Abstract
Musk odors have been used widely for fragrance and medicine for >2000 years because of their fascinating scent and physiological effects. Therefore, fragrance manufacturers have been eager to develop high-quality musk compounds that are safe and easily synthesized. We recently identified muscone-responsive olfactory receptors (ORs) MOR215-1 and OR5AN1 in mice and humans, respectively (Shirasu et al., 2014). In this study, we identified musk ORs that are evolutionarily closely related to MOR215-1 or OR5AN1 in various primates and investigated structure–activity relationships for various musk odorants and related compounds. We found that each species has one or two functional musk ORs that exhibit specific ligand spectra to musk compounds. Some of them, including the human OR5AN1, responded to nitro musks with chemical properties distinct from muscone. The ligand specificity of OR5AN1 reflects the perception of musk odors in humans. Genetic deletion of MOR215-1 in mice resulted in drastic reduction of sensitivity to muscone, suggesting that MOR215-1 plays a critical role in muscone perception. Therefore, the current study reveals a clear link between the identified OR and muscone perception. Moreover, the strategy established for screening ligands for the muscone OR may facilitate the development of novel and commercially useful musk odors.
SIGNIFICANCE STATEMENT The long-sought musk odor receptor family in mammals was discovered and found to be well conserved and narrowly tuned to musk odors. In mice, deletion of the most sensitive musk receptor resulted in drastic reduction in sensitivity to muscone, demonstrating a strong link between receptor and odor perception. In humans, we found one musk receptor that recognized both macrocyclic and nitro musks that had distinct chemical structures. The structure–activity relationships were in a good agreement with human sensory perception and therefore may be used to develop novel musk aroma in fragrance fields. Finally, identification of a natural ligand(s) for musk receptors in mammals other than musk deer would reveal an evolutionarily pivotal role in each species in the future.
Keywords: evolution, human, mouse, musk, olfactory, receptor
Introduction
Musk odors are considered to be “king” of the fragrance world because of their warm, sensual, fascinating scent. They have been used widely in commercial products such as perfumes and detergents. The first natural musk compound, muscone, was reported in 1906 by Walbaum as the major odorant in secretions of musk deer (Moschus moschiferus) (Walbaum, 1906). Muscone turned out to have a unique macrocyclic ketone structure with a 15-membered ring (Ruzicka et al., 1926). Males secrete muscone, which in turn attracts females, suggesting that muscone is a male pheromone in musk deer. Some reports showed that, in humans, testosterone and estradiol levels increased after muscone exposure to female subjects, suggesting that muscone may have physiological effects also on humans (Kato et al., 2004, Fukui et al., 2007). Other muscone-like macrocyclic ketones with a 15- to 17-membered ring such as civetone and cyclopentadecanone have been identified from the stink glands of civet cat (Viverra civetta), musk rat (Ondatra zibethicus), and musk shrew (Suncus murinus) (Asada et al., 2012).
Many compounds that mimic the aroma of muscone have been synthesized for perfumery applications (Matsuda et al., 2004). These include nitro musks (NMs) such as musk xylol and musk ketone and polycyclic musks (PCMs) such as galaxolide. The use of many of these compounds is now prohibited because of skin sensitivity (toxicity). Other macrocyclic musks (MCMs), such as muscenone and ethylene brassylate, have been developed as safe, biodegradable compounds with an excellent musk aroma, but low diffusibility and difficulty in synthesis continue to pose a challenge. Therefore, the development of new musk compounds remains an important goal of the cosmetics industry.
Although these compounds all smell musky, their structures differ considerably. Therefore, it is important to understand how these musk odor compounds are recognized at a receptor level in the human nose. In general, odorants are recognized in a combinatorial fashion by dozens of olfactory receptors (ORs) expressed in olfactory sensory neurons (Malnic et al., 1999; Touhara and Vosshall, 2009). However, muscone activates only a few glomeruli in the mouse olfactory bulb, suggesting that it is recognized by a small number of ORs in mice (Shirasu et al., 2014). Using a retrograde neural labeling technique, a muscone-responsive mouse OR, MOR215-1, was identified. MOR215-1 appeared to be tuned to several musk odorants (Shirasu et al., 2014). OR5AN1, the human OR most similar to MOR215-1 in amino acid sequence, was found to be a muscone receptor (Shirasu et al., 2014).
In this study, we looked for additional muscone ORs in mice and humans and their orthologs in various primate species. Our findings identified one or two muscone ORs in each species, although they differed with respect to ligand selectivity, sensitivity, and efficacy. To further examine the importance of the identified muscone ORs in recognizing musk odors, the effect of single muscone receptor deletion on muscone perception was examined in mice. The current studies provide insights into how each OR is involved in the perception of the cognate odor ligands. Moreover, these findings may be used to advance commercial development of novel musk odorants.
Materials and Methods
Odorants.
Musk odorants used in this study were kindly provided by Takasago International Corporation or were purchased from Wako or TCI. Eugenol was purchased from TCI. Propylene glycol was purchased from ADEKA. Mineral oil was purchased from Wako. The optical purity of l- and d-muscone is >98% enantiomeric excess (ee) and 96% ee, respectively. The musk odorants, musk analogs, and odorant mixtures used for the luciferase assay were prepared as 100 mm stock solutions in dimethyl sulfoxide. The compounds used for examining structure–activity relationships are grouped into five types: (1) MCMs, including 1. muscone, 2. muscenone, 3. cyclopentadecanone, 4. ambretone, 5. globanone, 6. cosmone, 7. 3-methylcyclotetradecanone, 8. ethylene brassylate, 9. ambrettolide, 10. exaltolide, and 11. habanolide; (2) NMs, including 12. musk xylol, and 13. musk ketone; (3) PCMs, including 14. galaxolide, 15. tonalide, 16. celestolide, and 17. cashmeran; (4) alicyclic musks (ACMs), including 18. helvetolide; and (5) others, including 19. cyclopentadecane, 20. cyclopentadecanol, 21. cyclohexanone, 22. cyclodecanone, 23. cycloundecanone, 24. 2-pentadecanone, and 25. 8-pentadecanone) (see Fig. 2A).
Figure 2.
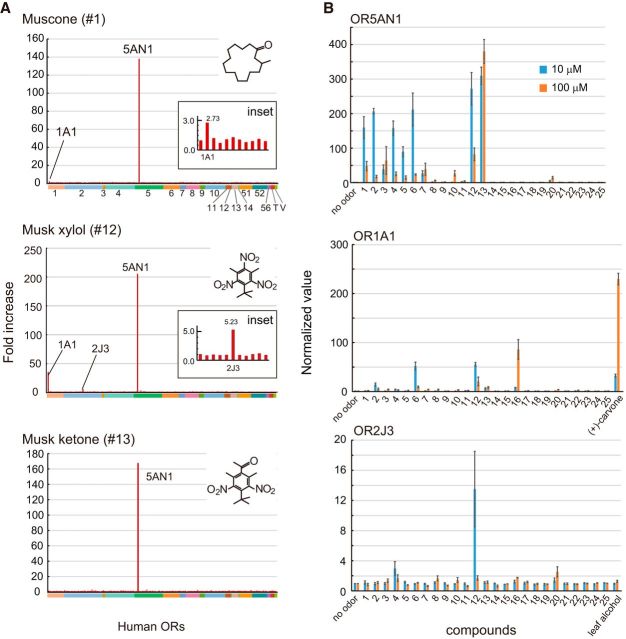
Structure–activity relationships of the muscone OR family for muscone-related compounds. A, Chemical structures of 18 musk odorants and seven related compounds used in structure–activity relationship studies grouped into five types: MCM (pink): 1. muscone [C16H30O], 2. muscenone [C16H28O], 3. cyclopentadecanone [C15H28O], 4. ambretone [C16H28O], 5. globanone [C16H28O], 6. cosmone [C15H26O], 7. 3-methylcyclotetradecanone [C15H28O], 8. ethylene brassylate [C15H26O4], 9. ambrettolide [C16H28O2], 10. exaltolide [C15H28O2], and 11. habanolide [C15H26O2]); NM (blue): 12. musk xylol [C12H15N3O6] and 13. musk ketone [C14H18N2O5]); PCM (yellow): 14. galaxolide [C18H26O], 15. tonalide [C18H26O], 16. celestolide [C17H24O], and 17. cashmeran [C14H22O]); ACM (orange): 18. helvetolide [C17H32O3]; and others (gray): 19. cyclopentadecane [C15H30], 20. cyclopentadecanol [C15H30O], 21. cyclohexanone [C6H10O], 22. cyclodecanone [C10H18O], 23. cycloundecanone [C11H20O], 24. 2-pentadecanone [C15H30O], and 25. 8-pentadecanone [C15H30O]). B, Response patterns of musk ORs to various musk-related compounds at 10 μm in the HEK293 luciferase assay. The upper dendrogram indicates the result of cluster analysis based on odor response patterns. The normalized values used for the cluster analysis were calculated by the following formula: (Fn−Fno odor) × 100/∑n=125Fn − Fno odor where Fn and Fno odor represent the fold increase for the nth odor and no odor, respectively. When Fn is smaller than Fno odor, the normalized value is regarded to be zero. Only positive values are shown. The empty boxes indicate that the value is <0.1.
Phylogenetic analyses.
Neighbor-joining phylogenetic trees (Saitou and Nei, 1987) were constructed using the program MEGA6 (Tamura et al., 2013) with Poisson correction distance and complete deletion option. MAFFT (http://mafft.cbrc.jp/alignment/software/) was used to make multiple alignments.
Cloning of ORs.
All ORs used in Figures 1, 2, 3, and 4B were amplified from human genomic DNA (Promega), mouse DNA (C57BL/6 that had been bred in our laboratory), or primate DNA and were extracted from tissues provided by the Primate Research Institute, Kyoto University. Cloned OR sequences of humans and mice are identical to those in GenBank (http://www.ncbi.nlm.nih.gov/genbank/). Cloned OR sequences of chimpanzees (Pan troglodytes), orangutans (Pongo abelii), macaques (Macaca mulatta), and marmosets (Callithrix jacchus) are identical to those identified in the previous study (Matsui et al., 2010). We inferred the nucleotide sequence of a functional version of the pseudogene OR5AN2P, OR5AN2, based on the sequence of the chimpanzee ortholog, PatrOR11R.6.1. We generated OR5AN2 by inserting a guanine nucleotide using Prime STAR Mutagenesis Basal Kit (TaKaRa).
Figure 1.

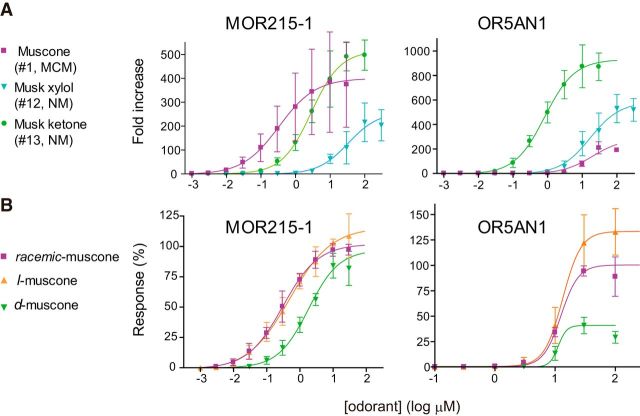
Phylogenetic analysis and responsiveness of the muscone OR family. A, Phylogeny and muscone responses of the muscone OR family in six mammalian species. Left, Phylogenetic tree of OR genes in three OGGs (Niimura et al., 2014). The scale bar indicates the number of amino acid substitutions per site. Bootstrap values obtained from 500 resamplings are shown on the tree. Right, Responses of ORs expressed in HEK293 cells to 100 μm muscone in the luciferase reporter gene assay. The ORs with colored letters showed relatively strong responses to muscone. Normalized value was calculated by dividing the fold increase upon stimulation with muscone in OR-expressing cells by the fold increase in empty vector control cells. The broken line indicates the value of empty vector control, 1.0. Error bars indicate SEM (n = 3). OR5AN1 and OR5AN2 (an inferred functional version of OR5AN2P, see Materials and Methods) in human; Patr_5AN2 (PatrOR11R.6.1) in chimpanzee; Poab_5AN1 (PoabORc2797.1) and Poab_5AN2 (PoabORc2797.5) in orangutan; Mamu_5AN1 (MamuOR14.1.9) and Mamu_5AN2 (MamuOR14.1.12) in macaque; Caja_5AN2 (CajaORc7026.1) in marmoset; MOR215-1 (Olfr1440), MOR214-1 (Olfr262), MOR214-2 (Olfr1436), MOR214-3 (Olfr235), MOR214-4 (Olfr1433/1434), MOR214-5 (Olfr1431), and MOR214-6 (Olfr1437) in mice. B, Mouse cluster on chromosome 19 corresponded to marmoset clusters on chromosome 11, macaque clusters on chromosome 14, orangutan clusters on chromosome 11, chimpanzee clusters on chromosome 11, and human clusters on chromosome 11. Each horizontal line represents a chromosome. The position of each OR gene is represented by a colored vertical bar above or below a horizontal line, the latter indicating the opposite transcriptional direction to the former. Each bar is colored according to the OGG to which the OR gene belongs. The genes in the same OGG are connected by a dashed line. C, Dose–response curves of muscone ORs. The squares show normalized luciferase values. The normalized luciferase value was calculated by the following formula: (Fn − Fno odor) * 100/(Fmax − Fno odor), where Fn represents the fold increase at a concentration of muscone, Fno odor represents the fold increase for no odor, and Fmax represents the fold increase for muscone at a concentration that showed a maximal response. EC50 and 95% confidential intervals (in parentheses) are shown in each graph. Error bars indicate SEM (n = 3).
Figure 3.
Dose–responses of MOR215-1 and OR5AN1 to NMs and muscone enantiomers. A, Dose–response curves of MOR215-1 and OR5AN1 to muscone, musk xylol, and musk ketone. The EC50 values (μm) of MOR215-1 to muscone were 0.50; to musk xylol, 34.9; and to musk ketone, 2.9. The EC50 values (μm) of OR5AN1 to muscone were 12.5; to musk xylol,16.7; and to musk ketone, 0.79. The calculation method of fold increase was described in the Materials and Methods. Error bars indicate SEM (n = 3). The data for muscone are the same as in Figure 1C. B, Dose–response curves of MOR215-1 and OR5AN1 to racemic-muscone, l-muscone, and d-muscone. The EC50 values (μm) of MOR215-1 to l-muscone were 0.63 and to d-muscone were 2.2. The EC50 values (μm) of OR5AN1 to l-muscone were 13.3 and to d-muscone were 11.3. Response (%) of y-axis is similar to the normalized luciferase value in Figure 1C, but Fmax represents the fold increase for racemic-muscone at a concentration that showed the maximal response. Error bars indicate SEM (n = 3). The data for racemic-muscone are the same as in Figure 1C.
Figure 4.
Screening of human ORs for muscone and NMs. A, Screening of musk receptors from the human chemosensory receptor repertoire. A total of 405 ORs, seven TAARs, and five VNRs expressed in HEK293 cells were stimulated with 100 μm muscone, musk xylol, or musk ketone, and the luciferase assay was performed. Numbers (#1–14, #51, #52, #56) with color bars on the x-axis indicate OR families. T, TAARs; V, VNRs. Fold increase was calculated using the same method as in Figure 3 (see Materials and Methods). Mean values of two screening replicates are shown. B, Structure–activity relationships of OR5AN1, OR1A1, and OR2J3 for musk-related compounds shown in Figure 2A. The blue and orange bars show the responses to 10 and 100 μm compounds, respectively. The normalized value was calculated for each compound by the following formula: Fn/Fempty vector, where Fn and Fempty vector represent the fold increase of each OR and empty vector, respectively. Error bars indicate SEM (n = 3).
ORs were amplified using Ex Taq (TaKaRa) with primers upstream (5′-TCAGGACGGACTCCCATCTGCATACA-3′ for OR5AN2P, 5′-GCGCGAATTCATGACTGGGGAAAGGAACAGTACAAG-3′ for PatrOR11R.6.1, 5′-GCGCGAATTCATGACTGGGGGAGGAAATATTACAG-3′ for PoabORc2797.1, 5′-GCGCGAATTCATGACTGGGGAAAGGAACAGTACAAG-3′ for PoabORc2797.5, 5′-GCGCGAATTCATGACTAGGGGAGGAAATATTACAG-3′ for MamuOR14.1.9, 5′-GCGCGAATTCATGACTGAGGAAAGGAACAGTACAAC-3′ for MamuOR14.1.12, 5′-GCGCGAATTCATGACTGGGGAAAGGAACAGTACAATTAC-3′ for CajaORc7026.1, 5′-GCGCGAATTCATGATTGCCAGGGGAAACAGCACAG-3′ for MOR214-1, 5′-GCGCAGATCTATGGAGGCAATGATTAAAGGAAAAAAC-3′ for MOR214-2, 5′-GCGCGAATTCATGATTGGGGAAAGAAATATTACCA-3′ for MOR214-3, 5′-GCGCATGATTGGGGGAAGAAATATTACCA-3′ for MOR214-4, 5′-GCGCGAATTCATGATTGGGGAAAGAAATATTACCA-3′ for MOR214-5, 5′-GCGCAGATCTATGATTAAGGAAAGAAACTTTAC-3′ for MOR214-6) and downstream (5′-GTAGTGGGAATAGTCCTGCCTGAGGG-3′ for OR5AN2P, 5′-GCGCCTCGAGTTATTTCATTCCATAACACCAGGAG-3′ for PatrOR11R.6.1, 5′-GCGCCTCGAGTCAGCAGCACCTTCTCTTTTGCAAC-3′ for PoabORc2797.1, 5′-GCGCCTCGAGTTATTTCATTCCATAACACCAGGAG-3′ for PoabORc2797.5, 5′-GCGCCTCGAGTCAGCAGCACCTTCTCTTTTGCAAC-3′ for MamuOR14.1.9, 5′-GCGCCTCGAGTTATTTCATTCCATAACACCAGGAG-3′ for MamuOR14.1.12, 5′-GCGCCTCGAGTTATTTCATTCCATAACACCAGGGG-3′ for CajaORc7026.1, 5′-GCGCCTCGAGCTAGTTGCAGATTGTCTTTTTCTGC-3′ for MOR214-1, 5′-GCGCCTCGAGTTAGCAGCATCCCAGCTTCTTCTGTAAC-3′ for MOR214-2, 5′-GCGCCTCGAGTTAGCTGCAAATTTTCTTTTTCTGC-3′ for MOR214-3, 5′-GCGCTTAGCTGCAGATTACCTTTTTCTGC-3′ for MOR214-4, 5′-GCGCCTCGAGTTAGTTGCAGATTTTCTTTTTCTGC-3′ for MOR214-5, 5′-GCCTCGAGTTAGCAGCATCCCAGCTTC-3′ for MOR214-6).
A polynucleotide encoding the first 20 aa of human rhodopsin was included as a tag in pME18S for expression in HEK293 cells.
Cell culture and the luciferase assay.
Cells were grown in a 37°C incubator containing 5% CO2. The luciferase reporter gene assay was performed as described previously (Zhuang and Matsunami, 2008; Shirasu et al., 2014). Dual-Glo Luciferase Assay System (Promega) was used to measure firefly and Renilla luciferase activities. cAMP response element promoter-containing firefly luciferase vector (CRE/luc2P-pGL4.29; Promega) was used to measure receptor activation. A thymidine kinase promoter-containing Renilla luciferase vector (TK/Rluc-pGL4.74; Promega) was used as an internal control for cell viability and transfection efficiency. A short form of RTP1 called RTP1S-pME18S vector was used to enhance functional expression of the ORs. HEK293 cells were cultured in poly-d-lysine-coated 96-well plates (BD BioCoat) and transfected with 50 ng of the tagged mouse OR vector, 10 ng of CRE/luc2P, 5 ng of TK/Rluc, and 10 ng of RTP1S-pME18S vector using 0.15 μl of Lipofectamine 2000 (Invitrogen) per well. Approximately 24 h after transfection, the medium was replaced with CD293 (Invitrogen) containing 20 μm l-glutamine in the presence or absence of an odorant and the cells were then incubated 3 h at 37°C and 5% CO2 (stimulation). Luminescence was measured using Centro LB960 plate reader (Berthold Technologies). Relative luciferase activity, L, was calculated as luminescence of firefly luciferase divided by luminescence of Renilla luciferase in a given well. Fold increase was calculated by the following formula: Ln/Lno odor, where Ln represents the relative luciferase activity in a given well and Lno odor represents the relative luciferase activity in a well not stimulated by any odorant. Methods for calculating normalized values in each figure are provided in figure legends. Each assay was done in triplicate. Mean values of three screening triplicates are defined as n = 1.
Data were analyzed with Microsoft Excel and GraphPad Prism 3. Cluster analysis of the OR ligand spectra in Figure 2 was by Ward's method using R 3.0.2.
Screening assay.
In the experiment screening for musk odor receptors (Fig. 4A), 405 human ORs, seven trace amine associated receptors (TAARs), and five vomeronasal receptors (VNRs) were cloned from human genomic DNA (Promega). All SNPs that were different from the reference sequences in GenBank (http://www.ncbi.nlm.nih.gov/genbank/) or HORDE (http://genome.weizmann.ac.il/horde/) were found in NCBI dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/).
To screen human ORs against muscone and musk xylol, HEK293 cells were cultured in 96-well plates and transfected with 75 ng of a FLAG-Rho-tagged OR vector, 30 ng of CRE/luc2P-pGL4.29 vector, 30 ng of CMV/hRluc-pGL4.75 vector, and 30 ng of human RTP1S-pME18S vector using 0.41 μl of Lipofectamine 2000 per well. Twenty-four to 27 h after transfection, cells were stimulated with muscone or musk xylol for 2.5 h. Only in the case of screening human ORs for musk ketone were HEK293 cells plated on poly-d-lysine-coated 384-well plates (BD BioCoat). The cells were transfected with a 25:10:19:1 mass ratio of FLAG-Rho-tagged OR vector, human RTP1S-pME18S vector, CRE/luc2P-pGL4.29 vector, and CMV/hRluc-pGL4.75 vector. The transfected cells were stimulated with DMEM in the presence or absence of musk ketone for 3 h. Luminescence from the 384-well plate was measured using EnVision (PerkinElmer). Each assay was done in duplicate. The mean (m) and the standard deviation (s) of fold increases among all of the examined receptors except for OR5AN1 were calculated. When a fold increase of a given receptor was larger than the value of m + 3s, the receptor was considered to be activated.
Behavioral assay.
C57BL/6 male and female mice (8–15 weeks old; CLEA Japan) MOR215-1 deletion male and female mice (8–29 weeks old; MOR103-1→MOR215-1-IRES tau-ECFP; Nakashima et al., 2013) were used. The housing room was maintained at a constant temperature (23 ± 1°C) and under a 12:12 h light:dark cycle. All animal experiments were approved by the Animal Care Committees of the University of Tokyo. Each animal was housed individually in its home cage with clean bedding. In the same home cage 2 d after isolation, odor-source-exploring experiments were conducted as described previously (Shirasu et al., 2014). A mouse was exposed for 5 min to the tip of a glass capillary that had been dipped in an odorant (odor source) and the time taken to begin actively sniffing the odor source (the tip of a capillary) was recorded. The tip of each capillary was carefully set so that the mouse could not reach it directly. Mice were exposed to a low concentration of odorant first. If the mouse did not find the odor source, then it was exposed to a higher concentration. The experiment was performed on each mouse until it found the odor source or up to three times a day. The experiment was repeated in the same animals >1 week later with other odor sources and concentrations. Tests were performed during dark periods and were designed in a double-blind manner. Mice were recorded with a digital video camera. The latency to find the odor source between wild-type (WT) and knock-out (KO) mice was compared statistically by performing the Scherier–Ray–Hare test, a nonparametric version of the repeated two-way ANOVA (Sokal and Rohlf, 2001; Márquez et al., 2015).
Results
Phylogenetic analysis
We reported previously that MOR215-1 and OR5AN1 are muscone receptors in mice and humans, respectively (Shirasu et al., 2014). To find muscone receptors in other mammals, we conducted phylogenetic analyses of OR genes using the orthologous gene groups (OGGs) among 13 placental mammals reported by Niimura et al. (2014), to which MOR215-1 and OR5AN1 were closely related in an OR gene tree. An OGG is defined as all extant descendant genes originated from a single gene in the most recent common ancestor of a given set of species (Niimura et al., 2014). MOR215-1 and OR5AN1 were found to belong to OGG2-417 and OGG2-59, respectively. OGG2-417 does not contain any primate OR genes. The most similar human OR gene to MOR215-1 was OR5AN2P belonging to OGG2-120, which contains several intact primate OR genes. It has been shown that other ORs closely related to OR5AN1 in humans (OR5A1 and OR5A2) and to MOR215-1 in mice (MOR215-2 and MOR215-3) do not respond to muscone (Shirasu et al., 2014). For these reasons, in this study, we focused on three OGGs, OGG2-59, OGG2-417, and OGG2-120, for further analysis (Fig. 1A). From these OGGs, we selected ORs from five primate species (marmoset, macaque, orangutan, chimpanzee, and human) and mice for analyzing muscone responsiveness (Fig. 1A). These OR genes are located on a syntenic chromosome among six species (Fig. 1B). Common non-OR genes, DTX4, MPEG, OSBP, and PATL1, are present near the OR gene cluster containing OGG2-417, OGG2-120, and OGG2-59 genes in all of the six species except for macaques.
Identification of the muscone receptor family in mammals
When we expressed each of the six ORs in the MOR214 subfamily in HEK293 cells, only MOR214-3 showed a strong response to 100 μm muscone in the luciferase reporter gene assay (Fig. 1A), consistent with the previous study (McClintock et al., 2014). This result suggests that most of these ORs have lost the ability to respond to muscone despite the mouse-specific OR gene expansion in OGG2-59. In the same functional assay, orangutan and macaque OR5AN1 orthologs Poab_5AN1 (PoabORc2797.1) and Mamu_5AN1 (MamuOR14.1.9) responded to muscone (the gene names in parentheses are from Niimura et al., 2014). A human OR pseudogene, OR5AN2P, belongs to OGG2-120. Among ORs in this OGG, orangutan and chimpanzee OR5AN2P orthologs, Poab_5AN2 (PoabORc2797.5) and Patr_5AN2 (PatrOR11R.6.1), exhibited strong responses to muscone, whereas macaque and marmoset orthologs, Mamu_5AN2 (MamuOR14.1.12) and Caja_5AN2 (CajaORc7026.1), in the same OGG weakly responded to muscone. Moreover, we constructed an inferred functional version of OR5AN2P based on the chimpanzee ortholog Patr_5AN2 by inserting a single guanine nucleotide after the 155th nucleotide in the OR5AN2P coding sequence (c.155_156 insG). The modified OR5AN2, however, did not respond to muscone. It is fair to say that OGG2-59, OGG2-417, and OGG2-120 represent orthologous gene families encoding muscone receptors in mammals.
We next investigated sensitivity of the ORs that strongly responded to muscone by applying various concentrations of muscone (Fig. 1C). The EC50 value for MOR215-1 was 0.5 μm and those of the other muscone receptors were between 5.4 and 19.4 μm. MOR215-1 appears to possess 10-fold greater sensitivity to muscone than other muscone receptors. It is notable that other typical ORs usually show EC50 values at μm levels, suggesting that MOR215-1 is a relatively sensitive OR.
Structure–activity relationships for various musk compounds
We examined the responsiveness of the muscone receptors to other musk odorants and related compounds, including MCMs, NMs, PCMs, and an ACM (Fig. 2A). Exposures of muscone or unsaturated MCM compounds at concentrations >100 μm often gave unreliable responses. This was likely due to cytotoxicity to HEK293 cells (Fig. 1C for muscone; data not shown for other compounds). Therefore, in this experiment, we used the 10 μm concentration, which was in the range of EC50 values.
Figure 2B shows relative responsiveness of each musk OR to 25 musk-related compounds. A cluster analysis based on the results of the structure–activity studies identified four clusters of musk ORs. Although phylogenetically close ORs tended to be in the same cluster, the clustering did not always reflect phylogenetic relationships. The first cluster of ORs (OR5AN1, Poab_5AN1, and Mamu_5AN1) showed responses to musk ketone (#13) most strongly and also to musk xylol (#12) and macrocyclic ketones with a 14- to 16-membered ring (#1–7), but very weakly to 17-membered rings (#8, #9). The second group (Caja_5AN2 and Mamu_5AN2) appeared to be relatively narrowly tuned ORs that responded only to macrocyclic ketones with a 15- or 16-membered ring and only very weak responses to some of other musk odorants. The third cluster (Patr_5AN2 and MOR215-1) showed relatively broad responses to musk odorants and related compounds. The fourth cluster (MOR214-3 and Poab_5AN2) responded to unsaturated macrocyclic ketones with a 15- or 16-membered ring (#2, #4, #5) more strongly than to muscone. The third and fourth clusters showed responses, not only to MCMs, a 15- to 17-membered ring with a ketone or lactone group (#1–11), but also to a hydroxyl group (#20). The oxygen, however, appears to be essential for the responsiveness, because C15 cyclic alkane (#19) did not induce responses.
Responsiveness of MOR215-1 and OR5AN1 to NMs and muscone enantiomers
We selected MOR215-1 and OR5AN1 as representatives from the two groups and measured their dose-dependent responses to muscone (MCM #1), musk xylol (NM #12), and musk ketone (NM #13; Fig. 3A). The dose–response curves for musk xylol and musk ketone were similar between MOR215-1 and OR5AN1, whereas their sensitivity to muscone was drastically different. The EC50 value of MOR215-1 for muscone (0.5 μm) was much lower than that of musk xylol (34.9 μm) and musk ketone (2.9 μm). In contrast, OR5AN1 responded to musk ketone (EC50 = 0.8 μm) much more strongly than to muscone (EC50 = 12.5 μm) and musk xylol (EC50 = 16.7 μm).
We reported previously that MOR215-1 has differential affinities to enantiomers of muscone in the Xenopus laevis oocyte expression system (Shirasu et al., 2014). We herein determined the sensitivity of MOR215-1 for muscone enantiomers in the luciferase assay (Fig. 3B). For MOR215-1, racemic-muscone (EC50 = 0.5 μm) and l-(R)-muscone (EC50 = 0.6 μm) showed higher affinity than d-(S)-muscone (EC50 = 2.2 μm), consistent with our previous results except that racemic- and l-muscone responses were similar in this study, likely due to the difference in the assay systems. The maximal responses were similar among the three musks. In contrast, OR5AN1 showed similar EC50 values for l-muscone (EC50 = 13.3 μm), racemic-muscone (EC50 = 12.5 μm), and d-muscone (EC50 = 11.3 μm), whereas the response intensities were drastically different: l-muscone, ∼300-fold increase; racemic-muscone, ∼200-fold increase; and d-muscone, ∼100-fold increase.
Screening musk odor receptors in the human OR repertoire
To further screen human musk ORs, HEK293 cells expressing each of 405 human ORs, seven TAARs, and five VNRs were stimulated with muscone, musk xylol, or musk ketone and the responsiveness was measured using the luciferase assay system. In addition to OR5AN1, which responded to all three musk compounds, OR1A1 was found to respond to muscone and musk xylol weakly, but not to musk ketone (Fig. 4A). We tested a series of musk compounds for OR1A1 and found that OR1A1 responded more to unsaturated C14–15 macrocyclic ketones, muscenone and cosmone (#2, #6) and a PCM, celestolide (#16; Fig. 4B). OR1A1 has been known to be a relatively nonselective OR that responds to a variety of odorants, including terpenes, aldehydes, alcohols, and ketones (Saito et al., 2009). Indeed, OR1A1 showed a robust response to a monoterpene (+)-carvone (Fig. 4B).
OR2J3 showed a relatively specific response to musk xylol (Fig. 4B). This OR is known as a cis-3-hexen-1-ol (leaf alcohol) receptor (McRae et al., 2012). However, we could not reproduce any responses to leaf alcohol in OR2J3. MOR256-17, having a sequence similarity to MOR256-18, a mouse ortholog to OR2J3, has been shown to respond to trinitrotoluene (Li et al., 2012), an explosive material, suggesting that the orthologous group containing OR2J3 includes a receptor for trinitrobenzene compounds.
All of these results indicate that OR5AN1 is the major musk receptor that recognizes muscone, musk xylol, and musk ketone in humans and that OR2J3, together with OR5AN1, may contribute to the sensation of musk xylol.
Odor-finding test for muscone in MOR215-1 KO mice
Our results so far demonstrated that each species has one or two muscone ORs, suggesting that the muscone OR genes have been evolutionarily and functionally well conserved. To examine the role of MOR215-1 in the perception of muscone in mice, we genetically deleted the MOR215-1 gene to test whether the KO mice could smell muscone. Although mice have two muscone ORs, MOR215-1 and MOR214-3, MOR215-1 has a muscone detection threshold that is two orders of magnitude smaller than that of MOR214-3 (Fig. 1C). Therefore, the deletion of MOR215-1 is expected to cause a shift of the threshold to higher concentrations or possibly lead to muscone anosmia.
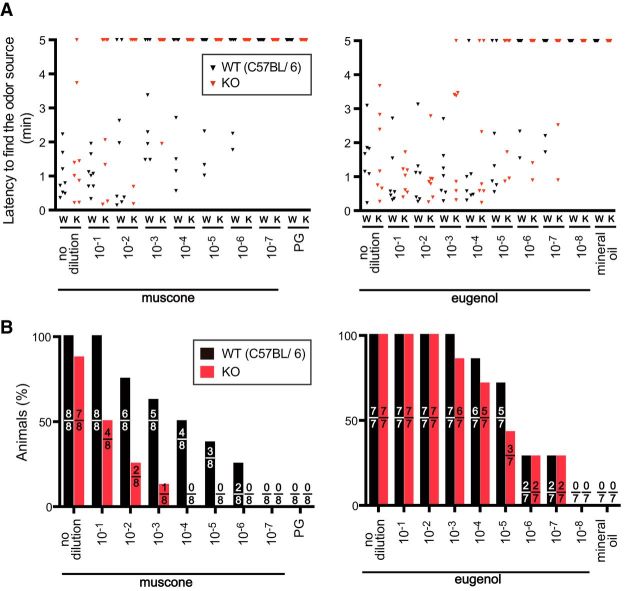
The MOR215-1 deletion (MOR215-1 KO) mice were subjected to the odor-finding test for muscone and eugenol (as a control) as described previously (Shirasu et al., 2014; see also Materials and Methods). More than half of the WT mice could find 10−4 dilution (0.01% volume per volume) muscone in 5 min, whereas none of the MOR215-1 KO mice could (Fig. 5). The latency to find the muscone odor sources is significantly different between WT and MOR215-1 KO mice (p < 0.001, the Scherier–Ray–Hare test statistics, H = 12.21). The detection threshold was two or three orders of magnitude higher in MOR215-1 KO mice than WT mice. In contrast, we did not find any significant difference for eugenol (p = 0.35, H = 0.88). These results indicate that, because of the presence of other receptors, including MOR214-3, MOR215-1 KO mice still possessed the ability to detect muscone albeit with much lower sensitivity. Therefore, MOR215-1 is crucial to detecting muscone in mice.
Figure 5.
Behavioral responses to muscone in MOR215-1 KO mice. A, Time to find the odor source in WT C57BL/6 mice (W) and MOR215-1 KO mice (K). Each triangle depicts a single individual. Animals that failed to find muscone (left) or eugenol (right) in 5 min were plotted at 5. PG, Propylene glycol. Muscone and eugenol were serially diluted from 10−1 (10%) to 10−7 (0.00001%) in PG and from 10−1 (10%) to 10−8 (0.000001%) in mineral oil. B, Summary of odor source-exploring experiments shown in Figure 5A. Vertical axis represents the percentage of the animals that found the odor sources within 5 min. The number of animals that found the odor sources and the number of total animals examined are shown inside each bar.
Discussion
In the early 1960s, Amoore (1963) proposed that thousands of odorants could be categorized into the seven primary odors and predicted the binding site structure for each primary odor. Musk odor was one of them and the shape of the musk binding site was estimated to be a disk-shaped column structure with approximately 10Å diameter. In this study, we have demonstrated that most of muscone receptors in primates and mice were tuned to both macrocyclic and nitro musks with chemical properties that are quite different. Lavine et al. (2003) showed that NMs and MCMs (called nitro-free musks in their paper) have common structural features that can be used to differentiate them from nonmusks. The space occupied by these musk compounds may be similarly circular with a diameter that fits into the disk shape as Amoore predicted. The present findings lead us to revisit Amoore's stereochemical theory of olfaction and confirm the important role of ORs in recognizing the shape of an odorant molecule.
Each of the six species examined in this study possesses one or two musk ORs, indicating that recognition of musk odors is specific and well conserved among species. Although we found similar structure–activity relationships in ORs across the OGGs, the ligand spectra were not necessarily similar among the muscone ORs within a single OGG. The structure–activity relationships suggest that the musk ORs can be categorized into four clusters (Fig. 2B). One type of OR showed strong responses to NMs and little response to macrocyclic lactones. Another two cluster types of ORs responded not only to macrocyclic ketones, but also to lactones, and thus appear to possess broader ligand spectra for macrocyclic compounds. The last type of ORs is the most specific and narrowly tuned to unsaturated macrocyclic ketones with a 15- or 16-membered ring. Unraveling the molecular basis for these unique structure–activity relationships in the musk receptor family will require solving the 3D structures of those receptors.
It is known that l-muscone possesses a stronger and higher-quality musk aroma than d-muscone, suggesting that muscone receptors are more responsive to l-muscone than to d-muscone (Kraft and Frater, 2001). Consistently, MOR215-1 responded to l-muscone more sensitively than to d-muscone, although the degree of the differences was somewhat different between the assay systems (Shirasu et al., 2014). For the human muscone receptor OR5AN1, no difference in EC50 values for l- and d-muscone was observed, but the response amplitude for l-muscone was ∼3-fold higher than that for d-muscone (Fig. 3B). This partial agonist characteristic may account for the weak and poor musk odor quality of d-muscone in human perception.
OR5AN1 also showed stronger responses to NMs than to muscone, consistent with the observation that human perception of NMs is more sensitive than muscone. Therefore, OR5AN1 may be the crucial musk receptor in humans. However, humans perceive macrocyclic lactones, PCMs, and ACMs as musk odors, even though these compounds do not activate at all or only slightly activate OR5AN1. This result suggests that other musk-responsive ORs participate in perception of these odorants. OR1A1 and OR2J3, which responded to muscone and musk xylol, have been known as broadly tuned ORs (Saito et al., 2009; McRae et al., 2012). These characteristics of OR1A1 and OR2J3 may explain the difference in scent properties of musk odorants.
In rodents, some expansion in the musk OR gene groups has occurred, resulting in, for example, a total of seven homologous ORs in mice. Of these, two musk ORs, MOR215-1 and MOR214-3, responded to musk odors in the assays using heterologous cells, consistent with the previous study (Fig. 1C; Shirasu et al., 2014; McClintock et al., 2014). MOR215-1 responded to musk ketone and musk xylol with ∼10- and 100-fold higher EC50 values, respectively, than to muscone. These differences may account for our observation that MOR215-1 did not respond to musk xylol and musk ketone during in vivo olfactory bulb imaging (Shirasu et al., 2014). The threshold concentration of muscone for MOR214-3 was ∼100-fold higher than that for MOR215-1, suggesting that MOR215-1 is the most sensitive muscone receptor. Indeed, genetic deletion of MOR215-1 resulted in drastic reduction in sensitivity to muscone in mice (Fig. 5), supporting the crucial role of MOR215-1 in mouse muscone perception.
Generally, odorants are recognized by multiple ORs among the 396 human ORs and 1130 ORs in mice. Therefore, typical ORs are functionally redundant, so single OR deletions generally do not affect olfaction. One exception so far is the case of deletion of TAAR4 (in the “other” class of chemosensory receptor) that caused anosmia to the cognate ligand β-phenylethylamine (Dewan et al., 2013). However, to our knowledge, this was the first case in which a single OR deletion affected the perception of the cognate ligand in mice. In humans, there are some cases in which genetic variation in a single receptor has a large influence on the intensity, sensitivity, or preference to the cognate odorant (Keller et al., 2007; Menashe et al., 2007; McRae et al., 2012; Jaeger et al., 2013; McRae, 2013; Mainland et al., 2014). These account for specific anosmia or hyperosmia for a particular odor. It has also been reported that some individuals cannot smell some musks (Whissell-Buechy and Amoore, 1973) and it is of interest whether this is due to a variation in the muscone OR.
In this study, we identified and described a conserved musk receptor family in mammals that is specific to musk odors but has different ligand spectra. Although a natural ligand(s) for these receptors in mammals other than musk deer remains to be elucidated, this family may play an evolutionarily pivotal role in each species. Indeed, our preliminary experiments suggest that there are unidentified compounds that activate MOR215-1 in mouse preputial gland extracts. The current study also reveals that, regardless of OR redundancy, deletion of a sensitive OR can affect the recognition threshold for the corresponding odorant dramatically, demonstrating a strong link between OR and odor perception. Finally, the structure–activity relationships of OR5AN1 appear to be in a good agreement with human sensory perception. This is an important observation that can inform future commercial development of useful musk aromas.
Footnotes
This work was supported by MEXT Japan (Grant-in-Aid for Scientific Research–S 24227003) and the Japanese Science and Technology Agency ERATO Touhara Chemosensory Signal Project. We thank Takasago International for providing highly pure musk odorants; Nagoya Higashiyama Zoo and Botanical Gardens for providing the orangutan sample (Baran, GAIN ID 0008) through the Great Ape Information Network and Primate Research Institute in Kyoto University; and H. Sakano for providing KO mice.
The authors declare no competing financial interests.
References
- Amoore JE. Stereochemical theory of olfaction. Nature. 1963;198:271–272. doi: 10.1038/198271a0. [DOI] [PubMed] [Google Scholar]
- Asada R, Kageyama K, Tanaka H, Saitoh Y, Miwa N. Antitumor and anti-invasive effects of diverse musk-fragrant macrocyclic ketones and their enhancement by hyperthermia. Mol Med Rep. 2012;5:148–152. doi: 10.3892/mmr.2011.585. [DOI] [PubMed] [Google Scholar]
- Dewan A, Pacifico R, Zhan R, Rinberg D, Bozza T. Non-redundant coding of aversive odours in the main olfactory pathway. Nature. 2013;497:486–489. doi: 10.1038/nature12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui H, Komaki R, Okui M, Toyoshima K, Kuda K. The effects of odor on cortisol and testosterone in healthy adults. Neuro Endocrinol Lett. 2007;28:433–437. [PubMed] [Google Scholar]
- Jaeger SR, McRae JF, Bava CM, Beresford MK, Hunter D, Jia Y, Chheang SL, Jin D, Peng M, Gamble JC, Atkinson KR, Axten LG, Paisley AG, Tooman L, Pineau B, Rouse SA, Newcomb RD. A Mendelian trait for olfactory sensitivity affects odor experience and food selection. Curr Biol. 2013;23:1601–1605. doi: 10.1016/j.cub.2013.07.030. [DOI] [PubMed] [Google Scholar]
- Kato M, Y Tanaka H. Study of the stimulation on the secretion of the female sex hormone by some perfumery raw material. Aroma Research. 2004;5:64–68. [Google Scholar]
- Keller A, Zhuang H, Chi Q, Vosshall LB, Matsunami H. Genetic variation in a human odorant receptor alters odour perception. Nature. 2007;449:468–472. doi: 10.1038/nature06162. [DOI] [PubMed] [Google Scholar]
- Kraft P, Fráter G. Enantioselectivity of the musk odor sensation. Chirality. 2001;13:388–394. doi: 10.1002/chir.1050. [DOI] [PubMed] [Google Scholar]
- Lavine BK, Davidson CE, Breneman C, Katt W, Sundling CM. Electronic van der Waals surface property descriptors and genetic algorithms for developing structure–activity correlations in olfactory databases. J Chem Inf Comput Sci. 2003;43:1890–1905. doi: 10.1021/ci030016j. [DOI] [PubMed] [Google Scholar]
- Li J, Haddad R, Chen S, Santos V, Luetje CW. A broadly tuned mouse odorant receptor that detects nitrotoluenes. J Neurochem. 2012;121:881–890. doi: 10.1111/j.1471-4159.2012.07740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainland JD, Keller A, Li YR, Zhou T, Trimmer C, Snyder LL, Moberly AH, Adipietro KA, Liu WL, Zhuang H, Zhan S, Lee SS, Lin A, Matsunami H. The missense of smell: functional variability in the human odorant receptor repertoire. Nat Neurosci. 2014;17:114–120. doi: 10.1038/nn.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96:713–723. doi: 10.1016/S0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- Márquez N, Martínez-Harms J, Vásquez RA, Mpodozis J. Early olfactory environment influences social behaviour in adult Octodon degus. PLoS One. 2015;10:e0118018. doi: 10.1371/journal.pone.0118018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda H, Watanabe S, Yamamoto K. New macrocyclic musk compounds. Chem Biodivers. 2004;1:1985–1991. doi: 10.1002/cbdv.200490152. [DOI] [PubMed] [Google Scholar]
- Matsui A, Go Y, Niimura Y. Degeneration of olfactory receptor gene repertories in primates: no direct link to full trichromatic vision. Mol Biol Evol. 2010;27:1192–1200. doi: 10.1093/molbev/msq003. [DOI] [PubMed] [Google Scholar]
- McClintock TS, Adipietro K, Titlow WB, Breheny P, Walz A, Mombaerts P, Matsunami H. In vivo identification of eugenol-responsive and muscone-responsive mouse odorant receptors. J Neurosci. 2014;34:15669–15678. doi: 10.1523/JNEUROSCI.3625-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRae JF, Mainland JD, Jaeger SR, Adipietro KA, Matsunami H, Newcomb RD. Genetic variation in the odorant receptor OR2J3 is associated with the ability to detect the “grassy” smelling odor, cis-3-hexen-1-ol. Chem Senses. 2012;37:585–593. doi: 10.1093/chemse/bjs049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRae JF, Jaeger SR, Bava CM, Beresford MK, Hunter D, Jia Y, Chheang SL, Jin D, Peng M, Gamble JC, Atkinson KR, Axten LG, Paisley AG, Williams L, Tooman L, Pineau B, Rouse SA, Newcomb RD. Identification of regions associated with variation in sensitivity to food-related odors in the human genome. Curr Biol. 2013;23:1596–1600. doi: 10.1016/j.cub.2013.07.031. [DOI] [PubMed] [Google Scholar]
- Menashe I, Abaffy T, Hasin Y, Goshen S, Yahalom V, Luetje CW, Lancet D. Genetic elucidation of human hyperosmia to isovaleric acid. PLoS Biol. 2007;5:e284. doi: 10.1371/journal.pbio.0050284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima A, Takeuchi H, Imai T, Saito H, Kiyonari H, Abe T, Chen M, Weinstein LS, Yu CR, Storm DR, Nishizumi H, Sakano H. Agonist-independent GPCR activity regulates anterior-posterior targeting of olfactory sensory neurons. Cell. 2013;154:1314–1325. doi: 10.1016/j.cell.2013.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Matsui A, Touhara K. Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals. Genome Res. 2014;24:1485–1496. doi: 10.1101/gr.169532.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka L, Stoll M, Schinz H. Zur Kenntnis des Kohlenstoffringes II. Synthese der carbocyclischen Ketone vom Zehner-bis zum Achtzehnerring. Helv Chim Acta. 1926;9:249–264. doi: 10.1002/hlca.19260090130. [DOI] [Google Scholar]
- Saito H, Chi Q, Zhuang H, Matsunami H, Mainland JD. Odor coding by a mammalian receptor repertoire. Sci Signal. 2009;2:ra9. doi: 10.1126/scisignal.2000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shirasu M, Yoshikawa K, Takai Y, Nakashima A, Takeuchi H, Sakano H, Touhara K. Olfactory receptor and neural pathway responsible for highly selective sensing of musk odors. Neuron. 2014;81:165–178. doi: 10.1016/j.neuron.2013.10.021. [DOI] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry. New York: W. H. Freeman; 2001. [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touhara K, Vosshall LB. Sensing odorants and pheromones with chemosensory receptors. Annu Rev Physiol. 2009;71:307–332. doi: 10.1146/annurev.physiol.010908.163209. [DOI] [PubMed] [Google Scholar]
- Walbaum H. Das natürliche Moschusaroma. J Prakt Chem. 1906;73:488–493. [Google Scholar]
- Whissell-Buechy D, Amoore JE. Odour-blindness to musk: simple recessive inheritance. Nature. 1973;242:271–273. doi: 10.1038/242271a0. [DOI] [PubMed] [Google Scholar]
- Zhuang H, Matsunami H. Evaluating cell-surface expression and measuring activation of mammalian odorant receptors in heterologous cells. Nat Protoc. 2008;3:1402–1413. doi: 10.1038/nprot.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]