Abstract
The generation of hair cells (HCs) from the differentiation of proliferating supporting cells (SCs) appears to be an ideal approach for replacing lost HCs in the cochlea and is promising for restoring hearing after damage to the organ of Corti. We show here that extensive proliferation of SCs followed by mitotic HC generation is achieved through a genetic reprogramming process involving the activation of β-catenin to upregulate Wnt signaling, the deletion of Notch1 to downregulate Notch signaling, and the overexpression of Atoh1 in Sox2+ SCs in neonatal mouse cochleae. We used RNA sequencing to compare the transcripts of the cochleae from control mice and from mice with β-catenin activation, Notch1 deletion, and β-catenin activation combined with Notch1 deletion in Sox2+ SCs. We identified the genes involved in the proliferation and transdifferentiation process that are either controlled by individual signaling pathways or by the combination of Wnt and Notch signaling. Moreover, the proliferation of SCs induced by Notch1 deletion disappears after deleting β-catenin in Notch1 knock-out Sox2+ cells, which further demonstrates that Notch signaling is an upstream and negative regulator of Wnt signaling.
SIGNIFICANCE STATEMENT We show here that the extensive proliferation of supporting cells (SCs) and the subsequent mitotic hair cell (HC) generation is achieved through a genetic reprogramming process involving activation of β-catenin to upregulate Wnt signaling, deletion of Notch1 to downregulate Notch signaling, and overexpression of Atoh1 in Sox2+ SCs in neonatal mice cochleae. By comparing the transcripts of the cochleae among controls, β-catenin activation, Notch1 deletion, and β-catenin activation combined with Notch1 deletion group, we identified multiple genes involved in the proliferation and transdifferentiation process that are either controlled by individual signaling pathways or by the combination of Wnt and Notch signaling. This provides a better understanding of the mechanisms behind mitotic HC generation and might provide new approaches to stimulating mitotic HC regeneration.
Keywords: Atoh1, hair cells, Notch, regeneration, reprogramming, Wnt
Introduction
Hair cell (HC) loss is the predominant cause for hearing and balance disorders. In mammals, in vivo HC regeneration has only been identified in neonatal cochleae and the number of regenerated HCs is quite low (Bramhall et al., 2014; Cox et al., 2014). Therefore, other strategies of generating HCs are needed for restoring hearing and balance function.
There is much evidence that cochlear HCs and supporting cells (SCs) are derived from a common progenitor cell population during development (Fekete et al., 1998). SCs can give rise to the new HCs with or without proliferation, which is the main source of HC regeneration in nonmammalian vertebrates (Raphael, 1992; Roberson et al., 2004). Therefore, SCs are considered to be a promising source for the generation of HCs (Burns and Corwin, 2013). However, direct transdifferentiation of SCs into new HCs exhausts the population of SCs, which are needed to maintain the survival of the newly regenerated HCs. Therefore, to regenerate the mosaic structure of the cochlea that is needed to restore hearing, SCs would need to be expanded before differentiate into HCs.
It has been reported that SC populations contain progenitor cells that can proliferate and regenerate new HCs in neonatal mammalian cochleae (Wang et al., 2006). Recently, these progenitor cells have been identified as Wnt-responsive Lgr5+ cells (Chai et al., 2012; Shi et al., 2012). Overexpressing β-catenin in Lgr5+ or Sox2+ cells can initiate the proliferation of cochlear progenitor cells, but only very limited numbers of SCs in BrdU+ foci differentiated into HCs (Shi et al., 2013). Our previous study reported that Notch inhibition led to the proliferation of progenitors in the postnatal cochlear sensory epithelium, which could serve as another approach to activating cochlear progenitor cell proliferation; however, the majority of generated HCs still came from direct transdifferentiation with the concomitant depletion of SCs (Li et al., 2015). Furthermore, the activation of progenitor cells was more obvious in the apical regions of cochlea, which is problematic because the majority of HC loss is in the basal region and the HC regeneration capability disappeared after postnatal day 7 (P7) (Shi et al., 2013; Cox et al., 2014). Therefore, there are significant challenges associated with generating HCs in the cochleae of older mice by manipulating a single pathway. To address these challenges, we investigated the coregulation of multiple signaling pathways.
The crosstalk between Wnt and Notch signaling has been shown to play an important role in cell fate decisions during otic placode formation (Jayasena et al., 2008); however, the interaction between these pathways remains largely uninvestigated in the postnatal mouse cochlea. Our previous study hinted that Notch signaling might act as a negative regulator for the progenitor proliferation induced by Wnt activation (Li et al., 2015), which was verified during the regeneration of HCs in the lateral line of zebrafish (Romero-Carvajal et al., 2015). Therefore, combining Wnt and Notch signaling in the cochlea might significantly increase the proliferation of progenitors and provide a new approach for HC generation in mammals.
In this study, by activating Wnt signaling and inhibiting Notch signaling, we genetically reprogrammed the Sox2+ SCs and observed extensive cell-cycle reentry in the neonatal mouse cochlea. We found significantly more proliferated SCs and more Myo7a+ HCs by simultaneous β-catenin overexpression and Notch1 inhibition in Sox2+ cells in vivo, suggesting the benefit of combining SC proliferation induced by β-catenin overexpression with transdifferentiation into HCs induced by Notch1 deletion. Moreover, we observed significantly more EdU+ Myo7a+ HCs through combined genetic programming with Notch inhibition and Wnt and Atoh1 overexpression. To further understand the mechanism behind the extensive proliferation of SCs and differentiation into HCs after the reprogramming process, we compared the gene transcripts of the cochleae among the control, β-catenin overexpression, Notch1 deletion, and β-catenin overexpression combined with Notch1 deletion mice. Last, the mechanism behind the interaction of Wnt and Notch signaling was further investigated in β-catenin and Notch1 double knock-out mice.
Materials and Methods
Mouse models and treatments.
Sox2CreERT2/+ (Stock 008875), Notch1flox/flox (Stock 007181), and β-cateninflox(exon2–6) (Stock 004152) mice were purchased from The Jackson Laboratory. β-cateninflox(exon3) mice were generously provided by Mark Taketo (Kyoto University). CAG-loxP-stop-loxP-Atoh1-HA+ mice were kind gifts from St. Jude Children's Research Hospital. Atoh1-nGFP mice were provided by Jane Johnson (University of Texas Southwestern Medical Center, Dallas).
Tamoxifen (Sigma-Aldrich) diluted in corn oil was injected intraperitoneally at P0 and P1 at doses of 0.20 mg/g bodyweight. Ten microliters of the thymidine analog 5-ethynyl-2′-deoxyuridine (EdU) (Click-iT EdU Imaging Kit; Invitrogen) were injected at 5 mg/ml intraperitoneally twice a day from P1 to P6. Male and female mice were used for experiments. All animal experiments were approved by the Institutional Animal Care and Use Committee of Fudan University.
Immunohistochemistry.
Cochleae were fixed in 4% PFA (Sigma-Aldrich) for 30 min and washed with 10 mm PBS. EdU was detected with Alexa Fluor azide using the Click-iT EdU Imaging Kits (Life Technologies) according to the manufacturer's protocol. The tissues were then blocked with 10% donkey serum in 10 mm PBS with 1% Triton X-100 for 1 h at room temperature and incubated with primary antibody overnight at 4°C. The primary antibodies were rabbit anti-MyosinVIIa (1:800 dilution; Proteus BioSciences), goat anti-Sox2 (1:300 dilution; Santa Cruz Biotechnology), chicken anti-EGFP (1:800 dilution; Abcam), guinea pig anti-VGLUT3 (1:1000 dilution; Millipore), goat anti-Prestin (1:400 dilution; Santa Cruz Biotechnology), and rat anti-HA (1:200 dilution; Roche). On the following day, the appropriate secondary Alexa Fluor-conjugated antibodies were incubated for 1–2 h at room temperature. HC bundles were labeled with phalloidin (Life Technologies) and nuclei were labeled with DAPI (1:800 dilution; Sigma-Aldrich).
RNA sequencing and analyses.
The apical and middle turns of at least six cochleae from each group were pooled to isolate the total RNA using the AllPrep DNA/RNA/Protein Mini Kit (QIAGEN) according to the manufacturer's protocol.
Preliminary processing of raw reads was performed using Casava 1.8 (Illumina). Subsequently, adapters and low-quality bases were trimmed using trimmomatic (version 0.23). Trimmed reads for each sample were aligned to the Mus musculus UCSC mm10 genome using the TopHat version 2.0.10 software package (Bowtie 2 version 2.2.1 software). These files were used as the input for the Cufflinks software, which is a complementary method used to generate assembled transcripts for each group, and the abundance was evaluated using read data. The fragments per kilobase per million mapped reads values were calculated for each gene to normalize the data. These assemblies were used with the Cuffdiff tools from the Cufflinks 2.2.1 package to calculate the differential expression levels and to evaluate the statistical significance of the detected alterations. A heat map of differentially expressed genes was generated with ClustVis using the default parameters.
Functional annotation.
DAVID (Database for Annotation, Visualization and Integrated Discovery) version 6.7 software (http://david.abcc.ncifcrf.gov/home.jsp) was used to determine the most functional annotation of significant genes in the datasets as described previously (Huang et al., 2009). The DAVID program calculates a modified Fisher's exact p-value to demonstrate gene ontology (GO) or molecular pathway enrichment. p-values < 0.05 were considered strongly enriched in the annotation category.
Image acquisition and cell counts.
Fluorescent images were acquired using a Leica SP8 confocal microscope. All of the images were digitally processed using ImageJ and Adobe Photoshop CS6. Cell counts from the confocal images were performed using ImageJ. Total numbers of HCs and Sox2+ EdU+ cells were counted separately in 100 μm regions of the cochlea in the apical, middle, and basal turns. For quantification of mitotic HCs, Myo7a+ EdU+ cells from all turns of the cochlea were counted.
Statistical analyses were performed using GraphPad Prism software. Cell counts were analyzed with one-way ANOVA.
Results
β-catenin overexpression and Notch1 deletion in Sox2+ cells led to intensive cell cycle reentry of SCs in the cochlea
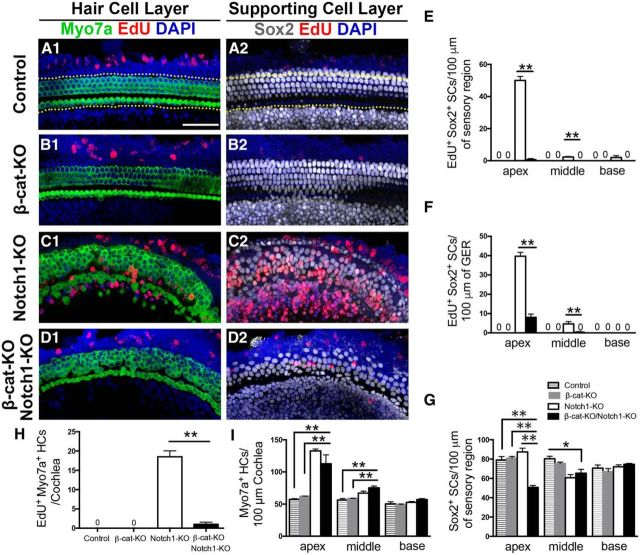
To test the effects of combining β-catenin overexpression and Notch1 deletion in Sox2+ SCs in neonatal mouse cochleae, we generated the Sox2CreERT2/+-β-cateninflox(exon3)-Notch1flox/flox (β-cat-OE/Notch1-KO) mouse in which constitutive activation of β-catenin and deletion of Notch1 in Sox2+ SCs was induced by tamoxifen administration at P0 and P1. The thymidine analog EdU was injected twice per day from P1 to P6 to label the cells entering the cell cycle, and the mice were killed at P7. Littermates lacking the Sox2CreER/T2+ allele were used as controls. In the control group, no EdU+Sox2+ cells were observed anywhere in the cochlea (Fig. 1A). Consistent with previous studies, we observed EdU+Sox2+ cell foci in the pillar cell region and greater epithelial ridge (GER) from Sox2CreER/T2-β-cateninflox(exon3) (β-cat-OE) cochleae (Fig. 1B,G,H, Table 1). We also observed EdU+Sox2+ cells in the Sox2CreER/T2-Notch1(flox/flox) (Notch1-KO) cochleae (Fig. 1C,G,H, Table 1). The number of EdU+ SCs decreased significantly from the apical to the basal turns of the cochlea. Compared with manipulating β-catenin or Notch1 signaling separately, more EdU+ SCs were identified in Sox2CreER/T2-β-cateninflox(exon3)-Notch1(flox/flox) (β-cat-OE/Notch1-KO) cochleae and these SCs extended beyond the pillar cell region and into the basal part of cochlea (Fig. 1D,G,H, Table 1). The total number of Sox2+ SCs in the sensory epithelium was determined to identify the effects of SC proliferation. In the cochlear cross-sections, we found significantly more Sox2+ cells stacked in multiple layers (Fig. 1F) and located in the auditory sensory epithelium region, as well as in the GER of β-cat-OE/Notch1-KO mice (Fig. 1I, Table 1).
Figure 1.
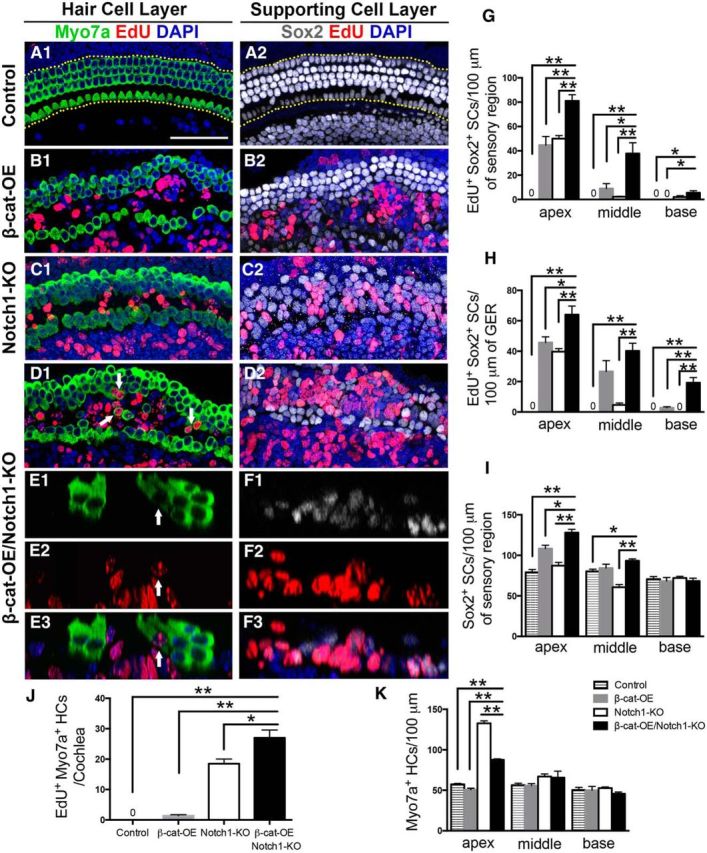
β-catenin overexpression (β-cat-OE) and Notch1 deletion (Notch1-KO) in Sox2+ cells induces extensive proliferation of SCs in the cochlea, but mitotic HC generation is limited. A, In control cochleae, no EdU+Sox2+ SC or EdU+Myo7a+ HCs were detected from P0 to P7. B, In β-cat-OE cochleae, numerous EdU+Sox2+ SCs were detected in the pillar cell region and GER of the SC layer (B2), but only a few EdU+Myo7a+ HCs were observed in the HC layer (B1). C, In Notch1-KO cochleae, most EdU+Sox2+ SCs were observed in the pillar cell region and GER in the SC layer (C2) and EdU+Myo7a+ HCs could only be observed in the sensory region of the HC layer (C1). D, In β-cat-OE/Notch1-KO cochleae, EdU+Sox2+ SCs were detected, not only in the pillar cell region and GER, but also in the Dieter's cells D2, Only limited numbers of EdU+ Myo7a+ HCs in the sensory region were detected. E, F, The tissue cross-section also shows the proliferation of SCs and the limited mitotic HC generation. Most of the EdU+Sox2+ cells were observed in the pillar cell region and Dieter's cell region and SCs in this region were stacked in multiple layers. G, H, The number of EdU+Sox2+ SCs was increased in β-cat-OE/Notch1-KO cochleae relative to the β-cat-OE and the Notch1-KO cochleae both in the sensory region and the GER. I, The number of Sox2+ SCs in the sensory region. J, Quantification and comparison of the number of EdU+Myo7a+ HCs from the apex to the base. The β-cat-OE/Notch1-KO cochleae had significantly more mitotic HCs. K, The number of Myo7a+ HCs. Data in G–K are presented as mean ± SEM. *p < 0.05, **p < 0.01. The dotted lines in A1 show the counting limits of SCs in the sensory region and GER. The arrows indicate the mitotic HCs. Scale bar, 50 μm.
Table 1.
Synergistic effect on β-catenin overexpression and Notch1 deletion on Sox2+ SCs proliferation and HCs generation
| Control (n = 4) | β-cat-OE (n = 3) | Notch1-KO (n = 4) | β-cat-OE Notch1-KO (n = 4) | |
|---|---|---|---|---|
| EdU+Sox2+ SCs/100 μm of sensory region | ||||
| Apex | 0 | 44.67 ± 7.13 | 50 ± 2.52 | 81.00 ± 5.12 |
| Mid | 0 | 9.00 ± 2.31 | 2.33 ± 0.33 | 37.75 ± 8.86 |
| Base | 0 | 0 | 2.00 ± 1.16 | 5.50 ± 1.71 |
| EdU+Sox2+ SCs/100 μm of GER | ||||
| Apex | 0 | 45.67 ± 3.76 | 39.67 ± 2.03 | 64.00 ± 5.770 |
| Mid | 0 | 26.67 ± 4.10 | 4.67 ± 1.20 | 40.25 ± 4.99 |
| Base | 0 | 2.67 ± 0.67 | 0 | 19.25 ± 3.40 |
| Sox2+ SCs/100 μm of sensory region | ||||
| Apex | 79.00 ± 2.08 | 108.30 ± 4.10 | 87.33 ± 3.93 | 128.00 ± 4.06 |
| Mid | 80.33 ± 2.60 | 84.33 ± 2.73 | 60.67 ± 3.28 | 93.25 ± 2.32 |
| Base | 70.67 ± 3.18 | 68.33 ± 4.41 | 72.00 ± 2.08 | 68.25 ± 3.54 |
| EdU+Myo7a+ HCs/cochlea | 0 | 1.33 ± 0.33 | 18.50 ± 1.56 | 27.00 ± 2.59 |
| Myo7a+ HCs/100 μm | ||||
| Apex | 57.33 ± 0.67 | 50.67 ± 1.85 | 132.67 ± 2.96 | 87.67 ± 1.20 |
| Mid | 56.33 ± 2.33 | 55.67 ± 1.45 | 67.00 ± 3.21 | 65.67 ± 7.97 |
| Base | 50.33 ± 3.18 | 50.00 ± 4.73 | 52.67 ± 1.33 | 45.67 ± 2.19 |
| EdU+Myo7a+/ EdU+ Sox2+ | ||||
| Apex | 0 | 1.30 ± 0.65% | 9.97 ± 0.02% | 3.86 ± 1.19% |
β-catenin overexpression and Notch1 deletion in Sox2+ cells led to limited mitotic HC generation
Previous reports have shown that the expansion of SCs can be achieved by upregulation of β-catenin or deletion of Notch1 in Sox2+ cells (Chai et al., 2012; Shi et al., 2013; Li et al., 2015) and that large numbers of SCs differentiated into HCs in the absence of Notch signaling (Yamamoto et al., 2006; Lin et al., 2011). We hypothesized that more proliferated SCs might differentiate into HCs by increasing SC proliferation through β-catenin upregulation and by increasing transdifferentiation into HCs by Notch1 deletion.
To test this hypothesis, we measured the ability of the proliferated SCs to differentiate into HCs by identifying EdU+Myo7a+ HCs. In control mice, no EdU+Myo7a+ cells were observed in any part of the cochlea (Fig. 1A). We found only a few EdU+Myo7a+ cells in the pillar cell region after upregulation of β-catenin (Fig. 1B,J, Table 1), and there was no significant increase in Myo7a+ HCs in the β-cat-OE cochleae (Fig. 1K, Table 1). However, many more EdU+Myo7a+ HCs were observed in the pillar cell region of the apical turn of the cochlea after Notch1 deletion (Fig. 1C,J, Table 1), and there were many more Myo7a+ HCs in the Notch1-KO cochleae (Fig. 1K, Table 1). In support of our hypothesis, we observed more EdU+Myo7a+ cells in the pillar cell region and the GER in the β-cat-OE/Notch1-KO cochleae (Fig. 1D,E,J, Table 1) and these results were further confirmed in Atoh1 reporter (Atoh1-nGFP) mice (data not shown). However, the number of Myo7a+ HCs was significantly greater in Notch1-KO mice than in β-cat-OE/Notch1-KO mice (Fig. 1K, Table 1). In the cochlear cross-sections of β-cat-OE/Notch1-KO mice, we observed multiple Myo7a+ HCs in both the inner and outer HC region and some of them were EdU+ (Fig. 1E). The differentiation ratios from proliferated SCs to HCs were calculated by counting the EdU+Sox2+ SCs and the EdU+Myo7a+ HCs from the apical turn of the cochlea (Table 1). Compared with the extensive proliferation of SCs induced by combining β-catenin overexpression and Notch1 deletion, only 3.86 ± 1.19% of the proliferated SCs in the apex differentiated into HCs, so the efficiency of mitotic HC generation is relatively low and needs to be improved. There was no significant difference between the control and β-cat-OE group in the number of Myo7a+ HCs in any part of the cochlea. However, compared with the control and β-cat-OE group, many more Myo7a+ HCs were observed in the apical region of the cochlea after Notch1 deletion with or without β-catenin overexpression, whereas the majority of HCs were EdU−, which suggests that the transdifferentiation might still be the main source of newly generated HCs after Notch1 deletion (Fig. 1A–D, J–K, Table 1).
Reciprocal interactions between the Wnt and Notch signaling pathways during reprogramming
Our previous studies revealed that Notch inhibition initiates the proliferation of Lgr5+ SCs and the mitotic generation of HCs in the neonatal mouse cochlea in vivo and in vitro, which hinted that Notch signaling might act as a negative regulator that inhibits the proliferation of Lgr5+ progenitors and maintains homeostasis of the cochlear sensory epithelium in terms of cell numbers. In the current study, this hypothesis was further confirmed by the cumulative effects of downregulating Notch1 and upregulating Wnt in controlling the proliferation of SCs. It has been reported that membrane-bound Notch associates physically with unphosphorylated (active) β-catenin in stem and colon cancer cells and that it negatively regulates posttranslational accumulation of the active β-catenin protein (Kwon et al., 2011). To further dissect the mechanism behind the interaction between Wnt and Notch1 signaling during the activation of progenitor cells in the neonatal mouse cochlea, we generated another mouse model, Sox2CreER/T2-β-cateninflox(exon2–6)-Notch1(flox/flox) (β-cat-KO/Notch1-KO), in which β-catenin and Notch1 were both knocked out in Sox2+ SCs after tamoxifen injection at P0 and P1. EdU was injected twice per day from P1 to P6 and these mice were killed at P7.
In the control group without the Sox2-CreER allele, there were no EdU+Sox2+ or EdU+Myo7a+ cells in any area of the cochlea (Fig. 2A,E,F, Table 2). Upon deleting β-catenin in SCs, no EdU+Sox2+ or EdU+Myo7a+ cells were observed anywhere in the cochlea (Fig. 2B,E,F, Table 2) and, compared with controls, there were no significant differences in the number of Myo7a+ HCs (Fig. 2I, Table 2). In Notch1-KO mice, EdSox2+ cells were identified both in the pillar cell region and the GER (Fig. 2C,E,F, Table 2), with a gradient from the apex to the base. Among the many new HCs generated after Notch1 deletion in SCs (Fig. 2I, Table 2), a few EdU+Myo7a+ HCs with Sox2 staining were identified in the pillar cell region (Fig. 2H, Table 2). When β-catenin was knocked out along with Notch1 deletion in Sox2+ cells, there was a significant decrease in EdU+Sox2+ cells, especially in the sensory epithelium. Only a few proliferated SCs were observed in the GER (Fig. 2D,F, Table 2) and very few EdU+Sox2+ cells could be identified in the apex of the sensory epithelium (Fig. 2E, Table 2). In addition, one or two EdU+Myo7a+ cells could occasionally be observed (Fig. 2D,H, Table 2). However, with or without β-catenin deletion, there was no significant effect on the overall increased Myo7a+ cell number after Notch1 deletion (Fig. 2I, Table 2).
Figure 2.
β-catenin knock-out (β-cat-KO) significantly reduces the proliferation of SCs and the generation of mitotic HCs in Notch1-deletion (Notch1-KO) cochleae. A, There were no EdU+Sox2+ SC or EdU+Myo7a+ HCs in the cochleae of the control mice. B, EdU+Sox2+ SCs and Myo7a+ EdU+ HCs were detected in both the sensory region and GER from the cochleae of the Notch1-KO mice. C, There were no EdU+Sox2+ SC or EdU+Myo7a+ HCs in the cochleae of β-cat-KO mice. D, The numbers of EdU+Sox2+ SCs and EdU+Myo7a+ HCs decreased significantly in the cochleae of β-cat-KO/Notch1-KO mice. E, F, The number of EdU+Sox2+ SCs in the sensory region and GER. G, The number of Sox2+ SCs in the sensory region. H, Compared with the cochleae of Notch1-KO mice, the number of EdU+Myo7a+ HCs decreased significantly in the cochleae of β-cat-KO/Notch1-KO mice. I, Quantification and comparison of the number of Myo7a+ HCs. There was no significant difference between Notch1-KO and β-cat-KO/Notch1-KO cochleae. Data in G–I are presented as mean ± SEM. *p < 0.05, **p < 0.01. The dotted lines in A1 show the counting limits of SCs in the sensory region and GER. Scale bar, 50 μm.
Table 2.
Synergistic effect of β-catenin and Notch1 double knockout on Sox2+ SCs proliferation and HCs generation
| Control (n =4) | β-cat-KO (n = 3) | Notch1-KO (n = 4) | β-cat-KO Notch1-KO (n = 3) | |
|---|---|---|---|---|
| EdU+Sox2+ SCs/100 μm of sensory region | ||||
| Apex | 0 | 0 | 50 ± 2.52 | 0.67 ± 0.67 |
| Mid | 0 | 0 | 2.33 ± 0.33 | 0 |
| Base | 0 | 0 | 2.00 ± 1.16 | 0 |
| EdU+Sox2+ SCs/100 μm of GER | ||||
| Apex | 0 | 0 | 39.67 ± 2/03 | 8.00 ± 1.73 |
| Mid | 0 | 0 | 4.67 ± 1.20 | 0.33 ± 0.33 |
| Base | 0 | 0 | 0 | 0 |
| Sox2+ SCs/100 μm of sensory region | ||||
| Apex | 79.00 ± 2.08 | 81.00 ± 1.73 | 87.33 ± 3.93 | 50.67 ± 2.02 |
| Mid | 80.33 ± 2.60 | 75.33 ± 0.88 | 60.67 ± 3.28 | 65.33 ± 4.06 |
| Base | 70.67 ± 3.18 | 67.00 ± 3.21 | 72.00 ± 2.08 | 74.67 ± 0.88 |
| EdU+Myo7a+ HCs/cochlea | 0 | 0 | 18.50 ± 1.56 | 1.00 ± 0.58 |
| Myo7a+ HCs/100 μm | ||||
| Apex | 57.33 ± 0.67 | 61.33 ± 0.88 | 132.67 ± 2.96 | 112.67 ± 13.86 |
| Mid | 56.33 ± 2.33 | 58.67 ± 0.33 | 67.00 ± 3.21 | 75.33 ± 2.67 |
| Base | 50.33 ± 3.18 | 49.00 ± 1.15 | 52.67 ± 1.33 | 57.00 ± 1.53 |
Our data provide direct evidence that Notch1 acts as a negative regulator of the proliferation of progenitor cells in the neonatal mouse cochlea through β-catenin activation and that Notch signaling serves as an upstream regulator of Wnt signaling for maintaining the quiescence of progenitor cells in the neonatal mouse cochlea.
Mechanism behind extensive cell proliferation after reprogramming
To assess the genome-wide gene expression profiles among controls, Notch1 deletion, β-catenin activation, and Notch1 deletion combined with β-catenin activation in Sox2+ SCs, we compared the transcripts of the cochleae (mainly from the apex and middle turns, where the greatest differences were seen for SC proliferation and HC generation) from each group using RNA sequencing. The cochleae were dissected from P7 mice that had been injected with tamoxifen at P0 and P1 to activate Cre and were examined under a microscope. Six to eight cochleae were pooled together for each RNA-sequencing experiment and three biological replicates were sequenced. We obtained between 98,000,000 and 189,000,000 reads for each sample, with 83–85% of the paired reads mapping correctly to the reference genome. The three biological replicates of each population showed high reproducibility. By comparing the transcripts among the three populations, we identified 1454 genes that had >2-fold differences in expression (p < 0.01). Gene expression clusters of the 1454 genes were generated and a heat map was created to aid in the visualization of the gene expression pattern (Fig. 3A). GO analysis was performed on clusters of genes using DAVID. Three gene clusters of particular interest are shown in Figure 3A.
Figure 3.
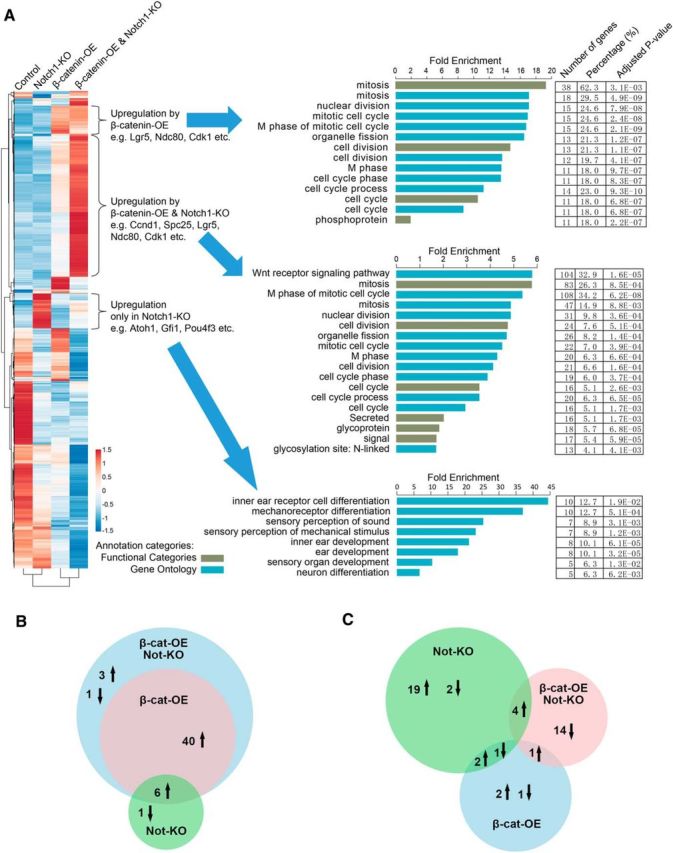
Global comparisons of gene expression among four different transgenic mouse lines and the expression pattern of genes by hierarchical clustering. A, Hierarchical clustering of the expression patterns of all differentially expressed genes. Red represents above-average expression levels and blue represents below-average levels. Each row represents one gene and each column represents one sample. Three gene clusters of interest were selected. B, Analysis of differentially expressed genes involved in the cell cycle and mitosis. C, Analysis of differentially expressed genes related to HC differentiation and function.
Genes in cluster 1 were upregulated in the β-cat-OE and the β-cat-OE/Notch1-KO group and GO analysis showed a significant number of genes involved in mitotic cell division and the cell cycle, including Lgr5, Cdk1, and Ndc80.
Genes in cluster 2 were upregulated significantly more in the β-cat-OE/Notch1-KO group and GO analysis showed that many of these genes are involved in cell cycle and mitotic cell division, especially the genes related to the Wnt receptor signaling pathway such as Ccnd1.
Genes in cluster 3 were upregulated in the Notch1-KO group and GO analysis revealed that many of these genes are associated with inner ear development and HC differentiation, such as Atoh1, Gfi1, Pou4f3, Ush2a, Jag2, Lhfpl5, Bsnd, Myo3a, Barhl1, Otof, Chrna 10, Ptprq, Miat, and Grk1, and that many of the genes are related to neuron differentiation, including Sall3, Ush2a, Lhfpl5, Jag2, Cdh4, Rasgrf1, Miat, Grk1, and Atoh1.
The differential expression of genes involved in the cell cycle and mitosis among the Notch1-KO group, β-cat-OE group, and β-cat-OE/Notch1-KO group were analyzed to better understand the targets of each individual pathway, as well as the cumulative effects on the proliferation of SCs by coregulating Notch and Wnt signaling. We found 40 genes related to the cell cycle and mitosis, including Ccnd1, Cdca5, and Cdca8, that were upregulated upon overexpression of β-catenin and the expression levels were significantly increased in the β-cat-OE/Notch1-KO group. There were another four genes exclusively regulated in the β-cat-OE/Notch1-KO group, including Chek1, Pim1, Rmp22, and Rad51. We also found that there were six cell cycle genes that were upregulated upon overexpression of β-catenin and were upregulated by inhibiting Notch signaling. Only one gene, S100a6, was exclusively downregulated by Notch inhibition (Fig. 3B).
Further, we found 25 genes related to differentiation and HC function that were upregulated by Notch1 deletion, but only two of them, Gjb3 and Coch, were upregulated upon overexpression of β-catenin. Four of them were also upregulated in the β-cat-OE/Notch1-KO group and 14 genes associated with HCs were exclusively downregulated in the β-cat-OE/Notch1-KO group, including Gjb6, Slc26a4, and Col4a3 (Fig. 3C). After the deletion of Notch1 in Sox2+ SCs, we found activation of Wnt downstream target genes in postnatal mice cochlea, including Cdkn2a (Wassermann et al., 2009) and Nanog (Yong et al., 2016).
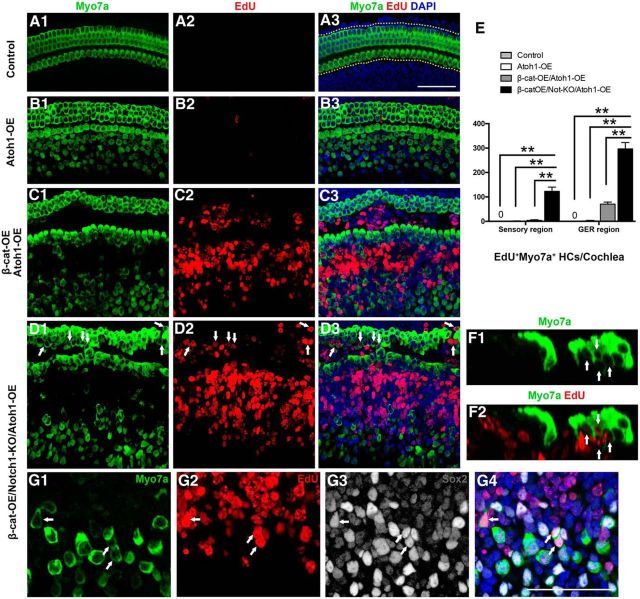
Mitotic HC generation through the overexpression of Atoh1 in Sox2+ cells
Contrary to our expectations, many of the proliferated SCs could not automatically differentiate into HCs after genetic programming with upregulation of β-catenin and downregulation of Notch1 and this raised the question of whether the proliferated cells could still react to Atoh1, an important transcriptional factor during the generation of HCs, and become HC-like cells.
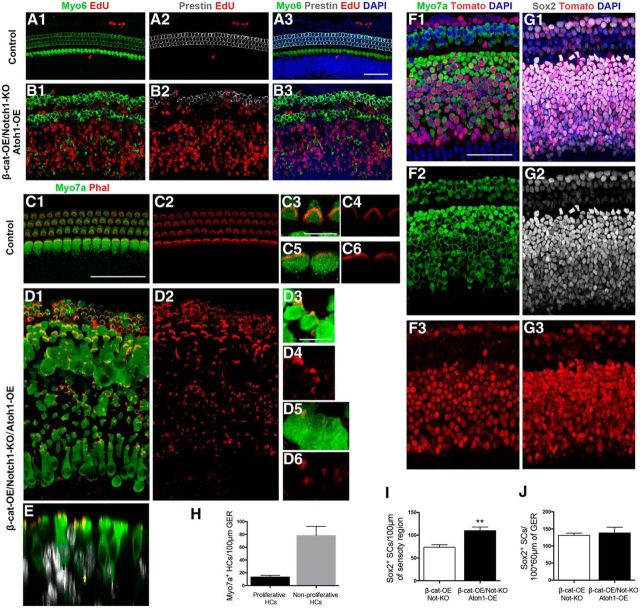
To test this hypothesis, we generated another transgenic mouse model in which Atoh1 is overexpressed in Sox2+ cells after tamoxifen administration at P0 and P1 with or without β-catenin activation and Notch1 deletion. The expression of transgene-Atoh1 was measured by the immunolabeling of HA tag from the cochlea of transgenic mice. We observed that the majority of Sox2+ SCs in the GER region (82.54 ± 5.44%), as well as in the sensory epithelial region (68.27 ± 4.69%), were upregulated with transgene after the injection of tamoxifen (see Fig. 5F,G). These mice were killed at P5 and, consistent with previous studies (Kelly et al., 2012), after the overexpression of Atoh1 in Sox2+ cells (Atoh1-OE), many of the SCs in the GER transdifferentiated into Myo7a+ cells, but very few EdU+Myo7a+ cells were identified (Fig. 4B,E, Table 3). Upon combining the activation of β-catenin, deletion of Notch1, and overexpression of Atoh1 in SCs (β-cat-OE/Notch1-KO/Atoh1-OE), we observed extensive proliferation of SCs in both the sensory epithelium and the GER and many of the proliferated cells differentiated into HCs (Fig. 4D–G, Table 3). In the sensory region, the efficiency was 13.85 ± 0.52%, which was much higher than the efficiency in β-cat-OE/Notch1-KO mice (3.86 ± 1.19%). In the GER, the number or proliferative HCs was 13.17 ± 1.17 and the number of nonproliferative HCs was 78.17 ± 5.83 (Fig. 5H, Table 3). To identify the effects of SC proliferation, we also counted the Sox2+ SCs in the sensory epithelium and GER of control and β-cat-OE/Notch1-KO/Atoh1-OE group. There were more Sox2+ SCs in the sensory region, but no significant difference in GER (Fig. 5I,J, Table 4).
Figure 5.
Characteristics of the generated HCs. A, There are no EdU+Myo6+ or EdU+Prestin+ HCs in the cochleae of control mice. B, In the cochleae of β-cat-OE/Notch1-KO/Atoh1-OE mice, the EdU+Myo7+ HCs could be labeled with Myo6, but only very few mitotic HCs in the sensory region could be stained with Prestin. Scale bar, 50 μm. C–E, The bundle structures of the new generated HC in the cochleae of control and β-cat-OE/Notch1-KO/Atoh1-OE mice. Arrows indicate the EdU+Myo7+ HCs. F, G, For showing the expression of transgene, the HA tag was labeled in the cochleae of Sox2CreER/T2/Atoh1-OE mice. H, Quantification of the EdU+Myo7+ and EdU−Myo7+ HCs in GER of β-cat-OE/Notch1-KO/Atoh1-OE mice. Data in H are presented as mean ± SEM. I, J, Quantification of Sox2+ EdU+ SCs in the sensory region and GER. Data in F and G are presented as mean ± SEM. *p < 0.01. Scale bars: A3, C1, I1, 50 μm; C3, 10 μm.
Figure 4.
Mitotic HC generation through the overexpression of Atoh1 (Atoh1-OE). A, In control mice, there are no ectopic HC or EdU+Myo7a+ HCs in the cochleae. B, Overexpression of Atoh1 in Sox2+ SCs resulted in numerous ectopic HCs in the GER, whereas few EdU+Myo7a+ HCs were identified in the cochlea. C, Only a limited number of proliferative SCs differentiated into HCs in the β-cat-OE/Atoh1-OE mice. D, E, In the β-cat-OE/Notch1-KO/Atoh1-OE mice, numerous EdU+Myo7a+ HCs were detected in both the sensory region and GER. E, Quantification and comparison of the number of EdU+Myo7a+ HCs in both the sensory region and GER among different groups. F, Cross-section of the cochleae from β-cat-KO/Notch1-KO/Atoh1-OE mice. G, EdU+Myo7a+ HCs in the GER at high magnification. Arrows indicate the EdU+Myo7a+ HCs. Data in E are presented as mean ± SEM. **p < 0.01. The dotted lines in A2 show the counting limits of HCs in the sensory region and GER. Scale bar, 50 μm.
Table 3.
Synergistic effect of β-catenin overexpression, Notch1 deletion, and Atoh1 overexpression on mitotic HCs generation
| Control (n = 4) | Atoh1-OE (n = 3) | β-cat-OE Atoh1-OE (n = 3) | β-cat-OE Notch1-KO Atoh1-OE (n = 3) | |
|---|---|---|---|---|
| EdU+Myo7a+ HCs/cochlea | ||||
| Sensory region | 0 | 0.67 ± 0.67 | 3.67 ± 1.67 | 122.67 ± 9.87 |
| GER | 0 | 1.67 ± 1.20 | 70.67 ± 4.67 | 296.67 ± 14.95 |
| Myo7a+HCs/100 μm GER | ||||
| Proliferative HCs | 13.17 ± 1.17 | |||
| Nonproliferative HCs | 78.17 ± 5.83 |
Table 4.
Comparison of the number of Sox2+ SCs between β-cat-OE/Notch1-KO and β-cat-OE/Notch1-KO/Atoh1-OE group
| β-cat-OE/Notch1-KO (n = 3) | β-cat-OE/Notch1-KO/Atoh1-OE (n = 3) | |
|---|---|---|
| Sox2+ SCs | ||
| 100 μm of sensory region | 73.67 ± 3.18 | 110.00 ± 4.62 |
| 100*60 μm of GER | 131.00 ± 3.79 | 138.00 ± 9.71 |
Mitotically generated Myo7a+ EdU+ cells are on the way to maturation
To further confirm the identity of the Myo7a+ EdU+ cells generated in our transgenic mouse models, we investigated the expression of Myo6, which is expressed at the onset of cochlear HC maturation, and measured the expression of Vglu3 (an inner HC marker) and Prestin (an outer HC marker) to determine the differentiation and maturation stages of the mitotically generated HCs. We found that all of the Myo7a+ EdU+ cells could be labeled with Myo6, only a few of the Myo7a+ EdU+ HCs in the sensory epithelium were labeled with Prestin, and no Myo7a+ EdU+ HCs in the GER could be labeled by Prestin (Fig. 5A,B). No EdU+Myo7a+ cells could be labeled by Vglu3 (data not shown). To identify the bundle structure of mitotically generated HCs (EdU+Myo7a+), we labeled the cochlea with phalloidin and observed that the EdU+Myo7a+ HCs had preliminary immature bundle structures, which clustered on the border area of new generated HCs. Compared with control, the bundle-like structures were relatively short and scattered (Fig. 5C,D). There was no significant difference in the bundle structures of the mitotically generated HCs from either the GER region or the sensory epithelial region.
Discussion
Wnt and Notch signaling are two fundamental pathways that play important roles in progenitor proliferation and cell fate determination in the inner ear (Murata et al., 2012; Munnamalai and Fekete, 2013; Hartman et al., 2010; Jacques et al., 2012; Shi et al., 2014). As previously reported, either activating canonical Wnt signaling or inhibiting Notch signaling can initiate the proliferation of SCs (Chai et al., 2012; Shi et al., 2013; Li et al., 2015). However, we found that the proliferation and transdifferentiation of SCs is mainly in the apical region, which is consistent with previous studies (Cox et al., 2014). The difference between the apical and middle turns of the cochlea in response to Notch downregulation and Wnt activation might be due to the differential expression of genes of Lgr5+ progenitors in the different regions of cochlea (Waqas et al., 2016). In our current study, we combined Wnt activation and Notch1 deletion in Sox2+ SCs and observed massive proliferation that extended from the apex to the base, demonstrating the synergistic effects on the proliferation of SCs by manipulating the two pathways. In addition, our method provides the first promising approach for activating HC generation in the basal region of the cochlea.
Although we did observe a few EdU+ Myo7a+ HCs generated by overexpressing β-catenin in the sensory region of cochlea, the majority of the proliferated SCs were trapped in the cell cycle and underwent continuous proliferation, which may due to that the proliferation takes precedence over cell fate determination at higher levels of β-catenin expression (Shi et al., 2013). We hypothesized that more proliferated SCs might differentiate into HCs by simultaneously inhibiting Notch and activating β-catenin. Compared with the extensive SC proliferation in β-cat-OE/Notch1-KO cochlea, only 3.86 ± 1.19% of the proliferated SCs differentiated into HCs. Most of the proliferated SCs could not automatically differentiate into HCs after the genetic programming, raising the question of whether the proliferated cells could still react to Atoh1, an important transcription factor for HC differentiation and maturation in the developing cochlea (Bermingham et al., 1999; Zheng and Gao, 2000), and become HC-like cells. We therefore combined the activation of β-catenin, deletion of Notch1, and overexpression of Atoh1. When all three pathways were manipulated, we observed significantly more mitotically generated HCs and this was the first successful use of a method to regenerate HCs through the manipulation of multiple genes.
A recent study showed that the proliferation of SCs and their differentiation into HCs were increased by coactivation of β-catenin and Atoh1 in neonatal cochlear Lgr5+ cells (Kuo et al., 2015) and the investigators attributed the synergistic effects to a repressive role of Atoh1 on Notch signaling, which was incidentally activated by β-catenin. However, HCs differentiated from proliferated SCs could not be evaluated clearly and only a few Myo7a+ EdU+ cells were identified in that study. In our current study, we used Wnt activation, Notch inhibition, and Atoh1 overexpression to induce significant numbers of proliferating SCs to differentiate into HCs.
We found that the generation of HCs in response to Wnt activation and Notch1 deletion did not occur at the expense of the SC population (based on counting Sox2+ cells in the sensory epithelium region), which provides a promising strategy that regenerates HCs while maintaining the SC population and delicate structure of the cochlea. However, due to the limitations of the transgenic mouse model, all of the genes are deleted or activated permanently after tamoxifen injection and their expressions cannot be controlled in an orderly manner as occurs during normal development. Therefore, in our study, the massive proliferation and differentiation of HCs went beyond the desired situation in which there should ideally be a balance in the ratio of sensory and nonsensory cell types for maintaining the structures. In addition, the generated HCs are not mature enough to express all of the HC markers, which is important for the transduction of hearing. Therefore, the mechanisms behind the production of HCs during development need to be better understood and new transgenic tools need to be developed for the transient and sequential regulation of gene expression.
The downstream target genes of Wnt signaling were upregulated after Notch1 deletion in Sox2+ SCs, whereas very few EdU+ SCs were identified, even in the apical region of sensory epithelium in Notch1 and β-catenin double knock-out transgenic mice (Fig. 2I). This demonstrates that the proliferation induced by Notch1 deletion is mediated by the activation of β-catenin and further suggests that Notch signaling may serve as an upstream and negative regulator of Wnt signaling in mouse cochlea. However, Notch inhibition induced the upregulation of Atoh1 and the following SC-to-HC transdifferentiation is Wnt independent, as we predicted in the previous study (Li et al., 2015).
Consistent with previous reports, GO analysis showed that a significant number of genes involved in inner ear development and HC differentiation were enriched after Notch1 deletion. The upregulated genes included Atoh1, Gfi1, and Pou4f3, which play important roles in HC formation and differentiation (Fig. 6C). Several genes related to congenital or progressive hearing loss, such as Esrrb (Weber et al., 2014), Bsnd (Rickheit et al., 2008), Ush2 a (Eudy et al., 1998), and Lhfpl5 (Xiong et al., 2012), were also upregulated, as well as other genes correlated with HC bundle formation, such as Ptprq (Shahin et al., 2010) and Xirp2 (Scheffer et al., 2015). This gene expression profile after Notch1 deletion will benefit the prediction and the investigation of specific gene functions. There were only three genes in the β-cat-OE group and four genes in β-cat-OE/Notch1-KO group that showed the same changes in the Notch1 deletion group, suggesting that there might be different targets behind the production of HCs by overexpressing β-catenin compared with Notch1 deletion.
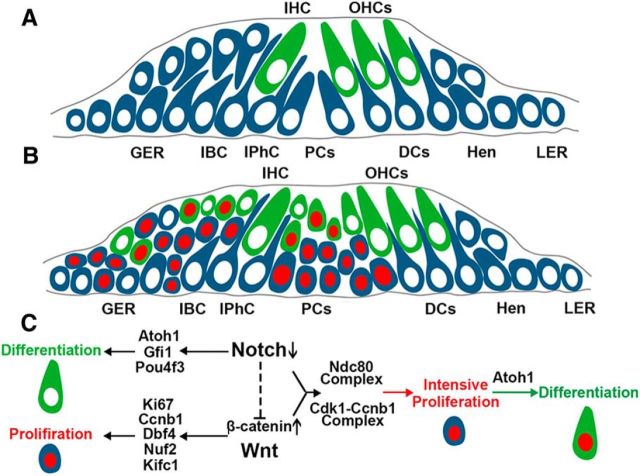
Figure 6.
Model of molecular mechanism during HC generation. A, Schematic cross-section of the organ of Corti of a neonatal mouse cochlea. B, Schematic cross-section of the organ of Corti of a neonatal mouse cochlea during HC generation. Blue cells indicate the Sox2+ cells, gray cells indicate Lgr5+ SCs, green cells indicate Myo7a+ HCs, and red nuclei indicate the EdU+ cells. C, Activation of Notch signaling inhibits the differentiation of HCs by inhibiting Atoh1, Gfi1, and Pou4f3. Notch signaling serves as a negative regulator for the proliferation of progenitors through inhibiting Wnt signaling. The activation of Wnt signaling promotes the proliferation of SCs through upregulating Ki67, Ccnb1, Dbf4, Nuf2, and Kifc1. In β-cat-OE/Notch1-KO mice, more cell-cycle-related genes are upregulated and lead to intensive proliferation of SCs, including the Ndc80 complex and Cdk1–cnb1. The proliferated SCs could still react to Atoh1 and differentiate into HCs. OHC, Outer HC; IHC, inner HC; IBC, inner border cell; IPhC, inner phalangeal cell; PC, pillar cell; DCs, Deiters' cells; Hen, Hensen's cells; LER, lesser epithelial ridge.
When comparing the gene expression profiles between β-cat-OE and β-cat-OE/Notch1-KO mice, we found that both groups showed enrichment of a significant number of genes related to the cell cycle and to mitotic cell division, including Ki67, which is a marker for proliferation (Scholzen and Gerdes, 2000); Dbf4, which regulates eukaryotic DNA replication (Yeeles et al., 2015); CCNB1 and CCNB2, which regulate chromosome segregation (Toit, 2014); and Stmn1, which is a member of a family of microtubule-destabilizing proteins (Niethammer et al., 2004). Compared with β-catenin activation, several genes involved in the cell cycle were further upregulated after the deletion of Notch1 and activation of β-catenin in Sox2+ SCs, such as Ndc80, Nuf2, Spc24, and Spc25, members of the Ndc80 complex, which is essential for kinetochore–microtubule interactions (McCleland et al., 2004; Cheerambathur et al., 2013) and Cdk1–Ccnb1 interactions, and its regulator Cdc25c, which is important for the onset of mitosis and successful completion of the M-phase (Takizawa and Morgan, 2000; Santos et al., 2012) (Fig. 6C). The significant activation of genes involved in cell proliferation and division on the molecular level is consistent with the extensive proliferation of SCs on the cellular level in the β-cat-OE/Notch1-KO group and an improved understanding of the mechanism behind the extensive proliferation might provide new approaches for promoting proliferative HC generation. Conversely, six of seven cell cycle genes upregulated by Notch1 deletion were also increased by the overexpression of β-catenin, which provides another piece of evidence that the proliferation of SCs induced by Notch inhibition is mainly through the activation of Wnt signaling.
β-catenin is a key factor involved in Wnt signaling and overexpression of β-catenin in Sox2+ SCs leads to the upregulation of some downstream targets or cooperative genes of Wnt signaling, such as Lgr5, Lgr6, Sp5, and Axin2 (Kuo et al., 2015), which is consistent with previous reports. Furthermore, when β-catenin overexpression and Notch1 deletion were combined in Sox2+ SCs, the genes that were upregulated in the β-cat-OE group were expressed at an even higher level and additional genes involved in Wnt signaling were unregulated, including Ccnb1, Axin2, Dkk1, Nkd1, and Nkd2. These results further demonstrated that Notch1 acts as a negative regulator of classical Wnt signaling, as we predicted in a previous study (Li et al., 2015). However, there are still multiple genes have not been fully described that were upregulated in β-cat-OE/Notch1-KO group, including Ccdc88c and Dact1. Further investigation into the mechanism behind the extensive proliferation of SCs might provide us with more practical strategies for generating HCs in the inner ear.
In summary, we have described the first successful demonstration of mitotic HC generation in the neonatal mouse cochlea by manipulating multiple signaling pathways in SCs (Fig. 6A,B) and we have shown that the increased proliferation of SCs and the mitotic generation of HCs occurred along with the increased expression of genes that are important for the development of the cochlea. We further conclude that Notch signaling serves as an upstream and negative regulator of Wnt signaling and that the proliferation, but not direct transdifferentiation, of SCs induced by Notch deletion requires the activation of β-catenin. In addition, the assessment of genome-wide gene expression profiles among the different groups might provide more hints for understanding the mechanisms behind mitotic HC generation and suggest new approaches to stimulate mitotic HC regeneration after HC loss.
Footnotes
This work was supported by grants from the Major State Basic Research Development Program of China (973 Program Grant 2015CB965000), the National Natural Science Foundation of China (Grants. 81400463, 81230019, 81470687, 81470692, 31500852, 31501194, and 81570911), the Jiangsu Province Natural Science Foundation (Grants BK20150022, BK20140620, and BK20150598), the Program of Leading Medical Personnel in Shanghai, the Fundamental Research Funds for the Central Universities (Grants 2242014R30022 and 021414380037), the Yingdong Huo Education Foundation, the Construction Program of the Shanghai Committee of Science and Technology (Grant 12DZ2251700), the Major Program of the Shanghai Committee of Science and Technology (Grants 14DJ1400203 and 11441901000), the Open Research Funds of the State Key Laboratory of Genetic Engineering of Fudan University (Grant SKLGE-1407), and the China Postdoctoral Science Foundation Funded Project (Grant 2014M551328). We thank Jin Li and Yalin Huang of the Institutes of Biomedical Sciences of Fudan University for providing technical support with the confocal microscope.
The authors declare no competing financial interests.
References
- Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zoghbi HY. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- Bramhall NF, Shi F, Arnold K, Hochedlinger K, Edge AS. Lgr5-positive supporting cells generate new hair cells in the postnatal cochlea. Stem Cell Reports. 2014;2:311–322. doi: 10.1016/j.stemcr.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JC, Corwin JT. A historical to present-day account of efforts to answer the question: “what puts the brakes on mammalian hair cell regeneration?”. Hear Res. 2013;297:52–67. doi: 10.1016/j.heares.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai R, Kuo B, Wang T, Liaw EJ, Xia A, Jan TA, Liu Z, Taketo MM, Oghalai JS, Nusse R, Zuo J, Cheng AG. Wnt signaling induces proliferation of sensory precursors in the postnatal mouse cochlea. Proc Natl Acad Sci U S A. 2012;109:8167–8172. doi: 10.1073/pnas.1202774109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur DK, Cheerambathur DK, Gassmann R, Gassmann R, Cook B, Cook B, Oegema K, Oegema K, Desai A. Crosstalk between microtubule attachment complexes ensures accurate chromosome segregation. Science. 2013;342:1239–1242. doi: 10.1126/science.1246232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox BC, Chai R, Lenoir A, Liu Z, Zhang L, Nguyen DH, Chalasani K, Steigelman KA, Fang J, Rubel EW, Cheng AG, Zuo J. Spontaneous hair cell regeneration in the neonatal mouse cochlea in vivo. Development. 2014;141:816–829. doi: 10.1242/dev.109421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Eudy JD, Weston MD, Yao S, Hoover DM, Rehm HL, Ma-Edmonds M, Yan D, Ahmad I, Cheng JJ, Ayuso C, Cremers C, Davenport S, Moller C, Talmadge CB, Beisel KW, Tamayo M, Morton CC, Swaroop A, Kimberling WJ, Sumegi J. Mutation of a gene encoding a protein with extracellular matrix motifs in Usher syndrome type IIa. Science. 1998;280:1753–1757. doi: 10.1126/science.280.5370.1753. [DOI] [PubMed] [Google Scholar]
- Fekete DM, Muthukumar S, Karagogeos D. Hair cells and supporting cells share a common progenitor in the avian inner ear. J Neurosci. 1998;18:7811–7821. doi: 10.1523/JNEUROSCI.18-19-07811.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman BH, Reh TA, Bermingham-McDonogh O. Notch signaling specifies prosensory domains via lateral induction in the developing mammalian inner ear. Proc Natl Acad Sci U S A. 2010;107:15792–15797. doi: 10.1073/pnas.1002827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques BE, Puligilla C, Weichert RM, Ferrer-Vaquer A, Hadjantonakis AK, Kelley MW, Dabdoub A. A dual function for canonical Wnt/ -catenin signaling in the developing mammalian cochlea. Development. 2012;139:4395–4404. doi: 10.1242/dev.080358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasena CS, Ohyama T, Segil N, Groves AK. Notch signaling augments the canonical Wnt pathway to specify the size of the otic placode. Development. 2008;135:2251–2261. doi: 10.1242/dev.017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly MC, Chang Q, Pan A, Lin X, Chen P. Atoh1 directs the formation of sensory mosaics and induces cell proliferation in the postnatal mammalian cochlea in vivo. J Neurosci. 2012;32:6699–6710. doi: 10.1523/JNEUROSCI.5420-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo BR, Baldwin EM, Layman WS, Taketo MM, Zuo J. In vivo cochlear hair cell generation and survival by coactivation of Catenin and Atoh1. J Neurosci. 2015;35:10786–10798. doi: 10.1523/JNEUROSCI.0967-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C, Cheng P, King IN, Andersen P, Shenje L, Nigam V, Srivastava D. Notch post-translationally regulates \beta -catenin protein in stem and progenitor cells. Nat Cell Biol. 2011;13:1244–1251. doi: 10.1038/ncb2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wu J, Yang J, Sun S, Chai R, Chen ZY, Li H. Notch inhibition induces mitotically generated hair cells in mammalian cochleae via activating the Wnt pathway. Proc Natl Acad Sci U S A. 2015;112:166–171. doi: 10.1073/pnas.1415901112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin V, Golub JS, Nguyen TB, Hume CR, Oesterle EC, Stone JS. Inhibition of Notch activity promotes nonmitotic regeneration of hair cells in the adult mouse utricles. J Neurosci. 2011;31:15329–15339. doi: 10.1523/JNEUROSCI.2057-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland ML, Kallio MJ, Barrett-Wilt GA, Kestner CA, Shabanowitz J, Hunt DF, Gorbsky GJ, Stukenberg PT. The vertebrate Ndc80 complex contains Spc24 and Spc25 homologs, which are required to establish and maintain kinetochore-microtubule attachment. Curr Biol. 2004;14:131–137. doi: 10.1016/j.cub.2003.12.058. [DOI] [PubMed] [Google Scholar]
- Munnamalai V, Fekete DM. Wnt signaling during cochlear development. Semin Cell Dev Biol. 2013;24:480–489. doi: 10.1016/j.semcdb.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata J, Ikeda K, Okano H. Notch signaling and the developing inner ear. Adv Exp Med Biol. 2012;727:161–173. doi: 10.1007/978-1-4614-0899-4_12. 2012. [DOI] [PubMed] [Google Scholar]
- Niethammer P, Bastiaens P, Karsenti E. Stathmin-tubulin interaction gradients in motile and mitotic cells. Science. 2004;303:1862–1866. doi: 10.1126/science.1094108. [DOI] [PubMed] [Google Scholar]
- Raphael Y. Evidence for supporting cell mitosis in response to acoustic trauma in the avian inner ear. J Neurocytol. 1992;21:663–671. doi: 10.1007/BF01191727. [DOI] [PubMed] [Google Scholar]
- Rickheit G, Maier H, Strenzke N, Andreescu CE, De Zeeuw CI, Muenscher A, Zdebik AA, Jentsch TJ. Endocochlear potential depends on Cl-channels: mechanism underlying deafness in Bartter syndrome IV. EMBO J. 2008;27:2907–2917. doi: 10.1038/emboj.2008.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson DW, Alosi JA, Cotanche DA. Direct transdifferentiation gives rise to the earliest new hair cells in regenerating avian auditory epithelium. J Neurosci Res. 2004;78:461–471. doi: 10.1002/jnr.20271. [DOI] [PubMed] [Google Scholar]
- Romero-Carvajal A, Navajas Acedo J, Jiang L, Kozlovskaja-Gumbrienė A, Alexander R, Li H, Piotrowski T. Regeneration of sensory hair cells requires localized interactions between the Notch and Wnt pathways. Dev Cell. 2015;34:267–282. doi: 10.1016/j.devcel.2015.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos SD, Wollman R, Meyer T, Ferrell JE, Ferrell JE., Jr Spatial positive feedback at the onset of mitosis. Cell. 2012;149:1500–1513. doi: 10.1016/j.cell.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer DI, Zhang DS, Shen J, Indzhykulian A, Karavitaki KD, Xu YJ, Wang Q, Lin JJ, Chen ZY, Corey DP. XIRP2, an actin-binding protein essential for inner ear hair-cell stereocilia. Cell Rep. 2015;10:1811–1818. doi: 10.1016/j.celrep.2015.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Shahin H, Rahil M, Abu Rayan A, Avraham KB, King MC, Kanaan M, Walsh T. Nonsense mutation of the stereociliar membrane protein gene PTPRQ in human hearing loss DFNB84. J Med Genet. 2010;47:643–645. doi: 10.1136/jmg.2009.075697. [DOI] [PubMed] [Google Scholar]
- Shi F, Kempfle JS, Edge AS. Wnt-responsive Lgr5-expressing stem cells are hair cell progenitors in the cochlea. J Neurosci. 2012;32:9639–9648. doi: 10.1523/JNEUROSCI.1064-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F, Hu L, Edge AS. Generation of hair cells in neonatal mice by β-catenin overexpression in Lgr5-positive cochlear progenitors. Proc Natl Acad Sci U S A. 2013;110:13851–13856. doi: 10.1073/pnas.1219952110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F, Hu L, Jacques BE, Mulvaney JF, Dabdoub A, Edge AS. β-Catenin is required for hair-cell differentiation in the cochlea. J Neurosci. 2014;34:6470–6479. doi: 10.1523/JNEUROSCI.4305-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa CG, Morgan DO. Control of mitosis by changes in the subcellular location of cyclin-B1-Cdk1 and Cdc25C. Curr Opin Cell Biol. 2000;12:658–665. doi: 10.1016/S0955-0674(00)00149-6. [DOI] [PubMed] [Google Scholar]
- Toit Du A. Cell cycle: Regulating chromosome segregation. Nat Rev Mol Cell Biol. 2014;15:364–365. doi: 10.1038/nrm3809. [DOI] [PubMed] [Google Scholar]
- Wang Z, Jiang H, Yan Y, Wang Y, Shen Y, Li W, Li H. Characterization of proliferating cells from newborn mouse cochleae. Neuroreport. 2006;17:767–771. doi: 10.1097/01.wnr.0000215781.22345.8b. [DOI] [PubMed] [Google Scholar]
- Waqas M, Guo L, Zhang S, Chen Y, Zhang X, Wang L, Tang M, Shi H, Bird PI, Li H, Chai R. Characterization of Lgr5+ progenitor cell transcriptomes in the apical and basal turns of the mouse cochlea. Oncotarget. 2016 doi: 10.18632/oncotarget.8636. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassermann S, Scheel SK, Hiendlmeyer E, Palmqvist R, Horst D, Hlubek F, Haynl A, Kriegl L, Reu S, Merkel S, Brabletz T, Kirchner T, Jung A. p16INK4a is a beta-catenin target gene and indicates low survival in human colorectal tumors. Gastroenterology. 2009;136:196–205.e2. doi: 10.1053/j.gastro.2008.09.019. [DOI] [PubMed] [Google Scholar]
- Weber ML, Hsin HY, Kalay E, BroŽková DS, Shimizu T, Bayram M, et al. Role of estrogen related receptor beta (ESRRB) in DFN35B hearing impairment and dental decay. BMC Med Genet. 2014;15:81. doi: 10.1186/1471-2350-15-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W, Grillet N, Elledge HM, Wagner TF, Zhao B, Johnson KR, Kazmierczak P, Müller U. TMHS is an integral component of the mechanotransduction machinery of cochlear hair cells. Cell. 2012;151:1283–1295. doi: 10.1016/j.cell.2012.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N, Tanigaki K, Tsuji M, Yabe D, Ito J, Honjo T. Inhibition of Notch/RBP-J signaling induces hair cell formation in neonate mouse cochleas. J Mol Med. 2006;84:37–45. doi: 10.1007/s00109-005-0706-9. [DOI] [PubMed] [Google Scholar]
- Yeeles JT, Deegan TD, Janska A, Early A, Diffley JF. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 2015;519:431–435. doi: 10.1038/nature14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong X, Tang B, Xiao YF, Xie R, Qin Y, Luo G, Hu CJ, Dong H, Yang SM. Helicobacter pylori upregulates Nanog and Oct4 via Wnt/β-catenin signaling pathway to promote cancer stem cell-like properties in human gastric cancer. Cancer Lett. 2016;374:292–303. doi: 10.1016/j.canlet.2016.02.032. [DOI] [PubMed] [Google Scholar]
- Zheng JL, Gao WQ. Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat Neurosci. 2000;3:580–586. doi: 10.1038/75753. [DOI] [PubMed] [Google Scholar]