), light sheet microscopy (Huisken et al., 2004; Planchon et al., 2011), and multifocal multiphoton microscopy (Bewersdorf et al., 1998; Bahlmann et al., 2007). Among these technologies, light-sheet microscopy is the most popular approach for observing living samples because this type of microscopy creates a thin slice of illuminated area and thereby provides a much faster imaging speed compared with confocal microscopy; light-sheet microscopy also minimizes unfocused background and premature photobleaching (Planchon et al., 2011). Recently, lattice light-sheet microscopy was developed to increase temporal and spatial resolution while also reducing phototoxic tissue damage (Chen et al., 2014).
Another limitation of conventional confocal microscopy is its spatial resolution, which does not allow imaging of individual synapses. Without the precise information regarding synapses, a detailed connectome will not be established. In the last two decades, the development of super-resolution microscopy, including stimulated emission depletion microscopy (Hell et al., 1994), localization microscopy (Betzig et al., 2006; Rust et al., 2006), and saturated structured illumination microscopy (Gustafsson, 2005), has achieved resolution at levels <100 nm, thereby enabling the possibility of reconstructing detailed dendritic structures, or even synapses, using optical approaches (Dani et al., 2010; Nägerl and Bonhoeffer, 2010). Furthermore, expansion microscopy has recently been developed as an alternative approach to high-resolution microscopy. In contrast to shrinkage-mediated imaging, expansion microscopy uses tissue enlargement by a water-absorbing gel that contains polyacrylate. Physical enlargement increases the optical resolution fourfold (Chen et al., 2015; Tillberg et al., 2016). Rapid development of innovative super-resolution imaging will soon enable direct visualization of synaptic connections in deep tissue.
Function
The aforementioned technological advances enable acquisition of high-resolution images for use in reconstructing a detailed connectome. However, to study brain functions, information is required regarding how diverse neurons differentially process signals, how signals flow between neurons within a responsive circuit, and how emergent properties generate behavior. Here, we discuss the technologies that are designed to classify neuron types, to identify synapses, to monitor cellular responses, to alter neural activity, and to control behaviors on a large scale.
Cell typing
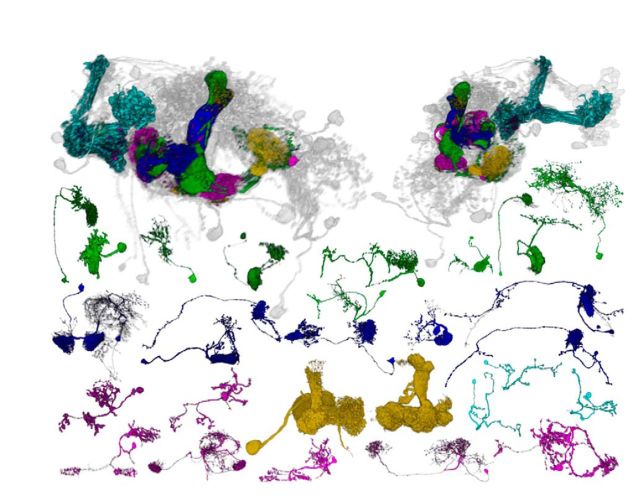
Structure determines function. Neurons are morphologically and functionally diverse, and categorizing them is therefore an important step toward systematic annotation for data management (van Pelt et al., 2001; Peng et al., 2015). Moreover, unlike EM, which can be used to reconstruct the connectome of one single individual, light-microscopy-based connectome has to be reconstructed by integrating images from a large number of individuals. The integration can be challenging considering the interindividual variation and hence crucially relies on proper cell-type categorization. Traditional neuronal taxonomy only considers neuron morphology; however, other properties related to function, such as spatial location, innervation sites, and connectivity, may also be useful considerations when designing tools for cell categorization. Initial attempts to differentiate neurons were based on innervations across different brain regions, with efforts to “bar code” each neuron for comparison (Chiang et al., 2011). Recently, more rigorous algorithms have been developed for clustering neuronal morphology by location and local geometry (Costa et al., 2016). In addition to morphology, the genes, molecular markers, or characteristic neural activity (Sharpee, 2014) may also be used to differentiate cell types (or subtypes). Using labeling with polarity makers, individual cells forming the mushroom body, which is the learning and memory center in the Drosophila brain, have been categorized into input, output, and modulatory neurons (Aso et al., 2014a, b) (Fig. 1). This comprehensive cell typing has led to the discovery of neurons responsible for individual smell preferences, which requires functional plasticity of the mushroom body output neurons acting collectively to register odor memories (Pai et al., 2013; Hige et al., 2015). Tracing mushroom body output neurons (as well as input and modulatory neurons) along their upstream and downstream neurons promises to reveal the circuit mechanisms of individual differences in odor memory. With the recent availability of large-scale neuron image databases for Drosophila, more cell-typing proposals are likely, which will greatly advance explorations of the connectome data.
Figure 1.
Diverse neurons forming mushroom bodies in the Drosophila brain. The mushroom body is comprised of three major types of intrinsic Kenyon cells (light blue), with dendrites forming calyx and axons forming γ, α′/β′, and α/β lobes. Two large modulatory neurons (yellow), anterior paired lateral and dorsal paired medial, innervate all mushroom body lobes. Each lobe is subdivided into consecutive domains, which are innervated by dopaminergic input neurons (magenta) output neurons (green), and some neurons with both axons and dendrites connecting between two mushroom body domains (dark blue). Assembling the intersections between Kenyon cells and other cells reveals an intricate mushroom body circuit (top). All neuron images were derived from FlyCircuit database.
Recordings of synaptic and neuronal activity
To study the flow of neural signals, we first need to identify whether two given neurons form synapses. In optical microscopy, synapses can be imaged using green fluorescence protein (GFP) reconstitution across synaptic partners (GRASP), which splits a GFP into two nonfluorescent fragments and expresses them separately in two different neurons. The GFP fragments may then reconstitute and emit fluorescence if two neurons contact closely within a distance similar to synaptic contact (Feinberg et al., 2008). GRASP has been demonstrated in C. elegans and Drosophila (Feinberg et al., 2008; Gordon and Scott, 2009), and a modified version, mGRASP, has also been developed for mammalian brains, using optimized transmembrane split-GFP carriers (Kim et al., 2012). However, from a functional perspective, it is more important to record synaptic activation than to simply identify the presence of synapses. Indeed, sensory signals can use different neural pathways under different environmental or behavioral conditions (H. H. Lin et al., 2013). Recoding synaptic activation can be achieved by a recently developed technique, X-RASP (Macpherson et al., 2015). X-RASP is a multicolor and activity-dependent GRASP that uses synaptobrevin, a small membrane protein found in secretory vesicles, as a carrier for the GFP1–10 fragment.
In addition to label-activated synapses, it is also informative to record the activity of individual neurons. Although calcium imaging has been widely used for observing neuronal activity, this technique requires online recording and is limited by a small field of view. These limitations are addressed by a new technique, calcium-modulated photoactivatable ratiometric integrator (CaMPARI), which uses the photoconvertible protein mEos2 as a probe for the calcium concentration. This technique allows a snapshot or recording of an episode of circuit activity within a precise time window and can be used for large volumes of brain tissue (Fosque et al., 2015).
Behavior manipulation
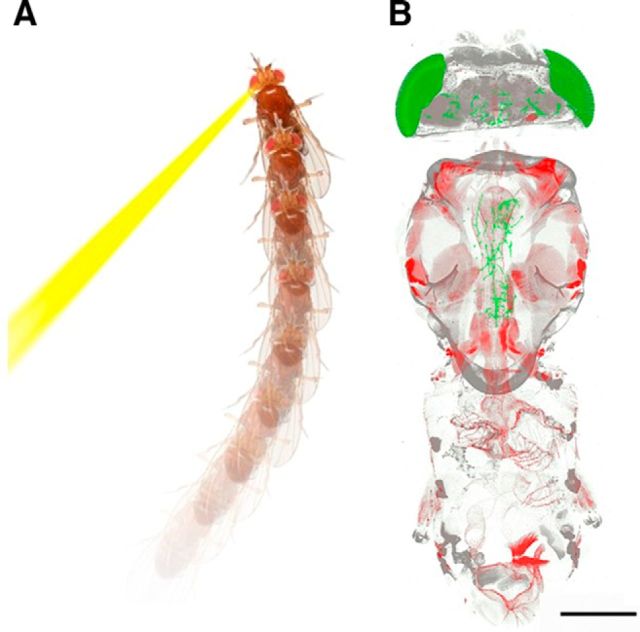
The ability to record synaptic and neuronal activity on a large scale facilitates systematic mapping of the neural correlates of behavior. However, correlation does not necessarily indicate a cause-and-effect relationship because the behavior and correlated neuronal activity may be elicited by a common source. To verify the cause-and-effect relationship, the behavior must be altered by activating or inhibiting the neurons of interest. This challenge is resolved by a number of recently developed technologies, including optogenetics (Boyden et al., 2005; Lima and Miesenböck, 2005; Nikolenko et al., 2007; Zhang et al., 2007; Hsiao et al., 2015), thermogenetics (Kitamoto, 2001; Peabody et al., 2009), chemogenetics (Armbruster et al., 2007; Becnel et al., 2013), magnetogenetics (Long et al., 2015; Wheeler et al., 2016), and sonogenetics (Ibsen et al., 2015). Most of these technologies express specific proteins (e.g., membrane receptors) in genetically targeted neurons and manipulate them by activating or silencing the proteins using physical (heat, light, magnetism, or ultrasound) or chemical means. Optogenetics, which uses light-sensitive opsins, is the most developed and adopted of these techniques. Optogenetics has been used to demonstrate a variety of neuron-behavior correlations, including locomotor control (Lima and Miesenböck, 2005; Aravanis et al., 2007; Kravitz et al., 2010; Leifer et al., 2011; Heiney et al., 2014; Y. Y. Lin et al., 2015), memory (Liu et al., 2012), and parallel pathway shunting (H. H. Lin et al., 2013). Despite its popularity, optogenetics has a major limitation: restricted light penetration depth. Fortunately, penetration depth can be improved by implementation of a red-orange light source with long-wavelength opsins (J. Y. Lin et al., 2013), or by multiphoton excitation (Rickgauer and Tank, 2009; Hsiao et al., 2015). The development of optogenetics and other neuron manipulation technologies enables possibilities for noninvasive neuron manipulation in free-moving animals (Leifer et al., 2011). In particular, because of its temporal and spatial precision benefits, optogenetics has been incorporated into a new behavioral apparatus, the Automated Laser Tracking and Optogenetic Manipulation System (ALTOMS), which integrates optogenetics, laser tracking, machine learning, and pattern recognition technologies (Wu et al., 2014). In ALTOMS, free-moving fruit flies are tracked by a video monitoring system and their behaviors are analyzed in real time. Targeted neurons in the flies can be activated or inhibited by a precisely positioned laser beam, using optogenetics, when preset criteria are met. ALTOMS offers opportunities to systematically map brain circuits that orchestrate specific behaviors, as well as testing predictions made by connectome analyses or computational models (Chiang et al., 2011; Shih et al., 2015). Neuroscientists will soon be able to construct integrated whole-body neuron-behavior maps in Drosophila using these advanced genetic tools to manipulate the activity of specific neurons (Bidaye et al., 2014) (Fig. 2).
Figure 2.
Optogenetic mapping of whole-body neural-behavioral circuits in Drosophila. A, Laser (593.5 nm) activation of a small group of brain neurons initiates backward walking behavior in a VT50660-Gal4>UAS-ReaChR fly fed with 100 μm all-trans-retinal. B, The whole-body anatomy (250 μm thick) reveals several brain neurons (green) extending axons to the thoracic muscles in the VT50660-Gal4>UAS-mko;z-disc::GFP fly. Red represents muscle. Gray represents body and tracheal cuticle stained by Congo red dye. Scale bar, 500 μm.
Informatics
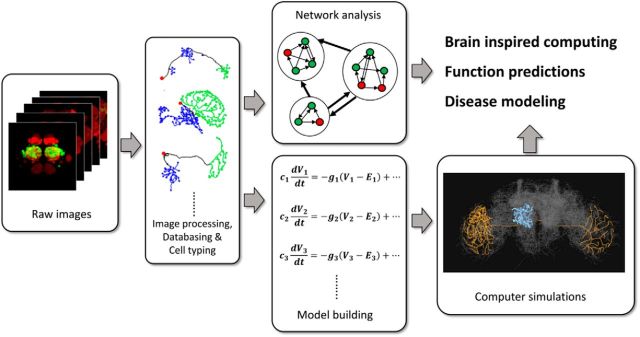
Large-scale connectome projects generate enormous quantities of 3D images, and each image requires complex processing before useful information can be extracted. Furthermore, advanced software tools are required for efficient analyses and management of the data. Here, we discuss the challenges and required informatics technologies in image processing, creating databases, and computer modeling (Fig. 3).
Figure 3.
Steps toward brain simulation. Raw images first undergo complex image processing, followed by tracing and segmentation of single neurons. The data are then analyzed and stored in a database, which contains information at the neuronal, circuitry, and system levels. The data can then be used for network analyses, as well as model construction, which form the foundation for large-scale brain simulations. These computer simulations and network analyses may inspire novel technologies in neuromorphic computing and provide insight into neural circuit mechanisms of brain functions and associated disorders.
Image processing
Typical postprocessing of optical brain images includes the following: (1) noise reduction; (2) segmentation and tracing, which extract the 3D structure of the target neurons from the background image; and (3) warping, which transforms and registers each neuronal image in standard brain coordinates. In a large-scale connectome project, such labor-intensive processes must be automated. Various computer tools for high-throughput image processing have been developed (Donohue and Ascoli, 2011; Peng et al., 2011a, b, 2015; Halavi et al., 2012; Helmstaedter and Mitra, 2012; Lee et al., 2012; Ostrovsky et al., 2013; Xiao and Peng, 2013). Next, the polarity (i.e., axon vs dendrite) of each neuronal arbor must be identified. Although the polarity can be accurately identified using immunolabeling, this information is often not available from connectome datasets. This issue is partially addressed by development of a computer algorithm, SPIN (skeleton-based polarity identification for neurons), which predicts the polarity of arbors purely based on morphological features extracted from neuronal skeletons (Lee et al., 2014).
Creating databases
A large-scale connectome project easily produces several terabytes or even petabytes of data. Although this amount of data is manageable with current database technology, the central challenges are as follows: (1) how to develop ontologies for efficient data management, (2) how to build a user-friendly web interface, and (3) how to integrate and transform the data to address scientific questions. Several large-scale databases have been established, which emphasize high-resolution neuron images and connectome for nematode Caenorhabditis elegans (Xu et al., 2013), fruit fly (Chiang et al., 2011; Shinomiya et al., 2011; Osumi-Sutherland et al., 2012; Parekh and Ascoli, 2013), mouse (Kuan et al., 2015), and other species (Peng et al., 2015). These databases provide invaluable information for connectomics research.
Network analysis and brain simulation
Connectomic data can be used to construct whole-brain or whole-body network models. By analyzing the network architecture with graph theory or other statistical tools, one can obtain insights into the functional organization of the brain (Sporns et al., 2005; Takemura et al., 2013; Shih et al., 2015). Additionally, the data can be applied to construction of detailed computational brain models. In the past decade, various large-scale modeling projects have been initiated to simulate the partial brain (Markram, 2006; Izhikevich and Edelman, 2008; Eliasmith et al., 2012). The whole-brain or whole-body connectome provides an opportunity for building computational models for a whole nervous system at the single-cell level. However, there are challenges that result from the enormous number of unknown parameters. Furthermore, if detailed neuronal and synaptic dynamics are simulated, the required computational power becomes incredibly large. Two lines of solutions are in development: (1) graphical processing unit-based computing (Ben-Shalom et al., 2013; Hoang et al., 2013; Givon and Lazar, 2016), and (2) neuromorphic chips (Schemmel et al., 2010; Indiveri et al., 2011; Benjamin et al., 2014; Furber et al., 2014; Merolla et al., 2014). Both solutions are much faster than the traditional CPU-based computations. However, coding for the graphical processing unit-based solution is relatively cumbersome and platform/hardware-dependent, whereas neuromorphic chips are limited by their inflexibility and inability to simulate biologically realistic models. Despite these challenges, several data-driven whole-brain or whole-body computational models at the single-neuron level are under development. OpenWorm is an international open science project with the aim to simulate the whole nervous system of C. elegans (Palyanov et al., 2011; Szigeti et al., 2014). The Flysim project aims to develop a brain-wide computational model for Drosophila, based on data from the FlyCircuit database (Huang et al., 2014). Additionally, NeuroKernel/NeuroArch is an innovative open source platform that provides graphical processing unit-based simulation for the Drosophila brain (Givon and Lazar, 2016).
Conclusion and perspective
Our ability to map the brains depends largely on the invention of connectomics neurotechnologies. In the coming decade, new technologies will allow us to identify abundant new types of neurons and to reveal the operation principles of information integration by the complex neural circuits, which hold the key to understanding our consciousness and intelligence. However, given the current microscopy technologies, it remains time-consuming to conduct whole-brain scanning at subcellular levels in large animals, which is even more burdensome for the whole-body connectome. Therefore, there is a need for another technology breakthrough that increases the image acquisition speed manyfold. Correspondingly, data processing must also be greatly improved because the scale of a connectome project is enormous.
It is reasonable to ask how society will benefit from the heavy investment currently made to the connectomics studies. The benefits can be explained by the following applications:
Basic science: Understanding the mind and consciousness is one of the most important questions underlying brain research. Various governments and agencies have invested tremendous resources to explore our universe; it is the same curiosity that motivates us to explore the universe within ourselves, which contains 86 billion neurons (Azevedo et al., 2009) and tens of thousand times more connections.
Public health: In the past few decades, considerable knowledge has been acquired regarding the genetic, molecular, and cellular mechanisms underlying various brain diseases. However, increasing evidence suggests relationships between circuitry-level abnormalities and several brain disorders, including epilepsy, autism, and schizophrenia (Yizhar et al., 2011; Lisman, 2012; Paz and Huguenard, 2015). Connectome data will facilitate studies on the circuit mechanisms of brain diseases and potentially result in improved treatments.
Industrial applications: Machine learning and artificial intelligence have made tremendous progress in recent years (Ghahramani, 2015; LeCun et al., 2015). Although artificial intelligence outperforms humans on certain tasks (e.g., playing Go), it still does not achieve human capabilities on most tasks, such as the speed of learning. Moreover, the nature of feedforward connections in most artificial neural networks is distinctly different from the strong feedback circuits that are present in every part of the human brain. Understanding detailed brain connectomes may reveal the core reasons for the fundamental differences between artificial intelligence and human brains, and may contribute to further advances in artificial intelligence.
In the era of rapidly developing biotechnologies, the ultimate goal of understanding brain activity and the resulting behavior is increasingly attainable. However, given the scale of a connectome project, no single laboratory, or even small group of laboratories, has the capacity to accomplish every aspect of a full connectome study from data acquisition to analyses. Hence, connectome projects should be conducted with publicly shared data, using a collaborative approach (Chiang et al., 2011; Milham, 2012; Burns et al., 2014; Poldrack and Gorgolewski, 2014; Peng et al., 2015). Data sharing would allow researchers from all disciplines to participate in generating new hypotheses and testing novel methods, thereby transforming connectomics research and accelerating progress toward understanding the brain.
Footnotes
We thank Hollis Cline for arranging this mini-review as a part of one of the four Presidential Special Lectures of Neuroscience 2016 Meeting at San Diego. We also thank Meimei Yang and our laboratory members Kuan-Lin Feng, Yan-Chih Ciou, Po-Yen Hsiao, and Chun-Yen Chen for assistance in preparing figures and illustrations.
The authors declare no competing financial interests.
References
- Aravanis AM, Wang LP, Zhang F, Meltzer LA, Mogri MZ, Schneider MB, Deisseroth K. An optical neural interface: in vivo control of rodent motor cortex with integrated fiberoptic and optogenetic technology. J Neural Eng. 2007;4:S143–S156. doi: 10.1088/1741-2560/4/3/S02. [DOI] [PubMed] [Google Scholar]
- Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci U S A. 2007;104:5163–5168. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y, Hattori D, Yu Y, Johnston RM, Iyer NA, Ngo TT, Dionne H, Abbott LF, Axel R, Tanimoto H, Rubin GM. The neuronal architecture of the mushroom body provides a logic for associative learning. eLife. 2014a;3:e04577. doi: 10.7554/eLife.04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y, Sitaraman D, Ichinose T, Kaun KR, Vogt K, Belliart-Guérin G, Plaçais PY, Robie AA, Yamagata N, Schnaitmann C, Rowell WJ, Johnston RM, Ngo TT, Chen N, Korff W, Nitabach MN, Heberlein U, Preat T, Branson KM, Tanimoto H, et al. Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila. eLife. 2014b;3:e04580. doi: 10.7554/eLife.04580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo FA, Carvalho LR, Grinberg LT, Farfel JM, Ferretti RE, Leite RE, Jacob Filho W, Lent R, Herculano-Houzel S. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J Comp Neurol. 2009;513:532–541. doi: 10.1002/cne.21974. [DOI] [PubMed] [Google Scholar]
- Bahlmann K, So PT, Kirber M, Reich R, Kosicki B, McGonagle W, Bellve K. Multifocal multiphoton microscopy (MMM) at a frame rate beyond 600 Hz. Opt Express. 2007;15:10991–10998. doi: 10.1364/OE.15.010991. [DOI] [PubMed] [Google Scholar]
- Becnel J, Johnson O, Majeed ZR, Tran V, Yu B, Roth BL, Cooper RL, Kerut EK, Nichols CD. DREADDs in Drosophila: a pharmacogenetic approach for controlling behavior, neuronal signaling, and physiology in the fly. Cell Rep. 2013;4:1049–1059. doi: 10.1016/j.celrep.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin BV, Gao P, McQuinn E, Choudhary S, Chandrasekaran AR, Bussat JM, Alvarez-Icaza R, Arthur JV, Merolla PA, Boahen K. Neurogrid: a mixed-analog-digital multichip system for large-scale neural simulations. Proc IEEE. 2014;102:699–716. doi: 10.1109/JPROC.2014.2313565. [DOI] [Google Scholar]
- Ben-Shalom R, Liberman G, Korngreen A. Accelerating compartmental modeling on a graphical processing unit. Front Neuroinformatics. 2013;7:4. doi: 10.3389/fninf.2013.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Bewersdorf J, Pick R, Hell SW. Multifocal multiphoton microscopy. Opt Lett. 1998;23:655–657. doi: 10.1364/OL.23.000655. [DOI] [PubMed] [Google Scholar]
- Bidaye SS, Machacek C, Wu Y, Dickson BJ. Neuronal control of Drosophila walking direction. Science. 2014;344:97–101. doi: 10.1126/science.1249964. [DOI] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Burns R, Vogelstein JT, Szalay AS. From cosmos to connectomes: the evolution of data-intensive science. Neuron. 2014;83:1249–1252. doi: 10.1016/j.neuron.2014.08.045. [DOI] [PubMed] [Google Scholar]
- Callaway EM. Transneuronal circuit tracing with neurotropic viruses. Curr Opin Neurobiol. 2008;18:617–623. doi: 10.1016/j.conb.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BC, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XS, Hammer JA, 3rd, Liu Z, English BP, Mimori-Kiyosue Y, Romero DP, Ritter AT, Lippincott-Schwartz J, Fritz-Laylin L, Mullins RD, Mitchell DM, Bembenek JN, Reymann AC, et al. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346:1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tillberg PW, Boyden ES. Expansion microscopy. Science. 2015;347:543–548. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang AS, Lin CY, Chuang CC, Chang HM, Hsieh CH, Yeh CW, Shih CT, Wu JJ, Wang GT, Chen YC, Wu CC, Chen GY, Ching YT, Lee PC, Lin CY, Lin HH, Wu CC, Hsu HW, Huang YA, Chen JY, et al. Three-dimensional reconstruction of brain-wide wiring networks in Drosophila at single-cell resolution. Curr Biol. 2011;21:1–11. doi: 10.1016/j.cub.2010.11.056. [DOI] [PubMed] [Google Scholar]
- Chiang AS, Liu YC, Chiu SL, Hu SH, Huang CY, Hsieh CH. Three-dimensional mapping of brain neuropils in the cockroach, Diploptera punctata. J Comp Neurol. 2001;440:1–11. doi: 10.1002/cne.1365. [DOI] [PubMed] [Google Scholar]
- Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, Pak S, Bernstein H, Ramakrishnan C, Grosenick L, Gradinaru V, Deisseroth K. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Manton JD, Ostrovsky AD, Prohaska S, Jefferis GS. NBLAST: rapid, sensitive comparison of neuronal structure and construction of neuron family databases. Neuron. 2016;91:293–311. doi: 10.1016/j.neuron.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini I, Ghobril JP, Di Giovanna AP, Allegra Mascaro AL, Silvestri L, Müllenbroich MC, Onofri L, Conti V, Vanzi F, Sacconi L, Guerrini R, Markram H, Iannello G, Pavone FS. A versatile clearing agent for multi-modal brain imaging. Sci Rep. 2015;5:9808. doi: 10.1038/srep09808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani A, Huang B, Bergan J, Dulac C, Zhuang X. Superresolution imaging of chemical synapses in the brain. Neuron. 2010;68:843–856. doi: 10.1016/j.neuron.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Carlos JA, Borrell J. A historical reflection of the contributions of Cajal and Golgi to the foundations of neuroscience. Brain Res Rev. 2007;55:8–16. doi: 10.1016/j.brainresrev.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2:e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt HU, Leischner U, Schierloh A, Jährling N, Mauch CP, Deininger K, Deussing JM, Eder M, Zieglgänsberger W, Becker K. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Methods. 2007;4:331–336. doi: 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- Donohue DE, Ascoli GA. Automated reconstruction of neuronal morphology: an overview. Brain Res Rev. 2011;67:94–102. doi: 10.1016/j.brainresrev.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliasmith C, Stewart TC, Choo X, Bekolay T, DeWolf T, Tang C, Rasmussen D. A large-scale model of the functioning brain. Science. 2012;338:1202–1205. doi: 10.1126/science.1225266. [DOI] [PubMed] [Google Scholar]
- Feinberg EH, Vanhoven MK, Bendesky A, Wang G, Fetter RD, Shen K, Bargmann CI. GFP reconstitution across synaptic partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron. 2008;57:353–363. doi: 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- Fosque BF, Sun Y, Dana H, Yang CT, Ohyama T, Tadross MR, Patel R, Zlatic M, Kim DS, Ahrens MB, Jayaraman V, Looger LL, Schreiter ER. Labeling of active neural circuits in vivo with designed calcium integrators. Science. 2015;347:755–760. doi: 10.1126/science.1260922. [DOI] [PubMed] [Google Scholar]
- Furber SB, Galluppi F, Temple S, Plana LA. The SpiNNaker Project. Proc IEEE. 2014;102:652–665. doi: 10.1109/JPROC.2014.2304638. [DOI] [Google Scholar]
- Ghahramani Z. Probabilistic machine learning and artificial intelligence. Nature. 2015;521:452–459. doi: 10.1038/nature14541. [DOI] [PubMed] [Google Scholar]
- Givon LE, Lazar AA. Neurokernel: an open source platform for emulating the fruit fly brain. PLoS One. 2016;11:e0146581. doi: 10.1371/journal.pone.0146581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon MD, Scott K. Motor control in a Drosophila taste circuit. Neuron. 2009;61:373–384. doi: 10.1016/j.neuron.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson MG. Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. Proc Natl Acad Sci U S A. 2005;102:13081–13086. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjieconomou D, Rotkopf S, Alexandre C, Bell DM, Dickson BJ, Salecker I. Flybow: genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster. Nat Methods. 2011;8:260–266. doi: 10.1038/nmeth.1567. [DOI] [PubMed] [Google Scholar]
- Halavi M, Hamilton KA, Parekh R, Ascoli GA. Digital reconstructions of neuronal morphology: three decades of research trends. Front Neurosci. 2012;6:49. doi: 10.3389/fnins.2012.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, Fukami K, Sakaue-Sawano A, Miyawaki A. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- Hama H, Hioki H, Namiki K, Hoshida T, Kurokawa H, Ishidate F, Kaneko T, Akagi T, Saito T, Saido T, Miyawaki A. ScaleS: an optical clearing palette for biological imaging. Nat Neurosci. 2015;18:1518–1529. doi: 10.1038/nn.4107. [DOI] [PubMed] [Google Scholar]
- Hampel S, Chung P, McKellar CE, Hall D, Looger LL, Simpson JH. Drosophila Brainbow: a recombinase-based fluorescence labeling technique to subdivide neural expression patterns. Nat Methods. 2011;8:253–259. doi: 10.1038/nmeth.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiney SA, Kim J, Augustine GJ, Medina JF. Precise control of movement kinematics by optogenetic inhibition of Purkinje cell activity. J Neurosci. 2014;34:2321–2330. doi: 10.1523/JNEUROSCI.4547-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19:780–782. doi: 10.1364/OL.19.000780. [DOI] [PubMed] [Google Scholar]
- Helmstaedter M, Mitra PP. Computational methods and challenges for large-scale circuit mapping. Curr Opin Neurobiol. 2012;22:162–169. doi: 10.1016/j.conb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hige T, Aso Y, Rubin GM, Turner GC. Plasticity-driven individualization of olfactory coding in mushroom body output neurons. Nature. 2015;526:258–262. doi: 10.1038/nature15396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang RV, Tanna D, Bray LC, Dascalu SM, Harris FC., Jr A novel CPU/GPU simulation environment for large-scale biologically realistic neural modeling. Front Neuroinformatics. 2013;7:19. doi: 10.3389/fninf.2013.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao PY, Tsai CL, Chen MC, Lin YY, Yang SD, Chiang AS. Non-invasive manipulation of Drosophila behavior by two-photon excited red-activatable channelrhodopsin. Biomed Opt Express. 2015;6:4344–4352. doi: 10.1364/BOE.6.004344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YC, Wang CT, Wang GT, Su TS, Hsiao PY, Lin CY, Hsieh CH, Chang HM, Lo CC. The Flysim project: persistent simulation and real-time visualization of fruit fly whole-brain spiking neural network model. Front. Neuroinform. Conference Abstract: Neuroinformatics 2014; 2014. [DOI] [Google Scholar]
- Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EH. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305:1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- Ibsen S, Tong A, Schutt C, Esener S, Chalasani SH. Sonogenetics is a non-invasive approach to activating neurons in Caenorhabditis elegans. Nat Commun. 2015;6:8264. doi: 10.1038/ncomms9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indiveri G, Linares-Barranco B, Hamilton TJ, van Schaik A, Etienne-Cummings R, Delbruck T, Liu SC, Dudek P, Häfliger P, Renaud S, Schemmel J, Cauwenberghs G, Arthur J, Hynna K, Folowosele F, Saighi S, Serrano-Gotarredona T, Wijekoon J, Wang Y, Boahen K. Neuromorphic silicon neuron circuits. Front Neurosci. 2011;5:73. doi: 10.3389/fnins.2011.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izhikevich EM, Edelman GM. Large-scale model of mammalian thalamocortical systems. Proc Natl Acad Sci U S A. 2008;105:3593–3598. doi: 10.1073/pnas.0712231105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson LA, Newsome WT, Anderson DJ, Bargmann CI, Brown EN, Deisseroth K, Donoghue JP, Hudson KL, Ling GS, MacLeish PR, Marder E, Normann RA, Sanes JR, Schnitzer MJ, Sejnowski TJ, Tank DW, Tsien RY, Ugurbil K, Wingfield JC. The BRAIN Initiative: developing technology to catalyse neuroscience discovery. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140164. doi: 10.1098/rstb.2014.0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER, Schwartz JH, Jessell TM, Siegelbaum SA, Hudspeth AJ, editors. Principles of neural science. Ed 5. New York: McGraw-Hill; 2012. p. 5. [Google Scholar]
- Ke MT, Fujimoto S, Imai T. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci. 2013;16:1154–1161. doi: 10.1038/nn.3447. [DOI] [PubMed] [Google Scholar]
- Kim J, Zhao T, Petralia RS, Yu Y, Peng H, Myers E, Magee JC. mGRASP enables mapping mammalian synaptic connectivity with light microscopy. Nat Methods. 2011;9:96–102. doi: 10.1038/nmeth.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamoto T. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J Neurobiol. 2001;47:81–92. doi: 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
- Kravitz AV, Freeze BS, Parker PR, Kay K, Thwin MT, Deisseroth K, Kreitzer AC. Regulation of parkinsonian motor behaviours by optogenetic control of basal ganglia circuitry. Nature. 2010;466:622–626. doi: 10.1038/nature09159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuan L, Li Y, Lau C, Feng D, Bernard A, Sunkin SM, Zeng H, Dang C, Hawrylycz M, Ng L. Neuroinformatics of the Allen mouse brain connectivity atlas. Methods. 2015;73:4–17. doi: 10.1016/j.ymeth.2014.12.013. [DOI] [PubMed] [Google Scholar]
- Kuwajima T, Sitko AA, Bhansali P, Jurgens C, Guido W, Mason C. ClearT: a detergent- and solvent-free clearing method for neuronal and non-neuronal tissue. Development. 2013;140:1364–1368. doi: 10.1242/dev.091844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Lee PC, Chuang CC, Chiang AS, Ching YT. High-throughput computer method for 3D neuronal structure reconstruction from the image stack of the Drosophila brain and its applications. PLoS Comput Biol. 2012;8:e1002658. doi: 10.1371/journal.pcbi.1002658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/S0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/S0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Lee YH, Lin YN, Chuang CC, Lo CC. SPIN: a method of skeleton-based polarity identification for neurons. Neuroinformatics. 2014;12:487–507. doi: 10.1007/s12021-014-9225-6. [DOI] [PubMed] [Google Scholar]
- Leifer AM, Fang-Yen C, Gershow M, Alkema MJ, Samuel AD. Optogenetic manipulation of neural activity in freely moving Caenorhabditis elegans. Nat Methods. 2011;8:147–152. doi: 10.1038/nmeth.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon WC, Pulver SR, Höckendorf B, McDole K, Branson K, Freeman J, Keller PJ. Whole-central nervous system functional imaging in larval Drosophila. Nat Commun. 2015;6:7924. doi: 10.1038/ncomms8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levoy M, Ng R, Adams A, Footer M, Horowitz M. Light field microscopy. ACM Trans Graph. 2006;25:924–934. doi: 10.1145/1141911.1141976. [DOI] [Google Scholar]
- Li A, Gong H, Zhang B, Wang Q, Yan C, Wu J, Liu Q, Zeng S, Luo Q. Micro-optical sectioning tomography to obtain a high-resolution atlas of the mouse brain. Science. 2010;330:1404–1408. doi: 10.1126/science.1191776. [DOI] [PubMed] [Google Scholar]
- Lichtman JW, Livet J, Sanes JR. A technicolour approach to the connectome. Nat Rev Neurosci. 2008;9:417–422. doi: 10.1038/nrn2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima SQ, Miesenböck G. Remote control of behavior through genetically targeted photostimulation of Neurons. Cell. 2005;121:141–152. doi: 10.1016/j.cell.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Lin CW, Lin HW, Chiu MT, Shih YH, Wang TY, Chang HM, Chiang AS. Automated in situ brain imaging for mapping the Drosophila connectome. J Neurogenet. 2015;26:57–168. doi: 10.3109/01677063.2015.1078801. [DOI] [PubMed] [Google Scholar]
- Lin HH, Chu LA, Fu TF, Dickson BJ, Chiang AS. Parallel neural pathways mediate CO2 avoidance responses in Drosophila. Science. 2013;340:1338–1341. doi: 10.1126/science.1236693. [DOI] [PubMed] [Google Scholar]
- Lin JY, Knutsen PM, Muller A, Kleinfeld D, Tsien RY. ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nat Neurosci. 2013;16:1499–1508. doi: 10.1038/nn.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YY, Wu MC, Hsiao PY, Chu LA, Yang MM, Fu CC, Chiang AS. Three-wavelength light control of freely moving Drosophila melanogaster for less perturbation and efficient social-behavioral studies. Biomed Opt Express. 2015;6:514–523. doi: 10.1364/BOE.6.000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman J. Excitation, inhibition, local oscillations, or large-scale loops: what causes the symptoms of schizophrenia? Curr Opin Neurobiol. 2012;22:537–544. doi: 10.1016/j.conb.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, Tonegawa S. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484:381–385. doi: 10.1038/nature11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YC, Chiang AS. High-resolution confocal imaging and three-dimensional rendering. Methods. 2003;30:86–93. doi: 10.1016/S1046-2023(03)00010-0. [DOI] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- Long X, Ye J, Zhao D, Zhang SJ. Magnetogenetics: remote non-invasive magnetic activation of neuronal activity with a magnetoreceptor. Sci Bull. 2015;60:2107–2119. doi: 10.1007/s11434-015-0902-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson LJ, Zaharieva EE, Kearney PJ, Alpert MH, Lin TY, Turan Z, Lee CH, Gallio M. Dynamic labelling of neural connections in multiple colours by trans-synaptic fluorescence complementation. Nat Commun. 2015;6:10024. doi: 10.1038/ncomms10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markram H. The Blue Brain Project. Nat Rev Neurosci. 2006;7:153–160. doi: 10.1038/nrn1848. [DOI] [PubMed] [Google Scholar]
- Mayerich D, Abbott L, McCormick B. Knife-edge scanning microscopy for imaging and reconstruction of three-dimensional anatomical structures of the mouse brain. J Microsc. 2008;231:134–143. doi: 10.1111/j.1365-2818.2008.02024.x. [DOI] [PubMed] [Google Scholar]
- Merolla PA, Arthur JV, Alvarez-Icaza R, Cassidy AS, Sawada J, Akopyan F, Jackson BL, Imam N, Guo C, Nakamura Y, Brezzo B, Vo I, Esser SK, Appuswamy R, Taba B, Amir A, Flickner MD, Risk WP, Manohar R, Modha DS. A million spiking-neuron integrated circuit with a scalable communication network and interface. Science. 2014;345:668–673. doi: 10.1126/science.1254642. [DOI] [PubMed] [Google Scholar]
- Micheva KD, Smith SJ. Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron. 2007;55:25–36. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milham MP. Open neuroscience solutions for the connectome-wide association era. Neuron. 2012;73:214–218. doi: 10.1016/j.neuron.2011.11.004. [DOI] [PubMed] [Google Scholar]
- Nägerl UV, Bonhoeffer T. Imaging living synapses at the nanoscale by STED microscopy. J Neurosci. 2010;30:9341–9346. doi: 10.1523/JNEUROSCI.0990-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A, Pfeiffer BD, Rubin GM. Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc Natl Acad Sci U S A. 2015;112:E2967–E2976. doi: 10.1073/pnas.1506763112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolenko V, Poskanzer KE, Yuste R. Two-photon photostimulation and imaging of neural circuits. Nat Methods. 2007;4:943–950. doi: 10.1038/nmeth1105. [DOI] [PubMed] [Google Scholar]
- Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, Mortrud MT, Ouellette B, Nguyen TN, Sorensen SA, Slaughterbeck CR, Wakeman W, Li Y, Feng D, Ho A, Nicholas E, et al. A mesoscale connectome of the mouse brain. Nature. 2014;508:207–214. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama T, Schneider-Mizell CM, Fetter RD, Aleman JV, Franconville R, Rivera-Alba M, Mensh BD, Branson KM, Simpson JH, Truman JW, Cardona A, Zlatic M. A multilevel multimodal circuit enhances action selection in Drosophila. Nature. 2015;520:633–639. doi: 10.1038/nature14297. [DOI] [PubMed] [Google Scholar]
- Osten P, Margrie TW. Mapping brain circuitry with a light microscope. Nat Methods. 2013;10:515–523. doi: 10.1038/nmeth.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrovsky A, Cachero S, Jefferis G. Clonal analysis of olfaction in Drosophila: image registration. Cold Spring Harb Protoc. 2013;2013:347–349. doi: 10.1101/pdb.prot071738. [DOI] [PubMed] [Google Scholar]
- Osumi-Sutherland D, Reeve S, Mungall CJ, Neuhaus F, Ruttenberg A, Jefferis GS, Armstrong JD. A strategy for building neuroanatomy ontologies. Bioinformatics. 2012;28:1262–1269. doi: 10.1093/bioinformatics/bts113. [DOI] [PubMed] [Google Scholar]
- Pai TP, Chen CC, Lin HH, Chin AL, Lai JS, Lee PT, Tully T, Chiang AS. Drosophila ORB protein in two mushroom body-output neurons is necessary for long-term memory formation. Proc Natl Acad Sci U S A. 2013;110:7898–7903. doi: 10.1073/pnas.1216336110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palyanov A, Khayrulin S, Larson SD, Dibert A. Towards a virtual C. elegans: a framework for simulation and visualization of the neuromuscular system in a 3D physical environment. In Silico Biol. 2011;11:137–147. doi: 10.3233/ISB-2012-0445. [DOI] [PubMed] [Google Scholar]
- Pan C, Cai R, Quacquarelli FP, Ghasemigharagoz A, Lourbopoulos A, Matryba P, Plesnila N, Dichgans M, Hellal F, Ertürk A. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat Methods. 2016;13:859–867. doi: 10.1038/nmeth.3964. [DOI] [PubMed] [Google Scholar]
- Parekh R, Ascoli GA. Neuronal morphology goes digital: a research hub for cellular and system neuroscience. Neuron. 2013;77:1017–1038. doi: 10.1016/j.neuron.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastrana E. Focus on mapping the brain. Nat Methods. 2013;10:481. doi: 10.1038/nmeth.2509. [DOI] [PubMed] [Google Scholar]
- Paz JT, Huguenard JR. Microcircuits and their interactions in epilepsy: is the focus out of focus? Nat Neurosci. 2015;18:351–359. doi: 10.1038/nn.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody NC, Pohl JB, Diao F, Vreede AP, Sandstrom DJ, Wang H, Zelensky PK, White BH. Characterization of the decision network for wing expansion in Drosophila using targeted expression of the TRPM8 channel. J Neurosci. 2009;29:3343–3353. doi: 10.1523/JNEUROSCI.4241-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Chung P, Long F, Qu L, Jenett A, Seeds AM, Myers EW, Simpson JH. BrainAligner: 3D registration atlases of Drosophila brains. Nat Methods. 2011a;8:493–500. doi: 10.1038/nmeth.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Long F, Myers G. Automatic 3D neuron tracing using all-path pruning. Bioinformatics. 2011b;27:i239–i247. doi: 10.1093/bioinformatics/btr237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Hawrylycz M, Roskams J, Hill S, Spruston N, Meijering E, Ascoli GA. BigNeuron: large-scale 3D neuron reconstruction from optical microscopy images. Neuron. 2015;87:252–256. doi: 10.1016/j.neuron.2015.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchon TA, Gao L, Milkie DE, Davidson MW, Galbraith JA, Galbraith CG, Betzig E. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat Methods. 2011;8:417–423. doi: 10.1038/nmeth.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack RA, Gorgolewski KJ. Making big data open: data sharing in neuroimaging. Nat Neurosci. 2014;17:1510–1517. doi: 10.1038/nn.3818. [DOI] [PubMed] [Google Scholar]
- Ragan T, Kadiri LR, Venkataraju KU, Bahlmann K, Sutin J, Taranda J, Arganda-Carreras I, Kim Y, Seung HS, Osten P. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat Methods. 2012;9:255–258. doi: 10.1038/nmeth.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-Lavigne M. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Richier B, Salecker I. Versatile genetic paintbrushes: Brainbow Technologies. Wiley Interdiscip Rev Dev Biol. 2015;4:161–180. doi: 10.1002/wdev.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickgauer JP, Tank DW. Two-photon excitation of channelrhodopsin-2 at saturation. Proc Natl Acad Sci U S A. 2009;106:15025–15030. doi: 10.1073/pnas.0907084106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schemmel J, Briiderle D, Griibl A, Hock M, Meier K, Millner S. A wafer-scale neuromorphic hardware system for large-scale neural modeling. Proc 2010 IEEE Int Symp Circuits Syst; 2010. pp. 1947–1950. [Google Scholar]
- Sharpee TO. Toward functional classification of neuronal types. Neuron. 2014;83:1329–1334. doi: 10.1016/j.neuron.2014.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih CT, Sporns O, Yuan SL, Su TS, Lin YJ, Chuang CC, Wang TY, Lo CC, Greenspan RJ, Chiang AS. Connectomics-based analysis of information flow in the Drosophila brain. Curr Biol. 2015;25:1249–1258. doi: 10.1016/j.cub.2015.03.021. [DOI] [PubMed] [Google Scholar]
- Shinomiya K, Matsuda K, Oishi T, Otsuna H, Ito K. Flybrain neuron database: a comprehensive database system of the Drosophila brain neurons. J Comp Neurol. 2011;519:807–833. doi: 10.1002/cne.22540. [DOI] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Kötter R. The Human Connectome: a structural description of the human brain. PLOS Comput Biol. 2005;1:e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szigeti B, Gleeson P, Vella M, Khayrulin S, Palyanov A, Hokanson J, Currie M, Cantarelli M, Idili G, Larson S. OpenWorm: an open-science approach to modeling Caenorhabditis elegans. Front Comput Neurosci. 2014;8:137. doi: 10.3389/fncom.2014.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tainaka K, Kubota SI, Suyama TQ, Susaki EA, Perrin D, Ukai-Tadenuma M, Ukai H, Ueda HR. Whole-body imaging with single-cell resolution by tissue decolorization. Cell. 2014;159:911–924. doi: 10.1016/j.cell.2014.10.034. [DOI] [PubMed] [Google Scholar]
- Takemura SY, Bharioke A, Lu Z, Nern A, Vitaladevuni S, Rivlin PK, Katz WT, Olbris DJ, Plaza SM, Winston P, Zhao T, Horne JA, Fetter RD, Takemura S, Blazek K, Chang LA, Ogundeyi O, Saunders MA, Shapiro V, Sigmund C, et al. A visual motion detection circuit suggested by Drosophila connectomics. Nature. 2013;500:175–181. doi: 10.1038/nature12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia JC, Kasthuri N, Hayworth KJ, Schalek R, Lichtman JW, Smith SJ, Buchanan J. High-contrast en bloc staining of neuronal tissue for field emission scanning electron microscopy. Nat Protoc. 2012;7:193–206. doi: 10.1038/nprot.2011.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillberg PW, Chen F, Piatkevich KD, Zhao Y, Yu CC, English BP, Gao L, Martorell A, Suk HJ, Yoshida F, DeGennaro EM, Roossien DH, Gong G, Seneviratne U, Tannenbaum SR, Desimone R, Cai D, Boyden ES. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat Biotechnol. 2016;34:987–992. doi: 10.1038/nbt.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Pelt J, van Ooyen A, Uylings HB. The need for integrating neuronal morphology databases and computational environments in exploring neuronal structure and function. Anat Embryol. 2001;204:255–265. doi: 10.1007/s004290100197. [DOI] [PubMed] [Google Scholar]
- Wheeler MA, Smith CJ, Ottolini M, Barker BS, Purohit AM, Grippo RM, Gaykema RP, Spano AJ, Beenhakker MP, Kucenas S, Patel MK, Deppmann CD, Güler AD. Genetically targeted magnetic control of the nervous system. Nat Neurosci. 2016;19:756–761. doi: 10.1038/nn.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Wickersham IR, Lyon DC, Barnard RJ, Mori T, Finke S, Conzelmann KK, Young JA, Callaway EM. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Chu LA, Hsiao PY, Lin YY, Chi CC, Liu TH, Fu CC, Chiang AS. Optogenetic control of selective neural activity in multiple freely moving Drosophila adults. Proc Natl Acad Sci U S A. 2014;111:5367–5372. doi: 10.1073/pnas.1400997111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Peng H. APP2: automatic tracing of 3D neuron morphology based on hierarchical pruning of a gray-weighted image distance-tree. Bioinformatics. 2013;29:1448–1454. doi: 10.1093/bioinformatics/btt170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Jarrell TA, Wang Y, Cook SJ, Hall DH, Emmons SW. Computer assisted assembly of connectomes from electron micrographs: application to Caenorhabditis elegans. PLoS One. 2013;8:e54050. doi: 10.1371/journal.pone.0054050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen CK, Lubeck E, Shah S, Cai L, Gradinaru V. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell. 2014;158:945–958. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Prigge M, Schneider F, Davidson TJ, O'Shea DJ, Sohal VS, Goshen I, Finkelstein J, Paz JT, Stehfest K, Fudim R, Ramakrishnan C, Huguenard JR, Hegemann P, Deisseroth K. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang LP, Brauner M, Liewald JF, Kay K, Watzke N, Wood PG, Bamberg E, Nagel G, Gottschalk A, Deisseroth K. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L. Mosaic analysis with double markers in mice. Cell. 2005;121:479–492. doi: 10.1016/j.cell.2005.02.012. [DOI] [PubMed] [Google Scholar]