Abstract
MicroRNAs (miRNAs) are small, noncoding RNAs that posttranscriptionally regulate gene expression in many tissues. Although a number of brain-enriched miRNAs have been identified, only a few specific miRNAs have been revealed as critical regulators of synaptic plasticity, learning, and memory. miR-9-5p/3p are brain-enriched miRNAs known to regulate development and their changes have been implicated in several neurological disorders, yet their role in mature neurons in mice is largely unknown. Here, we report that inhibition of miR-9-3p, but not miR-9-5p, impaired hippocampal long-term potentiation (LTP) without affecting basal synaptic transmission. Moreover, inhibition of miR-9-3p in the hippocampus resulted in learning and memory deficits. Furthermore, miR-9-3p inhibition increased the expression of the LTP-related genes Dmd and SAP97, the expression levels of which are negatively correlated with LTP. These results suggest that miR-9-3p-mediated gene regulation plays important roles in synaptic plasticity and hippocampus-dependent memory.
SIGNIFICANCE STATEMENT Despite the abundant expression of the brain-specific microRNA miR-9-5p/3p in both proliferating and postmitotic neurons, most functional studies have focused on their role in neuronal development. Here, we examined the role of miR-9-5p/3p in adult brain and found that miR-9-3p, but not miR-9-5p, has a critical role in hippocampal synaptic plasticity and memory. Moreover, we identified in vivo binding targets of miR-9-3p that are involved in the regulation of long-term potentiation. Our study provides the very first evidence for the critical role of miR-9-3p in synaptic plasticity and memory in the adult mouse.
Keywords: hippocampus, long-term potentiation, memory, microRNA
Introduction
Long-term memory formation requires de novo synthesis of mRNAs and proteins (Martin et al., 2000). Gene expression can be regulated at multiple stages such as transcriptional initiation, posttranscription, and translational levels. Recent studies have shown that microRNAs (miRNAs) posttranscriptionally regulate gene expression (Bartel, 2004; He and Hannon, 2004). miRNAs are short nucleotides (∼22 nt) regulating the stability and translation of target mRNAs via complementary binding to 3′-untranslated regions (3′ UTRs), which fine-tunes the expression of various target genes and orchestrates diverse biological processes, including proliferation, development, metabolism, apoptosis, and cell fate decision (Bartel, 2004; He and Hannon, 2004; Marcuzzo et al., 2015). There is emerging evidence suggesting that miRNAs play essential roles in synaptic plasticity and memory in both invertebrates and vertebrates (Schratt, 2009). For example, forebrain-specific disruption of Dicer1, a key RNase III enzyme for miRNA biogenesis, affected the expression of a set of brain-specific miRNAs and improved performance in learning and memory tasks in adult mice (Konopka et al., 2010). miR-134, which is specifically expressed in the brain, was shown to regulate the size of dendritic spines, excitatory synaptic transmission, and synaptic plasticity (Schratt et al., 2006; Gao et al., 2010). The role of another brain-specific miRNA, miR-124, was studied in the marine snail Aplysia californica (Rajasethupathy et al., 2009). It is exclusively expressed in the sensory neuron and regulates serotonin-mediated synaptic plasticity through the regulation of CREB (Rajasethupathy et al., 2009). In addition to these miRNAs, several miRNAs are reported to be highly enriched in the brain (Sempere et al., 2004), but their roles in synaptic plasticity, learning, and memory are still largely unknown.
A single miR-9 precursor produces two mature miRNAs, miR-9-5p and miR-9-3p, which are abundantly expressed in both developing and adult brain (Yuva-Aydemir et al., 2011). miR-9-5p is evolutionarily highly conserved across vertebrate species and shows brain-specific expression (Sempere et al., 2004; He et al., 2012). miR-9-5p was shown to be involved in neurogenesis (Krichevsky et al., 2006; Leucht et al., 2008; Shibata et al., 2011), proliferation and differentiation of neural progenitor cells (Zhao et al., 2009; Delaloy et al., 2010), and axon development (Otaegi et al., 2011; Dajas-Bailador et al., 2012). The complementary strand miR-9-3p was also suggested to be involved in neural development (Yoo et al., 2009; Yoo et al., 2011; Wei et al., 2013). Moreover, the expression levels of miR-9-3p are decreased in neurodegenerative diseases such as Huntington's and Alzheimer's disease (Cogswell et al., 2008; Packer et al., 2008). Interestingly, the seed sequence of miR-9-3p is identical to that of the invertebrate-specific miRNAs miR-4 and miR-79, which implies the functional importance of miR-9-3p across the species (Lai et al., 2004). However, the role of miR-9-5p/3p in the adult brain, especially in synaptic plasticity and memory, remains to be elucidated.
In the present study, we demonstrate that miR-9-3p, but not miR-9-5p, is crucially involved in hippocampal synaptic plasticity and memory. Inhibition of miR-9-3p activity using a competitive inhibitor impaired hippocampal long-term potentiation (LTP) and hippocampus-dependent memory. Furthermore, miR-9-3p inhibition altered the expression of LTP-related genes, suggesting that miR-9-3p-mediated gene regulation is critical for synaptic plasticity and memory.
Materials and Methods
Mice.
Male C57BL/6N mice were used for all experiments. The electrophysiology and behavioral experiments used 4- to 5-week-old and 8-week-old mice, respectively, purchased from Orient Bio. Animals were housed on a 12/12 h light/dark cycle in standard laboratory cages with ad libitum access to food and water and were cared for in accordance with the regulations and guidelines of Institutional Animal Care and Use Committees of Seoul National University.
Construction of sponge plasmid and adeno-associated virus (AAV) production.
Oligonucleotides (Integrated DNA Technologies) containing six bulged binding sites for miR-9-3p and seven bulged binding sites for miR-9-5p were subcloned into pEGFP-C1 with BglII and HinDIII. EGFP-miR-9-3p and EGFP-miR-9-5p sponge was subcloned into an AAV vector with the human synapsin promoter (hSYN) using NheI and HindIII. For the EGFP-CXCR4 control sponge, a CMV-d2EGFP-CXCR4 plasmid was purchased from Addgene (plasmid 21967) and subcloned into a pEGFP-C1 vector and then an AAV vector in the same manner as the EGFP-miR-9-3p sponge. The EGFP-CXCR4 control sponge has seven bulged binding sites for artificial miRNA based on a sequence from the CXCR4 gene. AAVs expressing each sponge were produced as described previously (Choi et al., 2014). Briefly, human embryonic kidney (HEK)-293T cells were transfected with an AAV vector expressing each sponge, AAV2/1, and pAdDeltaF6 using calcium phosphate. AAVs were harvested and purified using an iodixanol (Axis-Shield) gradient.
Stereotaxic virus injection.
Mice were deeply anesthetized with a ketamine/xylazine mixture and mounted in a stereotaxic apparatus (Stoelting). Their eyes were protected by ophthalmic gel. The virus was injected using a 33-gauge metal needle (Plastics One) connected to a 10 μl Hamilton syringe at a rate of 0.1 μl/min. After the injection, the needle was left for an additional 7 min before withdrawal. For 3-week-old male mice, 0.5 μl of virus was injected into the CA1 [stereotaxic coordinates: −1.9 mm anteroposterior (AP), ±1.25 mm mediolateral (ML), −1.5 mm dorsoventral (DV)]. For 8-week-old mice, two sites in the CA1 of both hippocampi were targeted 0.5 μl of virus injected into each site (site one: −1.6 mm AP, ±1.3 mm ML, −1.6 mm DV; site two: −2.3 AP, ±2.0 mm ML, −1.7 mm DV).
Immunohistochemistry.
The AAV-miR-9-3p or AAV-control sponge-infused mice were perfused with 4% paraformaldehyde (PFA) in PBS. The brains were removed and kept in 4% PFA overnight at 4°C. Brains were sectioned with a cryostat at a thickness of 40 μm. Sections were incubated in a blocking solution (2% goat serum, 0.2% Triton X-100 in PBS) for 1 h and then incubated with the NeuN (1:1000; Millipore) antibody in the blocking solution overnight at 4°C. Sections were then incubated with anti-mouse Alexa Fluor 555 IgG (1:300; Invitrogen) in blocking solution for 2 h at room temperature (RT). Sections were then imaged with a fluorescent microscope (IX51; Olympus).
Target gene prediction of brain-enriched miRNAs.
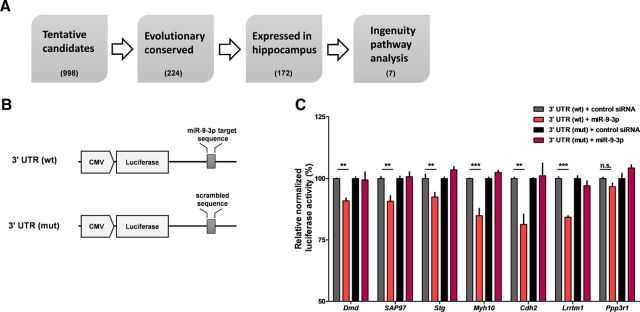
To predict the target genes of miR-9-3p in the mouse hippocampus, a series of bioinformatics analyses were performed. Among the putative candidates genes predicted by TargetScan version 6.2, the top 25% were selected based on the context + score (Garcia et al., 2011) and genes with only 6mer sites in their 3′ UTRs were removed. To identify genes with highly conserved target sequences, target sequences including ≥1 nucleotide were removed with a <0.5 PhyloP conservation score (phyloP60way, mm10, UCSC genome browser, http://genome.ucsc.edu; Kent et al., 2002). After further removing genes in which mRNA expression in the mouse hippocampus did not fall in the top 10,000 highly expressed genes, the target genes of brain-enriched miRNAs were obtained. Next, gene ontology and network analysis was performed using Ingenuity Pathway Analysis (QIAGEN) to identify the number of genes with functional annotations related to excitatory synapses, LTP, and learning and memory (see Fig. 5A). Among the target genes of miR-9-3p, genes with loss or gain of function that induced any changes in LTP were obtained by searching previously published literature and >1 PhyloP conservation scores. Seven final target genes of miR-9-3p were obtained (see Table 1).
Figure 5.
miR-9-3p regulates the expression of LTP-related genes. A, Flow chart of bioinformatic analyses. The TargetScan algorithm predicted tentative target genes of miR-9-3p. Further analyses were performed based on evolutionary conservation of miR-9-3p target sequence and reliable expression in hippocampus. Finally, seven candidate target genes were identified via Ingenuity Pathway Analysis. Numbers in parentheses indicate the number of counted target genes via each bioinformatics analysis. B, Schematic diagrams of the reporter constructs used for the luciferase assays. C, miR-9-3p suppresses the expression of the luciferase reporters containing the 3′ UTRs of selected LTP-related genes [n = 4–6 for each group, 2-way ANOVA, miR-9-3p × mutation; Dmd, F(1,20) = 5.91, p = 0.0245; SAP97, F(1,20) = 9.59, p = 0.0057; Stg, F(1,20) = 14.97, p = 0.001; Myh10, F(1,20) = 31.39, p < 0.0001; Cdh2, F(1,12) = 8.95, p = 0.0112; Lrrtm1, F(1,12) = 27.5, p = 0.0002; Ppp3r1, F(1,12) = 7.06, p = 0.0209; Bonferroni's post tests, 3′ UTR (wt) + control siRNA versus 3′ UTR (wt) + miR-9-3p, **p < 0.01, ***p < 0.001]. Data are mean ± SEM.
Table 1.
List of final candidate target genes of miR-9-3p
| Gene symbol | RefSeq ID | Description | Typea | Context+ score | Min PhyloPb | LTP change by gene manipulation |
Reference(s) | |
|---|---|---|---|---|---|---|---|---|
| Gain of function | Loss of function | |||||||
| Dmd | NM_007868 | Dystrophin, muscular dystrophy | 7 mer-m8, 7 mer-m8 | −0.2611 | 1.42, 1.45 | ND | ↑ | (Vaillend et al., 1999, 2004) |
| Dlg1 (SAP97) | NM_001252435 | Discs, large homolog 1 | 7 mer-m8 | −0.19174 | 1.95 | ↓ | Not affected | (Nakagawa et al., 2004; Howard et al., 2010; Li et al., 2011) |
| Myh10 | NM_175260 | Myosin, heavy polypeptide 10, non-muscle | 8 mer | −0.181 | 3.25 | ND | (Rex et al., 2010) | |
| Cacng2 (Stargazin) | NM_007583 | Calcium channel, voltage-dependent, gamma subunit 2 | 8 mer | −0.18095 | 3.36 | Serine 9 phosphorylation-dependent bidirectional synaptic plasticity | (Tomita et al., 2005) | |
| Lrrtm1 | NM_028880 | Leucine rich repeat transmembrane neuronal 1 | 7 mer-m8 | −0.17026 | 1.53 | ND | ↓ | (Soler-Llavina et al., 2013) |
| Cdh2 | NM_007664 | Cadherin 2 | 7 mer-A1, 7 mer-A1 | −0.16819 | 1.20, 4.29 | ND | ↓ | (Tang et al., 1998; Bozdagi et al., 2000) |
| Ppp3r1 | NM_024459 | Protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type I) | 7 mer-A1 | −0.13017 | 2.84 | ND | Threshold shift | (Zeng et al., 2001) |
aType of mirna target sequences depending on seed matching (Bartel, 2009).
bMinimum PhyloP conservation score of target sequences.
ND, Not determined.
Luciferase assay.
To validate the miR-9-3p target genes, the 3′ UTR was cloned from each of the putative target genes downstream of the firefly luciferase coding region in modified pGL3 luciferase reporter vectors (pGL3-UC). The following primers were used for cloning: Dmd, forward 5′-GCCGTGTAAT TCTAGA GACGGGCTGAAATGTCTAA-3′, reverse 5′-TATCCCGCGG GAATTC CACTTTAATACTTGATGGTTAT-3′; SAP97, forward 5′-GCCGTGTAAT TCTAGA ATGCAAAATTGTAAAA-3′, reverse 5′-TATCCCGCGG GAATTC ACTAAATCACTGATTA-3′; Myh10, forward 5′-GCCGTGTAAT TCTAGA CATTCAGTGACAGCAAATCA-3′, reverse 5′-TATCCCGCGG GAATTC ACAGCCAGTACAATTCATAC-3′; Stargazin, forward 5′-GCCGTGTAAT TCTAGA TAATAAGTGAATTTAC-3′, reverse 5′-TATCCCGCGG GAATTC TTGTCTTTACAAGTTC-3′; Lrrtm1, forward 5′-GCCGTGTAAT TCTAGA CAGCGGGCACCAGGACT-3′, reverse 5′-TATCCCGCGG GAATTC GATTATGGGGTGGGGTTTT-3′; Cdh2, forward 5′-GCCGTGTAAT TCTAGA ACGGCAGGACGGACTTG-3′, reverse 5′-TATCCCGCGG GAATTC GCGTATCACATTTAACTTTC-3′; Ppp3r1, forward 5′-GCCGTGTAAT TCTAGA ACAAGAAAGTATCTATATATACA-3′, reverse 5′-TATCCCGCGG GAATTC GTGAAATAAAAGTAAGGATGG-3′. The vector was a generous gift from Dr. V. Narry Kim (Seoul National University). The mutation of target sequence was produced by mutagenesis kit (Enzynomics). HEK-293T cells were transfected in 12-well plates with 300 ng of firefly luciferase reporter, 200 ng of pcDNA3-renila luciferase, and a 20 nm concentration of the synthetic miRNA duplexes or negative control duplexes (Bioneer) using Lipofectamine 2000 (Invitrogen). Dual-Glo luciferase assays (Promega) were performed 24 h after transfection according to the manufacturer's instructions. To generate the miR-9-5p/3p expression construct, the miR-9-3p gene was amplified from the genomic locus of mouse miR-9-3p and cloned into a pcDNA3 vector (Invitrogen).
Electrophysiology.
Animals (4- to 5-week-old male mice) were deeply anesthetized with isoflurane, decapitated, and coronal hippocampal slices (300 μm thick) were prepared using a vibratome (VT1200S; Leica). Slices were allowed to recover for at least 1 h in a recovery chamber at RT with oxygenated artificial CSF (ACSF) containing the following (in mm): 124 NaCl, 2.5 KCl, 1 NaH2PO4, 25 NaHCO3, 10 glucose, 2 CaCl2, and 2 MgSO4. After recovery, the CA3 region was removed from the slice and the remaining hippocampal tissue was transferred to a recording chamber and maintained at RT with oxygenated ACSF. For experiments that examine LTP and basal synaptic properties, the patch-clamp recording pipettes (3∼5 MΩ) were filled with an internal solution containing the following (in mm): 145 K-gluconate, 5 NaCl, 10 HEPES, 1 MgCl2, 0.2 EGTA, 2 MgATP, and 0.1 Na3GTP, 280–300 mOsm adjust to pH 7.2 with KOH. Picrotoxin (100 μm) was added to the ACSF to block the GABA-R-mediated currents. EPSCs were evoked at 0.05 Hz and three successive EPSCs were averaged and expressed relative to the normalized baseline. Because the LTP was measured using whole-cell recording, the baseline responses were only recorded for 5 min to minimize the washout effect. To induce theta-burst LTP, theta-burst stimulations (TBS; five trains of burst with four pulses at 100 Hz at a 200 ms interval) were repeated four times at 10 s intervals. For experiments measuring long-term depression (LTD) and NMDAR-mediated currents, internal solution containing the following (in mm): 130 CsMeSO4, 5 NaCl, 10 HEPES, 1 MgCl2, 0.5 EGTA, 4 MgATP, 0.3 Na3GTP, 5 QX-314, 280–300 mOsm adjusted to pH 7.2 with CsOH was used. LTD was induced with a 1 Hz, 5 min train of stimulation at −40 mV in the presence of picrotoxin (100 μm). To measure NMDAR-mediated currents, CNQX (20 μm) and picrotoxin (100 μm) were added to the ACSF. The input–output relationship of NMDA-receptor-mediated EPSC was recorded at −30 mV. Experiments measuring miniature EPSCs (mEPSCs) and spontaneous EPSCs (sEPSCs) were performed with internal solution containing the following (in mm): 100 Cs-gluconate, 5 NaCl, 10 HEPES, 10 EGTA, 20 TEA-Cl, 3 QX-314, 4 MgATP, and 0.3 Na3GTP, 280–300 mOsm pH adjusted to 7.2 with CsOH. Both mEPSCs and sEPSCs were recorded in the presence of picrotoxin (100 μm). For the mIPSC and sIPSC recordings, an internal solution containing the following (in mm): 145 KCl, 5 NaCl, 10 HEPES, 10 EGTA, 10 QX-314, 4 MgATP, and 0.3 Na3GTP 280–300 mOsm pH adjusted to 7.2 with KOH was used. Both mIPSCs and sIPSCs were recorded in the presence of kynurenic acid (2 μm). For the mEPSC and mIPSC recordings, 1 μm tetrodotoxin was added. Hippocampal neurons were voltage-clamped at −70 mV using an Axopatch 200B (Molecular Devices). Only cells with a change in access resistance <20% were included in the analysis. AAV-GFP-sponge expression was confirmed by a cooled CCD camera (ProgRes MF cool; Jenoptik) and fluorescence microscope (BX51WI; Olympus). The MiniAnalysis program (Synaptosoft) was used for mEPSC, sEPSC, mIPSC, and sIPSC analysis.
Morris water maze.
The water maze was made of an opaque gray Plexiglas, 140 cm in diameter and 100 cm in height. The maze was filled with water (22–23°C) to a depth of 30 cm. The water was kept opaque by adding a small amount of white paint. The hidden platform (a circle 10 cm in diameter) was placed at the center of the target quadrant and submerged ∼1.5 cm below the surface of the water. For acquisition of spatial memory, mice were placed in water and trained to find the hidden platform. The training consisted of 4 60 s trials per day for 5 consecutive days. The starting point was randomized for the four trials so that they were equally distributed among the four maze quadrants. If the mice failed to find the platform within 60 s, they were guided manually to the platform by an experimenter and allowed to remain on the platform for 20 s. When the mice found and mounted on the platform, they were allowed to remain on the platform for 20 s. The intertrial interval (ITI) was 60 s between trials. The probe trial was performed on day 6 to measure spatial memory, which consisted of free swimming (60 s) without the platform. For the probe test, mice were placed in the center of the maze.
Object location memory task.
The object location memory task was performed as described previously (Lee et al., 2014). All experiments were performed under dim light. Each mouse was handled for 5 min per day for 4 consecutive days. For habituation, mice were exposed to the arena, an acrylic box (33 × 33 × 30 cm), for 15 min for 2 d without the objects. A black, triangle-shaped sticker was placed on one side of the arena to provide a visual anchor. During the training session, mice were allowed to explore two identical objects for 10 min. For the test session, 24 h after training, one of the objects was placed at the same location as the training session (nondisplaced object), but the other was moved to a new location (displaced object) and then mice were placed back into the arena for 5 min. Switching the placement of the objects was randomly counterbalanced. Training and test sessions of the task were recorded with a digital camera placed above the arena. The exploration time was manually counted to be the amount of time the mice spent sniffing or exploring the object within 1 cm of the object. The discrimination index was calculated as ([displaced object exploration time/total object exploration time] × 100). All sessions of the task and analysis were performed blindly. Mice that showed >65% preference for any object during the training session were excluded from the analysis.
Trace fear conditioning.
Trace fear conditioning was performed as described previously (Lim et al., 2014). White noise (15 s, 3 kHz, 75 dB) was used as the conditioned stimulus (CS) and electrical foot shock (0.5 s, 0.5 mA) was used as the unconditioned stimulus (US). For trace training, mice were exposed to the conditioning chamber for 60 s and subjected to seven CS-trace–US–ITI trials (trace, 30 s; ITI, 210 s). The next day, mice were exposed to the novel context, which was changed to octagonal walls, a different grid, vanilla scent, and red light. The memory test consisted of a 60 s acclimation period followed by seven CS-ITI trials (ITI, 210 s). All behavioral tests were performed in the conventional conditioning chamber and analyzed by the FreezeFrame system (Coulbourn Instruments).
Open-field task.
Mice were allowed to explore the arena, an opaque white box (40 × 40 × 40 cm), under dim light. The movement of each mouse in the central (within 10 × 10 cm and 20 × 20 cm) and peripheral area was recorded for 10 min. All data were recorded and analyzed with EthoVision 3.1 (Noldus).
Elevated zero maze task.
The elevated zero maze is a modified version of the elevated plus maze that avoids the ambiguity of time spent in the central square of the elevated plus maze. The maze was made of white Plexiglas. Two opposite open and closed quadrants (15 cm walls enclosed two opposing closed quadrants) were placed on circular platform (50 cm diameter, 5 cm width). The platform was elevated 65 cm above ground level. Mice were placed in the center of the closed quadrants and the time spent in each quadrant was recorded for 5 min under fluorescent lights with EthoVision 3.1 (Noldus) software.
Western blots.
The expression of the AAV-miR-9-3p, AAV-miR-9-5p or AAV-control sponge was confirmed by fluorescence microscopy and only the CA1 regions expressing EGFP were collected and frozen in liquid nitrogen for Western blot analysis. Samples were homogenized using the TissueLyser LT (QIAGEN) with homogenization buffer (320 mm sucrose, 10 mm HEPES, pH 7.4) containing protease inhibitor mixture (Roche). The homogenized suspension was centrifuged at 1000 × g for 10 min at 4°C. The supernatant was centrifuged at 12,000 × g for 20 min at 4°C to obtain the pellet (the crude synaptosomal membrane). The pellet was resuspended in RIPA buffer (20 mm HEPES, 0.15 mm NaCl, 1% Triton X-100, 1% sodium deoxycholate, 1% SDS, 1 mm dithiothreitol, pH 7.5) containing protease inhibitor mixture, followed by rotation (1 h, 4°C) and centrifugation (10,000 × g 15 min, 4°C). The supernatant was collected and its protein level was measured by a BCA protein assay (Pierce). Equal amounts of protein were subjected to SDS-PAGE and Western blots with the following antibodies: Dmd (1:200, sc-73592; Santa Cruz Biotechnology), SAP97 (1:1000, 75-030; Neuromab), and pan-cadherin (1:20,000, sc-59876; Santa Cruz Biotechnology).
Results
Inhibition of miR-9-3p impairs LTP
LTP at the hippocampal Shaffer collateral-CA1 (SC-CA1) synapses is thought to be a key mechanism underlying hippocampus-dependent long-term memory (Bliss and Collingridge, 1993; Lee, 2014). We investigated the role of endogenous miR-9-5p/3p in LTP at the SC-CA1 synapses using the miRNA sponge, which consists of a transcript containing multiple tandem miRNA-binding sites and works as a competitive decoy target for the endogenous miRNA (Ebert et al., 2007).
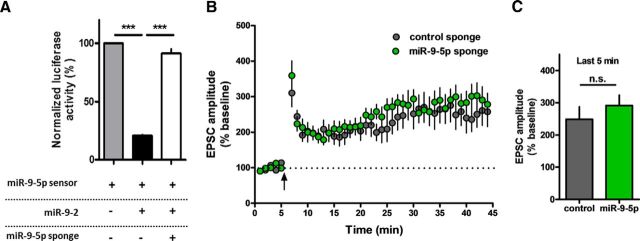
First, we designed an miR-9-3p sponge expressing the EGFP genes with 3′ UTR containing six bulged miR-9-3p-binding sites. A sponge construct containing the EGFP genes with 3′ UTR containing seven bulged artificial miRNA-binding sites that are not target sequences of any known miRNA was used as a control. To validate the efficacy of miRNA sponges, we performed a luciferase assay in HEK-293T cells. miR-9-3p sensor expresses firefly luciferase, which can be regulated via the miRNA target sequence in its 3′ UTR. Among three genetic loci encoding miR-9-5p/3p in mammals (miR-9-1, miR-9-2, and miR-9-3), we cloned miR-9-2 due to its most abundant expression in mouse brain (Shibata et al., 2011). Overexpression of miR-9-2 reduced the activity of the firefly luciferase, which was reversed by cotransfection of miR-9-3p sponge (n = 5 per group, one-way ANOVA, F(2,12) = 86, p < 0.0001; Fig. 1A). These data show clearly that the miR-9-3p sponge suppresses the activity of miR-9-3p. We generated AAV vectors encoding the miR-9-3p sponge and the control sponge under the control of synapsin promoter (Fig. 1B), which were stably expressed in dorsal hippocampus (Fig. 1C). Then, we investigated the role of miR-9-3p in hippocampal LTP using AAV-miRNA sponge. Two weeks after injection of the AAV-miRNA sponges into the dorsal CA1 region of hippocampus, we performed whole-cell patch-clamp recordings. miR-9-3p sponge expression significantly reduced the level of theta-burst-induced LTP compared with the control sponge or naive group (Fig. 1D). Statistical analysis of the EPSC amplitude of the last 5 min of recording showed significant impairment of LTP in miR-9-3p sponge-expressing group (last 5 min of recording: control, n = 6 cells from 3 mice; miR-9-3p, n = 9 cells from 6 mice; naive, n = 7 cells from 5 mice; one-way ANOVA, F(2,19) = 9.514, p = 0.0014; Tukey's multiple-comparison test, control vs miR-9-3p, miR-9-3p vs naive, p < 0.01; Fig. 1E). In addition to LTP, we also investigated whether miR-9-3p sponge expression affects LTD in the CA3–CA1 synapse of the hippocampus (Fig. 1F). We found that inhibition of miR-9-3p has no significant effect on LTD (last 5 min of recording, control, n = 9 cells from 4 mice, miR-9-3p, n = 7 cells from 3 mice, unpaired two-tailed t test, t(14) = 0.6813, p = 0.5068; Fig. 1G). Next, we examined the role of miR-9-5p in hippocampal LTP. Luciferase assay confirmed that miR-9-5p sponge inhibited miR-9-5p activity (n = 4 per group, one-way ANOVA, F(2,9) = 376.2, p < 0.0001; Fig. 2A). Unlike miR-9-3p, inhibition of miR-9-5p had no effect on the level of theta-burst LTP (last 5 min of recording, control, n = 12 cells from 7 mice, miR-9-5p, n = 14 cells from 6 mice, unpaired two-tailed t test, t(24) = 0.8454, p = 0.4062; Fig. 2B,C), demonstrating that only miR-9-3p, not miR-9-5p, is involved in hippocampal LTP.
Figure 1.
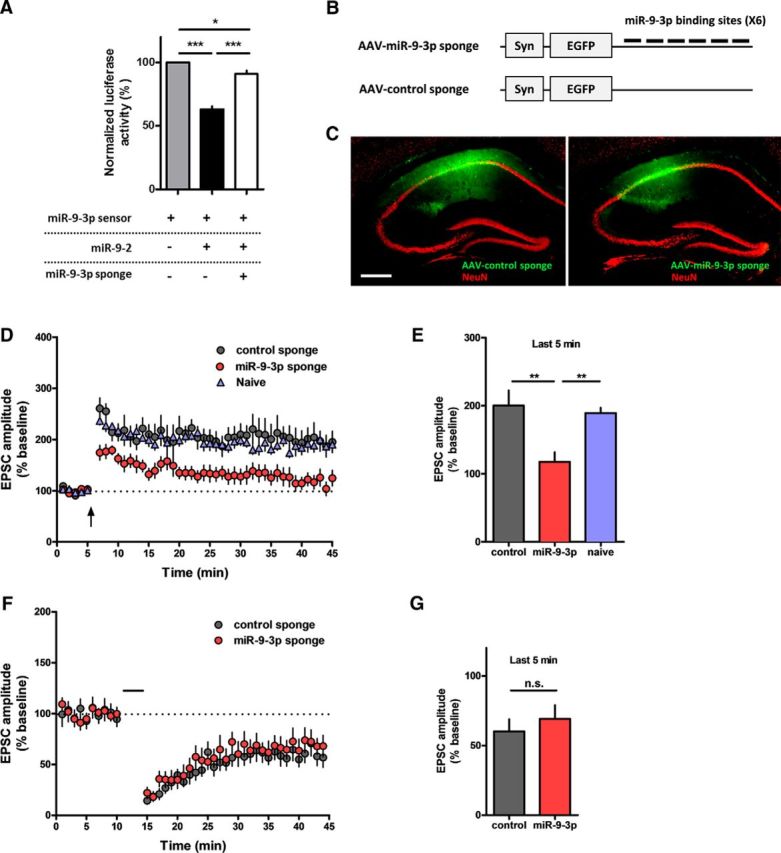
AAV-miR-9-3p sponge-mediated miR-9-3p inhibition blocks hippocampal LTP. A, Luciferase assay showing that the miR-9-3p sponge specifically suppresses the activity of miR-9-3p (*p < 0.05, ***p < 0.001, n = 5 per group, Tukey's multiple-comparisons test after significant 1-way ANOVA). The expression of miR-9-2, a genetic locus encoding miR-9-5p/3p, inhibited the expression of the miR-9-3p sensor, which rescued by cotransfection of the miR-9-3p sponge. B, Schematic diagrams of AAV-miR-9-3p-sponge. Six bulged miR-9-3p-binding sites were contained in the 3′ UTR of the EGFP gene. C, Representative fluorescence images showing EGFP expression in hippocampus 4 weeks after injection of AAV-miR-sponge. Scale bar, 300 μm. D, Theta-burst LTP recordings were significantly impaired in miR-9-3p sponge-expressing neurons. E, Summary graph representing the average EPSC amplitudes of the last 5 min of recording in naive, control sponge, and miR-9-3p sponge groups (last 5 min of recording, control, n = 6 cells from 3 mice, miR-9-3p, n = 9 cells from 6 mice, naive, n = 7 cells from 5 mice, **p < 0.01, Tukey's multiple-comparisons test after 1-way ANOVA). F, Hippocampal LTD was comparable between control and miR-9-5p sponge-expressing neurons. G, Summary graph representing the average EPSC amplitudes of the last 5 min recording in control sponge and miR-9-3p sponge groups (last 5 min of recording, control, n = 9 cells from 4 mice, miR-9-3p, n = 7 cells from 3 mice, unpaired two-tailed t test, t(14) = 0.6813, p = 0.5068). Data are mean ± SEM.
Figure 2.
AAV-miR-9-5p sponge-mediated miR-9-5p inhibition has no effect on hippocampal LTP. A, Luciferase assay showing that the miR-9-5p sponge specifically suppresses the activity of miR-9-5p (***p < 0.001, n = 4 per group, Tukey's multiple-comparisons test after significant 1-way ANOVA). B, Theta-burst LTP was comparable between control and miR-9-5p sponge. C, Summary graph represents the average EPSC amplitudes of the last 5 min recording in control and miR-9-5p sponge groups (control, n = 12 cells from 7 mice, miR-9-5p, n = 14 cells from 6 mice, unpaired 2-tailed t test, t(24) = 0.8454, p = 0.4062). Data are mean ± SEM.
Inhibition of miR-9-3p does not affect membrane excitability and basal synaptic transmission
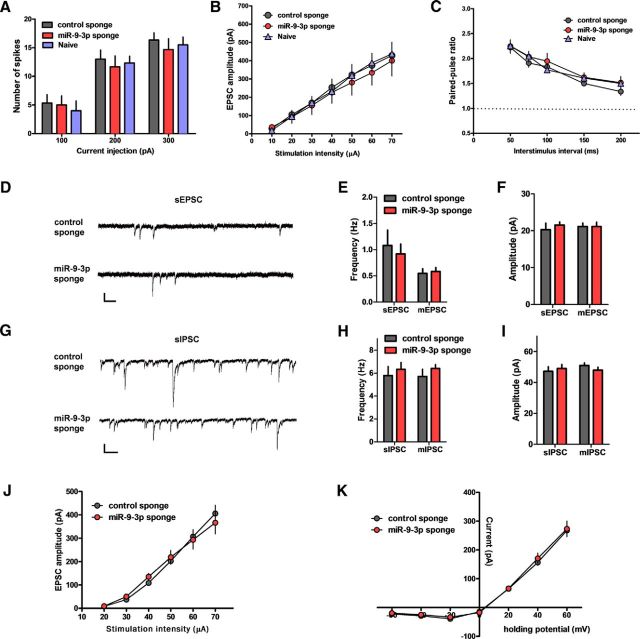
We examined the effect of blocking miR-9-3p activity on intrinsic membrane excitability. Action potentials were measured after applying step current pulses in CA1 pyramidal neurons. There were no significant differences in the number of action potentials between groups, suggesting that miR-9-3p sponge expression does not affect the membrane excitability (n = 6 cells from 3 mice per group, two-way ANOVA, F(4,45) = 0.11, p = 0.9786; Fig. 3A). Next, we examined basal synaptic strength and presynaptic transmission by measuring the input–output relationship and paired-pulse facilitation ratio, respectively. We found that inhibition of miR-9-3p affect neither the input–output relationship nor the paired-pulse facilitation ratios (input–output relationship, control, n = 8 from 3 mice, miR-9-3p, n = 6 from 3 mice, naive, n = 7 from 3 mice, two-way ANOVA, F(12,126) = 0.07, p = 1.0000; paired-pulse facilitation ratios, n = 6 cells from 3 mice per group, two-way ANOVA, F(8,75) = 0.29, p = 0.9680; Fig. 3B,C). We also measured sEPSCs, sIPSCs, mEPSCs, and mIPSCs to test whether blocking miR-9-3p activity affects excitatory or inhibitory synaptic transmission in CA1 pyramidal neurons. However, we found that the frequencies and amplitudes of sEPSCs, sIPSCs, mEPSCs, and mIPSCs were comparable among neurons expressing miR-9-3p sponge or control sponge, suggesting that inhibition of miR-9-3p activity has no effect on excitatory and inhibitory synaptic transmission (sEPSC, control, n = 6 cells from 3 mice, miR-9-3p, n = 11 cells from 5 mice; mEPSC, control, n = 7 cells from 3 mice, miR-9-3p, n = 9 from 5 mice; sIPSC, control, n = 10 cells from 5 mice, miR-9-3p, n = 9 cells from 4 mice; mIPSC, control, n = 11 cells from 5 mice, miR-9-3p, n = 6 cells from 4 mice; Fig. 3D–I). In addition, we investigated whether the miR-9-3p sponge expression affects NMDA receptor function, which is known to be critical for LTP induction (Collingridge et al., 1983; Tsien et al., 1996). We found that inhibition of miR-9-3p did not affect the input–output relationship (control, n = 8 cells from 3 mice, miR-9-3p, n = 11 cells from 4 mice; two-way ANOVA, F(5,102) = 0.4, p = 0.8482; Fig. 3J) and I–V plot (control, n = 11 cells from 3 mice, miR-9-3p, n = 12 cells from 4 mice; two-way ANOVA, F(6,147) = 0.11, p = 0.995; Fig. 3K) of NMDA receptor-mediated EPSC in the presence of CNQX, suggesting that blocking miR-9-3p does not affect NMDA receptor function.
Figure 3.
AAV-miR sponge-mediated inhibition of miR-9-3p activity has no effects on basal synaptic properties and NMDAR-mediated synaptic transmission. A, Number of action potentials generated by current injection (n = 6 for each group). B, Input–output relationship (n = 7, naive; n = 8, control sponge; n = 6, miR-9-3p sponge). C, Paired-pulse ratio at the indicated interstimulus intervals (n = 6 for each group). D–F, sEPSCs and mEPSCs from CA1 pyramidal neurons expressing control sponge or miR-9-3p sponge. D, Representative sEPSC recording traces. Scale bars, 25 pA and 200 ms. Graphs show sEPSC (n = 6, control sponge; n = 11, miR-9-3p sponge) and mEPSC (n = 7, control sponge; n = 9, miR-9-3p sponge) frequency (E) and amplitude (F). G–I, sIPSCs and mIPSCs from CA1 pyramidal neurons expressing control sponge or miR-9-3p sponge. G, Representative sIPSC recording traces. Scale bars, 50 pA and 200 ms. Graphs show sIPSC (n = 10, control sponge; n = 9, miR-9-3p sponge) and mIPSC (n = 11, control sponge; n = 6, miR-9-3p sponge) frequency (H) and amplitude (I). J, Input–output relationship of NMDAR-mediated EPSCs (n = 8, control sponge; n = 11, miR-9-3p sponge). K, NMDAR-mediated I–V plot (n = 11, control sponge; n = 12, miR-9-3p sponge). Data are mean ± SEM.
Inhibition of miR-9-3p activity impairs hippocampus-dependent memory
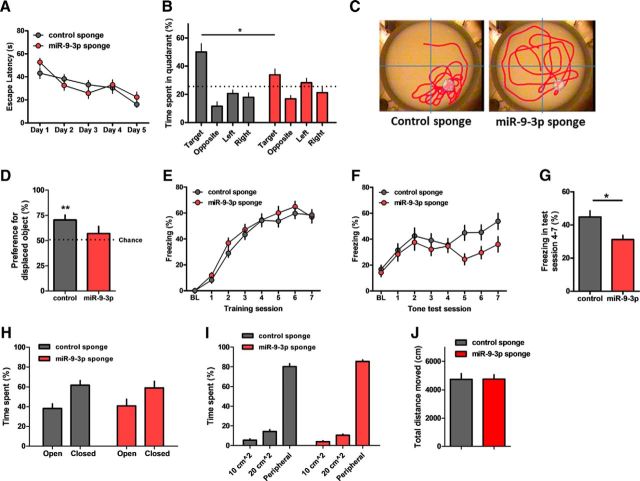
Next, we examined the behavioral effects of inhibiting miR-9-3p activity via AAV-miRNA sponge infection in the dorsal CA1 region of the adult mouse hippocampus. We first tested mice in the Morris water maze task to examine hippocampus-dependent spatial memory. Control or miR-9-3p sponge-expressing mice showed similar escape latencies during the training sessions (control, n = 9 mice, miR-9-3p, n = 10 mice, repeated-measures two-way ANOVA, the effect of sponge, F(1,68) = 0.15, p = 0.7062; Fig. 4A). However, in the probe test in which the platform is removed from the pool, miR-9-3p sponge-expressing mice spent significantly less time in the target quadrant than the control sponge-expressing mice (two-way ANOVA, sponge × quadrant, F(3,68) = 4.44, p = 0.0066; Bonferroni's post test, target quadrant, control vs miR-9-3p, p < 0.05; Fig. 4B,C), suggesting that blocking miR-9-3p activity impairs spatial memory. Second, the mice were examined in the object-location recognition task, which exploits the innate nature of mice to explore spatially novel objects and is known to be hippocampus dependent (Oliveira et al., 2010). The miR-9-3p sponge-expressing mice failed to recognize the displaced object, whereas the control sponge-expressing mice showed a significant preference for the displaced object (control, n = 8 mice, one-sample paired t test, t(7) = 4.069, p = 0.0048; miR-9-3p, n = 9 mice, one-sample paired t test, t(8) = 0.9769, p = 0.3572; Fig. 4D). Finally, we examined the effect of miR-9-3p inhibition on fear memory in trace fear-conditioning task, a more demanding hippocampus-dependent behavioral learning paradigm requiring temporal processing and attention (Huerta et al., 2000; Zhao et al., 2005; Lim et al., 2014). Control and miR-9-3p sponge-expressing mice displayed similar freezing levels during the trace fear-conditioning training sessions (control, n = 20 mice, miR-9-3p, n = 19 mice, repeated-measures two-way ANOVA, sponge × day, F(7,259) = 0.72, p = 0.656; Fig. 4E). When trace memory was measured 24 h after trace fear conditioning, however, the miR-9-3p sponge group showed significantly less freezing compared with the control sponge group during test sessions (control, n = 20 mice, miR-9-3p, n = 19 mice, repeated-measures two-way ANOVA, sponge × day, F(7,259) = 2.02, p = 0.0526; last 4 test sessions, sessions 4–7, unpaired two-tailed t test, t(6) = 2.998, p = 0.0241; Fig. 4F,G). AAV-miR-9-3p-expressing mice showed significantly impaired memory in all of three different hippocampus-dependent tasks. These results clearly indicate that miR-9-3p has a critical role in hippocampus-dependent memory. Importantly, miR-9-3p sponge expression did not alter either anxiety level or locomotor activity (Fig. 4H–J).
Figure 4.
Inhibition of miR-9-3p activity impairs hippocampus-dependent memories. A–C, AAV-miR-9-3p sponge-expressing mice showed impaired spatial memory in the Morris water maze test (n = 9, AAV-control sponge; n = 10, AAV-miR-9-3p sponge). Escape latencies to the platform were similar in control and miR-9-3p sponge groups (repeated-measures 2-way ANOVA, the effect of sponge, F(1,68) = 0.15, p = 0.7062) (A). Probe test examining time spent in the target quadrant on day 6 showed significant spatial memory deficits in miR-9-3p sponge group (2-way ANOVA, sponge × quadrant, F(3,68) = 4.44, p = 0.0066; Bonferroni's post tests, *p < 0.05) (B). C, Representative images showing the swimming traces of mice during the probe trial. Dashed circle indicates the location of removed platform. D, Object location memory was significantly impaired in the miR-9-3p sponge group (n = 8, AAV-control sponge; n = 9, AAV-miR-9-3p sponge, 1-sample paired t test, **p < 0.01). The percentage preference for the displaced objects in the testing session is reported and the dotted line indicates chance (50%) preference. E–G, Trace fear conditioning was impaired in the miR-9-3p sponge group (n = 20, AAV-control sponge; n = 19, AAV-miR-9-3p sponge). The average freezing duration during training sessions was similar in control and miR-9-3p sponge groups (repeated-measures 2-way ANOVA, sponge × day, F(7,259) = 0.72, p = 0.6560) (E). Average freezing duration during testing sessions (repeated-measures 2-way ANOVA, sponge × day, F(7,259) = 2.02, p = 0.0526) (F). Average freezing duration during testing sessions 4–7 (unpaired 2-tailed t test, *p < 0.05) (G). H–J, Basal anxiety level and locomotor activity were intact in both AAV-sponge groups. The elevated zero maze test (n = 10, control sponge; n = 11, miR-9-3p sponge) (H). Open-field test (n = 10, control sponge, n = 11, miR-9-3p sponge) (I, J). Data are mean ± SEM.
miR-9-3p suppresses the expression of LTP-related genes
Combining results from the electrophysiological and behavioral experiments strongly suggest that miR-9-3p has a critical role in regulating gene expression related to LTP and memory. To identify molecular targets of miR-9-3p, we performed a series of bioinformatic analyses (Fig. 5A). We first predicted 998 tentative target genes of miR-9-3p using TargetScan algorithm. Among the predicted targets, mRNAs harboring evolutionarily conserved miR-9-3p target sequence (224 genes) and expressed in the hippocampus (172 genes) were subjected to the Ingenuity Pathway Analysis to identify the genes with functional annotations related to LTP and memory. We found seven genes as final candidate target genes of miR-9-3p (Table 1). A comprehensive literature search also revealed that the gain or loss of function of each candidate target gene affects LTP (see Table 1, LTP change by gene manipulation).
We validated that miR-9-3p suppresses the expression of luciferase reporter constructs containing the 3′ UTRs of candidate target genes in in vitro luciferase assays [n = 4–6 for each group, two-way ANOVA, miR-9-3p × mutation; Dmd, F(1,20) = 5.91, p = 0.0245; SAP97, F(1,20) = 9.59, p = 0.0057; Stg, F(1,20) = 14.97, p = 0.001; Myh10, F(1,20) = 31.39, p < 0.0001; Cdh2, F(1,12) = 8.95, p = 0.0112; Lrrtm1, F(1,12) = 27.5, p = 0.0002; Ppp3r1, F(1,12) = 7.06, p = 0.0209; Bonferroni's post tests, 3′ UTR (wt) + control siRNA versus 3′ UTR (wt) + miR-9-3p, **p < 0.01, ***p < 0.001; Fig. 5B,C]. For six of the seven candidate target genes, miR-9-3p suppressed the translation of luciferase reporter. The mutation of target sequence within 3′ UTRs diminished the effect of miR-9-3p, indicating the specific binding of miR-9-3p to the 3′ UTR of candidate target genes.
Dmd and SAP97 are novel molecular targets of miR-9-3p
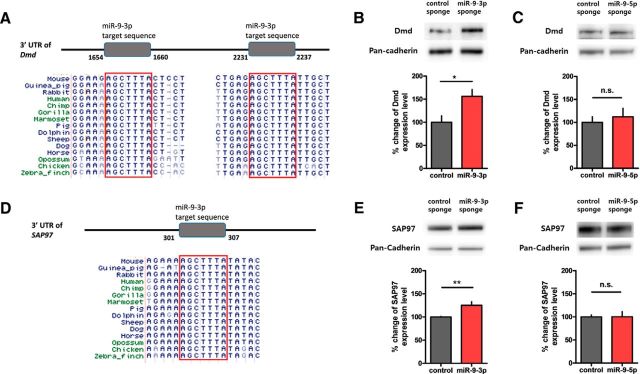
Based on the role of miRNAs in translational repression, inhibition of miR-9-3p via miRNA sponge should increase the expression of target gene. The increased expression level of target gene may impair hippocampal LTP. Therefore, we hypothesized that the genuine targets of miR-9-3p were negatively correlated with LTP. Among the seven candidate target genes, Dmd (also known as dystrophin) and SAP97 (also known as Dlg1) showed negative correlation with LTP. Therefore, we focused on Dmd and SAP97 as molecular targets of miR-9-3p.
Bioinformatic analysis identified that both Dmd and SAP97 3′ UTRs contain a highly conserved miR-9-3p target sequence (Fig. 6A,D). To validate whether Dmd and SAP97 are genuine targets of miR-9-3p in vivo, we expressed control and miR-9-3p sponge in the dorsal CA1 region for 4 weeks and performed Western blots with only the CA1 regions expressing EGFP. We confirmed that the protein levels of Dmd and SAP97 in the hippocampus were significantly increased by inhibiting miR-9-3p (Dmd, control, 100.0 ± 14.0%, n = 7 hippocampi, miR-9-3p, 156.2 ± 14.9%, n = 8 hippocampi, unpaired two-tailed t test, t(13) = 2.724, p = 0.0173; SAP97, control, 100.0 ± 1.2%, n = 5 hippocampi, miR-9-3p, 125.2 ± 7.8%, n = 4 hippocampi, unpaired two-tailed t test, t(7) = 3.606, p = 0.0087; Fig. 6B,E). Conversely, inhibition of miR-9-5p did not alter the protein levels of Dmd and SAP97 (Dmd, control, 100.0 ± 12.4%, n = 4 hippocampi, miR-9-5p, 112.1 ± 18.6%, n = 4 hippocampi, unpaired two-tailed t test, t(6) = 0.5428, p = 0.6068; SAP97, control, 100.0 ± 4.1%, n = 4 hippocampi, miR-9-5p, 100.2 ± 11.2%, n = 4 hippocampi, unpaired two-tailed t test, t(6) = 0.01657, p = 0.9873; Fig. 6C,F), suggesting that miR-9-3p specifically regulates the expression of Dmd and SAP97. Together, our results suggest that miR-9-3p regulates LTP and memory by maintaining optimal expression levels of LTP-related genes such as Dmd and SAP97.
Figure 6.
Dmd and SAP97 are novel targets of miR-9-3p. A, Bioinformatic analyses identified highly conserved miR-9-3p target sequences within Dmd 3′ UTR. B, The Dmd protein level was significantly increased in the hippocampi overexpressing miR-9-3p sponge (n = 7, AAV-control sponge; n = 8, AAV-miR-9-3p sponge, unpaired 2-tailed t test, *p < 0.05). C, The Dmd protein level was not changed by miR-9-5p inhibition (n = 4, AAV-control sponge; n = 4, AAV-miR-9-5p sponge, unpaired 2-tailed t test). D, Bioinformatic analyses identified highly conserved miR-9-3p target sequence within SAP97 3′ UTR. E, The SAP97 protein level was significantly increased in hippocampi overexpressing the miR-9-3p sponge (n = 5, AAV-control sponge; n = 4, AAV-miR-9-3p sponge, unpaired 2-tailed t test, **p < 0.01). F, The SAP97 protein level was not changed by miR-9-5p inhibition (n = 4, AAV-control sponge; n = 4, AAV-miR-9-5p sponge, unpaired 2-tailed t test). Data are mean ± SEM.
Discussion
Our results demonstrate that miR-9-3p is critically involved in hippocampal LTP and long-term memory. The miR-9-5p/3p gene is ancient in evolution and well conserved. Drosophila has five genes corresponding to miR-9-5p/3p and most vertebrate species have several copies of this gene, which implies the functional importance of miR-9-5p/3p (Wheeler et al., 2009). So far, most of functional studies on miR-9 have highlighted their roles in neuronal development (Leucht et al., 2008; Otaegi et al., 2011; Shibata et al., 2011; Dajas-Bailador et al., 2012). For example, miR-9-2/miR-9-3 double knock-out mice in which both miR-9-5p and miR-9-3p are reduced showed defects in neurogenesis and abnormal telencephalic structures (Shibata et al., 2011). However, miR-9-5p and miR-9-3p are abundantly expressed, not only in neural progenitors, but also in postmitotic neurons (Yuva-Aydemir et al., 2011; Liu et al., 2012). Interestingly, the miR-9 level was shown to be decreased in the postmortem brains of Huntington's disease patients (Packer et al., 2008) and Alzheimer's disease patients (Cogswell et al., 2008), suggesting that miR-9 might be involved in normal brain functions in adults. Recently, Giusti et al. (2014) showed that inhibition of miR-9-5p using sponge technique impairs dendritic growth and excitatory synaptic transmission. Interestingly, the miR-9-3p sponge had no effect on dendritic growth of cultured neurons (Giusti et al., 2014), which is consistent with our finding that the miR-9-3p sponge did not affect basal synaptic transmission.
miR-9-3p was shown to control the transition from neural progenitor to postmitotic neurons by switching the chromatin remodeling complexes from neural-progenitor-specific BAF (npBAF) to neuron-specific BAF. Furthermore, suppressing miR-9-3p activity in postmitotic neurons induced the expression of BAF53a, which is an npBAF (Yoo et al., 2009; Yoo et al., 2011), suggesting that miR-9-3p is critical for neuronal differentiation. In the present study, we provide the first evidence for the adult function of miR-9-3p in mammalian brain using multiple approaches. First, we used an AAV sponge vector to inhibit miR-9-3p in vivo and confirmed that miR-9-3p is critical for LTP, but not for LTD induction. Second, we showed that inhibition of miR-9-3p in vivo impairs hippocampus-dependent memory in multiple behavioral tasks. Finally, we showed that miR-9-3p, but not miR-9-5p, suppresses the expression of the well known LTP-related genes Dmd and SAP97. It is interesting that the expression levels of Dmd and SAP97 have been shown to be negatively correlated with the LTP level. Dmd-deficient mice showed enhanced LTP (Vaillend et al., 2004) and SAP97 overexpression impaired LTP (Nakagawa et al., 2004; Li et al., 2011).
Absence or mutation in Dmd gene causes membrane instability in muscles, thereby inducing Duchenne muscular dystrophy (DMD), the symptoms of which are severe muscle degradation and progressive muscular weakness. In addition to the features of DMD in skeletal muscle, DMD patients also show nonprogressive cognitive impairments, which implies critical roles of Dmd in brain. The animal model of DMD, the mdx mouse, showed impairment of memory consolidation in certain behavior tasks (Sicinski et al., 1989; Muntoni et al., 1991; Vaillend et al., 1995; Vaillend et al., 2004), supporting the importance of Dmd in cognitive function. Although it remains unknown whether the miR-9-3p level is altered in DMD patients, our finding showing downregulation of Dmd by miR-9-3p may provide new insights for the development of treatments for cognitive impairment in DMD patients.
SAP97 is the only member of PSD-95-like membrane associated guanylate kinases (PSD-MAGUKs) that binds directly to the C-terminus of GluA1 and is involved in AMPA receptor (AMPAR) trafficking (Leonard et al., 1998; Cai et al., 2002). For example, Nash et al. (2010) showed that the SAP97–myosin VI complex plays a critical role in trafficking of AMPARs to synapses; however, its role in excitatory synaptic transmission and AMPAR trafficking is controversial. Some researchers have reported that overexpressing SAP97 enhances mEPSC frequency or evoked EPSC amplitude (Rumbaugh et al., 2003; Nakagawa et al., 2004), whereas others have shown that overexpressing SAP97 has no effect (Schnell et al., 2002; Schlüter et al., 2006). The effects of SAP97 overexpression on synaptic transmission seem to be influenced by many factors, such as isoforms of SAP97 (Schlüter et al., 2006; Waites et al., 2009), compensation with other PSD-MAGUKs (Schlüter et al., 2006; Howard et al., 2010), or matureness of synapse (Howard et al., 2010). In the present study, SAP97 was increased by miR-9-3p inhibition, but we could not detect any changes in excitatory synaptic transmission (Fig. 2). This result is consistent with a previous result showing no effect of SAP97 overexpression on synaptic transmission in mature neurons (Howard et al., 2010). Conversely, the effect of overexpressing SAP97 on LTP is clear: when pairing-LTP or chemical LTP stimulus was given, SAP97 overexpressed neurons showed LTP deficits (Nakagawa et al., 2004; Li et al., 2011), which is consistent with our LTP results. Furthermore, we showed that miR-9-3p inhibition did not affect NMDAR function, suggesting that miR-9-3p is involved in LTP induction, very likely through regulating AMPAR trafficking. Depending on the stimulation paradigms, LTP can be divided into two forms: a PKA and protein-synthesis-dependent form and a PKA and protein-synthesis-independent form (Park et al., 2016). The induction of PKA-sensitive LTP requires multiple episodes of stimuli with proper interepisode intervals (Woo et al., 2003; Kim et al., 2010; Park et al., 2014; Park et al., 2016). In our experiments, LTP was induced by TBS with relatively short intervals, which may not be sensitive to protein synthesis and PKA inhibitors. Considering that miRNAs regulate protein translation, it would be of interest to determine whether miR-9-3p is also involved in the protein-synthesis-dependent form of LTP.
In conclusion, our results demonstrate the critical role of miR-9-3p in the normal adult mouse brain; miR-9-3p was shown to have functions beyond regulating neuronal differentiation and dendritic development. In addition, our study may provide an insight into understanding the molecular etiology of cognitive disorders in which the expression of miR-9-5p/3p is downregulated, such as Huntington's disease and Alzheimer's disease.
Footnotes
This work was supported by the National Research Foundation of Korea funded by the Korean government (Grant 2012R1A3A1050385 to B.-K.K.) and the Ministry of Health and Welfare of the Republic of Korea (Grant HI14C-1922-010014 to Y.-S.L.). We thank Dr. V. Narry Kim for helpful discussion.
The authors declare no competing financial interests.
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- Bozdagi O, Shan W, Tanaka H, Benson DL, Huntley GW. Increasing numbers of synaptic puncta during late-phase LTP: N-cadherin is synthesized, recruited to synaptic sites, and required for potentiation. Neuron. 2000;28:245–259. doi: 10.1016/S0896-6273(00)00100-8. [DOI] [PubMed] [Google Scholar]
- Cai C, Coleman SK, Niemi K, Keinänen K. Selective binding of synapse-associated protein 97 to GluR-A α-amino-5-hydroxy-3-methyl-4-isoxazole propionate receptor subunit is determined by a novel sequence motif. J Biol Chem. 2002;277:31484–31490. doi: 10.1074/jbc.M204354200. [DOI] [PubMed] [Google Scholar]
- Choi JH, Yu NK, Baek GC, Bakes J, Seo D, Nam HJ, Baek SH, Lim CS, Lee YS, Kaang BK. Optimization of AAV expression cassettes to improve packaging capacity and transgene expression in neurons. Mol Brain. 2014;7:17. doi: 10.1186/1756-6606-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, Prinjha RK, Richardson JC, Saunders AM, Roses AD, Richards CA. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- Collingridge GL, Kehl SJ, McLennan H. Excitatory amino acids in synaptic transmission in the Schaffer collateral-commissural pathway of the rat hippocampus. J Physiol. 1983;334:33–46. doi: 10.1113/jphysiol.1983.sp014478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajas-Bailador F, Bonev B, Garcez P, Stanley P, Guillemot F, Papalopulu N. microRNA-9 regulates axon extension and branching by targeting Map1b in mouse cortical neurons. Nat Neurosci. 2012 doi: 10.1038/nn.3082. In press. [DOI] [PubMed] [Google Scholar]
- Delaloy C, Liu L, Lee JA, Su H, Shen F, Yang GY, Young WL, Ivey KN, Gao FB. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell. 2010;6:323–335. doi: 10.1016/j.stem.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wang WY, Mao YW, Gräff J, Guan JS, Pan L, Mak G, Kim D, Su SC, Tsai LH. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giusti SA, Vogl AM, Brockmann MM, Vercelli CA, Rein ML, Trumbach D, Wurst W, Cazalla D, Stein V, Deussing JM, Refojo D. MicroRNA-9 controls dendritic development by targeting REST. eLife. 2014:3. doi: 10.7554/eLife.02755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- He M, Liu Y, Wang X, Zhang MQ, Hannon GJ, Huang ZJ. Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron. 2012;73:35–48. doi: 10.1016/j.neuron.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard MA, Elias GM, Elias LA, Swat W, Nicoll RA. The role of SAP97 in synaptic glutamate receptor dynamics. Proc Natl Acad Sci U S A. 2010;107:3805–3810. doi: 10.1073/pnas.0914422107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta PT, Sun LD, Wilson MA, Tonegawa S. Formation of temporal memory requires NMDA receptors within CA1 pyramidal neurons. Neuron. 2000;25:473–480. doi: 10.1016/S0896-6273(00)80909-5. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Huang T, Abel T, Blackwell KT. Temporal sensitivity of protein kinase a activation in late-phase long term potentiation. PLoS Comput Biol. 2010;6:e1000691. doi: 10.1371/journal.pcbi.1000691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka W, Kiryk A, Novak M, Herwerth M, Parkitna JR, Wawrzyniak M, Kowarsch A, Michaluk P, Dzwonek J, Arnsperger T, Wilczynski G, Merkenschlager M, Theis FJ, Köhr G, Kaczmarek L, Schütz G. MicroRNA loss enhances learning and memory in mice. J Neurosci. 2010;30:14835–14842. doi: 10.1523/JNEUROSCI.3030-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell–derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC, Wiel C, Rubin GM. Complementary miRNA pairs suggest a regulatory role for miRNA: miRNA duplexes. RNA. 2004;10:171–175. doi: 10.1261/rna.5191904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS. Genes and signaling pathways involved in memory enhancement in mutant mice. Mol Brain. 2014;7:43. doi: 10.1186/1756-6606-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Ehninger D, Zhou M, Oh JY, Kang M, Kwak C, Ryu HH, Butz D, Araki T, Cai Y, Balaji J, Sano Y, Nam CI, im HK, Kaang BK, Burger C, Neel BG, Silva AJ. Mechanism and treatment for learning and memory deficits in mouse models of Noonan syndrome. Nat Neurosci. 2014;17:1736–1743. doi: 10.1038/nn.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard AS, Davare MA, Horne MC, Garner CC, Hell JW. SAP97 is associated with the α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor GluR1 subunit. J Biol Chem. 1998;273:19518–19524. doi: 10.1074/jbc.273.31.19518. [DOI] [PubMed] [Google Scholar]
- Leucht C, Stigloher C, Wizenmann A, Klafke R, Folchert A, Bally-Cuif L. MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat Neurosci. 2008;11:641–648. doi: 10.1038/nn.2115. [DOI] [PubMed] [Google Scholar]
- Li D, Specht CG, Waites CL, Butler-Munro C, Leal-Ortiz S, Foote JW, Genoux D, Garner CC, Montgomery JM. SAP97 directs NMDA receptor spine targeting and synaptic plasticity. J Physiol. 2011;589:4491–4510. doi: 10.1113/jphysiol.2011.215566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CS, Hoang ET, Viar KE, Stornetta RL, Scott MM, Zhu JJ. Pharmacological rescue of Ras signaling, GluA1-dependent synaptic plasticity, and learning deficits in a fragile X model. Genes Dev. 2014;28:273–289. doi: 10.1101/gad.232470.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DZ, Ander BP, Tian Y, Stamova B, Jickling GC, Davis RR, Sharp FR. Integrated analysis of mRNA and microRNA expression in mature neurons, neural progenitor cells and neuroblastoma cells. Gene. 2012;495:120–127. doi: 10.1016/j.gene.2011.12.041. [DOI] [PubMed] [Google Scholar]
- Marcuzzo S, Bonanno S, Kapetis D, Barzago C, Cavalcante P, D'Alessandro S, Mantegazza R, Bernasconi P. Up-regulation of neural and cell cycle-related microRNAs in brain of amyotrophic lateral sclerosis mice at late disease stage. Mol Brain. 2015;8:5. doi: 10.1186/s13041-015-0095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SJ, Grimwood PD, Morris RG. Synaptic plasticity and memory: an evaluation of the hypothesis. Annu Rev Neurosci. 2000;23:649–711. doi: 10.1146/annurev.neuro.23.1.649. [DOI] [PubMed] [Google Scholar]
- Muntoni F, Mateddu A, Serra G. Passive avoidance behaviour deficit in the mdx mouse. Neuromusc Disord. 1991;1:121–123. doi: 10.1016/0960-8966(91)90059-2. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Futai K, Lashuel HA, Lo I, Okamoto K, Walz T, Hayashi Y, Sheng M. Quaternary structure, protein dynamics, and synaptic function of SAP97 controlled by L27 domain interactions. Neuron. 2004;44:453–467. doi: 10.1016/j.neuron.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Nash JE, Appleby VJ, Corrêa SA, Wu H, Fitzjohn SM, Garner CC, Collingridge GL, Molnár E. Disruption of the interaction between myosin VI and SAP97 is associated with a reduction in the number of AMPARs at hippocampal synapses. J Neurochem. 2010;112:677–690. doi: 10.1111/j.1471-4159.2009.06480.x. [DOI] [PubMed] [Google Scholar]
- Oliveira AM, Hawk JD, Abel T, Havekes R. Post-training reversible inactivation of the hippocampus enhances novel object recognition memory. Learn Mem. 2010;17:155–160. doi: 10.1101/lm.1625310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaegi G, Pollock A, Hong J, Sun T. MicroRNA miR-9 modifies motor neuron columns by a tuning regulation of FoxP1 levels in developing spinal cords. J Neurosci. 2011;31:809–818. doi: 10.1523/JNEUROSCI.4330-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park P, Volianskis A, Sanderson TM, Bortolotto ZA, Jane DE, Zhuo M, Kaang B-K, Collingridge GL. NMDA receptor-dependent long-term potentiation comprises a family of temporally overlapping forms of synaptic plasticity that are induced by different patterns of stimulation. Philos Trans R Soc B Biol Sci. 2014;369:20130131. doi: 10.1098/rstb.2013.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park P, Sanderson TM, Amici M, Choi SL, Bortolotto ZA, Zhuo M, Kaang BK, Collingridge GL. Calcium-permeable AMPA receptors mediate the induction of the protein kinase A-dependent component of long-term potentiation in the hippocampus. J Neurosci. 2016;36:622–631. doi: 10.1523/JNEUROSCI.3625-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasethupathy P, Fiumara F, Sheridan R, Betel D, Puthanveettil SV, Russo JJ, Sander C, Tuschl T, Kandel E. Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron. 2009;63:803–817. doi: 10.1016/j.neuron.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex CS, Gavin CF, Rubio MD, Kramar EA, Chen LY, Jia Y, Huganir RL, Muzyczka N, Gall CM, Miller CA, Lynch G, Rumbaugh G. Myosin IIb regulates actin dynamics during synaptic plasticity and memory formation. Neuron. 2010;67:603–617. doi: 10.1016/j.neuron.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbaugh G, Sia GM, Garner CC, Huganir RL. Synapse-associated protein-97 isoform-specific regulation of surface AMPA receptors and synaptic function in cultured neurons. J Neurosci. 2003;23:4567–4576. doi: 10.1523/JNEUROSCI.23-11-04567.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüter OM, Xu W, Malenka RC. Alternative N-terminal domains of PSD-95 and SAP97 govern activity-dependent regulation of synaptic AMPA receptor function. Neuron. 2006;51:99–111. doi: 10.1016/j.neuron.2006.05.016. [DOI] [PubMed] [Google Scholar]
- Schnell E, Sizemore M, Karimzadegan S, Chen L, Bredt DS, Nicoll RA. Direct interactions between PSD-95 and stargazin control synaptic AMPA receptor number. Proc Natl Acad Sci U S A. 2002;99:13902–13907. doi: 10.1073/pnas.172511199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G. microRNAs at the synapse. Nat Rev Neurosci. 2009;10:842–849. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J Neurosci. 2011;31:3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicinski P, Geng Y, Ryder-Cook AS, Barnard EA, Darlison MG, Barnard PJ. The molecular basis of muscular dystrophy in the mdx mouse: a point mutation. Science. 1989;244:1578–1580. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- Soler-Llavina GJ, Arstikaitis P, Morishita W, Ahmad M, Südhof TC, Malenka RC. Leucine-rich repeat transmembrane proteins are essential for maintenance of long-term potentiation. Neuron. 2013;79:439–446. doi: 10.1016/j.neuron.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Hung CP, Schuman EM. A role for the cadherin family of cell adhesion molecules in hippocampal long-term potentiation. Neuron. 1998;20:1165–1175. doi: 10.1016/S0896-6273(00)80497-3. [DOI] [PubMed] [Google Scholar]
- Tomita S, Stein V, Stocker TJ, Nicoll RA, Bredt DS. Bidirectional synaptic plasticity regulated by phosphorylation of stargazin-like TARPs. Neuron. 2005;45:269–277. doi: 10.1016/j.neuron.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Tsien JZ, Huerta PT, Tonegawa S. The essential role of hippocampal CA1 NMDA receptor-dependent synaptic plasticity in spatial memory. Cell. 1996;87:1327–1338. doi: 10.1016/S0092-8674(00)81827-9. [DOI] [PubMed] [Google Scholar]
- Vaillend C, Rendon A, Misslin R, Ungerer A. Influence of dystrophin-gene mutation onmdx mouse behavior. I. Retention deficits at long delays in spontaneous alternation and bar-pressing tasks. Behav Genet. 1995;25:569–579. doi: 10.1007/BF02327580. [DOI] [PubMed] [Google Scholar]
- Vaillend C, Ungerer A, Billard JM. Facilitated NMDA receptor-mediated synaptic plasticity in the hippocampal CA1 area of dystrophin-deficient mice. Synapse. 1999;33:59–70. doi: 10.1002/(SICI)1098-2396(199907)33:1<59::AID-SYN6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Vaillend C, Billard JM, Laroche S. Impaired long-term spatial and recognition memory and enhanced CA1 hippocampal LTP in the dystrophin-deficient Dmdmdx mouse. Neurobiol Dis. 2004;17:10–20. doi: 10.1016/j.nbd.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Waites CL, Specht CG, Härtel K, Leal-Ortiz S, Genoux D, Li D, Drisdel RC, Jeyifous O, Cheyne JE, Green WN, Montgomery JM, Garner CC. Synaptic SAP97 isoforms regulate AMPA receptor dynamics and access to presynaptic glutamate. J Neurosci. 2009;29:4332–4345. doi: 10.1523/JNEUROSCI.4431-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Li H, Miao J, Liu B, Zhan Y, Wu D, Zhang Y, Wang L, Fan Y, Gu H, Wang W, Yuan Z. miR-9*-and miR-124a-Mediated switching of chromatin remodelling complexes is altered in rat spina bifida aperta. Neurochem Res. 2013;38:1605–1615. doi: 10.1007/s11064-013-1062-8. [DOI] [PubMed] [Google Scholar]
- Wheeler BM, Heimberg AM, Moy VN, Sperling EA, Holstein TW, Heber S, Peterson KJ. The deep evolution of metazoan microRNAs. Evol Dev. 2009;11:50–68. doi: 10.1111/j.1525-142X.2008.00302.x. [DOI] [PubMed] [Google Scholar]
- Woo NH, Duffy SN, Abel T, Nguyen PV. Temporal spacing of synaptic stimulation critically modulates the dependence of LTP on cyclic AMP-dependent protein kinase. Hippocampus. 2003;13:293–300. doi: 10.1002/hipo.10086. [DOI] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuva-Aydemir Y, Simkin A, Gascon E, Gao FB. MicroRNA-9. RNA Biol. 2011;8:557–564. doi: 10.4161/rna.8.4.16019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Chattarji S, Barbarosie M, Rondi-Reig L, Philpot BD, Miyakawa T, Bear MF, Tonegawa S. Forebrain-specific calcineurin knockout selectively impairs bidirectional synaptic plasticity and working/episodic-like memory. Cell. 2001;107:617–629. doi: 10.1016/S0092-8674(01)00585-2. [DOI] [PubMed] [Google Scholar]
- Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol. 2009;16:365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MG, Toyoda H, Ko SW, Ding HK, Wu LJ, Zhuo M. Deficits in trace fear memory and long-term potentiation in a mouse model for fragile X syndrome. J Neurosci. 2005;25:7385–7392. doi: 10.1523/JNEUROSCI.1520-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]