Abstract
The synaptic cleft is the space through which neurotransmitters convey neural information between two synaptic terminals. This space is presumably filled with extracellular matrix molecules involved in synaptic function or differentiation. However, little is known about the identities of the matrix components, and it remains unclear how these molecules organize the matrix in synaptic clefts. In this study, we identified Hasp, a Drosophila secretory protein containing CCP and WAP domains. Molecular genetic analysis revealed that Hasp diffuses extracellularly and is predominantly captured at synaptic clefts of cholinergic synapses. Furthermore, Hasp regulates levels of DLG and the nAChR subunits Dα6 and Dα7 at postsynaptic terminals. Hasp is required for trapping of another matrix protein, Hig, which is also secreted and diffused in the brain, at synaptic clefts of cholinergic synapses; however, Hig is dispensable for localization of Hasp at synaptic clefts. In addition, in the brains of triple mutants for the nAChR subunits Dα5, Dα6, and Dα7, the level of Hig, but not Hasp, was markedly reduced in synaptic regions, indicating that these nAChR subunits are required to anchor Hig to synaptic clefts. High-resolution microscopy revealed that Hasp and Hig exhibit segregated distribution within individual synaptic clefts, reflecting their differing roles in synaptogenesis. These data provide insight into how Hasp and Hig construct the synaptic cleft matrix and regulate the differentiation of cholinergic synapses, and also illuminate a previously unidentified architecture within synaptic clefts.
SIGNIFICANCE STATEMENT The synapse has been extensively studied because it is essential for neurotransmission. By contrast, the space between the synaptic terminals, the synaptic cleft, is still an undeveloped research area despite its ubiquity in synapses. In fruit fly brains, we obtained evidence that the matrix protein Hasp and the previously identified Hig, both of which are secreted extracellularly, localize predominantly to synaptic clefts of cholinergic synapses, and modulate the levels of nAChR subunits on postsynaptic membranes. However, Hasp and Hig play differential roles in matrix formation and exhibit segregated distribution within synaptic clefts. These results reveal the molecular mechanisms of synaptic matrix construction and illuminate a molecular architecture within synaptic clefts previously unrevealed in any animal species.
Keywords: acetylcholine, drosophila, Hasp, Hig, matrix, synapse
Introduction
The synapse comprises presynaptic and postsynaptic terminals that are separated by a very narrow space, the synaptic cleft. Neurotransmitters traverse this extracellular space to convey neural information between the two terminals, a process that is essential for various neural functions. The synaptic cleft, which also serves as an interface that regulates the differentiation of synapses, is not simply an empty space; instead, it is filled with matrix proteins forming a scaffold that organizes membrane molecules on the synaptic terminals. To date, the matrix components in synaptic clefts have not been thoroughly identified, especially in the CNS.
Because synaptic function largely relies on neurotransmitter receptors localized at the postsynaptic membranes, the local density and efficiency of neurotransmitter receptors are critical for proper control of synaptic function. Previous studies showed that several proteins secreted into the extracellular space regulate clustering of neurotransmitter receptors (Broadie et al., 2011; Nakayama and Hama, 2011). Agrin, found in vertebrates, is a proteoglycan that clusters AChR at neuromuscular junctions (NMJs) (McMahan, 1990). Multiple studies have investigated how Agrin released by motor neurons transmits the signal to various cytoplasmic proteins and eventually to AChR (Wu et al., 2010; Barros et al., 2011). In Caenorhabditis elegans, LEV-9 and OIG-4, which are released by muscles, promote clustering of AChR at NMJs (Gendrel et al., 2009; Rapti et al., 2011). The long isoform of C. elegans Punctin/MADD-4, secreted by cholinergic motor neurons, clusters AChRs, whereas its short isoform, released by GABAergic motor neurons, clusters GABAA receptors at the NMJs (Pinan-Lucarré et al., 2014; Tu et al., 2015). In Drosophila NMJs, which are mostly glutamatergic, clustering of glutamate receptors depends on the secreted protein Mind-the-Gap (Rohrbough et al., 2007; Kim et al., 2012). In mice, Cbln1, which links Neurexin to the glutamate receptor GluD2 at cerebellar synapses (Matsuda et al., 2010; Uemura et al., 2010), induces GluD2 clustering in culture cells (Matsuda et al., 2010). Thus, several secretory proteins involved in clustering receptors have been studied in cholinergic, GABAergic, and glutamatergic NMJs, as well as in glutamatergic synapses in the CNS. However, the molecular mechanisms underlying the differentiation of other types of synapses remain to be revealed. In addition, it remains unclear how the secreted proteins distribute and organize a matrix within an individual synaptic cleft.
We previously identified the hikaru genki (hig) gene in a genetic screen for Drosophila mutants that exhibited reduced locomotor behavior (Hoshino et al., 1993). Hig, a secretory protein (Hoshino et al., 1999) with one Ig domain and a maximum of five complement control protein (CCP) domains, localizes to the synaptic clefts of mature and nascent synapses in the brain (Hoshino et al., 1996). Subsequent analyses revealed that Hig localizes predominantly at synaptic clefts of cholinergic synapses in the CNS and regulates the levels of nAChR subunits and DLG, a Drosophila PSD-95 family member (Funke et al., 2005), in the postsynaptic terminals (Nakayama et al., 2014). Hig does not simply diffuse over the entire space of the synaptic cleft but, instead, is juxtaposed with the area of nAChR on the postsynaptic membrane. During synaptogenesis, Hig secreted from cholinergic or noncholinergic neurons or even from glia cells is captured in synaptic clefts of cholinergic synapses, suggesting that a specific mechanism is responsible for anchoring Hig to synaptic clefts.
In this study, we identified Hasp (Hig-anchoring scaffold protein), a CCP domain-containing synaptic matrix protein predominantly localized at synaptic clefts of cholinergic synapses in the Drosophila brain. Hasp has a domain organization resembling that of LEV-9 of Caenorhabditis elegans (Briseño-Roa and Bessereau, 2014). The data show that Hasp is required for the synaptic localization of Hig and nAChR subunits; however, Hig and nAChR subunits are not reciprocally required for Hasp localization. High-resolution microscopy revealed that Hig and Hasp are nonuniformly distributed in individual synaptic clefts, suggesting the presence of functionally distinct matrix compartments.
Materials and Methods
Drosophila strains.
The haspD1 mutant was generated from GS7260 (Toba et al., 1999) by imprecise P-element excision. higdd37 is a null mutation caused by a deficiency in the hig gene (Hoshino et al., 1993). The mutant strain nAcRα-30DDAS1 (hereafter Dα6DAS1) (Watson et al., 2010), GAL4 driver lines (elavc155, Cha, VGluT, Eaat1, Repo, 5HTR1B, Ple, da, and OK307-GAL4), and UAS-mCD8-GFP were obtained from the Bloomington Stock Center. Ddc-Gal4, Dα7PΔEY6, and UAS-Dα7-EGFP were obtained from J. Hirsh (Li et al., 2000), H. J. Bellen (Fayyazuddin et al., 2006), and S. J. Sigrist (Leiss et al., 2009), respectively. The RNAi lines for AChR subunits used in this study are as follows: Dα1 (Bloomington stock #28688), Dα2 (#27493), Dα3 (#27671), Dα4 (#31985), Dα5 (#25943), Dβ1 (#31883), Dβ2 (#28038), and Dβ3 (#25927).
To generate UAS-hasp-HA, hasp was amplified by PCR from RE03726 (Stapleton et al., 2002), which contains the longest isoform of hasp cDNA. The resultant fragment, whose stop codon was removed, was cloned into pBluescript II containing 3× hemagglutinin (HA). C-terminally HA-tagged hasp (hasp-HA) was recloned into the KpnI/XbaI sites of pUASTattB (Bischof et al., 2007). Transgenic fly lines expressing UAS-hasp-HA were generated by Phi-C31-mediated site-specific integration at attP2.
Antibody production.
To generate anti-Hasp antibody, a fragment of hasp cDNA encoding the N-terminal fragment (amino acids 108-443) was amplified by PCR using RE03726 as a template and cloned into pET15b (Novagen). Hexahistidine fusion proteins were expressed in Escherichia coli BL21 (DE3), purified, and injected into guinea pigs and rats. The anti-Hasp antisera were purified by affinity binding to the corresponding antigens.
Immunohistochemistry.
The brains of adult male flies were dissected in PBS, fixed with 4% PFA in PBS on ice for 1 h, and stained with the following antibodies: anti-Hasp (1:1000), anti-Hig (1:1000), anti-Bruchpilot (nc82, 1:20, DSHB) (Wagh et al., 2006), anti-ChAT (ChAT4B1, 1:20, DSHB) (Takagawa and Salvaterra, 1996), anti-DVGLUT (1:500; (Mahr and Aberle, 2006), anti-Rdl (1:1000) (Enell et al., 2007), anti-DLG (4F3, 1:100, DSHB) (Parnas et al., 2001), anti-DN-cadherin (Ex#7, 1:20) (Iwai et al., 1997), or anti-GFP (1:500) (Invitrogen). AlexaFluor (405, 488, 546, or 647)-conjugated antibodies (1:200, Invitrogen) were used as secondaries. For staining with anti-Dα7 (1:1000, a gift from H. J. Bellen) and rabbit anti-Dα6 (1:1000) (Nakayama et al., 2014), dissected brains were fixed with 4% PFA in PBS for 10 min on ice. Samples were observed by a sequential scan on an SP2 or SP8 confocal microscope (Leica). Quantification of confocal images was performed, using the Leica confocal software, and fluorescence intensity was compared with that of control samples stained simultaneously in the same tube. Unless otherwise indicated, at least seven samples were examined for the purpose of statistical analysis. Each optical section was obtained from a different brain. For quantification of the levels of Hig, Hasp, Dα6, and Dα7 (see Figs. 3, 4, 6, 7,), the mean fluorescence staining intensity of each antibody in the corresponding mutants (higdd37, haspD1, Dα6DAS1, and Dα7PΔEY6) was measured and subtracted from the levels of the test samples. For DLG (see Fig. 3), background levels were measured in the brains stained only for DN-cadherin, with simultaneous application of the same secondary antibodies used for the test samples, but with no DLG antibody. The mean background level was subtracted from the DLG levels of the test samples. For quantification of synaptic proteins in pupal brains (see Fig. 10), the background levels of Hig, Hasp, and Dα6 were measured in brains stained for Brp, with application of the same secondary antibodies used for the test samples, but with no other primary antibodies. The background levels of Brp in the pupal brains were measured in the visual field outside the brains of the test samples and then subtracted from the Brp levels. For structured illumination microscopy (SIM) analysis using an ELYRA S.1 microscope (Zeiss), PFA-fixed brains were infused with 30% sucrose in PBS by gradually increasing the concentration of sucrose (5%, 10%, and 20%), embedded in Tissue-Tek OCT compound (Sakura Finetek), and frozen in liquid nitrogen. Frozen sections (10 μm) were placed on 3-aminopropyltrietoxysilane-coated cover glasses and stained with antibodies. Channels of SIM images were aligned with channel alignment tool of the ZEN software (Zeiss) using the data of TetraSpeck fluorescent microspheres (Invitrogen). Three-dimensional images were generated using the Imaris software (Zeiss).
Figure 3.
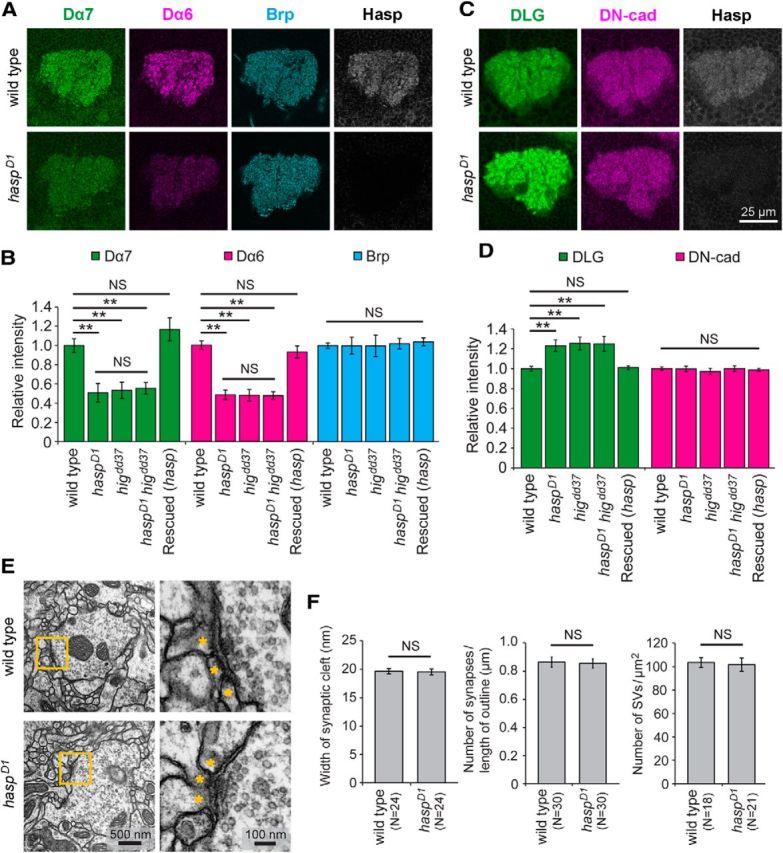
The levels of nAChR subunits and DLG are altered in the MB calyx of haspD1 adult brains. A, Reduced accumulation of nAChR subunits Dα7 and Dα6. The levels of nAChR subunits Dα7 and Dα6 decreased, whereas presynaptic active-zone protein Brp was maintained at the wild-type level in haspD1. B, Quantitative analysis of fluorescence intensities of Dα7, Dα6, and Brp signals in the calyces of wild-type (N = 39), haspD1 (N = 11), higdd37 (N = 7), haspD1 higdd37 (N = 10), and rescued flies (elav-Gal4/Y; haspD1; UAS-hasp-HA/+) (N = 23). The altered levels of Dα7 and Dα6 did not significantly differ among haspD1, higdd37, and haspD1 higdd37. C, Increased accumulation of DLG. The level of DLG increased in haspD1, whereas DN-cadherin was maintained at the normal level. D, Quantitative analysis of DLG and DN-cadherin signals in wild-type (N = 32), haspD1 (N = 11), higdd37 (N = 9), haspD1 higdd37 (N = 9), and rescued flies (N = 10). The altered levels of DLG did not significantly differ among haspD1, higdd37, and haspD1 higdd37. Each optical section was obtained from a distinct brain. Error bars indicate ± SEM. **p < 0.01, relative to wild-type (Steel–Dwass test). NS, Not significantly different. E, Structure of synaptic terminals in the calyx microglomeruli of wild-type and haspD1. Yellow-line square is magnified in the right panel to show the synaptic structure, including the active zone. The dendritic endings are indicated by asterisks. F, Quantification of the synaptic cleft width, number of synapses per unit of length of presynaptic terminal outline, and number of synaptic vesicles (SVs) per unit of area of presynaptic terminal. None of these parameters significantly differed between wild-type and haspD1. The brain sections were prepared from three flies for each genotype. The number of microglomeruli examined is indicated in parentheses.
Figure 4.
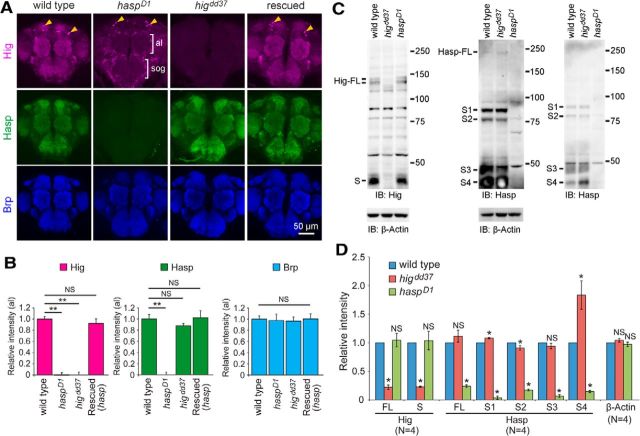
Hasp localizes Hig to synaptic clefts of cholinergic synapses. A, Localization of Hig in synaptic regions of haspD1 brains. Hig signals disappeared throughout the synaptic regions in haspD1 (indicated by brackets) but were present in neuronal cell bodies (arrowheads). Loss of Hig in synaptic regions was restored in rescued flies (elav-Gal4/Y; haspD1; UAS-hasp-HA/+). The level of Hasp was unchanged in higdd37 brains, including synaptic regions. Anti-Brp antibody was used to label the synaptic regions. al, Antenna lobe; sog, subesophageal ganglion. B, Quantifications of Hig, Hasp, and Brp in antennal lobes (al). The number of samples examined: wild-type, N = 22; haspD1, N = 12; higdd37, N = 8; Rescued, N = 10. Each optical section was obtained from a distinct brain. Error bars indicate ± SEM. **p < 0.01, relative to wild-type (Steel–Dwass test). NS, Not significantly different. C, Western blotting of Hig and Hasp proteins. Extracts from adult heads of wild-type, higdd37, and haspD1 flies were probed with antibodies for Hig (left) or Hasp (middle and right). Right blot, One-fourth of each sample used in the middle blot was loaded to clarify the patterns of S3 and S4 bands at the cost of full-length (FL) bands. β-Actin was used as a loading control. All Hig bands (two FLs and S) in haspD1 flies and most Hasp bands (FL and S1–3) in higdd37 flies were present nearly at wild-type levels. The level of Hasp S4 was elevated in higdd37 (see right blot). The two FL bands of Hig may have been produced by splicing (Hoshino et al., 1993). Several Hasp bands are likely to be generated by either splicing or cleavage (Fig. 1A; see below). Molecular markers (kDa) are indicated on the right of each blot. D, Quantification of Hig and Hasp bands in wild-type, higdd3, and haspD1 flies. The levels of Hasp S1 and S2 were slightly changed in higdd37. The signal intensity of each band was compared with the corresponding wild-type band in each of four blots. Error bars indicate ± SEM. *p < 0.05, relative to wild-type (Steel–Dwass test). NS, Not significantly different.
Figure 6.
Hasp diffuses extracellularly and is trapped by cholinergic synapses. A, Rescue of the hasp phenotype by ectopic expression of Hasp-HA. The indicated GAL4 lines were used to drive Hasp-HA expression in cholinergic, glutamatergic, serotonergic, or dopaminergic neurons, or glia, in the hasp mutant background. The genotype of the control is +/Y; haspD1; UAS-hasp-HA/+. Unless stated otherwise, a single copy of UAS-hasp-HA on the third chromosome was used for these experiments. The genotype of Ple (Hasp-HA × 2) was +/Y; haspD1; Ple-Gal4, UAS-hasp-HA/UAS-hasp-HA. B, Expression patterns of GAL4 drivers used in rescue experiments. GFPnls were used to label nuclei but occasionally diffused into cell processes. The synaptic regions were stained for Brp (magenta). C, D, Rescue of synaptic distribution of Hasp and Hig. Hasp-HA secreted from Ple-GAL4 neurons in haspD1 brains localized to the central brain (C) and MB calyx (D). Consequently, endogenous Hig appeared in the same area as in the wild-type. Levels of both Hig and Hasp-HA in synaptic regions depended on the dose of UAS-hasp-HA. Cell bodies of Ple-GAL4 neurons are indicated by arrows. al, Antennal lobe; sog, subesophageal ganglion. E, Quantification of the levels of Hasp-HA and Hig in the MB calyx of rescued flies in the haspD1 background. Each level was normalized to the corresponding level of Brp. Each optical section was prepared from a different brain. Error bars indicate ± SEM.
Figure 7.
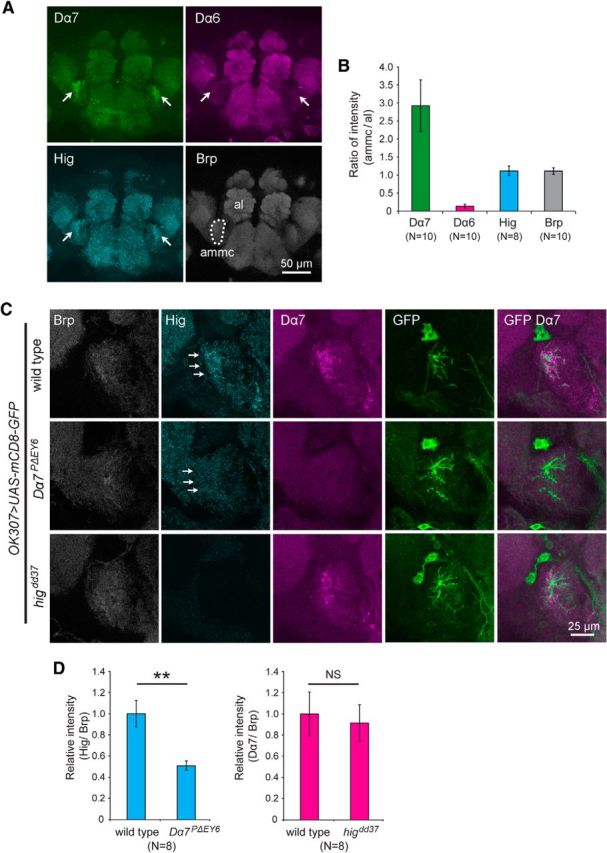
Synaptic Hig is present at reduced levels in the antennal mechanosensory and motor center (ammc) of Dα7PΔEY6 mutant brains. A, Staining patterns of Dα7, Dα6, Hig, and Brp in wild-type brains. Accumulation of Dα7 and Hig was observed in the ammc (arrows). However, Dα6 signal was faint in ammc. The dotted area of ammc was used for quantification of signal levels. B, Quantification of ratios of fluorescence intensities of Dα7, Dα6, Hig, and Brp per unit area of ammc to those of antennal lobes (al). C, Reduction of Hig at ammc in Dα7PΔEY6 brains. PLDs of the giant fiber neuron, which innervate ammc, were labeled with mCD8-GFP driven by OK307-GAL4. The Hig level was reduced in ammc of Dα7PΔEY6 brains (arrows), whereas the Dα7 level was not reduced in higdd37. D, Quantification of levels of Hig in Dα7PΔEY6 and Dα7 in higdd37 relative to those in wild-type. Fluorescence intensities were measured in PLD areas labeled with GFP. Each level is normalized to the corresponding Brp levels. Error bars indicate ± SEM. **p < 0.01 (Mann–Whitney U test). NS, Not significantly different.
Figure 10.
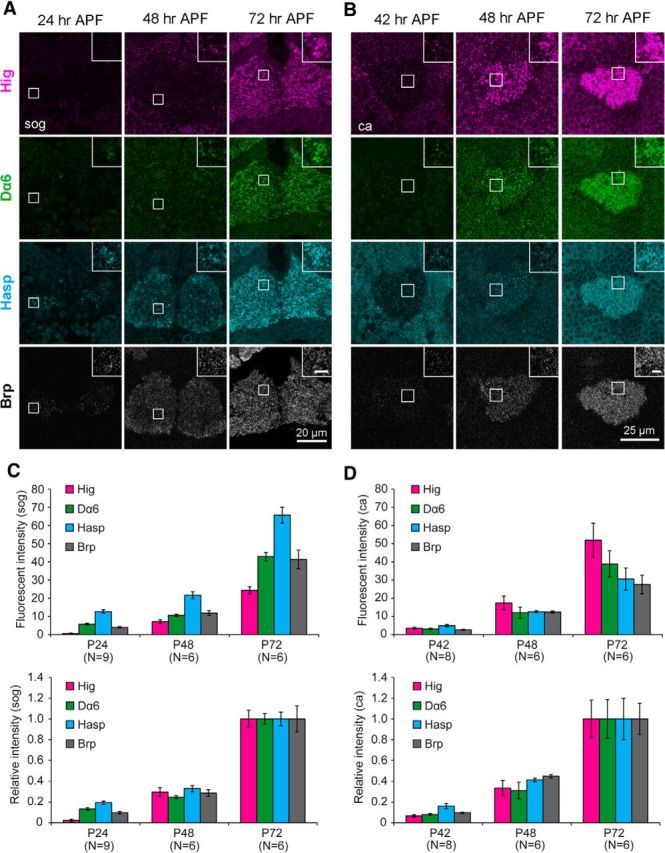
Developmental profile of synaptic localization of Hasp, Hig, Dα6, and Brp during pupal stages. A, Immunolabeling of developing subesophageal ganglion (sog). At 24 h APF, Hasp and Dα6 signals are clearly detectable at the synaptic regions labeled with Brp, but the Hig signal is very faint, and absent in most Hasp puncta (inset). The signals for all these proteins increased at 48 h APF and increased further at 72 h APF. B, Immunolabeling of developing mushroom body calyx (ca). At 42 h APF, signals of Hig, Dα6, Hasp, and Brp were weakly detected at similar levels in synaptic regions. All synaptic signals increased at 48 h APF and at 72 h APF. Squares are magnified in the insets. Scale bars: insets, 2 μm. C, D, Quantification of fluorescence signals of Hig, Dα6, Hasp, and Brp in synaptic regions of sog (C) and ca (D) during synaptogenesis. Lower graphs indicate the signal intensities of the four proteins relative to the values at 72 h APF. The number of optical sections examined is indicated below the bars.
Colocalization analysis.
To measure Manders' colocalization coefficient (MCC), coloc2 function in ImageJ/Fiji was used. First, a single-channel image was converted into a grayscale image, and ROI of the synaptic regions, including antennal lobes (al) and subesophageal ganglion (sog) in Figure 1 and a single microglomerulus (square area of 2.6 × 2.6 μm2) in Figure 9, were set. The contrast was adjusted using the Minimum/Maximum contrast function such that the mean background levels measured in areas with no tissues was set to 0 and the maximum intensity in the ROI was set to 255. Then, a threshold was set excluding all pixels with intensity <51 (and 101 and 151 for Fig. 9D,E), and the processed image was transformed into a binary image by applying the threshold. Binary images of two different channels were used to test the colocalization analysis using the coloc2 function. MCC M1 or M2 indicates the ratio of the pixels with signals in both the magenta and green channels to the pixels with signals in the magenta or green channel, respectively. For the analysis of microglomeruli (Fig. 9E), a total of 48 sections (six sequential z-sections with 0.2 μm intervals of eight independent microglomeruli) were measured.
Figure 1.
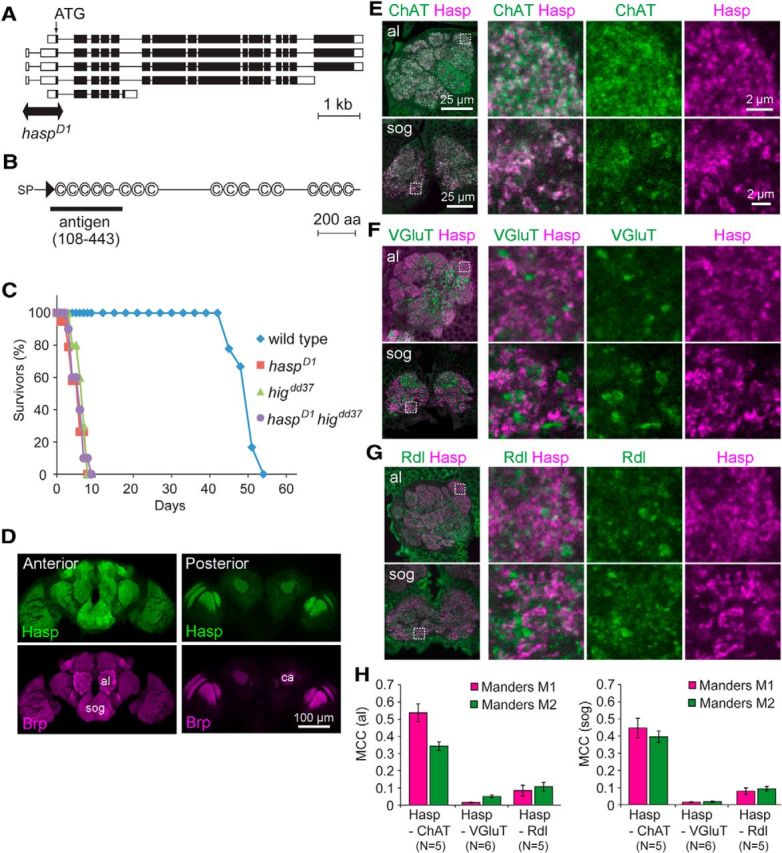
Hasp localizes at cholinergic synapses in the brain. A, Schematic structures of the five splicing isoforms of hasp (CG10186) mRNA. Double arrow spans the genomic region deleted in haspD1 mutants. Open boxes represent untranslated regions. Filled boxes represent coding regions. Lines indicate introns. B, Domain structure of Hasp, which includes a signal peptide (SP), a WAP domain (triangle), and 17 CCP domains (C). Black bar represents the amino acid sequence used as an antigen to generate anti-Hasp antibody. C, Reduced longevity of adult male mutant flies. The survival curves of higdd37, haspD1, and double-mutant higdd37 haspD1 flies are similar. D, Distribution of Hasp in the brain. Frontal sections of the anterior and posterior brain regions are shown. Hasp is present in the entire synaptic region labeled with Brp. al, Antennal lobe; sog, subesophageal ganglion; ca, calyx of the mushroom body. E–G, Double immunolabeling of synapses with antibodies against ChAT and Hasp (E), VGLUT and Hasp (F), and Rdl and Hasp (G) in the indicated synaptic regions. Dotted squares are magnified in the right three panels. H, MCCs for the two indicated proteins in the antennal lobe (left) and subesophageal ganglion (right). Both M1 and M2 of Hasp-ChAT were much higher than those of Hasp-VGluT and Hasp-Rdl.
Figure 9.
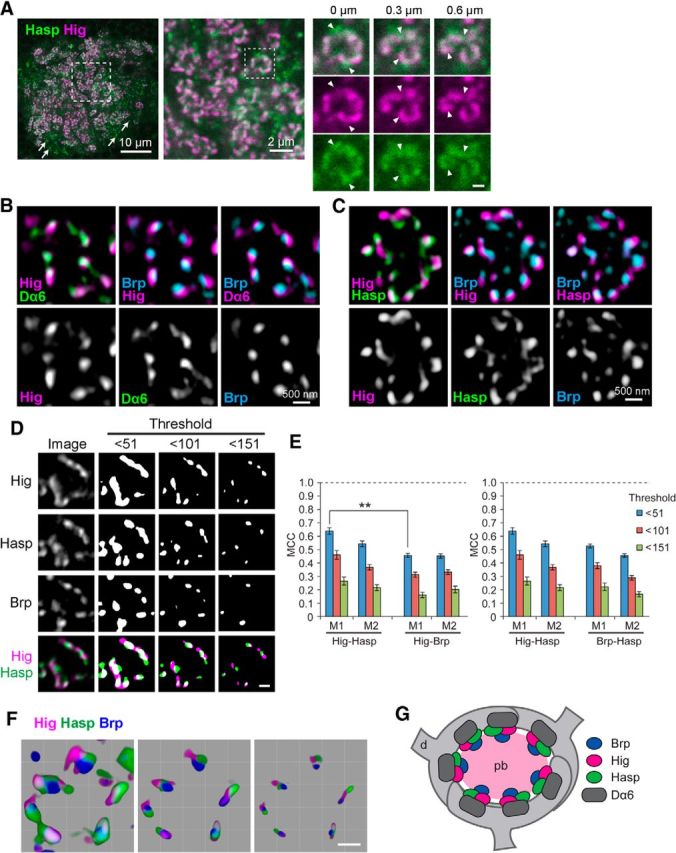
Hig and Hasp form matrix compartments in synaptic clefts. A, Confocal images of MB calyx. Both Hasp (green) and Hig (magenta) are closely associated in microglomeruli, but only Hasp is visible in nonmicroglomerular synapses (arrows); Hig is detectable only in overexposures. Dotted square in leftmost panel is magnified in the middle panel, and the region of a single microglomerulus (dotted square in the middle panel) is enlarged and shown as three sets of serial optical sections with Z intervals of 300 nm. Scale bar: serial image, 500 nm. Hasp does not completely overlap with Hig, as indicated by triangles. B, C, High-resolution images of single microglomeruli obtained by structured illumination microscopy. Optical sections triply immunolabeled for Hig, Dα6, and Brp (B) or Hig, Hasp, and Brp (C) are shown with two colors for each image. Hig was closely associated with presynaptic Brp and postsynaptic Dα6 (B). Hig and Hasp were located in close proximity but occupy distinct areas in the same synaptic cleft associated with Brp puncta (C). D, Binary images of a single microglomerulus triply labeled for Hig, Hasp, and Brp. The original image is shown in the leftmost column. Merged figures are shown at the bottom. Nonuniform distribution of the three proteins becomes clearer as the signal threshold is set higher. Binary images were processed after removal of signals with intensities lower than the threshold indicated at the top of each column. E, MCCs. As the threshold is increased, a similar reduction in MCC is observed for Hig-Hasp, Hig-Brp, and Hasp-Brp. Error bars indicate ± SEM. **p < 0.01, relative to M1 of Hig-Hasp with the same threshold (Mann–Whitney U test). F, Three-dimensional reconstruction images of a single microglomerulus. The threshold of image was arbitrarily changed, with the lowest at the left and the highest at the right. Scale bars: D, F, 500 nm. G, Schematic model for the distribution of Brp, Hig, Hasp, and Dα6 in a microglomerulus. Hig and Hasp localize to synaptic clefts. pb, Presynaptic bouton of olfactory projection neuron; d, dendrite of MB Kenyon cell.
Immunoelectron microscopy.
The brains of adult male flies were dissected in PBS and fixed with periodate-lysine-PFA solution on ice for 1 h. The fixed samples were infused with 30% sucrose/PBS and embedded in OCT as described above, and the frozen sections (10 μm) were placed on 3-aminopropyltrietoxysilane-coated glass slides. After removal of OCT by washing with PBS, samples were treated with 150 mm glycine/PBS to quench the remaining aldehydes, blocked with 5% normal goat serum/0.1% saponin/PBS, and stained with anti-Hasp antibody (1:1000) overnight at 4°C. After washing with PBS, the samples were incubated with Nanogold-conjugated anti-rat IgG (1:40, Nanoprobes) overnight at 4°C. After washing with PBS, the samples were fixed with 1% glutaraldehyde/PBS for 10 min, washed with distilled water, and silver-enhanced for 6 min at 20°C using the HQ SILVER kit (Nanoprobes). The samples were washed with distilled water, fixed with 1% OsO4 for 15 min, dehydrated, and embedded in Epon 812 (Taab). Finally, thin sections were stained with uranyl acetate and Reynolds' lead solution.
Electron microscopy.
The brains of adult male flies were fixed with 2% PFA/2.5% glutaraldehyde/0.1 m sodium cacodylate, pH 7.4, overnight at 4°C. The fixed samples were washed with 3% sucrose/0.1 m sodium cacodylate at 4°C, fixed with 1% OsO4/0.1 m sodium cacodylate for 1 h on ice, dehydrated, and embedded in Epon 812 (Taab).
Quantification of Western blotting.
The intensity of protein bands obtained by Western blotting was quantified by “Gels” analysis in ImageJ. To subtract background, the baseline of a peak of Hig or Hasp was adjusted to the baseline measured in the corresponding region in the lane of higdd37 or haspD1, respectively.
Immunoprecipitation.
Hasp-HA, Hig-GFP, or Dα7-GFP was expressed ubiquitously by da-GAL4. Adult heads were dissected in PBS, and soluble proteins were extracted in HBST [10 mm HEPES, pH 7.4, 150 mm NaCl, 0.5% (w/v) Triton X-100 and protease inhibitors (Roche, #04693159001)]. Anti-HA-tag mAb or anti-GFP mAb magnetic beads (MBL, M132-9, and D153-9) were used for immunoprecipitation analysis. For immunoprecipitation of Dα6 and Dα7-GFP, membrane proteins were extracted from Drosophila heads in membrane solubilization buffer (0.2 m NaCl, 1.8% (w/v) Triton X-100, 0.6% (w/v) sodium deoxycholate, 10% (v/v) glycerol, 10 mm Tris-HCl, pH 8.5, and protease inhibitors) as previously described (Schloss et al., 1988). To examine the interaction of Hig with Dα6 and Dα7-GFP (Fig. 5C), the membrane fraction was incubated with rabbit anti-Dα6 antibody and Protein G Mag Sepharose (GE Healthcare Life Sciences) and with anti-GFP mAb magnetic beads, respectively. After washed with HBST, the beads were incubated with a soluble fraction extracted from wild-type heads in HBST. For immunoblotting of Dα6, guinea pig anti-Dα6 antibody (1:1000) was used.
Figure 5.
Hig forms protein complexes with Hasp, Dα6, and Dα7. A, B, Coimmunoprecipitation of Hasp and Hig. Hasp-HA (A) or Hig-GFP (B) was expressed ubiquitously by da-GAL4, and brain extracts were immunoprecipitated with anti-HA beads or anti-GFP beads, respectively. Hasp-HA and endogenous Hig (A), and Hig-GFP and endogenous Hasp (B) in the immunoprecipitated (IP) fractions were detected with the corresponding antibodies. Full-length Hig (two bands) and Hasp were coimmunoprecipitated with Hasp-HA and Hig-GFP, respectively (A,B, right blots). Multiple bands derived from Hasp-HA and Hig-GFP indicate that these tagged proteins were cleaved (A,B, left blots). Extract from wild-type heads was used as a control (right lanes) for each experiment. I, Input; IB, immunoblot. Hasp-HA and Hig-GFP contain the tags at the C terminus, whereas anti-Hasp and anti-Hig antibodies recognize the 108-443 amino acids of Hasp and the 35–295 amino acids of Hig, respectively. Predicted molecular weights of full-length proteins: Hig, 107.0 kDa; Hig-GFP, 133.9 kDa; Hasp, 183.5 kDa; Hasp-HA, 187.3 kDa. The observed molecular weights of these proteins were higher than predicted, suggesting that these proteins are glycosylated. C, Coimmunoprecipitation of Hig and Dα6. Membrane proteins extracted from wild-type or Dα6DAS1 were immunoprecipitated with anti-Dα6 (left blot). The precipitants were further incubated with the soluble fraction extracted from wild-type and examined whether Hig was coimmunoprecipitated with Dα6 (right blot). D, Coimmunoprecipitation of Hig and Dα7-GFP. Membrane proteins extracted from wild-type or flies expressing Dα7-GFP were immunoprecipitated with anti-GFP (left blot). The precipitants were further incubated with the soluble fraction extracted from wild-type, and examined whether Hig was coimmunoprecipitated with Dα7-GFP (right blot). C, D, A short form of Hig (S) was detectable in the IP fraction. SFr represents input of a soluble fraction extracted from wild-type or higdd37. The molecular weight of Dα6 is lower than predicted (59.1 kDa), possibly because of its hydrophobic property. Filled triangles represent the bands of immunoprecipitated polypeptides detected with the antibodies for the tags or proteins shown below each blot. Open triangles represent the absence of the corresponding bands.
Results
The matrix protein Hasp predominantly localizes at the synaptic clefts of cholinergic synapses in the Drosophila brain
The synaptic cleft matrix protein Hig contains a maximum of five CCP domains. Therefore, to identify a novel matrix protein filling synaptic clefts, we searched the SMART database (Schultz et al., 1998) for other proteins with such domains. Among the CCP-containing proteins in the database, we focused our attention on Hasp (encoded by CG10186 on the Drosophila genome), which has 17 CCP domains and a WAP domain preceded by a signal sequence at its N terminus (Fig. 1B). According to the FlyAtlas database (Robinson et al., 2013), the hasp mRNA is selectively expressed in the CNS. To investigate the function of the encoded protein, we generated a deficiency mutant of hasp by imprecise P-element excision. The haspD1 mutant genome lacks the region spanning the transcription start site and second exon, which contains the initiation codon (Fig. 1A), strongly suggesting that this allele is a null. haspD1 homozygotes were largely inviable, and the rare surviving adult flies were short-lived (Fig. 1C) with severely reduced locomotion. Similar phenotypes were observed in higdd37 null mutants and haspD1higdd37 double mutants (Fig. 1C). In addition, the protein structure and expression data of Hasp resembled those of hig, suggesting that Hasp and Hig share a role in the process of brain development or function.
Immunostaining with anti-Hasp antibodies revealed that Hasp was localized in most synaptic regions labeled with the presynaptic active-zone protein Bruchpilot (Brp) (Wagh et al., 2006) in the adult brain (Fig. 1D). These signals completely disappeared in the brains of haspD1 mutants (see Figs. 3A,C, 4A), demonstrating the specificity of the Hasp antibody used in this study. Double immunolabeling for Hasp and specific synaptic markers revealed that synaptic Hasp was closely associated with choline acetyltransferase (ChAT), a presynaptic marker of cholinergic neurons (Fig. 1E). The MCCs between Hasp and ChAT were 0.54 ± 0.05 (M1) and 0.34 ± 0.03 (M2) in the antennal lobe (al), and 0.45 ± 0.06 (M1) and 0.40 ± 0.03 (M2) in the subesophageal ganglion (sog) (Fig. 1H). However, Hasp signals were mostly excluded from areas containing vesicular glutamate transporter (VGLUT), a presynaptic marker of glutamatergic neurons (Fig. 1F,H; M1 = 0.016 ± 0.002 and M2 = 0.050 ± 0.008 in al, and M1 = 0.015 ± 0.002 and M2 = 0.018 ± 0.003 in sog). The GABA receptor Rdl and Hasp were segregated in most synaptic regions, although they overlapped in a larger fraction of synapses than VGLUT and Hasp (Fig. 1G,H; M1 = 0.084 ± 0.032 and M2 = 0.11 ± 0.03 in al, and M1 = 0.079 ± 0.019 and M2 = 0.093 ± 0.012 in sog). This overlap may be due to inhibitory connections of GABAergic neurons to cholinergic synaptic terminals, as revealed by electron microscopy in the insect brains (Distler and Boeckh, 1997; Yasuyama et al., 2002). Collectively, these results indicate that Hasp is predominantly distributed in cholinergic synapses in the brain.
Because Hasp was predicted to be a secretory protein, we investigated whether it was localized extracellularly at the periphery of synapses or synaptic clefts. The microglomerulus of the mushroom body (MB) calyx, a specialized synaptic complex comprising a single axon terminal of the olfactory projection neuron (PN) at its center and a few surrounding dendritic terminals of MB Kenyon cells, predominantly forms multiple cholinergic synaptic connections (Fig. 2A,B,F) (Yasuyama et al., 2002). Hasp signals were detected in the surrounding regions of each PN axon terminal labeled for ChAT (Fig. 2C). Immunoelectron microscopy revealed several clusters of Hasp signals in the microglomeruli at synaptic clefts associated with electron-dense postsynaptic structures (Fig. 2D,E), whereas these signals were rarely detected in the sections from haspD1 brains (Fig. 2E). Therefore, we conclude that Hasp localizes at synaptic clefts of cholinergic synapses in the Drosophila brain (Fig. 2F).
Figure 2.
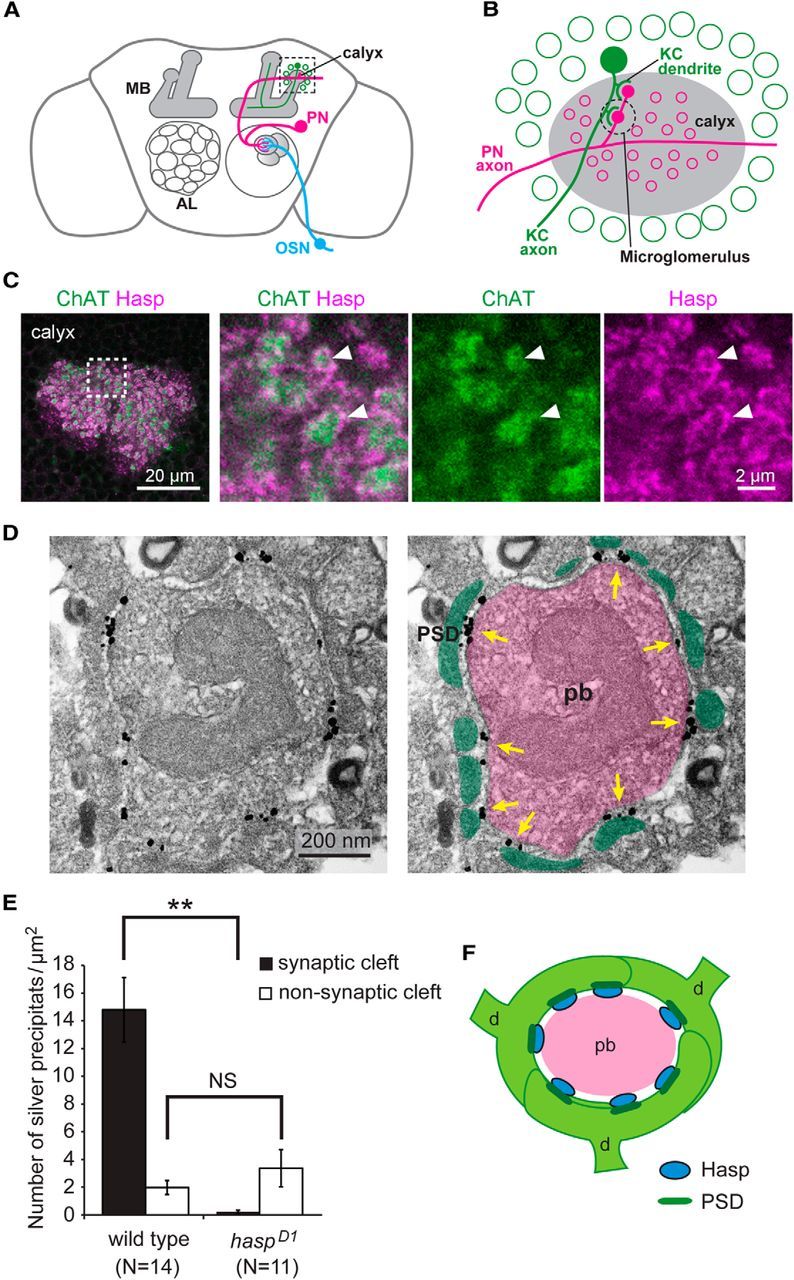
Hasp localizes at synaptic clefts. A, Schematic representation of the olfactory circuit. The axon of olfactory sensory neuron (OSN) targets one of the glomeruli in the antennal lobe (AL), where the dendrite of PN forms synapses with OSNs. The PN axon extends to the calyx of the MB. B, Structure of the calyx. A, Dotted square is enlarged to show the calyx structure (gray), where synaptic connections occur between the PN axon and the dendrites of Kenyon cells (KC) to form a specialized synaptic complex called the microglomerulus. The cell bodies of KCs (green circles) are located in the region surrounding the calyx. C, Localization of Hasp at the microglomeruli of the calyx. Dotted square is magnified in the right three panels. Arrowheads indicate microglomeruli. Discrete Hasp signals surround the ChAT-positive presynaptic PN axon terminal in each of microglomeruli. D, Localization of Hasp, revealed by immunoelectron microscopy. Left, Preembedding gold labeling of Hasp in a single microglomerulus. Silver-enhanced signals of Hasp are detected in clusters at the synaptic cleft. Right, The picture shown at left is marked to indicate the presynaptic bouton (pb) of PN axon (pink), the postsynaptic densities (PSDs) of KC dendritic endings (green), and clusters of Hasp signals (yellow arrows). E, Quantification of Hasp signals at microglomeruli. Silver precipitates per unit area of the PN terminal were counted within and outside synaptic clefts. The precipitates were frequently detected at synaptic clefts in the wild-type but were rarely detected there in haspD1. Error bars indicate ± SEM. **p < 0.01, relative to wild-type (Mann–Whitney U test). NS, Not significantly different. F, Structure of a single microglomerulus. The microglomerulus is formed by a single presynaptic bouton (pb) of PN axon and a few claw-like dendritic endings (d) of KCs. Each dendritic ending usually contains a few PSDs (dark green), where AChRs are localized. Hasp (blue) localizes at the synaptic cleft close to PSDs.
Hasp regulates the synaptic distribution of Dα6, Dα7, and DLG
The localization of Hasp at cholinergic synapses suggested that this protein is involved in a mechanism related to differentiation of cholinergic synapses. To test this idea, we examined the distribution of nAChR subunits Dα7 and Dα6 in the MB calyx in haspD1 brains. The levels of both Dα7 and Dα6 were significantly reduced to 51% and 48% of the wild-type level, respectively (Fig. 3A,B), whereas the presynaptic active-zone protein Brp (Fig. 3A,B) and the cell adhesion protein DN-cadherin (Fig. 3C,D) were present at wild-type levels. We also examined the distribution of DLG, a Drosophila homolog of PSD-95 family proteins, which serve as cytoplasmic scaffolds for neurotransmitter receptors. In contrast to Dα7 and Dα6, the level of DLG increased by 23% in haspD1 brains (Fig. 3C,D). The altered synaptic accumulation of Dα7, Dα6, and DLG in haspD1 was restored to wild-type levels by pan-neuronal expression of Hasp-HA (Fig. 3B,D). Thus, Hasp modulates the accumulation of the synaptic membrane proteins as well as the cytoplasmic protein at the postsynaptic terminals. Phenotypes similar to those of haspD1 were observed in both higdd37 and higdd37haspD1 brains (Fig. 3B,D), further corroborating the idea that Hasp and Hig act in the same pathway of synaptic differentiation.
The altered accumulation of synaptic molecules at postsynaptic terminals suggested that synaptic structures may be defective in haspD1 brains. To test this idea, we examined the ultrastructure of microglomeruli in MB calyx (Fig. 3E). However, no apparent alterations were detected in the morphology of postsynaptic densities. In addition, the distance between the presynaptic and postsynaptic terminals was not significantly changed by the loss of Hasp (Fig. 3F), indicating that other matrix or membrane proteins are sufficient to maintain the size of synaptic clefts. We also counted active zones and synaptic vesicles in the presynaptic PN axon terminals, but they were not significantly different in the mutant brains (Fig. 3F).
Hasp is essential for the synaptic localization of Hig
The extensive similarities between the haspD1 and higdd37 phenotypes led us to hypothesize that Hasp and Hig interact closely with each other to control synaptic differentiation. Notably, immunostaining revealed that Hig was absent throughout the synaptic regions of haspD1 brains (Fig. 4A,B), but present in neuronal cell bodies (Fig. 4A). Furthermore, the soluble fraction extracted from the heads of haspD1 flies contained two forms of full-length Hig, as well as the short form, at wild-type levels (Fig. 4C,D). This indicates that production of Hig is normal in haspD1, and that its localization to synaptic clefts is regulated by Hasp. On the other hand, the synaptic distribution of Hasp was not reciprocally altered in higdd37 brains (Fig. 4A,B). Full-length Hasp and its short forms (S1-S3) were detected mostly at normal levels in head extracts of higdd37 flies, although the amount of S4 was elevated (Fig. 4C,D). Thus, Hasp is required for the localization of Hig at the synaptic clefts of cholinergic synapses, but Hig is dispensable for localization of Hasp to the synaptic clefts.
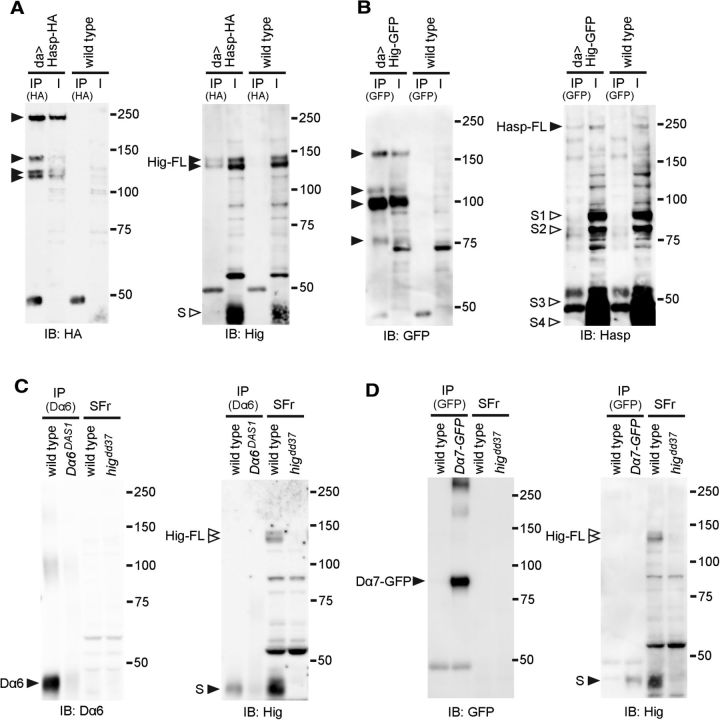
To determine whether Hasp physically interacts with Hig, we performed immunoprecipitation analysis, using transgenic flies expressing Hasp-HA (Fig. 5A) or Hig-GFP (Fig. 5B), each of which contains the tag at its C terminus. Two forms of full-length endogenous Hig coprecipitated with ectopically expressed Hasp-HA pulled down with HA antibody (Fig. 5A, right blot); reciprocally, a full length of endogenous Hasp also coprecipitated with ectopic Hig-GFP pulled down with GFP antibody (Fig. 5B, right blot). Notably, both Hasp-HA and Hig-GFP produced four forms of polypeptides containing the tags, indicating that these proteins were cleaved at three or more sites (Fig. 5A,B, left blots). These results indicate that Hasp and Hig (or their cleaved fragments) form a protein complex, providing molecular evidence of capturing Hig by Hasp at synaptic clefts of cholinergic synapses.
Hasp diffuses extracellularly and is trapped by cholinergic synapses
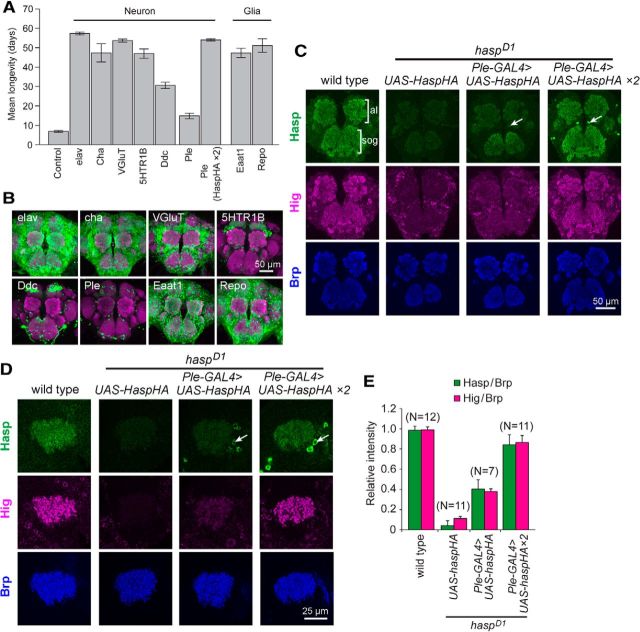
Because Hasp predominantly localized to the synaptic clefts of cholinergic synapses, we predicted that transgenic Hasp expression in cholinergic neurons would rescue the mutant phenotype of haspD1. To investigate this possibility, we expressed Hasp-HA in cholinergic or noncholinergic neurons in the haspD1 background under the control of several GAL4 drivers (Fig. 6A,B). As expected, the reduction in longevity of haspD1 mutants was restored by expression of Hasp-HA in all neurons (elav-Gal4) or cholinergic neurons (Cha-Gal4). However, the longevity phenotype was also rescued by Hasp-HA expression in glutamatergic neurons (VGluT-Gal4) and neurons producing serotonin receptor 1B (5HTR1B-Gal4). In addition, the mutant phenotype was partially rescued by Hasp-HA expression in subsets of dopaminergic and serotonergic neurons (Ddc-Gal4) (Pech et al., 2013; Riemensperger et al., 2013), which are less abundant than cholinergic and glutamatergic neurons (Fig. 6B). When Hasp-HA was driven by ple-Gal4 in a small number of dopaminergic neurons (Fig. 6B) that produce tyrosine hydroxylase (Friggi-Grelin et al., 2003), the longevity phenotype of haspD1 was only partially rescued, whereas Ple-Gal4 fully rescued the mutant phenotype when the expression level was increased by an additional copy of hasp-HA (Fig. 6A). Notably, even Hasp-HA expression in glial cells (Eaat1-Gal4 and Repo-Gal4) rescued the mutant phenotype (Fig. 6A,B). Thus, Hasp does not need to be expressed in cholinergic neurons to perform its function, so long as a sufficient amount of Hasp is secreted into the brain. These data suggest that ectopically expressed Hasp-HA is diffused extracellularly and trapped in the synaptic clefts of cholinergic synapses and that Hasp-HA localizes Hig to the synaptic sites.
To confirm this, we examined the distribution of Hasp-HA driven by Ple-Gal4 and endogenous Hig in haspD1 brains. Hasp-HA secreted from a small number of dopaminergic neurons was detected across the whole synaptic region (Fig. 6C) and MB calyx microglomeruli (Fig. 6D,E), which are predominantly cholinergic (Yasuyama et al., 2002). Accordingly, endogenous Hig appeared in these structures in a pattern indistinguishable from the wild-type, although the staining intensity of both Hasp-HA and Hig proteins depended on the dose of Hasp-HA transgene (Fig. 6C–E). Therefore, we conclude that, after secretion from cholinergic or noncholinergic neurons, Hasp diffuses into the extracellular spaces and is trapped in the synaptic clefts of cholinergic synapses, where it is responsible for localizing Hig.
nAChR subunits localize Hig but not Hasp to the synaptic clefts of cholinergic synapses
The findings described above reveal the relationships between Hasp, Hig, and nAChR subunits in terms of regulation of their synaptic localization: Hasp affects the localization of Hig and nAChR subunits, and Hig regulates the level of nAChR subunits but not Hasp. To elucidate the roles of nAChR subunits in the synaptic localization of Hasp and Hig, we examined the distribution of those two proteins in nAChR subunit mutants. In Dα7PΔEY6, levels of Hasp and Hig were not altered in most synaptic regions in the brain (see Fig. 8C; and data not shown). However, in the posterior lateral dendrites (PLDs) of giant fiber neurons, in which both Hig and Dα7 were normally abundant but Dα6 was barely detectable (Fig. 7A,B), the level of Hig was clearly reduced in Dα7PΔEY6 (Fig. 7C,D). By contrast, the level of Dα7 was not reciprocally altered in the dendrites of higdd37 (Fig. 7C,D). In Dα6DAS1 brains, no obvious changes in the levels of Hasp and Hig were detected throughout the synaptic regions, including the PLDs of giant fiber neurons (Fig. 8C; and data not shown). However, in the Dα7PΔEY6Dα6DAS1double mutant, Hig fluorescence was significantly reduced in the entire synaptic region (Fig. 8A), and MB calyx contained ∼66% of the wild-type level (Fig. 8B,C); however, the Hasp level in the mutant brain was unchanged (Fig. 8A–C). The Drosophila genome encodes seven α and three β nAChR subunits. Hence, we knocked down each of these genes in the Dα7PΔEY6Dα6DAS1double mutant using RNAi constructs. Among the combinations of these mutations, knockdown of Dα5 in the double-mutant background decreased the level of Hig throughout the synaptic regions (Fig. 8A) and retained ∼19% of the wild-type level in MB calyx but did not affect the level of Hasp (Fig. 8A–C). In addition, knockdown of Dα5 in the wild-type background did not significantly change the levels of both Hig and Hasp (Fig. 8C). Thus, Dα5, Dα6, and Dα7 are redundantly required for localization of Hig to synaptic clefts of cholinergic synapses, but not for localization of Hasp.
Figure 8.
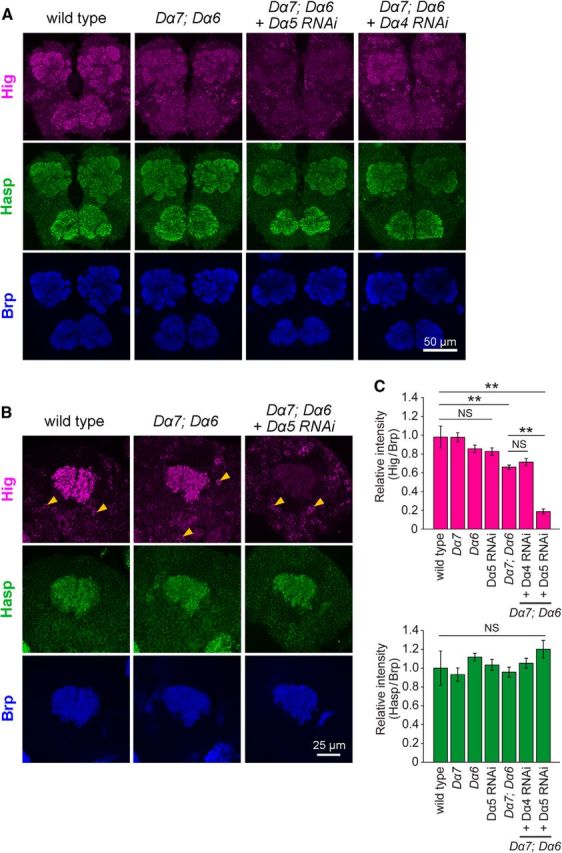
nAChR subunits are required for synaptic localization of Hig, but not Hasp. A, B, Triple immunolabeling for Brp, Hasp, and Hig in anterior brain regions (A) and MB calyx (B); genotypes are indicated above the images. The level of Hig decreased throughout the synaptic regions, including MB calyx, in the Dα7PΔEY6; Dα6DAS1 double mutant. This reduction was enhanced by knockdown of Dα5 (A, B), but not by knockdown of Dα4 (A). C, Levels of Hig (top) and Hasp (bottom) in MB calyx of mutant brains. Each level is normalized to the corresponding Brp levels and shown as a ratio relative to the wild-type. The number of samples examined: wild-type, N = 88; Dα7PΔEY6, N = 18; Dα6DAS1, N = 11; Dα5 RNAi, N = 15; Dα7PΔEY6 Dα6DAS1, N = 12; Dα7PΔEY6 Dα6DAS1 Dα4 RNAi, N = 20; Dα7PΔEY6 Dα6DAS1 Dα5 RNAi, N = 14. Error bars indicate ± SEM. The Steel–Dwass test was used for statistical analysis. **p < 0.01 relative to wild-type. NS, Not significantly different.
The reduction in levels of Hig in the triple nAChR subunit mutants and the decrease in levels of Dα6 and Dα7 in higdd37 mutants suggest that Hig and those subunits may physically interact to form a protein complex. To examine this idea, we performed immunoprecipitation experiments, adding the soluble fraction extracted from wild-type to the membrane fraction immunoprecipitated with Dα6 antibody (Fig. 5C) or to the membrane fraction of Dα7-GFP flies immunoprecipitated with GFP antibody (Fig. 5D). The short form of Hig (S) was coprecipitated with both Dα6 and Dα7-GFP. These results indicate that Hig forms a protein complex with Dα6 and Dα7, consistent with the genetic interaction for their localization at synapses.
Hig and Hasp display nonuniform distribution within individual synaptic clefts
The Hasp-mediated capture of Hig in cholinergic synapses suggests that Hasp and Hig colocalize within synaptic clefts. To determine how these proteins are distributed in this structure, we examined their immunostaining patterns in the microglomeruli of the MB calyx. A previous study showed that in each microglomerulus, Brp, nAChR subunits, and Hig are localized in several discrete synaptic regions (Fig. 9G) (Nakayama et al., 2014). In the microglomeruli, signals for Hasp were colocalized with those of Hig (Fig. 9A), but in synapses outside the microglomeruli (Fig. 9A, arrows), Hasp was associated with Dα6 and a faint Hig signal that was detectable only in overexposure (data not shown). Therefore, the relative levels of Hig and Hasp can vary depending on the type of cholinergic synapses in the MB calyx. Within individual microglomeruli, although Hig and Hasp were closely associated in the synaptic clefts, they did not completely overlap with each other (Fig. 9A). To examine the spatial relationships between Hig, Hasp, Brp, and nAChR subunits in more detail, we used SIM, which provides optical images with resolution of 100–130 nm along the x- and y-axes and 250–340 nm along the z-axis (Schermelleh et al., 2010; Maglione and Sigrist, 2013); these resolutions are smaller than the widths of synaptic clefts (19.6 ± 0.4 nm; Fig. 3F) in the CNS. Nevertheless, SIM provides information about protein localization that cannot be obtained by conventional confocal microscopy. Hig in the synaptic cleft, Dα6 on the postsynaptic membrane, and Brp in the presynaptic terminal were closely juxtaposed (Fig. 9B). This high-resolution image was consistent with the mutual dependency of Hig and AChR subunits for synaptic localization. Notably, in a single synaptic cleft associated with a dot of Brp signal, Hig and Hasp, in various patterns, occupied distinct areas with some overlap (Fig. 9C). When colocalization coefficients of the binary images were calculated with a low signal threshold (<51), M1 and M2 for Hig and Hasp fluorescence signals (M1 = 0.64 ± 0.03, M2 = 0.54 ± 0.02) tended to be larger than the respective coefficients for Hig and Brp signals (M1 = 0.46 ± 0.02, M2 = 0.45 ± 0.02) and for Brp and Hasp signals (M1 = 0.53 ± 0.02, M2 = 0.46 ± 0.01), although statistical analysis only indicated that M1 for Hig and Hasp signals was significantly different from that for Hig and Brp signals (Fig. 9D,E). These data may reflect specific positions of the three proteins within synaptic structures. Because Brp in the presynaptic cytoplasm should not colocalize with Hig and Hasp, both of which are confined to synaptic clefts, the coefficients for Brp and Hig, and Brp and Hasp would theoretically be zero; therefore, the molecular colocalization between Hig and Hasp should be, if not zero, much lower than the value estimated from the colocalization coefficients. In addition, as the signal threshold of SIM images was increased, areas with fluorescent signals became smaller and gradually lost their original shapes. The colocalization coefficients for Hig and Hasp in these processed images were reduced similarly to those for Hig and Brp, or Hasp and Brp (Fig. 9D,E). Three-dimensional images generated from optical sections with altered thresholds consistently showed segregated distribution of Hig, Hasp, and Brp (Fig. 9F). These data indicate that large fractions of Hig and Hasp regions do not overlap with each other and that the proteins largely occupy distinct spaces within individual synaptic clefts (Fig. 9G).
Developmental profile of Hasp, Hig, and nAChR subunits during differentiation of cholinergic synapses
To further understand the molecular mechanism of matrix formation during synaptogenesis, we examined the developmental profile of synaptic localization of Hasp, Hig, AChR subunit Dα6, and Brp during the pupal stages, when immature synapses undergo active differentiation. In the subesophageal ganglion (Fig. 10A), at 24 h after puparium formation (APF), both Dα6 and Hasp were present in a small number of fluorescent puncta in synaptic regions labeled with Brp, whereas the Hig signal was much fainter (Fig. 10A,C) and barely detectable at most puncta of Hasp signals (Fig. 10A, insets). The number of fluorescent signals for all of these proteins increased as development proceeded. At 72 h APF, Hig, Dα6, Hasp, and Brp proteins exhibited strong signals; Hasp was most intense and Hig was least intense in the synaptic regions (Fig. 10A,C). In MB calyx, the developmental profile was different from that in subesophageal ganglion (Fig. 10B). The signals of all four synaptic proteins were weak at 42 h APF, increased similarly during later developmental stages, and could easily be detected in the synaptic regions at 48 h APF (Fig. 10B,D). All four proteins exhibited strong fluorescent signals in pupae at 72 h APF; Hig signals were the most intense (Fig. 10B,D). A developmental profile resembling that in MB calyx was also observed in the optic lobes (data not shown). These data indicate that Hasp localizes to cholinergic synaptic clefts before appearance of Hig in subesophageal ganglion, and arrives no later than Hig in the other brain regions we examined.
Discussion
We previously identified Hig as a matrix protein predominantly localized at the synaptic cleft of cholinergic synapses in the Drosophila brain. In this study, we revealed that Hasp, another matrix component occupies cholinergic synaptic clefts. Both proteins contain multiple CCP domains, and the loss of either protein causes similar behavioral and molecular phenotypes, suggesting that both proteins are involved in the same process of synaptic development or function. Consistent with this, Hasp and Hig localize close to each other at cholinergic synapses. However, high-resolution imaging revealed that these proteins occupy distinct areas within synaptic clefts. These results provide novel insight into the molecular architecture of the synaptic cleft matrix in the CNS and suggest that each of the areas containing Hig or Hasp plays a distinct role in synaptogenesis.
Genetic analysis revealed that the roles of Hasp and Hig proteins in synaptic differentiation are not identical: although both proteins similarly affect the levels of nAChR subunits and DLG, Hasp is required for Hig to localize at the synaptic cleft, whereas Hig is dispensable for the synaptic localization of Hasp. These functional relationships raise the possibility that Hasp directly regulates the levels of nAChR subunits, as well as those of DLG, and simultaneously mediates anchoring of Hig at synapses. Alternatively, Hasp may only be involved in capture of Hig and regulates the distribution of the synaptic proteins as a secondary consequence of its main function. Our data indicate that the altered levels of AChR subunits Dα6, Dα7, and DLG in hasp and hig single mutants and hasp hig double mutants are quantitatively similar, strongly suggesting that the primary role of Hasp is localizing Hig to the synaptic clefts. The close interaction between Hig and nAChR subunits was corroborated by genetic data showing that Dα5, Dα6, and Dα7 are redundantly required for localization of Hig, but not Hasp at synaptic clefts, and also by coimmunoprecipitation of Hig with Dα6 and Dα7. Thus, Hig and the nAChR subunits mutually interact for their synaptic distribution, and the physiologically important role of Hasp is localizing Hig at synaptic clefts.
In C. elegans, LEV-9, a Hasp homolog, LEV-10, a transmembrane protein containing CUB domains, and Oig-4, a secretory protein containing an Ig domain, are required for LAChR clustering; the absence of any of these proteins, including LAChR, causes the loss of all the other proteins on NMJs (Rapti et al., 2011). In Drosophila, however, Hasp is localized normally at the synaptic cleft in the CNS when Hig or a subset of nAChR subunits is missing. This difference between the mechanisms underlying synaptic localization of LEV-9 and Hasp could be explained simply by evolutionary diversification among species, or alternatively by differences in synaptic architecture between NMJ and CNS synapses.
We have not yet determined how Hasp localizes Hig at synaptic clefts. Hasp may either trap extracellularly diffusing Hig or prevent degradation of Hig localized at synaptic clefts. Hasp contains a WAP domain, which has been implicated in protease inhibition (Bingle and Vyakarnam, 2008), implying that Hasp stabilizes Hig by preventing its degradation. However, immunoblot analysis indicated that the amounts of full-length and short form Hig polypeptides were unchanged in extracts from hasp mutants, suggesting instead that Hasp recruits Hig at synaptic clefts. Hasp and Hig occupy their respective areas, which may be completely separate or partly overlap with each other. This regional distribution suggests that a single Hasp molecule may not be sufficient to trap Hig. Rather, a number of Hasp molecules may construct a Hasp compartment, which could serve as a scaffold for Hig or a Hig-based compartment maintained within synaptic clefts. A previous study showed that C. elegans LEV-9 must be processed into fragments to cluster AChR at NMJs (Briseño-Roa and Bessereau, 2014). Consistent with this, Hasp and Hig are processed to produce truncated polypeptides. Therefore, the patterns of Hig and Hasp staining observed in this study may represent the distribution of a mixture of Hig and Hasp fragments containing their respective N-terminal amino acid-sequences (the antigens used to raise the antibodies) and may not reflect the entire fragment distribution. Further studies are required to reveal the details of Hig and Hasp cleavage, as well as the distribution of the processed fragments in synaptic clefts.
Hig could regulate the accumulation of nAChR on postsynaptic membranes via either of two mechanisms. Hig has an Ig domain and a maximum of five CCP domains in its C-terminal half and the residual N-terminal half contains an RGD sequence, a putative integrin binding site (Campbell and Humphries, 2011). This domain organization can be used to form a scaffold complex that may physically interact with nAChR subunits and thereby either maintain clustering of the receptors on postsynaptic membranes or prevent their degradation. Alternatively, Hig may transduce signals through transmembrane proteins into the cytoplasm of postsynaptic terminals and induce clustering of nAChRs that move laterally on the membrane, as reported for Agrin-mediated AChR clustering (Dai et al., 2000).
Our mutant analysis revealed that loss of Hig or Hasp resulted in an increase in the level of DLG, as well as a reduction in the levels of Dα6 and Dα7, indicating that Hig also affects the accumulation of cytoplasmic proteins in postsynaptic terminals. It is notable that PSD-95 family members in vertebrates are present at cholinergic synapses, where they function as scaffolds for AChR (Conroy et al., 2003; Parker et al., 2004), as they do for glutamate receptors at glutamatergic synapses (Xu, 2011; Opazo et al., 2012). Moreover, synaptic PSD-95 accumulation is increased by reduced synaptic activity and decreased by elevated activity via regulation of phosphorylation or palmitoylation in glutamatergic synapses (Kim et al., 2007; Noritake et al., 2009). The increase of DLG in hasp mutant brains may reflect similar homeostatic regulation in the Drosophila cholinergic synapses: the reduced synaptic activity caused by the decrease in Dα6 and Dα7 levels may activate a compensatory mechanism by which DLG accumulates to a greater extent on postsynaptic membranes.
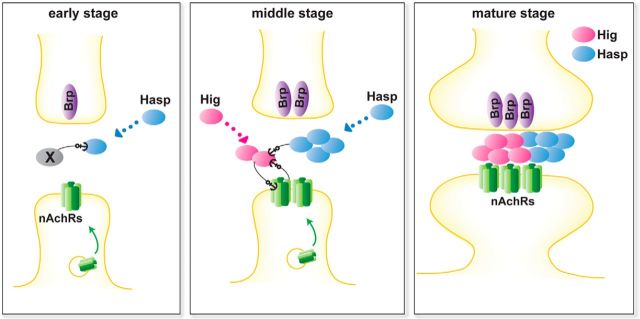
On the basis of our data, we propose a model that illustrates how the synaptic cleft matrix is constructed during synaptogenesis (Fig. 11). During the early stages of synaptogenesis, when synaptic structures are immature, Hasp is secreted extracellularly, diffused, and trapped by an unknown molecule, occupying a particular space in the synaptic clefts of cholinergic synapses. The molecule involved in trapping Hasp may be a secretory or membrane protein localized specifically to the cholinergic synapses. During this and later stages, the Hasp-containing scaffold increases its volume by incorporating new Hasp molecules, and nAChR subunits start to accumulate on postsynaptic membranes. Following Hasp localization, secreted Hig molecules are continuously captured in the differentiating matrix architecture containing the Hasp scaffold, as well as maintained by nAChR subunits, thereby increasing the volume of the Hig-containing scaffold. Reciprocally, the Hig scaffold stabilizes nAChR subunits on the postsynaptic membranes by a physical interaction in synaptic clefts or signaling into the cytoplasm of postsynaptic terminals. In mature cholinergic synapses, the two scaffolding complexes divide synaptic clefts into compartments, reflecting their distinct roles in synaptic differentiation. To further understand the entire process of matrix construction, it will be important to identify other matrix components in the Hasp and Hig scaffold complexes, and especially the Hasp-anchoring molecules.
Figure 11.
Schematic model for the construction of matrix by Hasp, Hig, and Dα6 during synaptogenesis. Early stage: An unidentified factor X, which may be either a secretory or membrane protein, initially anchors diffusing Hasp at the cholinergic synaptic clefts. nAChRs may be exocytosed at this stage to localize on the postsynaptic membrane. Middle stage: Hasp forms a scaffold complex, incorporating new Hasp and other molecules. This complex, together with AChR subunits, captures diffusing Hig and support construction of Hig-based scaffold complex, which in turn maintains the levels of AChR subunits on the postsynaptic membranes. Both Hasp and Hig can be secreted from either the closest synaptic terminals or from distant cells. Mature stage: Hasp and Hig form a large scaffold complex and establish matrix compartments in the synaptic cleft of cholinergic synapses. Both proteins are still abundantly expressed and supplied at this stage. Both Hasp and Hig produce truncated polypeptides, but it remains unclear how the polypeptides are distributed within synaptic clefts. Therefore, this model pertains to the distribution of full-length and truncated Hasp and Hig polypeptides recognized by the antibodies used in this study. An “anchor” does not necessarily refer to direct binding between the two proteins. For clarity, factor X is not depicted at the middle and mature stages. For additional details, see Discussion.
The specific localization of both Hig and Hasp at cholinergic synapses suggests that the molecular composition of synaptic matrix may be related to the type of synapse and the distinct complement of neurotransmitters and receptors. In mice, >30 genes encoding predicted CCP proteins are expressed in the CNS (see Allen Brain Atlas). One of these proteins, SRPX2, regulates the formation of glutamatergic synapses in the brain (Sia et al., 2013). Further work should attempt to elucidate how these CCP proteins participate in synaptogenesis and how their combinatorial repertoire is involved in the diversification of synaptic properties. Because synaptic clefts are the space through which neurotransmitters disperse, the molecular composition of the matrix may also affect the behavior of neurotransmitters, thereby influencing synaptic plasticity and the efficiency of neurotransmission. Further studies focusing on the matrix architecture of synaptic clefts may reveal novel aspects of synaptic differentiation and function.
Footnotes
This work was supported by Japan Society for the Promotion of Science KAKENHI Grants 23650170 and 15K06721 to C.H. and Grant 15K21489 to M.N. We thank H.J. Bellen, U. Thomas, H. Aberle, D.R. Nässel, T. Uemura, and the Developmental Studies Hybridoma Bank for antibodies; Berkeley Drosophila Genome Project for cDNA; S.J. Sigrist, H.J. Bellen, the Bloomington Stock Center, and the Drosophila Genetic Resource Center at Kyoto Institute of Technology for fly stocks; and I. Takahashi, K. Watase, and Y. Wada for assistance with data processing.
The authors declare no competing financial interests.
References
- Barros CS, Franco SJ, Müller U. Extracellular matrix: functions in the nervous system. Cold Spring Harb Perspect Biol. 2011;3:a005108. doi: 10.1101/cshperspect.a005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingle CD, Vyakarnam A. Novel innate immune functions of the whey acidic protein family. Trends Immunol. 2008;29:444–453. doi: 10.1016/j.it.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briseño-Roa L, Bessereau JL. Proteolytic processing of the extracellular scaffolding protein LEV-9 is required for clustering acetylcholine receptors. J Biol Chem. 2014;289:10967–10974. doi: 10.1074/jbc.C113.534677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadie K, Baumgartner S, Prokop A. Extracellular matrix and its receptors in Drosophila neural development. Dev Neurobiol. 2011;71:1102–1130. doi: 10.1002/dneu.20935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell ID, Humphries MJ. Integrin structure, activation, and interactions. Cold Spring Harb Perspect Biol. 2011;3:piia004994. doi: 10.1101/cshperspect.a004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy WG, Liu Z, Nai Q, Coggan JS, Berg DK. PDZ-containing proteins provide a functional postsynaptic scaffold for nicotinic receptors in neurons. Neuron. 2003;38:759–771. doi: 10.1016/S0896-6273(03)00324-6. [DOI] [PubMed] [Google Scholar]
- Dai Z, Luo X, Xie H, Peng HB. The actin-driven movement and formation of acetylcholine receptor clusters. J Cell Biol. 2000;150:1321–1334. doi: 10.1083/jcb.150.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distler PG, Boeckh J. Synaptic connections between identified neuron types in the antennal lobe glomeruli of the cockroach, Periplaneta americana: I. Uniglomerular projection neurons. J Comp Neurol. 1997;378:307–319. doi: 10.1002/(SICI)1096-9861(19970217)378:3<307::AID-CNE1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Enell L, Hamasaka Y, Kolodziejczyk A, Nässel DR. gamma-Aminobutyric acid (GABA) signaling components in Drosophila: immunocytochemical localization of GABA(B) receptors in relation to the GABA(A) receptor subunit RDL and a vesicular GABA transporter. J Comp Neurol. 2007;505:18–31. doi: 10.1002/cne.21472. [DOI] [PubMed] [Google Scholar]
- Fayyazuddin A, Zaheer MA, Hiesinger PR, Bellen HJ. The nicotinic acetylcholine receptor Dalpha7 is required for an escape behavior in Drosophila. PLoS Biol. 2006;4:e63. doi: 10.1371/journal.pbio.0040063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friggi-Grelin F, Coulom H, Meller M, Gomez D, Hirsh J, Birman S. Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J Neurobiol. 2003;54:618–627. doi: 10.1002/neu.10185. [DOI] [PubMed] [Google Scholar]
- Funke L, Dakoji S, Bredt DS. Membrane-associated guanylate kinases regulate adhesion and plasticity at cell junctions. Annu Rev Biochem. 2005;74:219–245. doi: 10.1146/annurev.biochem.74.082803.133339. [DOI] [PubMed] [Google Scholar]
- Gendrel M, Rapti G, Richmond JE, Bessereau JL. A secreted complement-control-related protein ensures acetylcholine receptor clustering. Nature. 2009;461:992–996. doi: 10.1038/nature08430. [DOI] [PubMed] [Google Scholar]
- Hoshino M, Matsuzaki F, Nabeshima Y, Hama C. hikaru genki, a CNS-specific gene identified by abnormal locomotion in Drosophila, encodes a novel type of protein. Neuron. 1993;10:395–407. doi: 10.1016/0896-6273(93)90329-P. [DOI] [PubMed] [Google Scholar]
- Hoshino M, Suzuki E, Nabeshima Y, Hama C. Hikaru genki protein is secreted into synaptic clefts from an early stage of synapse formation in Drosophila. Development. 1996;122:589–597. doi: 10.1242/dev.122.2.589. [DOI] [PubMed] [Google Scholar]
- Hoshino M, Suzuki E, Miyake T, Sone M, Komatsu A, Nabeshima Y, Hama C. Neural expression of hikaru genki protein during embryonic and larval development of Drosophila melanogaster. Dev Genes Evol. 1999;209:1–9. doi: 10.1007/s004270050221. [DOI] [PubMed] [Google Scholar]
- Iwai Y, Usui T, Hirano S, Steward R, Takeichi M, Uemura T. Axon patterning requires DN-cadherin, a novel neuronal adhesion receptor, in the Drosophila embryonic CNS. Neuron. 1997;19:77–89. doi: 10.1016/S0896-6273(00)80349-9. [DOI] [PubMed] [Google Scholar]
- Kim MJ, Futai K, Jo J, Hayashi Y, Cho K, Sheng M. Synaptic accumulation of PSD-95 and synaptic function regulated by phosphorylation of serine-295 of PSD-95. Neuron. 2007;56:488–502. doi: 10.1016/j.neuron.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Bao H, Bonanno L, Zhang B, Serpe M. Drosophila Neto is essential for clustering glutamate receptors at the neuromuscular junction. Genes Dev. 2012;26:974–987. doi: 10.1101/gad.185165.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiss F, Koper E, Hein I, Fouquet W, Lindner J, Sigrist S, Tavosanis G. Characterization of dendritic spines in the Drosophila central nervous system. Dev Neurobiol. 2009;69:221–234. doi: 10.1002/dneu.20699. [DOI] [PubMed] [Google Scholar]
- Li H, Chaney S, Roberts IJ, Forte M, Hirsh J. Ectopic G-protein expression in dopamine and serotonin neurons blocks cocaine sensitization in Drosophila melanogaster. Curr Biol. 2000;10:211–214. doi: 10.1016/S0960-9822(00)00340-7. [DOI] [PubMed] [Google Scholar]
- Maglione M, Sigrist SJ. Seeing the forest tree by tree: super-resolution light microscopy meets the neurosciences. Nat Neurosci. 2013;16:790–797. doi: 10.1038/nn.3403. [DOI] [PubMed] [Google Scholar]
- Mahr A, Aberle H. The expression pattern of the Drosophila vesicular glutamate transporter: a marker protein for motoneurons and glutamatergic centers in the brain. Gene Expr Patterns. 2006;6:299–309. doi: 10.1016/j.modgep.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Miura E, Miyazaki T, Kakegawa W, Emi K, Narumi S, Fukazawa Y, Ito-Ishida A, Kondo T, Shigemoto R, Watanabe M, Yuzaki M. Cbln1 is a ligand for an orphan glutamate receptor δ2, a bidirectional synapse organizer. Science. 2010;328:363–368. doi: 10.1126/science.1185152. [DOI] [PubMed] [Google Scholar]
- McMahan UJ. The agrin hypothesis. Cold Spring Harb Symp Quant Biol. 1990;55:407–418. doi: 10.1101/SQB.1990.055.01.041. [DOI] [PubMed] [Google Scholar]
- Nakayama M, Hama C. Modulation of neurotransmitter receptors and synaptic differentiation by proteins containing complement-related domains. Neurosci Res. 2011;69:87–92. doi: 10.1016/j.neures.2010.11.006. [DOI] [PubMed] [Google Scholar]
- Nakayama M, Matsushita F, Hama C. The matrix protein Hikaru genki localizes to cholinergic synaptic clefts and regulates postsynaptic organization in the Drosophila brain. J Neurosci. 2014;34:13872–13877. doi: 10.1523/JNEUROSCI.1585-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noritake J, Fukata Y, Iwanaga T, Hosomi N, Tsutsumi R, Matsuda N, Tani H, Iwanari H, Mochizuki Y, Kodama T, Matsuura Y, Bredt DS, Hamakubo T, Fukata M. Mobile DHHC palmitoylating enzyme mediates activity-sensitive synaptic targeting of PSD-95. J Cell Biol. 2009;186:147–160. doi: 10.1083/jcb.200903101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo P, Sainlos M, Choquet D. Regulation of AMPA receptor surface diffusion by PSD-95 slots. Curr Opin Neurobiol. 2012;22:453–460. doi: 10.1016/j.conb.2011.10.010. [DOI] [PubMed] [Google Scholar]
- Parker MJ, Zhao S, Bredt DS, Sanes JR, Feng G. PSD93 regulates synaptic stability at neuronal cholinergic synapses. J Neurosci. 2004;24:378–388. doi: 10.1523/JNEUROSCI.3865-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnas D, Haghighi AP, Fetter RD, Kim SW, Goodman CS. Regulation of postsynaptic structure and protein localization by the Rho-type guanine nucleotide exchange factor dPix. Neuron. 2001;32:415–424. doi: 10.1016/S0896-6273(01)00485-8. [DOI] [PubMed] [Google Scholar]
- Pech U, Pooryasin A, Birman S, Fiala A. Localization of the contacts between Kenyon cells and aminergic neurons in the Drosophila melanogaster brain using SplitGFP reconstitution. J Comp Neurol. 2013;521:3992–4026. doi: 10.1002/cne.23388. [DOI] [PubMed] [Google Scholar]
- Pinan-Lucarré B, Tu H, Pierron M, Cruceyra PI, Zhan H, Stigloher C, Richmond JE, Bessereau JL. C. elegans Punctin specifies cholinergic versus GABAergic identity of postsynaptic domains. Nature. 2014;511:466–470. doi: 10.1038/nature13313. [DOI] [PubMed] [Google Scholar]
- Rapti G, Richmond J, Bessereau JL. A single immunoglobulin-domain protein required for clustering acetylcholine receptors in C. elegans. EMBO J. 2011;30:706–718. doi: 10.1038/emboj.2010.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemensperger T, Issa AR, Pech U, Coulom H, Nguỹên MV, Cassar M, Jacquet M, Fiala A, Birman S. A single dopamine pathway underlies progressive locomotor deficits in a Drosophila model of Parkinson disease. Cell Rep. 2013;5:952–960. doi: 10.1016/j.celrep.2013.10.032. [DOI] [PubMed] [Google Scholar]
- Robinson SW, Herzyk P, Dow JA, Leader DP. FlyAtlas: database of gene expression in the tissues of Drosophila melanogaster. Nucleic Acids Res. 2013;41:D744–D750. doi: 10.1093/nar/gks1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrbough J, Rushton E, Woodruff E, 3rd, Fergestad T, Vigneswaran K, Broadie K. Presynaptic establishment of the synaptic cleft extracellular matrix is required for post-synaptic differentiation. Genes Dev. 2007;21:2607–2628. doi: 10.1101/gad.1574107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Heintzmann R, Leonhardt H. A guide to super-resolution fluorescence microscopy. J Cell Biol. 2010;190:165–175. doi: 10.1083/jcb.201002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P, Hermans-Borgmeyer I, Betz H, Gundelfinger ED. Neuronal acetylcholine receptors in Drosophila: the ARD protein is a component of a high-affinity alpha-bungarotoxin binding complex. EMBO J. 1988;7:2889–2894. doi: 10.1002/j.1460-2075.1988.tb03146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia GM, Clem RL, Huganir RL. The human language-associated gene SRPX2 regulates synapse formation and vocalization in mice. Science. 2013;342:987–991. doi: 10.1126/science.1245079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton M, Liao G, Brokstein P, Hong L, Carninci P, Shiraki T, Hayashizaki Y, Champe M, Pacleb J, Wan K, Yu C, Carlson J, George R, Celniker S, Rubin GM. The Drosophila gene collection: identification of putative full-length cDNAs for 70% of D. melanogaster genes. Genome Res. 2002;12:1294–1300. doi: 10.1101/gr.269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagawa K, Salvaterra P. Analysis of choline acetyltransferase protein in temperature sensitive mutant flies using newly generated monoclonal antibody. Neurosci Res. 1996;24:237–243. doi: 10.1016/0168-0102(95)00999-X. [DOI] [PubMed] [Google Scholar]
- Toba G, Ohsako T, Miyata N, Ohtsuka T, Seong KH, Aigaki T. The gene search system: a method for efficient detection and rapid molecular identification of genes in Drosophila melanogaster. Genetics. 1999;151:725–737. doi: 10.1093/genetics/151.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu H, Pinan-Lucarré B, Ji T, Jospin M, Bessereau JL. C. elegans Punctin clusters GABA(A) receptors via neuroligin binding and UNC-40/DCC recruitment. Neuron. 2015;86:1407–1419. doi: 10.1016/j.neuron.2015.05.013. [DOI] [PubMed] [Google Scholar]
- Uemura T, Lee SJ, Yasumura M, Takeuchi T, Yoshida T, Ra M, Taguchi R, Sakimura K, Mishina M. Trans-synaptic interaction of GluRδ2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010;141:1068–1079. doi: 10.1016/j.cell.2010.04.035. [DOI] [PubMed] [Google Scholar]
- Wagh DA, Rasse TM, Asan E, Hofbauer A, Schwenkert I, Dürrbeck H, Buchner S, Dabauvalle MC, Schmidt M, Qin G, Wichmann C, Kittel R, Sigrist SJ, Buchner E. Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron. 2006;49:833–844. doi: 10.1016/j.neuron.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Watson GB, Chouinard SW, Cook KR, Geng C, Gifford JM, Gustafson GD, Hasler JM, Larrinua IM, Letherer TJ, Mitchell JC, Pak WL, Salgado VL, Sparks TC, Stilwell GE. A spinosyn-sensitive Drosophila melanogaster nicotinic acetylcholine receptor identified through chemically induced target site resistance, resistance gene identification, and heterologous expression. Insect Biochem Mol Biol. 2010;40:376–384. doi: 10.1016/j.ibmb.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Wu H, Xiong WC, Mei L. To build a synapse: signaling pathways in neuromuscular junction assembly. Development. 2010;137:1017–1033. doi: 10.1242/dev.038711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W. PSD-95-like membrane associated guanylate kinases (PSD-MAGUKs) and synaptic plasticity. Curr Opin Neurobiol. 2011;21:306–312. doi: 10.1016/j.conb.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuyama K, Meinertzhagen IA, Schürmann FW. Synaptic organization of the mushroom body calyx in Drosophila melanogaster. J Comp Neurol. 2002;445:211–226. doi: 10.1002/cne.10155. [DOI] [PubMed] [Google Scholar]