Abstract
To define the cell populations that drive joint inflammation in rheumatoid arthritis (RA), we applied single-cell RNA sequencing (scRNA-seq), mass cytometry, bulk RNA-seq and flow cytometry to T cells, B cells, monocytes and fibroblasts from 51 samples of synovial tissue from patients with RA or osteoarthritis. Utilizing an integrated strategy based on canonical correlation analysis of 5,265 scRNA-seq profiles, we identified 18 unique cell populations. Combining mass cytometry and transcriptomics together revealed cell states expanded in RA synovia: THY1(CD90)+HLA-DRAhi sublining fibroblasts, IL1B+ pro-inflammatory monocytes, ITGAX+TBX21+ autoimmune-associated B cells and PDCD1+ T peripheral helper (Tph) and T follicular helper (Tfh). We defined distinct subsets of CD8+ T cells characterized by a GZMK+, GZMB+ and GNLY+ phenotype. We mapped inflammatory mediators to their source cell populations; for example, we attributed IL6 expression to THY1+HLA-DRAhi fibroblasts, and IL1B production to pro-inflammatory monocytes. These populations are potentially key mediators of RA pathogenesis.
Rheumatoid arthritis (RA) is an autoimmune disease with chronic inflammation in the synovium of the joint tissue1–3. This inflammation leads to joint destruction, disability and shortened life span4. Defining key cellular subsets and their activation states in the inflamed tissue is a critical step to define new therapeutic targets for RA. CD4+ T cell5,6 B cells7, monocytes8,9, and fibroblasts10,11 have established relevance to RA pathogenesis. Here, we use single cell technologies to view all of these cell types simultaneously across a large collection of samples from inflamed joints. We believe a global single-cell portrait of how different cell types work together would advance our understanding of therapeutics.
Application of transcriptomic and cellular profiling technologies to whole synovial tissue has already identified specific cell populations associated with RA3,12–14. However, most studies have focused on a pre-selected cell type, surveyed whole tissues rather than disaggregated cells, or used only a single technology platform. The latest advances in single-cell technologies offer an opportunity to identify disease-associated cell subsets in human tissues at high resolution in an unbiased fashion15–17. These technologies have already been used to discover roles for T peripheral helper (Tph) cells18 and HLA-DR+CD27− cytotoxic T cells19 in RA pathogenesis. Studies using scRNA-seq have defined myeloid cell heterogeneity in human blood20 and identified overabundance of PDPN+CD34−THY1+ (THY1, also known as CD90) fibroblasts in RA synovial tissue15,21.
To generate high-dimensional multi-modal single-cell data from synovial tissue samples collected across a collaborative network of research sites, we developed a robust pipeline22 in the Accelerating Medicines Partnership Rheumatoid Arthritis and Lupus (AMP RA/SLE) consortium. We collected and disaggregated tissue samples from patients with RA and osteoarthritis (OA), and then subjected constituent cells to scRNA-seq, sorted-population bulk RNA-seq, mass cytometry, and flow cytometry. We developed a unique computational strategy based on canonical correlation analysis (CCA) to integrate multi-modal transcriptomic and proteomic profiles at a single cell level. A unified analysis of single cells across data modalities can precisely define contributions of specific cell subsets to pathways relevant to RA and chronic inflammation.
RESULTS
Generation of parallel mass cytometric and transcriptomic data from synovial tissue
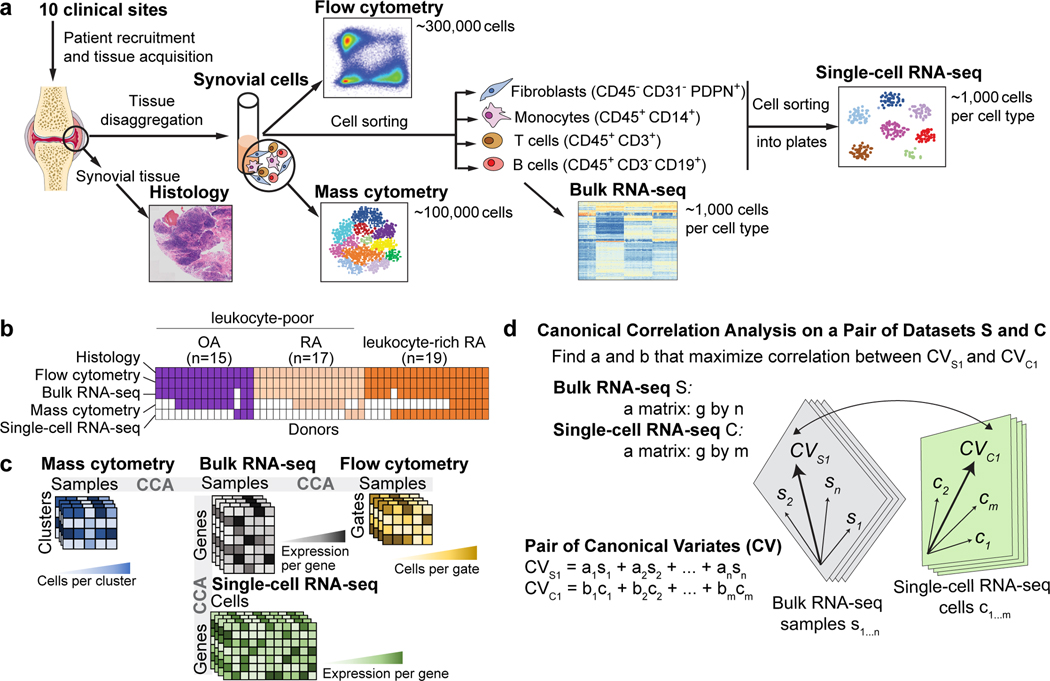
In phase 1 of AMP RA/SLE, we recruited 36 patients with RA that met the 1987 American College of Rheumatology (ACR) classification criteria and 15 patients with OA from 10 clinical sites over 16 months (Supplementary Table 1) and obtained synovial tissues from ultrasound-guided biopsies or joint replacements (Methods, Fig. 1a). We required that all tissue samples included had synovial lining documented by histology. Synovial tissue disaggregation yielded an abundance of viable cells for downstream analyses (362,190 +/− 7,687 (mean +/− SEM) cells per tissue). We used our validated strategy for cell sorting22 (Fig. 1a) to isolate B cells (CD45+CD3−CD19+), T cells (CD45+CD3+), monocytes (CD45+CD14+), and stromal fibroblasts (CD45−CD31−PDPN+) (Supplementary Fig. 1a). We applied bulk RNA-seq to all four sorted subsets for all 51 samples. For samples with sufficient cell yield (Methods), we also measured single-cell protein expression using a 34-marker mass cytometry panel (n=26, Supplementary Table 2), and single-cell RNA expression in sorted cell populations (n=21, Fig. 1b).
Figure 1.
Overview of synovial tissue workflow and pairwise analysis of high-dimensional data. a. We acquired synovial tissue, disaggregated the cells, sorted them into four gates representing fibroblasts (CD45−CD31−PDPN+), monocytes (CD45+CD14+), T cells (CD45+CD3+), and B cells (CD45+CD3−CD19+). We profiled these cells with mass cytometry, flow cytometry, sorted low-input bulk RNA-seq, and single-cell RNA-seq. Here, we use Servier Medical Art by Servier for the joint picture. b. Presence and absence of five different data types for each tissue sample. c. Schematic of each dataset and the shared dimensions used to analyze each of the three pairs of datasets with canonical correlation analysis (CCA). d. CCA finds a common mapping for two datasets. For bulk RNA-seq and single-cell RNA-seq, we first find a common set of g genes present in both datasets. Each bulk sample si gets a coefficient ai and each cell ci gets a coefficient bi. The linear combination of all samples s1…n arranges bulk genes along the canonical variate CVs1 and the linear combination of all cells c1…m arranges single-cell genes along CVc1. CCA finds the coefficients a1…n and b1…m that arrange the genes from the two datasets in such a way that the correlation between CVs1 and CVc1 is maximized. After CCA finds the first pair of canonical variates, the next pair is computed on the residuals, and so on.
Summary of computational data integration strategy to define cell populations
To confidently define RA-associated cell populations, we integrated multiple data modalities (Fig. 1b, c). We use bulk RNA-seq data as the reference point because it was available for all of the donors and most of the cell types, it had the highest dimensionality and least sensitive to technical artifacts (Fig. 1b).
Integrating scRNA-seq with bulk RNA-seq data ensures robust discovery of cell populations. Here, we used CCA to find linear combinations of bulk RNA-seq samples and scRNA-seq cells (Fig. 1c, d) to create gene expression profiles that were maximally correlated. These linear combinations captured sources of shared variation between the two datasets and allowed us to identify individual cell populations that drive variation in the bulk RNA-seq data. We analyzed the scRNA-seq data by using the canonical variate coefficients for each cell to compute a nearest neighbor network, identifying clusters with a community detection algorithm, and evaluating the separation between clusters with Silhouette analysis (Methods, Supplementary Fig. 2b).
We identified cell clusters in mass cytometry data with density-based clustering23. Next, we used CCA to identify linear combinations of bulk RNA-seq genes and mass cytometry cluster abundances that maximize correlation across patients. These canonical variates offer a way to visualize genes and mass cytometry clusters together. We then queried this CCA result with the best marker genes from scRNA-seq to establish a relationship between each scRNA-seq cluster and each mass cytometry cluster (Methods). We also used CCA to associate bulk gene expression in each sample with proportions of cells in different flow cytometry gates.
Flow cytometry features define a set of RA synovia that are leukocyte-rich
Histology of RA synovial tissues revealed heterogeneous tissue composition with variable lymphocyte and monocyte infiltration (Fig. 2a,b, Supplementary Fig. 2c,d). This heterogeneity was expected, because variation in tissue immune cell infiltration reflects local disease activity in the source joint. Consequently, we employed a data-driven approach to separate samples based on flow cytometry of lymphocyte and monocyte infiltration in each tissue sample (Supplementary Fig. 1b,c). We calculated a multivariate normal distribution of these parameters based on OA samples as a reference, and for each RA sample we calculated the Mahalanobis distance from OA24. We defined the maximum OA distance (4.5) as the threshold for defining leukocyte-rich RA (>4.5, n=19) or leukocyte-poor RA (<4.5, n=17) samples (Methods, Supplementary Fig. 1d). Whereas leukocyte-rich RA tissues had significant infiltration of synovial T cells and B cells, leukocyte-poor RA tissues had cellular compositions more similar to OA (Fig. 2c). Synovial monocyte abundances were similar between RA and OA (Fig. 2c).
Figure 2.
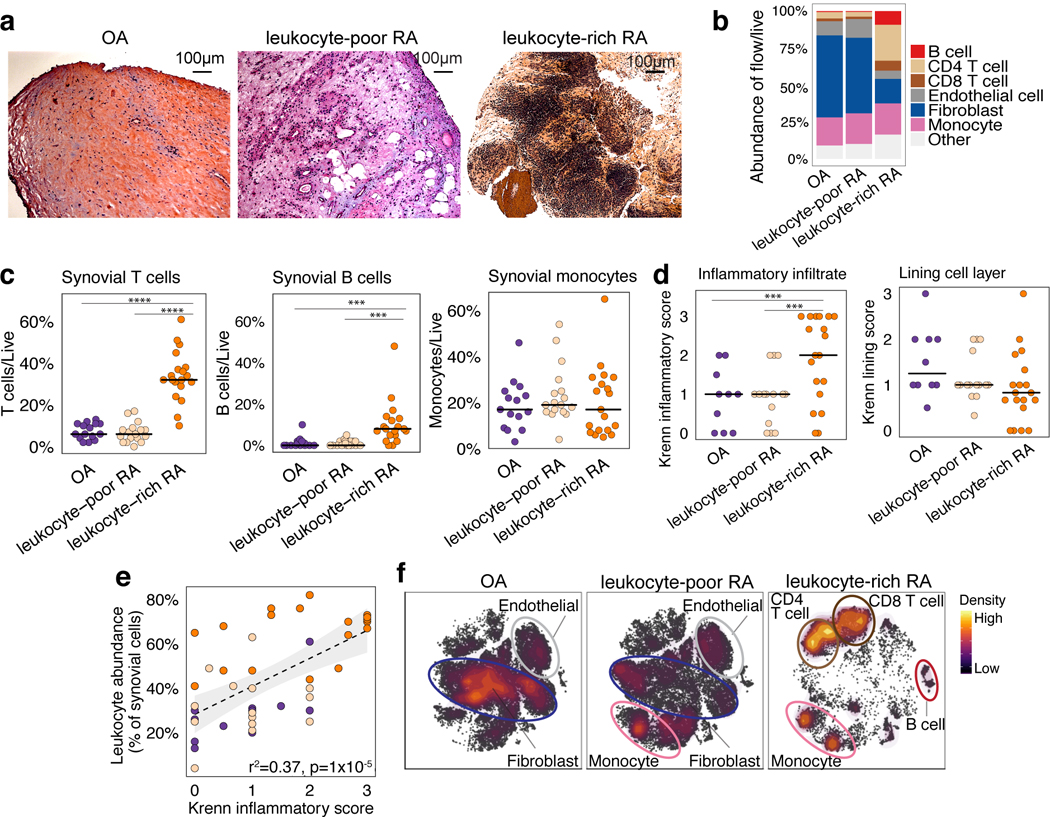
Distinct cellular composition in synovial tissue from OA, leukocyte-poor RA, and leukocyte-rich RA patients. a. Histological assessment of synovial tissue derived from OA (n = 15 independent tissue samples), leukocyte-poor RA (n = 17 independent tissue samples), and leukocyte-rich RA (n = 19 independent tissue samples). b. Cellular composition of major synovial cell types by flow cytometry. c. Synovial T cells, B cells, and monocytes by flow cytometry in samples from OA (n = 15), leukocyte-poor RA (n = 17), and leukocyte-rich RA (n = 19). Leukocyte-rich RA tissues were significantly higher infiltrated in synovial T cells (Student’s one-sided t-test P = 4×10−9, t-value = 8.92, df = 22.27) compared to leukocyte-poor RA and OA. Leukocyte-rich RA tissues were significantly higher infiltrated in synovial B cells (Student’s one-sided t-test P = 1×10−3, t-value = 3.50, df = 20.56) compared to leukocyte-poor RA and OA. Center value is mean. Statistical significance levels: ****P<1×10−4 and ***P<1×10−3. d. Quantitative histologic inflammatory scoring of both sublining cell layer and lining layer. Leukocyte-rich RA samples (n = 19) exhibited higher (Student’s one-sided t-test P = 1×10−3, t-value = 3.21, df = 30.66) Krenn inflammation scores than leukocyte-poor RA (n=15) and OA tissues (n = 10) samples. Center value is mean. e. Correlation between leukocyte infiltration assessed by cytometry with histologic inflammation score (n = 44 biologically independent samples). Student’s one-sided t-test P = 3×10−09, t-value = 7.15, df = 46.51. f. tSNE visualization of synovial cell types in OA, leukocyte-poor RA, and leukocyte-rich RA by mass cytometry density plot.
To test if our classification indicates inflammation, we assessed tissue histology and assigned each sample a Krenn inflammation score25. Samples we classified as leukocyte-rich RA had a significantly higher Krenn inflammation score than leukocyte-poor RA or OA (Fig. 2d). In contrast, synovial lining membrane hyperplasia was not significantly different between leukocyte-rich RA, leukocyte-poor RA, and OA samples (Fig. 2d). We observed significant correlation between synovial leukocyte infiltration measured by flow cytometry and the histological Krenn inflammation score (Fig. 2e). Mass cytometry in 26 synovial tissues was consistent with flow cytometry and histology. OA and leukocyte-poor RA samples were characterized by high abundance of fibroblasts and endothelial cells; while leukocyte-rich RA tissues were characterized by high abundance of CD4 T, CD8 T, and B cells (Fig. 2f, Supplementary Fig. 3a).
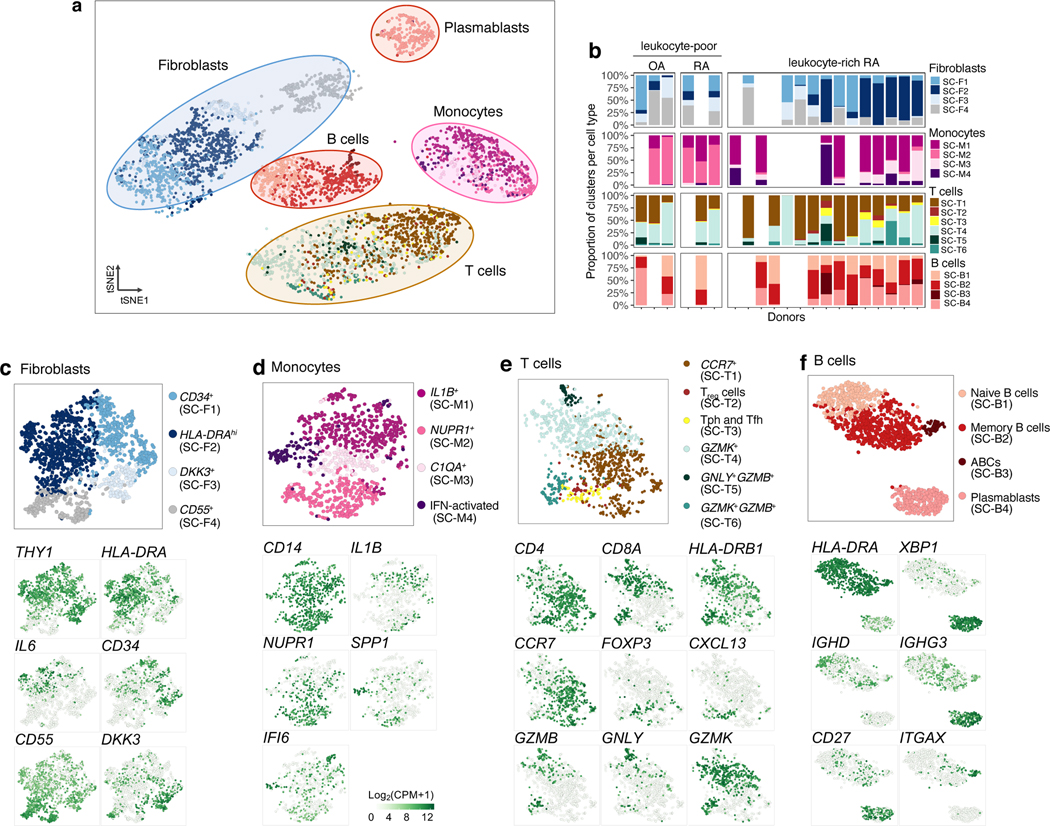
Single-cell RNA-seq analysis reveals distinct cell subpopulations
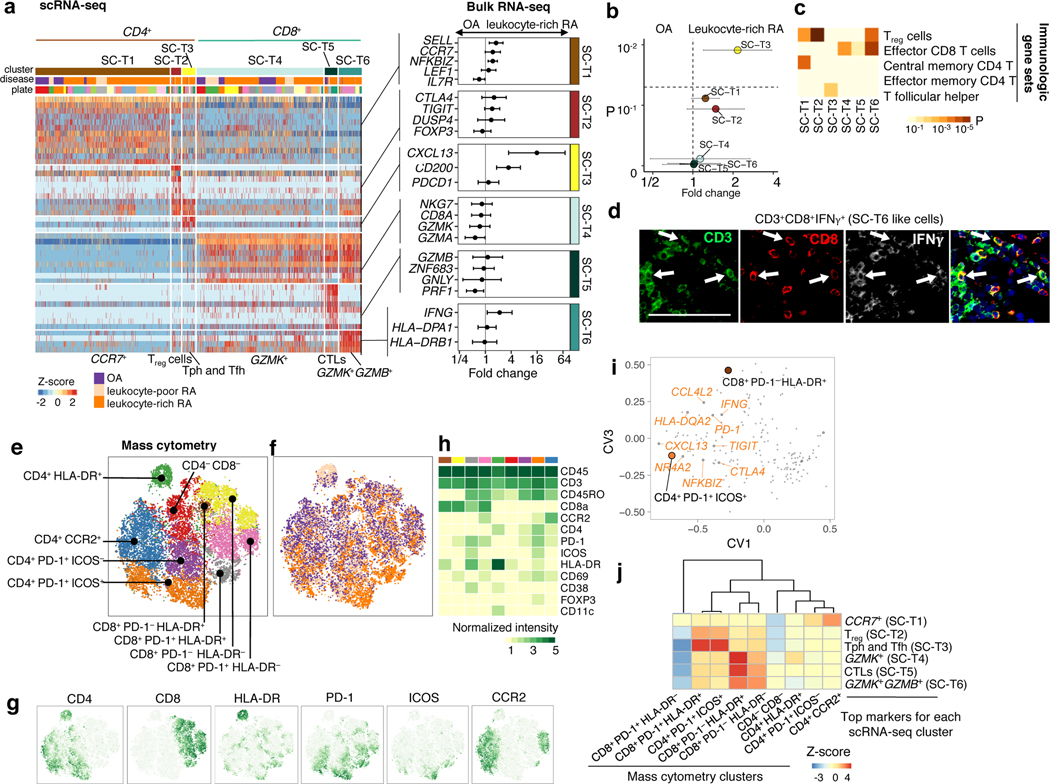
Next, we analyzed 5,265 scRNA-seq profiles passing quality control (Methods), including 1,142 B cells, 1,844 fibroblasts, 750 monocytes, and 1,529 T cells. We used canonical variates (from CCA with bulk RNA-seq) to define 18 cell clusters that were independent of donor (n=21) and technical plate (n=24) effects (Fig. 3a–bb, Supplementary Fig. 2c, Supplementary 4a). In contrast, conventional PCA-based clustering led to clusters that were confounded by batch effects (Supplementary Fig. 4b). All of the clusters in the PCA-based clustering, excluding clusters confounded by batch, were identified in CCA-based clustering. Next, we compared expression values between cells in the cluster and all other cells to select cluster marker genes (Methods, Supplementary Table 4). For selected genes, we show expression values in each cell positioned in a t-distributed Stochastic Neighbor Embedding (tSNE26) (Fig. 3c–f). Among fibroblasts, we identified four putative subpopulations (Fig. 3c): CD34+ sublining fibroblasts (SC-F1), HLA-DRAhi sublining fibroblasts (SC-F2), DKK3+ sublining fibroblasts (SC-F3), and CD55+ lining fibroblasts (SC-F4). In monocytes (Fig. 3d), we identified IL1B+ pro-inflammatory monocytes (SC-M1), NUPR1+ monocytes (SC-M2), C1QA+ monocytes (SC-M3), and interferon (IFN) activated monocytes (SC-M4). In T cells (Fig. 3e), we identified three CD4+ clusters: CCR7+ T cells (SC-T1), FOXP3+ regulatory T cells (Treg cells) (SC-T2), and PDCD1+ Tph and T follicular helper (Tfh) (SC-T3); and three CD8+ clusters: GZMK+ T cells (SC-T4), GNLY+GZMB+ cytotoxic lymphocytes (CTLs) (SC-T5), and GZMK+GZMB+ T cells (SC-T6). Within B cells (Fig. 3f), we identified four cell clusters, including naive IGHD+CD27− (SC-B1) and IGHG3+CD27+ memory B cells (SC-B2). We identified an autoimmune-associated B cells (ABCs) cluster (SC-B3) with high expression of ITGAX (also known as CD11c) and a plasmablast cluster (SC-B4) with high expression of immunoglobulin genes and XBP1, a transcription factor for plasma cell differentiation27.
Figure 3.
High-dimensional transcriptomic scRNA-seq clustering reveals distinct cell type subpopulations. a. 18 clusters across 5,265 cells from all cell types on a tSNE visualization. b. Cluster abundances across donors. c. Fibroblasts: three types of THY1+ sublining fibroblasts (SC-F1, SC-F2, and SC-F3) and CD55+ lining fibroblasts (SC-F4). d. Monocytes: two activated cell states of IL1B+ pro-inflammatory (SC-M1) and IFN-activated (SC-M4) monocytes. e. T cells: CD4+ subsets: SC-T1, SC-T2, SC-T3, and CD8+ subsets: SC-T4, SC-T5, and SC-T6. f. B cells: HLA+ (SC-B1, SC-B2, and SC-B3) and plasmablasts (SC-B4). The cluster colors in c-f are consistent with (a).
We assessed protein fluorescence measurements of typical cell type markers, which were consistent with our identified scRNA-seq clusters (Supplementary Fig. 2e). Cell density quantified from 10 histology samples was correlated with the lymphocyte flow cytometric cell yields, suggesting that samples with the most single cell measurements are those with the best yields and the most inflammation (Supplementary Fig. 5).
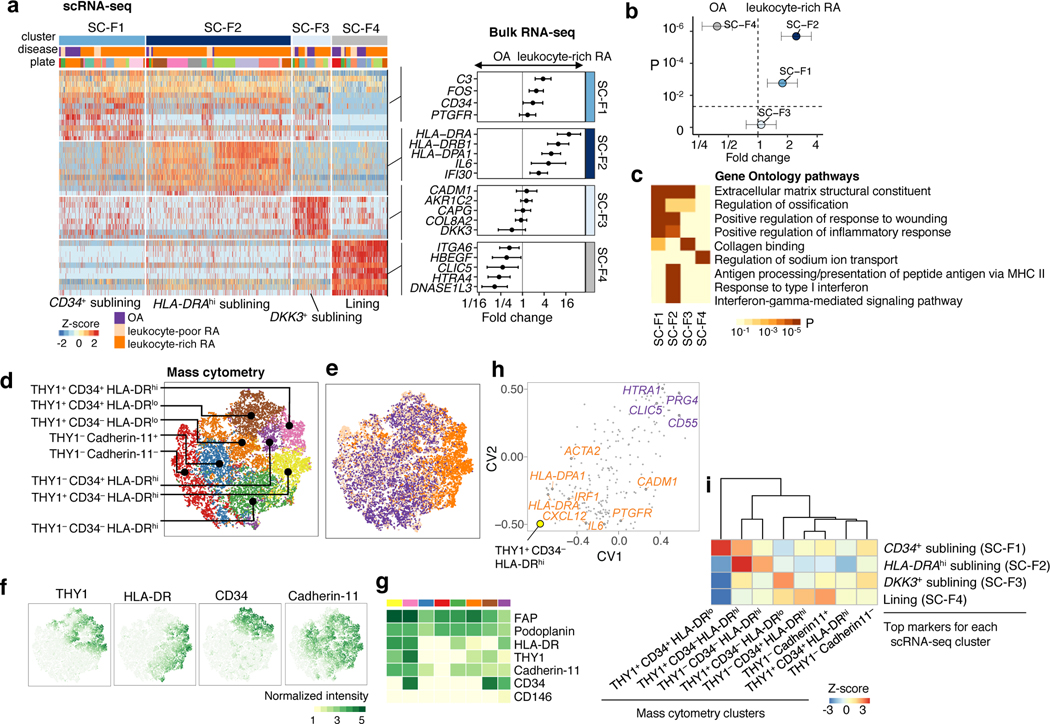
Distinct synovial fibroblasts defined by cytokine activation and MHC II expression
To identify the fibroblast subpopulations overabundant in leukocyte-rich RA synovia, we selected marker genes for each cluster and assessed their expression levels in bulk RNA-seq from sorted fibroblasts (CD45−PDPN+) from RA and OA patients. For example, genes associated with HLA-DRAhi (SC-F2) fibroblasts were more highly expressed in bulk RNA-seq samples from leukocyte-rich RA than OA (t-test p<1×10−3 for HLA-DRA, IFI30, and IL6) (Fig. 4a). Since the expression profile of a bulk tissue sample is an aggregate of the expression profiles of its constituent cell populations, this result suggests expansion of HLA-DRAhi (SC-F2) fibroblasts in RA tissues. Genes associated with CD55+ fibroblasts (SC-F4) were significantly more highly expressed in bulk RNA-seq samples from OA than leukocyte-rich RA (t-test p<1×10−3 for HBEGF, CLIC5, HTRA4, and DNASE1L3) (Fig. 4a). CD55+ fibroblasts (SC-F4) were the most transcriptionally distinct subset from the three THY1+ clusters (SC-F1–3), including the highest expression of lubricin (PRG4), suggesting that these cells represent synovial lining fibroblasts and THY1+ fibroblasts (SC-F1–3) represent sublining (Fig. 4a). Next, we use the averaged expression level of the best marker genes for each scRNA-seq cluster (AUC>0.7) and tested for differential expression in bulk RNA-seq fibroblast samples from leukocyte-rich RA and OA synovia. The gene averages for HLA-DRAhi sublining fibroblasts (SC-F2) and CD34+ sublining fibroblasts (SC-F1) were higher in leukocyte-rich RA compared to OA (t-test p=2×10−6 and p=2×10−3, respectively), while the gene averages for CD55+ lining fibroblasts (SC-F4) were higher in OA than leukocyte-rich RA (t-test p=5×10−7) (Fig. 4b).
Figure 4.
Distinct synovial fibroblast subsets defined by cytokine activation and MHC II expression. a. scRNA-seq analysis identified three sublining subsets, CD34+ (SC-F1), HLAhi (SC-F2), and DKK3+ (SC-F3) and one lining subset (SC-F4). Differential analysis between leukocyte-rich RA (n = 16) and OA (n = 12) bulk RNA-seq fibroblast samples shows marker genes upregulated or downregulated in leukocyte-rich RA. Fold changes with 95% confidence interval (CI). b. By querying the leukocyte-rich RA (n = 16) and OA (n = 12) fibroblast bulk RNA-seq samples, scRNA-seq cluster HLA-DRAhi (SC-F2) and CD34+ (SC-F1) fibroblasts are significantly overabundant (two-sided Student’s t-test P=2×10−6, t-value=6.2, df = 23.91 and P=2×10−3, t-value = 3.20, df = 25.41, respectively) in leukocyte-rich RA relative to OA. Lining fibroblasts (SC-F4) are overabundant (two-sided Student’s t-test P=5×10−7, t-value=−5.31, df =21.97) in OA samples. Fold changes with 95% CI. c. Pathway enrichment analysis for each cluster. Two-sided Kolmogorov-Smirnov test with 105 permutations; Benjamini-Hochberg FDR is shown. d-e. Identified subpopulations from fibroblasts (n = 25,161) and disease status from 6 leukocyte-rich RA, 9 leukocyte-poor RA, and 8 OA by mass cytometry on the same gating with scRNA-seq. f-g. Normalized intensity of distinct protein markers shown in tSNE visualization and averaged for each cluster heatmap. h. CCA projections of mass cytometry clusters and bulk RNA-seq genes. First two canonical variates (CVs) separated genes upregulated in leukocyte-rich RA from genes upregulated in OA. HLAhi genes are highly associated with THY1+CD34−HLA-DRhi by mass cytometry. i. Integration of mass cytometry clusters with scRNA-seq clusters based on the top markers (AUC > 0.7) for each scRNA-seq cluster using top 10 canonical variates in the low-dimensional CCA space. We computed the spearman correlation between each pair of scRNA-seq cluster and mass cytometry cluster in the CCA space and performed permutation test 104 times. Z-score is calculated based on permutation p-value. We observed HLAhigh sublining fibroblasts by scRNA-seq are strongly correlated with THY1+CD34−HLA-DRhi fibroblasts by mass cytometry.
Consistent with the role of synovial fibroblasts in matrix remodeling, the sublining fibroblast subsets (SC-F1–3) expressed genes encoding extracellular matrix constituents (Fig. 4c). HLA-DRAhi sublining fibroblasts (SC-F2) expressed genes related to MHC class II presentation and the interferon gamma-mediated signaling pathway (IFI30) (Fig. 4a,c), suggesting upregulation of MHC class II in response to interferon-gamma signaling in these cells. We identified a novel sublining fibroblast subtype (SC-F3) that is characterized by high expression of DKK3, CADM1 and COL8A2 (Fig. 4a).
To independently confirm the presence of four fibroblast subpopulations discovered by scRNA-seq, we analyzed CD45−PDPN+ cells in mass cytometry data, and found eight putative cell clusters with differential protein levels of THY1, HLA-DR, CD34, and Cadherin-11 without obvious batch effects (Fig. 4d–g, Supplementary Fig. 3b). CCA revealed that greater abundance of THY1+CD34−HLA-DRhi fibroblasts measured by mass cytometry is associated with higher expression of IL6, CXCL12, and HLA-DRA in bulk RNA-seq of the same samples, suggesting these cells are in an active cytokine-producing state (Fig. 4h). CCA allowed us to place mass cytometry clusters in the same space as bulk RNA-seq genes, so we could query the positions of scRNA-seq genes within this space to find the correspondence between scRNA-seq clusters and mass cytometry clusters (Fig. 4i, Methods). We found HLA-DRAhi sublining fibroblasts (SC-F2) correspond to THY1+CD34−HLA-DRhi fibroblasts (z-score=2.8), and CD34+ sublining fibroblasts (SC-F1) correspond to THY1+CD34+HLA-DRlo fibroblasts (z-score=2.7) (Table 1). Consistent with differential expression analysis of bulk RNA-seq, we found that THY1+CD34−HLA-DRhi cells in the mass cytometry data were overabundant in leukocyte-rich RA relative to leukocyte-poor RA and OA controls (36% versus 2% of fibroblasts, MASC OR = 33.8 (95% CI: 11.7–113.1), one-sided MASC p=1.9×10−5) (Table 1).
Table 1.
Connection between cell populations determined by mass cytometry and scRNA-seq clusters and disease associations. Bold mass cytometry clusters are significantly enriched in leukocyte-rich RA (one-sided Benjamini-Hochberg FDR q value < 0.05). Two significant digits are given to the one-sided F-tests conducted on nested models with MASC. 95% confidence interval (CI) for the odds ratio (OR) is given for each mass cytometry cluster. Where possible, we have identified the most similar scRNA-seq clusters for each cluster found by mass cytometry. The mass cytometry analysis is performed on downsampled datasets of 25,161 fibroblasts from 23 patients, 15,298 monocytes from 26 patients, 19,985 T cells from 26 patients, and 8,179 B cells from 23 patients.
| leukocyte-poor | leukocyte-rich | One-sided | leukocyte-rich | ||
|---|---|---|---|---|---|
| scRNA-seq cluster | mass cytometry cluster | RA and OA | RA | MASC p value | OR (CI) |
| THY1− Cadherin-11− | 21% | 4% | 1.00 | 0.04 (0–0.2) | |
| Lining (SC-F4) | THY1− Cadherin-11+ | 18% | 2% | 1.00 | 0.1 (0–0.3) |
| THY1− CD34+ HLA-DRhi | 7% | 3% | 0.87 | 0.5 (0.3–1.2) | |
| THY1− CD34− HLA-DRhi | 17% | 15% | 0.48 | 1.2 (0.3–4.4) | |
| HLAhi sublining (SC-F2) | THY1+ CD34− HLA-DRhi | 2% | 36% | 1.9×10−5 | 33.8 (11.7–113.1) |
| DKK3+ sublining (SC-F3) | THY1+ CD34− HLA-DRlow | 16% | 15% | 0.66 | 0.8 (0.3–1.8) |
| CD34+ sublining (SC-F1) | THY1+ CD34+ HLA-DRlow | 18% | 4% | 1.00 | 0.2 (0.1–0.4) |
| THY1+ CD34+ HLA-DRhi | 2% | 21% | 1.6×10−4 | 25.5 (7.5–101.8) | |
| NUPR1+ (SC-M2) | CD11c− | 30% | 4% | 1.00 | 0.1 (0–0.4) |
| IL1B+ (SC-M1), IFN-activated (SC-M4) | CD11c+ CCR2+ | 34% | 40% | 0.23 | 1.6 (0.7–3.6) |
| CD11c+ CD38− | 13% | 2% | 1.00 | 0.1 (0–0.3) | |
| CD11c+ CD38− CD64+ | 13% | 3% | 0.93 | 0.3 (0.1–1) | |
|
IL1B+ (SC-M1), IFN-activated (SC-M4), C1QA+ (SC-M3) |
CD11c+ CD38+ | 15% | 51% | 6.7×10−5 | 7.8 (3.6–17.2) |
| CD4− CD8− | 15% | 9% | 0.95 | 0.6 (0.3–1) | |
| CCR7+(SC-T1) | CD4+ CCR2+ | 26% | 13% | 1.00 | 0.4 (0.2–0.7) |
| CD4+ HLA-DR+ | 6% | 2% | 0.83 | 0.7 (0.2–4.1) | |
| CD4+ PD-1+ ICOS− | 13% | 12% | 0.81 | 0.9 (0.5–1.6) | |
| Tph and Tfh (SC-T3) | CD4+ PD-1+ ICOS+ | 11% | 25% | 2.7×10−4 | 3.0 (1.7–5.2) |
| CD8+ PD-1− HLA-DR− | 14% | 9% | 0.76 | 0.7 (0.3–1.5) | |
|
GZMK+GZMB+(SC-T6), GZMK+ (SC-T4),
CTLs (SC-T5) |
CD8+ PD-1− HLA-DR+ | 2% | 1% | 0.64 | 0.9 (0.4–2.2) |
| CD8+ PD-1+ HLA-DR− | 13% | 14% | 0.40 | 1.1 (0.6–1.9) | |
| Tph and Tfh (SC-T3) | CD8+ PD-1+ HLA-DR+ | 1% | 15% | 9.2×10−5 | 11.8 (4.9–34.2) |
| plasmablasts (SC-B4) | CD38++ CD20− IgM− IgD− | 6% | 12% | 0.01 | 3.3 (1.2–10.5) |
| CD38++ CD20− IgM+ HLA-DR+ | 1% | 3% | 0.01 | 6.9 (1.3–83.1) | |
| Memory B cells (SC-B2) | IgM− IgD− HLA-DR− | 27% | 2% | 1.00 | 0.1 (0–0.3) |
| CD38+ HLA-DR++ CD20− CD11c+ | 19% | 6% | 0.56 | 0.9 (0.1–6.7) | |
| ABCs (SC-B3) | IgM− IgD− HLA-DR++ CD20+ CD11c+ | 4% | 12% | 2.7×10−3 | 5.7 (1.8–22.3) |
| IgM− IgD− HLA-DR+ | 32% | 20% | 0.98 | 0.4 (0.2–1) | |
| IgA+ IgM− IgD− | 5% | 4% | 0.68 | 0.9 (0.5–1.6) | |
| Naïve B cells (SC-B1) | IgM+ IgD− | 22% | 11% | 0.97 | 0.5 (0.2–1) |
| IgM+ IgD+ CD11c− | 12% | 26% | 0.02 | 4.0 (1.3–12.0) | |
| IgM+ IgD+ CD11c+ | 4% | 7% | 0.14 | 2.2 (0.74 – 7.7) |
To validate that the protein surface markers from mass cytometry were capturing the same transcriptional populations from scRNA-seq, we isolated fibroblasts from 10 synovial tissue samples based on surface protein levels of THY1 and HLA-DR and applied bulk RNA-seq (Supplementary Fig. 6a). We trained a linear discriminant analysis (LDA) classifier on fibroblast scRNA-seq data and used it to determine the most similar scRNA-seq cluster for each bulk RNA-seq sample. The sorted THY1+HLA-DR+ fibroblast population was similar to THY1+HLA-DRAhi (SC-F2) and the THY1− HLA-DR− population was similar to THY1− (SC-F4) (Supplementary Fig. 7a–d). Genes upregulated in the sorted THY1+HLA-DR+ fibroblasts included the interleukin IL6 and the chemokine CXCL12, consistent with the scRNA-seq data.
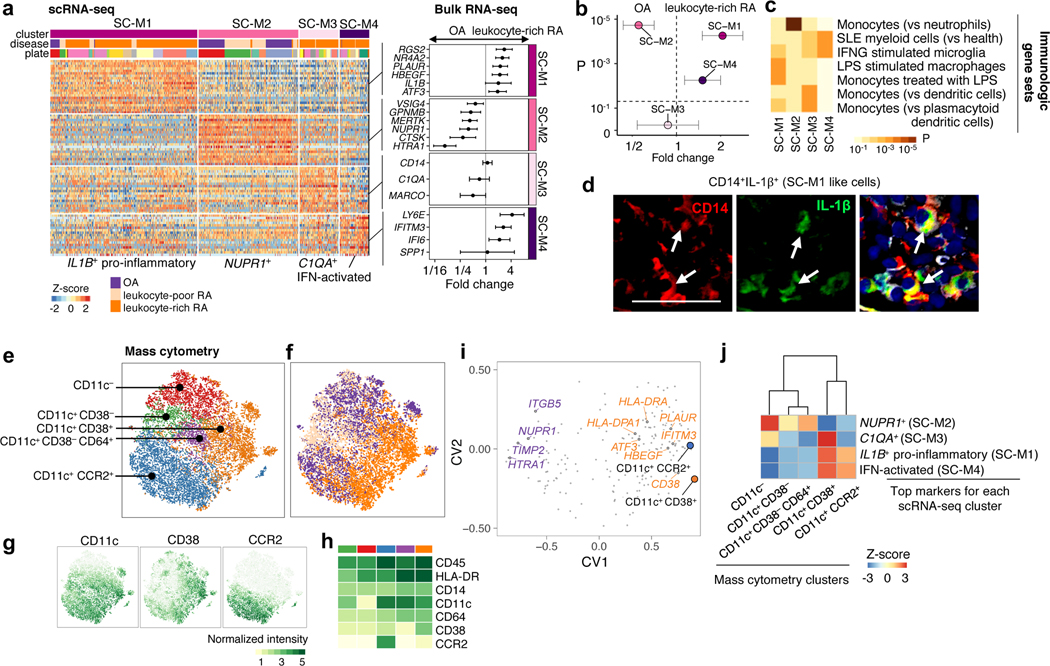
Activation states define heterogeneity among synovial monocytes
We identified four transcriptionally distinct monocyte subsets in the scRNA-seq data: IL1B+ pro-inflammatory monocytes (SC-M1), NUPR1+ monocytes (SC-M2), C1QA+ monocytes (SC-M3) and IFN-activated SPP1+ monocytes (SC-M4) (Fig. 5a). In bulk RNA-seq monocyte samples from leukocyte-rich RA and OA donors, we found that genes associated with IL1B+ monocytes (SC-M1), including NR4A2, HBEGF, PLAUR and the IFN-activated gene IFITM3 were significantly upregulated in leukocyte-rich RA samples (t-test p<1×10−4). In contrast, marker genes associated with NUPR1+ monocytes (SC-M2) were downregulated in leukocyte-rich RA relative to OA (Fig. 5a). Next, we took the average of the top marker genes (AUC>0.7) for each monocyte scRNA-seq subset and tested for differential expression of these averages in the bulk RA versus OA RNA-seq data. This analysis suggests that leukocyte-rich RA synovia have a greater abundance of IL1B+ monocytes (t-test p=6×10−5) and IFN-activated monocytes (t-test p=6×10−3), but lower abundance of NUPR1+ monocytes (t-test p=2×10−5) (Fig. 5b). These data suggest that cytokine activation drives expansion of unique monocyte populations in active RA synovia.
Figure 5.
Unique activation states define synovial monocytes heterogeneity. a. scRNA-seq analysis identified four subsets: IL1B+ pro-inflammatory monocytes (SC-M1), NUPR1+ monocytes (SC-M2) with a mixture of leukocyte-poor RA and OA cells, C1QA+ (SC-M3), and IFN-activated monocytes (SC-M4). Differential analysis by bulk RNA-seq on leukocyte-rich RA samples (n = 17) and OA samples (n = 13) revealed upregulation/downregulation of cluster marker genes. Effect sizes with 95% CI are given. b. By querying the bulk RNA-seq, we found scRNA-seq cluster IL1B+ pro-inflammatory monocytes (two-sided Student’s t-test P=6×10−5, t-value=4.56, df =26.33) and IFN-activated monocytes (two-sided Student’s t-test P=6×10−3, t-value=3.28, df =23.68) are upregulated in leukocyte-rich RA (n = 17) compared to OA (n = 13), while SC-M2 is depleted (two-sided Student’s t-test P=2×10−5, t-value=−5.62, df=26.81) in leukocyte-rich RA. Error bars indicate mean and 95% CI. c. Pathway enrichment analysis indicates the potential pathways for each subset. Two-sided Kolmogorov-Smirnov test with 105 times permutation was performed; Benjamini-Hochberg was used to control the FDR of multiple tests. The standard names for the immunological gene sets from up to bottom are: Genes down-regulated in neutrophils versus monocytes (GSE22886); Genes down-regulated in healthy myeloid cells versus SLE myeloid cells (GSE10325); Genes down-regulated in control microglia cells versus those 24 h after stimulation with IFNG (GSE1432); Genes down-regulated in unstimulated macrophage cells versus macrophage cells stimulated with LPS (GSE14769); Genes up-regulated monocytes treated with LPS versus monocytes treated with control IgG (GSE9988); Genes up-regulated in monocytes versus myeloid dendritic cells (mDC) (GSE29618); Genes up-regulated in monocytes versus plasmacytoid dendritic cells (pDC) (GSE29618). d. Detection of pro-inflammatory IL-1β in inflamed synovium by multicolor immunofluorescent staining with antibodies CD14 (red), IL-1β (green), and counterstained with DAPI (blue) identified CD14+IL-1β+ cells (white arrow). The experiment was repeated > 5 times with staining of 6 independent leukocyte-rich RA samples with similar results. Image was acquired at 200 magnification. Scale bar is 50 μm. e–f. Identified subpopulations from monocytes (n = 15,298) and disease status from 6 leukocyte-rich RA, 9 leukocyte-poor RA, and 11 OA by mass cytometry on the same gating with scRNA-seq. g-h. Normalized intensity of distinct protein markers by tSNE visualization and averaged for each cluster in heatmap. i. Integration of identified mass cytometry clusters with bulk RNA-seq reveals genes that are associated with CD11c+CD38+ and CD11c+CCR2+, like IFITM3, CD38, HBEGF, ATF3, and HLA+ genes. j. Integration of mass cytometry clusters and scRNA-seq clusters revealed that CD11c+CD38+ by mass cytometry are significantly associated with IL1B+ pro-inflammatory (SC-M1) monocytes.
With GSEA, we tested MSigDB immunologic gene sets and found IL1B+ monocytes (SC-M1) have relatively high expression levels of genes defining the LPS response in monocytes and macrophages (Fig. 5b). This suggests IL1B+ monocytes (SC-M1) are similar to TLR-activated IL-1-producing pro-inflammatory monocytes. Among Gene Ontology gene sets, we found SPP1+ monocytes (SC-M4) express genes induced by type I and II IFN (Supplementary Fig. 8a), including IFITM3 and IFI6 (Fig. 5a). The transcriptional profiles of monocytes in SC-M2 and SC-M3 do not align with known activation states, possibly indicating that these clusters represent cell phenotypes tailored to the unique homeostatic needs of the synovium. Immunofluorescence staining confirmed the presence of CD14 and IL-1β positive cells in 6 tissue samples, consistent with an enrichment of the IL1B+ pro-inflammatory monocytes (SC-M1) phenotype in RA synovium (Fig. 5d, Supplementary Fig. 9a,b).
In the mass cytometry data, we identified five CD14+ monocyte clusters (Fig. 5e–h, Supplementary Fig. 3c). Using CCA to integrate mass cytometry and bulk RNA-seq data, we found that samples with a greater abundance of CD11c+CCR2+ and CD11c+CD38+ using mass cytometry also had a higher expression of IFITM3, PLAUR, CD38, and HLA genes (Fig. 5i). This was consistent with a correspondence between the CD11c+CD38+ mass cytometry cluster and the activated monocyte scRNA-seq cluster IL1B+ (SC-M1) and SPP1+ (SC-M4) (z-score=2.3 and 2.3, respectively) (Fig. 5j, Table 1). Supporting this finding, we confirmed that CD11c+CD38+ monocytes are significantly expanded in leukocyte-rich RA (OR = 7.8 (95% CI: 3.6–17.2), one-sided MASC p=6.7×10−5) (Table 1). Conversely, NUPR1+ monocytes (SC-M2) correspond to CD11c− monocytes in mass cytometry and are inversely correlated with inflammatory monocyte populations (z-score=2.7) (Fig. 5j, Table 1).
To confirm that putative populations from mass cytometry correspond to those identified by scRNA-seq clusters, we sorted CD14+ monocytes from 4 synovial tissue samples using CD11c and CD38 protein markers and assayed them with RNA-seq (Supplementary Fig. 6c). Importantly, we found that CD14+ synovial cells had high expression of both CD11c and CD38 particularly in the RA samples. The CD14+CD11c+++CD38+++ and CD14+CD11c+CD38− sorted cells were consistent with IL1B+ pro-inflammatory (SC-M1) and NUPR1+ (SC-M2) cells, respectively (Supplementary Fig. 7e–h). These data, alongside the mass cytometry data, support the findings of greater abundance of IL1B+ pro-inflammatory (SC-M1) monocytes and lower abundance of NUPR1+ (SC-M2) monocytes in leukocyte-rich RA samples.
Heterogeneity in synovial CD4 and CD8 T cells defined by effector functions
We found three CD4+ and three CD8+ T cell subsets in the scRNA-seq data (Fig. 6a). CCR7+ T cells (SC-T1) expressed genes in the MSigDB immunologic gene set for central memory T cells (Fig. 6a, c). The two other CD4+ populations, FOXP3+ Treg cells and PDCD1+ Tph and Tfh cells, were marked by high expression of FOXP3 (SC-T2) and CXCL13 (SC-T3) by examining differentially expressed genes between these two clusters18 (Supplementary Fig. 8c). CXCL13, a chemokine expressed by Tph cells, was upregulated in bulk-sorted T cells (CD45+CD14−CD3+) from leukocyte-rich RA compared to OA (t-test p=1.2×10−4) (Fig. 6a). We found that the average of marker genes for Tph and Tfh cells (SC-T3) (AUC>0.7) was higher in leukocyte-rich RA than OA samples (t-test p=0.01) (Fig. 6b), suggesting greater abundance of Tph and activated T cells in RA than OA. We identified three CD8 T cell subsets characterized by distinct expression patterns of effector molecules GZMK, GZMB, GZMA and GNLY (Fig. 6a). We defined these populations as GZMK+ (SC-T4), GNLY+GZMB+ cytotoxic T lymphocytes (CTLs) (SC-T5), and GZMK+GZMB+ T cells (SC-T6). GZMK+GZMB+ T cells (SC-T6) also expressed HLA-DPA1 and HLA-DRB1, and other genes suggestive of an effector phenotype (Fig. 6a,c).
Figure 6.
Synovial T cells display heterogeneous CD4 and CD8 T cell subpopulations in RA synovium. a. scRNA-seq analysis identified three CD4+ subsets: CCR7+ (SC-T1), Treg cells (SC-T2), and Tph and Tfh (SC-T3); and three CD8+ subsets: GZMK+ (SC-T4), CTLs (SC-T5), and GZMK+GZMB+ (SC-T6). Differential expression analysis on leukocyte-rich RA (n = 18) comparing with OA (n = 13) on sorted T cell bulk RNA-seq samples revealed that CXCL13 is most significantly enriched in leukocyte-rich RA compared to OA. Effect sizes with 95% CI are given. b. Disease association of scRNA-seq clusters by aggregating top markers (AUC>0.7) by comparing leukocyte-rich RA (n = 18) with OA (n = 13) using bulk RNA-seq. Tph and Tfh cells (SC-T4) are upregulated (two-sided Student’s t-test p=0.01, t-value=2.73, df =29.00) in leukocyte-rich RA. Error bars indicate mean and 95% CI. c. Pathway analysis based on immunologic gene set enrichment indicates the potential enriched T cell states pathways. Two-sided Kolmogorov-Smirnov test with 105 times permutation was performed; Benjamini-Hochberg was used to control the FDR of multiple tests. The brief description of the standard names from up to bottom are: Genes up-regulated in CD4 high cells from thymus: Treg versus T conv (GSE42021); Genes up-regulated in comparison of effector CD8 T cells versus memory CD8 T cells (GOLDRATH); Genes down-regulated in comparison of effector memory T cells versus central memory T cells from peripheral blood mononuclear cells (PBMC) (GSE11057); Genes up-regulated in comparison of effective memory CD4 T cells versus Th1 cells (GSE3982); Genes up-regulated in comparison of T follicular helper (Tfh) cells versus Th17 cells (GSE11924). d. Detection of CD3+CD8+IFNγ+ (white arrow) in inflamed RA synovium by multicolor immunofluorescent staining with antibodies CD3 (green), CD8 (red), IFNγ (white), and counterstained with DAPI (blue). The experiment was repeated > 5 times with staining of 6 independent leukocyte-rich RA samples with similar results. Image was acquired at 200 magnification. Scale bar is 50 μm. e-f. Identified subpopulations from T cells (n = 19,985) and disease status from 6 leukocyte-rich RA, 9 leukocyte-poor RA, and 11 OA by mass cytometry. g-h. Distinct patterns of protein markers by tSNE and heatmap that define these clusters. i. Integration of identified mass cytometry clusters with bulk RNA-seq using CCA reveals bulk genes that are associated with CD4+PD-1+ICOS+ and CD8+PD-1−HLA-DR+ by mass cytometry. j. Integration of mass cytometry clusters with scRNA-seq clusters on the top markers (AUC>0.7) for each scRNA-seq cluster in the top 10 canonical variates. Z-score based on permutation test reveals that CD4+PD-1+ICOS+ and CD8+PD-1+HLA-DR+ by mass cytometry are highly associated with Tph and Tfh (SC-T3) by scRNA-seq; CD8+PD-1−HLA-DR+ T cells by mass cytometry are highly associated with CD8+ T cells (SC-T4, SC-T5, and SC-T6).
To confirm these findings, we applied intracellular staining to tissues from RA samples and RNA-seq to sorted CD8 T cells. Intracellular staining of GZMK and GZMB proteins in disaggregated tissue samples from patients with RA revealed that the majority of CD8 T cells in synovial tissue express GZMK (Supplementary Fig. 10a). Furthermore, we found that most HLA-DR+ CD8 T cells express both GZMB and GZMK by intracellular protein staining (Supplementary Fig. 10b). In a comparison of 7 synovial tissue samples, CD8 T cells had higher proportion of IFNγ+ cells than CD4 T cells from the same sample (Supplementary Fig. 10c,d). We also applied immunofluorescence to 6 synovial tissue samples and found that IFNγ+CD3+CD8+ T cells were more frequent in RA than OA (Fig. 6d, Supplementary Fig. 9c,d). Overall, these results closely mirror the findings from the scRNA-seq clusters.
Using mass cytometry, we identified nine putative T cell clusters among the synovial T cells (CD45+CD14−CD3+) (Fig. 6e–h, Supplementary Fig. 3d). By integrating bulk RNA-seq with mass cytometry cluster abundances, we found that higher gene expression of CXCL13 and inhibitory receptors TIGIT and CTLA4 was associated with greater abundance of the CD4+PD-1+ICOS+ mass cytometry cluster. Greater abundance of CD8+ PD-1−HLA-DR+ cells was associated with greater expression of IFNG (Fig. 6i). We found correspondence between Tph and Tfh cells (SC-T3) and CD4+PD-1+ICOS+ T cells (z-score = 3.4). CD8+ subsets including GZMK+GZMB+ (SC-T6), CTLs (SC-T5), and GZMK+ (SC-T4) tracked with CD8+PD-1−HLA-DR+ T cells by mass cytometry (Fig. 6j, Table 1). In addition, CD4+PD-1+ICOS+ cells were significantly overabundant in leukocyte-rich RA (MASC OR = 3 (95% CI: 1.7–5.2), one-sided MASC p=2.7×10−4) (Table 1).
Autoimmune-associated B cells expanded in RA synovium by single-cell RNA-seq
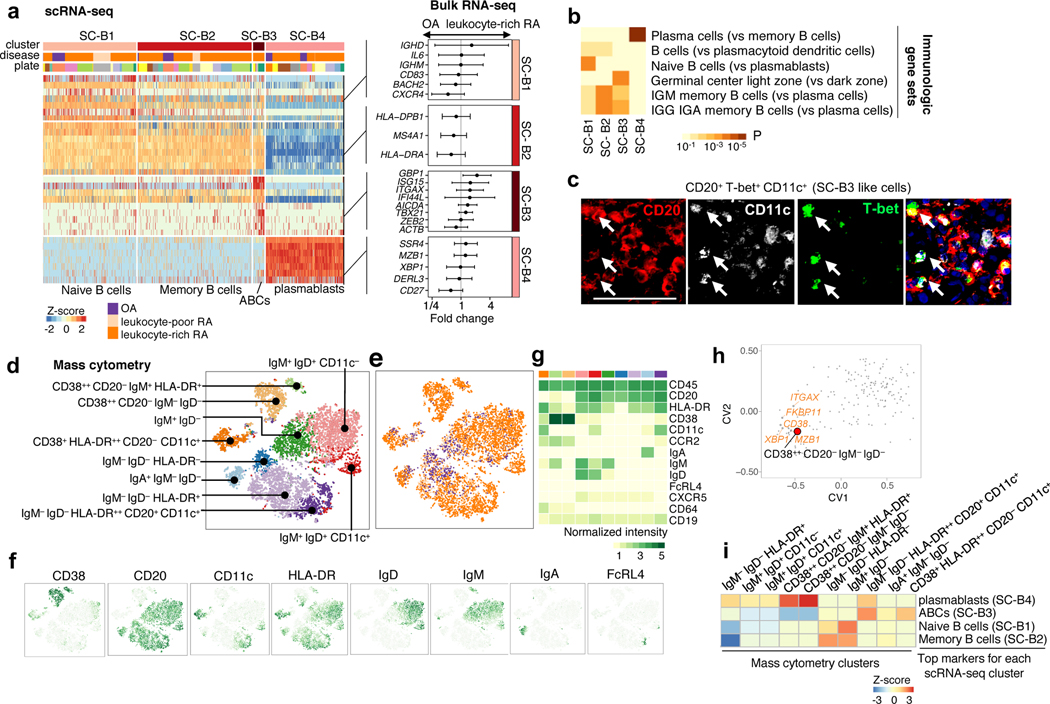
We identified four synovial B cell clusters with scRNA-seq: naive B cells (SC-B1), memory B cells (SC-B2), ITGAX+ ABC cells (SC-B3), and plasmablasts (SC-B4) (Fig. 7a). GSEA with Gene Ontology pathways suggested that SC-B1, SC-B2, and SC-B3 clusters represent activated B cells (Supplementary Fig. 8b). GSEA with MSigDB immunological gene sets revealed that SC-B1 cells express naive B cell genes, while SC-B2 and SC-B3 cells express IgM and IgG memory B cell genes (Fig. 7b). SC-B3 cells express high levels of ITGAX and TBX21 (T-bet), which are markers of autoimmunity-associated B cells (Fig. 3f and Fig. 7a)28,29, as well as markers of recently activated B cells including ACTB30. High expression of AICDA is consistent with the recently reported transcriptomic analysis of CD11c+ B cells from SLE peripheral blood31. Interferon stimulated genes (GBP1 and ISG15) are also expressed in ABCs (SC-B3) and upregulated in leukocyte-rich RA (Fig. 7a). While ABCs (SC-B3) constitute a relatively small proportion of all B cells, they are almost exclusively derived from two patients with leukocyte-rich RA (Fig. 3b). To confirm the presence of ABCs in human tissues, we applied immunofluorescence staining to 6 synovial tissue samples. RA synovium had increased numbers of CD20+T-bet+ CD11c+ B cells compared to OA synovium. Specifically, we observed ABC cells in tissue sections from the same inflamed tissue samples that had a high proportion of ABCs by scRNA-seq analysis (Fig. 7c, Supplementary Fig. 9e, f).
Figure 7.
Synovial B cells display heterogeneous subpopulations in RA synovium. a. scRNA-seq analysis identified naive B cells (SC-B1), memory B cells (SC-B2), autoimmune-associated B cells (ABCs) (SC-B3), and plasmablasts (SC-B4). Differential expression analysis is given by comparing leukocyte-rich RA (n = 16) with OA (n = 7) using bulk RNA-seq B cell samples. Effect size with 95% CI are given. b. Pathway enrichment analysis using immunologic gene sets indicates the distinct enriched pathways for each scRNA-seq cluster. Two-sided Kolmogorov-Smirnov test with 105 times permutation was performed; Benjamini-Hochberg was used to control the FDR of multiple tests. The standard names for the immunological gene sets from up to bottom are: Genes up-regulated in plasma cells versus memory B cells (GSE12366); Genes up-regulated in comparison of B cells versus plasmacytoid dendritic cells (pDC) (GSE29618); Genes up-regulated in B lymphocytes: naive versus plasmablasts (GSE42724); Genes up-regulated in B lymphocytes: human germinal center light zone versus dark zone (GSE38697); Genes up-regulated in comparison of memory IgM B cells versus plasma cells from bone marrow and blood (GSE22886); Genes up-regulated in comparison of memory IGG and IGA B cells versus plasma cells from bone marrow and blood (GSE22886). c. Detection of CD20+T-bet+CD11c+ (white arrow) in inflamed synovium by multicolor immunofluorescence. Immunofluorescent staining with antibodies CD20 (red), CD11c (white), T-bet (green), and counterstained with DAPI (blue). The experiment was repeated > 5 times with staining of 6 independent leukocyte-rich RA samples with similar results. Image was acquired at 200 magnification. Scale bar is 50 μm. d-e. Identified subpopulations of B cells (n = 8,179) and disease status from 6 leukocyte-rich RA, 9 leukocyte-poor RA, and 8 OA by mass cytometry. f-g. Distinct expression patterns of protein markers by tSNE and averaged for each cluster in heatmap. h. Integrating mass cytometry clusters with bulk RNA-seq data using CCA shows that CD38+CD20−Ig− (plasmablasts) population is highly associated with gene expression of plasma cells makers, like XBP1. i. Integration of mass cytometry clusters with scRNA-seq clusters suggested that CD38++CD20−IgM+HLA-DR+ and CD38++CD20−IgM−IgD− are significantly associated with plasmablast (SC-B4); IgM−IgD−HLA-DR++CD20+CD11c+ B cells are associated with ABCs (SC-B3).
We identified 10 putative B cell clusters in the mass cytometry data (CD45+CD3−CD14−CD19+) (Fig. 7d–g, Supplementary Fig. 3e). CCA analysis showed that samples with higher gene expression of CD38, MZB1, and plasma cell differentiation factor XBP1 had greater abundance of CD38++CD20−IgM−IgD− plasmablasts (Fig. 7h). Plasmablasts (SC-B4) corresponded with CD38++CD20−IgM−IgD− B cells (z-score=2.7) (Fig. 7i, Table 1). ABCs (SC-B3) corresponded with the IgM− IgD− HLA-DR++ CD20+ CD11c+ mass cytometry cluster (z-score=1.6), which is significantly overabundant in leukocyte-rich RA (OR = 5.7 (95% CI: 1.8–22.3), one-sided MASC p=2.7×10−3) (Fig. 7i, Table 1). Mass cytometry analysis further identified three putative subsets within CD11c+ cells: IgM−IgD−HLA-DR++CD20+CD11c+, CD38+HLA-DR++CD20−CD11c+, and IgM+IgD+CD11c+, which is suggestive of additional heterogeneity within ABCs.
To demonstrate that CD19+CD11c+ cells by surface protein markers correspond to SC-B3 (ABCs), we flow-sorted CD19+CD11c+ cells from an independent cohort of 6 RA synovial samples and applied RNA-seq (Supplementary Fig. 6b). We show that these RNA-seq profiles are most consistent with ABC cells (Supplementary Fig. 7i–k). In these sorted samples, we found more putative marker genes (e.g. ZEB2 and CIITA) and interferon-induced genes (IFITM3 and IFI27) for the ABC population (Supplementary Fig. 7l).
Inflammatory pathways and effector modules revealed by global single cell profiling
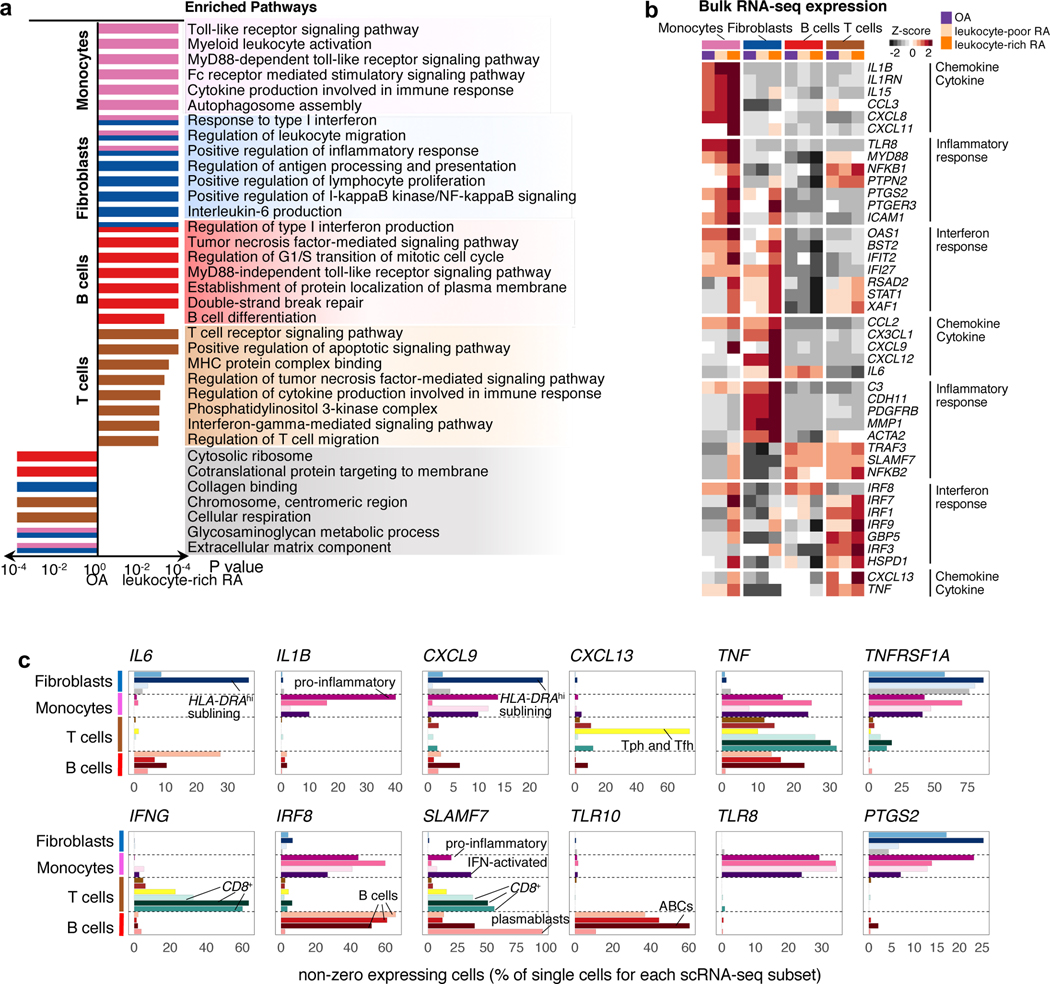
We used bulk and single cell transcriptomes of sorted synovial cells to examine pathologic molecular signal pathways. First, principal component analysis (PCA) on post-QC OA and RA bulk RNA-seq samples (Supplementary Fig. 11a,b) showed that cell type accounted for most of the data variance. Each cell type expressed specific marker genes, PDGFRA for fibroblasts, C1QA for monocytes, CD3D for T cells, and CD19 for B cells (Supplementary Fig. 11c). Within each cell type, PCA showed that leukocyte-rich RA samples separated from OA and leukocyte-poor RA samples (Supplementary Fig. 11d–g). Differential gene expression analysis between leukocyte-rich RA and OA (FC>2 and FDR<0.01) revealed genes upregulated in leukocyte-rich RA tissues: 173 in fibroblasts, 159 in monocytes, 10 in T cells, and 5 in B cells. To define the pathways relevant to leukocyte-rich RA, we used GSEA weighted by gene effect sizes on Gene Ontology pathways and identified type I interferon response and inflammatory response (monocytes and fibroblasts) (Supplementary Fig. 11h–i), Fc receptor signaling (monocytes), NF-kappa B signaling (fibroblasts), and interferon gamma (T cells) (Fig. 8a). Leukocyte-rich RA samples had significantly higher expression of some genes in fibroblasts and monocytes: inflammatory response genes (PTGS2, PTGER3, and ICAM1), interferon response genes (IFIT2, RSAD2, STAT1, and XAF1), and chemokine or cytokine genes (CCL2 and CXCL9) (Fig. 8b), consistent with a coordinated chemotactic response to interferon activation. T cells had upregulation of interferon regulatory factors (IRFs), including IRF7 and IRF9, and monocytes had upregulation of IRF7, IRF8 and IRF9. Taken together, pathway analysis suggests crosstalk between immune and stromal cells in leukocyte-rich RA synovia. Inflammatory response genes upregulated in leukocyte-rich RA had comparable expression levels between leukocyte-poor RA and OA synovial cells (Fig. 8b)
Figure 8.
Transcriptomic profiling of synovial cells reveals upregulation of inflammatory pathways in RA synovium. a. Pathway enrichment using bulk RNA-seq identified shared and unique inflammatory response pathways for each cell type. Two-sided Kolmogorov-Smirnov test with 105 permutations was performed on 18 leukocyte-rich RA, 17 leukocyte-poor RA, and 14 OA. b. Bulk RNA-seq profiling of genes obtained from the significantly enriched pathways from (a) shows the averaged gene expression from each group (18 leukocyte-rich RA, 17 leukocyte-poor RA, and 14 OA) normalized across all cell type samples. c. scRNA-seq profiling resolved that inflammatory cytokines/chemokines, interferon responsive, and inflammatory responsive genes were driven by a global upregulation within a synovial cell type or discrete cell states.
Next, we asked whether inflammatory cytokines upregulated in leukocyte-rich RA are driven by global upregulation within a single synovial cell type, or specific upregulation within a discrete cell subset defined by scRNA-seq. Whereas TNF was produced at a high level by multiple monocyte, B cell and T cell populations; IL6 expression was restricted to HLA-DRAhi sublining fibroblasts (SC-F2) and a subset of B cells (SC-B1) (Fig. 8c); CD8 T cells, rather than CD4 T cells, were the dominant source of IFNG transcription in leukocyte-rich synovia.
We also observed cell subset-specific responses to inflammatory pathways. Toll-like receptor signaling pathway was enriched in B cells and monocytes in leukocyte-rich RA tissues (Fig. 8a). At the single cell level, we observed that TLR10 was only expressed by activated B cells, indicating that TLR10 has a functional role within the B cell lineage. In contrast, TLR8 was elevated in all RA monocyte subsets. The hematopoietic cell-specific transcription factor IRF8 was expressed in a significant fraction of monocytes and B cells that cooperatively regulate differentiation of monocytes and activated B cells in RA synovium. SLAMF7 is highly expressed by pro-inflammatory monocytes (SC-M1), IFN-activated monocytes (SC-M4), CD8 T cells, and plasmablasts (SC-B4).
Furthermore, mass cytometry analysis across all identified cell clusters revealed that leukocyte-rich RA patients show high cell abundances of HLA-DRhi fibroblast populations, Tph cells, CD11c+CD14+ monocytes, and CD11c+ B cell populations (Supplementary Fig. 3f).
DISCUSSION
Using multi-model, high-dimensional synovial tissue data we defined stromal and immune cell populations overabundant in RA and described their transcriptional contributions to essential inflammatory pathways. Recognizing the considerable variation in disease duration and activity, treatment types, and joint histology scores32, we elected to use a molecular parameter, based on percent leukocytes of the total cellularity, to classify our samples at the local tissue level. We note that differences in leukocyte enrichment of joint replacement samples and biopsy samples were best explained by leukocyte infiltration and not by the histological scores (Supplementary Fig. 1, Supplementary Fig. 11d–g).
This study and a previous study33 have highlighted sublining fibroblasts as a potential therapeutic target in RA. Sublining fibroblasts are a major source of pro-inflammatory cytokines such as IL6 (Fig. 4), and a specific subset of sublining fibroblasts expressing MHC II (SC-F2, THY1+CD34-HLA-DRhi) was >15 fold expanded in RA tissues. Further studies are needed to define molecular mechanisms that regulate sublining fibroblast expansion in RA. T cells, B cells, and monocyte proportions track with expression of individual fibroblast genes (Supplementary Fig. 11j). We found DNASE1L3, a gene whose loss of function is associated with RA34 and systemic lupus erythematosus35 to be highly expressed in CD55+ lining fibroblasts (SC-F4) (Fig. 4a). We identified a novel fibroblast subset (SC-F3) with high expression of DKK3+ (Fig. 4), encoding Dickkopf3, a protein upregulated in OA that prevents cartilage degradation in vitro36.
Transcriptional heterogeneity in the synovial monocytes indicated that distinct RA-enriched subsets are driven by inflammatory cytokines and interferons (Fig. 5). This suggests monocytes may be differentially polarized by unique cytokine combinations in local microenvironments. These newly identified inflammatory phenotypes align with RA therapeutic targets, including anti-TNF therapies and interferon pathway JAK kinase inhibitors37. The NUPR1+ (SC-M2) monocytes were inversely correlated with tissue inflammation, and expressed high levels of monocyte tissue remodeling factors such as MERTK (Fig. 5)38. Alternatively, NUPR1+ markers such as osteoactivin (GPNMB) and cathepsin K (CTSK) may indicate a subset of osteoclast progenitors that control bone remodeling (Fig. 5)37,39. Furthermore, spatial studies—particularly focused on lining versus sublining, perivascular and lymphocyte aggregate-associated monocytes—will help understand the functional roles of these subsets.
Single cell classification of T cell subsets in RA synovium demonstrated CD4+ T cell heterogeneity that is consistent with distinction between the homing capacity and effector functions of these subsets. Consistent with previous studies, we observed expansion of PDCD1+CD4+ Tph cells (SC-T3) within leukocyte-rich RA. We also found CD8 T cell subsets (SC-T4–6) characterized by a distinct granzyme expression pattern (Fig. 6a). A larger study may be better powered to differentiate the relative expansion of individual subpopulations.
A critical unmet need in RA is identifying therapeutic targets for patients failing to respond to disease-modifying antirheumatic drugs (DMARDs)41. We observed upregulation of chemokines (CXCL8, CXCL9, and CXCL13), cytokines (IFNG and IL1542,43), and surface receptors (PDGFRB and SLAMF7) in distinct immune and stromal cell populations, suggesting potential novel targets. This study was enabled by advances in the statistical integration of single-cell data and our recent work optimizing robust methodologies for disaggregation of synovial tissue22.
We developed advanced strategies to integrate multiple molecular datasets by modulating technical artifact from single cell technologies44, while emphasizing biological signals. CCA has been successfully employed in other contexts to integrate high-dimensional biological data45,46. Our CCA-based strategy analyzed scRNA-seq data using canonical variates that capture variance that are present in both single-cell and bulk RNA-seq data. The shared variances likely represent biological trends, and not technical factors that would likely be uncorrelated in these two independent datasets. We further confirmed that the identified scRNA-seq clusters are well correlated with the bulk RNA-seq data and also the mass cytometry data (Supplementary Fig. 12, 13).
The two single cell modalities used in this study, mass cytometry and scRNA-seq, complement each other. Single-cell RNA-seq captures expression of thousands of genes, but at the cost of sparse data47. Mass cytometry captures hundreds of thousands of individual cells, but measures a limited number (~40)48 of pre-selected markers. However, since markers are backed with decades of experimental experience they can be effective at defining cellular heterogeneity49. To make the analysis consistent, we gated mass cytometry cells on the same markers upon which the scRNA-seq was gated. Combining mass cytometry with the extended dimensionality of scRNA-seq enables quantification of well-established cell populations and discovery of novel cell states, such as the CD8 T cell states noted here. As an ongoing AMP phase 2 study, we are examining larger numbers of ungated cell populations from ~100 synovial tissue patients with RA by capturing mRNA and protein expression simultaneously50 with detailed clinical data and ultrasound score evaluation of synovitis. We anticipate that this larger study will enable us to not only discover additional subpopulations, but to better define their link to clinical sub-phenotypes.
It is essential to interrogate the tissue infiltration of diseases other than RA, including SLE, type I diabetes, psoriasis, multiple sclerosis and other organ targeting conditions. Application of multiple single-cell technologies together can help define key novel populations, thereby providing new insights about etiology and potential therapies.
Methods
Study design and patient recruitment
The study was performed in accordance with protocols approved by the institutional review board. A multicenter, cross-sectional study of individuals undergoing elective surgical procedures and a prospective observational study of synovial biopsy specimens from patients with RA ≥ age 18, with at least one inflamed joint, recruited from 10 contributing sites in the network. Synovial tissues were obtained from joint replacement procedures or ultrasound-guided biopsies, followed by cryopreservation in cryopreservation media Cryostor CS10 (Sigma-Aldrich) and transit to a central technology site.
Histological assessment of synovial tissue and quality control
Synovial tissue quality and grading of synovitis were evaluated in formalin-fixed, paraffin-embedded sections by histologic analysis (H and E staining). Specimens were identified as synovium by the presence of a lining layer or by characteristic histologic features of synovium, including the presence of loose fibrovascular or fatty tissue lacking a lining layer. Samples consisting of dense fibrous tissue, joint capsule or other tissues were determined not to be synovium. For each histological and molecular analysis, we generated pooled data from 6–8 separate fragments from different sites in the same joint. Thus, this should be representative of the whole tissue and mitigate much of the biopsy site-to-site variability. Krenn lining scores (0–3) and inflammation scores (0–3) for each tissue sample were determined independently by three pathologists25.
Tissue disaggregation for mass cytometry and RNA-sequencing
For pipeline analysis, synovial tissue samples stored in cryovials were disaggregated into single cell suspension as describe. Briefly, synovial tissue fragments were separated mechanically and enzymatically in digestion buffer (Liberase TL (Sigma-Aldrich) 100 ug/mL and DNase I (New England Biolabs) 100 ug/ml in RPMI) in a 37°C water bath for 30 minutes. Single cell suspensions from disaggregated synovial tissues were assessed for cell quantity and cell viability by trypan Blue. For samples with more than 200,000 viable synovial cells, 50% of all synovial cells were allocated for analysis by mass cytometry and the remaining cells were allocated for RNA-seq. For samples with less than 200,000 viable synovial cells, all synovial cells were utilized for RNA-seq analysis.
Synovial cell sorting strategy for RNA sequencing
Synovial T cells, B cells, monocytes, and fibroblasts were isolated from disaggregated synovial tissue, as described22. Briefly, disaggregated synovial cells were stained with antibodies against CD45 (HI30), CD90 (5E10), podoplanin (NZ1.3), CD3 (UCHT1), CD19 (HIB19), CD14 (M5E2), CD34 (4H11), CD4 (RPA-T4), CD8 (SK1), CD31 (WM59), CD27 (M-T271), CD235a (KC16), using human TruStain FcX in 1% BSA in Hepes-Buffered Saline (HBS,20 mM HEPES, 137 mM NaCl, 3mM Kcl, 1mM CaCl2) for 30 minutes. 1000 viable (PI-) T cells (CD45+, CD3+, CD14−), monocytes (CD45+, CD3−, CD14+), B cells (CD45+, CD3−, CD14−, CD19+), and synovial fibroblasts (CD45−, CD31−, PDPN+) were collected by fluorescence-activated cell sorting (BD FACSAria Fusion) directly in buffer RLT (Qiagen) for bulk RNA-seq. For single cell RNA-seq, live cells of each population were re-sorted into 384-well plates single cells with a maximum of 144 cells for each cell type, per patient sample.
Flow sorting strategy for bulk RNA-seq experimental validation
For bulk RNA-seq validation experiments, RA and OA synovial tissue were disaggregated and synovial cells were stained with cell-type specific antibody panels. For each cell subset, up to 1000 cells were collected directly into buffer TCL (Qiagen). Antibody panels used to define cell subsets are fibroblasts: CD90 (5E10), podoplanin (NZ1.3), HLA-DR (G46–6); B cell subsets: HLA-DR (G46–6), CD11c (3.9), CD19 (SJ25C1), CD27 (M-T271), IgD (IA6–2), CD3 (UCHT1), CD14 (M5E2), CD38 (HIT2); Monocyte subsets: CD14-BV421 (M5E2), CD38-APC (HB-7), and CD11c-PECy7 (B-ly6). Immediately prior to sorting, DAPI or LIVE/DEAD viability dye was added to cell suspensions and cells were passed through a 100μm filter. Synovial cell subsets were sorted based on flow cytometry gating schema shown in Supplementary Fig. 6. In all, we sorted THY1− DR− populations from 4 OA samples, THY1+DR− population from 4 OA and 6 RA samples, and THY1+ DR+ population from 6 RA samples. For monocytes, we sorted CD14+CD11c+++CD38+++ population from 2 RA samples and CD14+CD11c+ CD38− population from 2 OA samples. For B cells, we sorted CD11c−IgD−CD27+ population from 6 RA samples, CD11c−IgD+CD27− population from 3 RA samples, CD19+CD11c+ population from 3 RA samples, and plasma cells from 3 RA samples.
To validate the identified single-cell populations using bulk RNA-seq, we fit an LDA (Linear Discriminant Analysis) classifier on the scRNA-seq cell clusters and then classified each flow sorted bulk RNA-seq sample. For each cell type, 1) we trained an LDA model on the scRNA-seq clusters with the top 500 marker genes for each cluster; 2) Next, we applied this LDA model to classify each sample of bulk sorted cells and estimated the maximum posterior probability for each sample. In summary, we tested if we could sort new cells from new, independent samples and see the same gene expression profiles in the new bulk samples as the original scRNA-seq samples.
Multicolor immunofluorescent staining of paraffin synovial tissue
Briefly, 5 mm thick formalin fixed paraffin sections were incubated in a 60°C oven to melt paraffin. Slides were quickly transferred to xylenes to completely dissolve the paraffin and after 5 minutes transferred to absolute ethanol. Slides were left in absolute ethanol for 5 minutes and then transferred to 95% ethanol. At the end of the 5 minutes immersion in 95% ethanol, slides were rinsed several times with distilled water and transfer to a plastic coplin jar filled with 1× DAKO retrieval solution (S1699, Dakocytomation). Antigens were unmasked by immersing of plastic coplin jar in boiling water for 30 minutes. Slides were let cool down for 10 minutes at room temperature and washed several times with distilled water. Non-specific binding was blocked with 5% normal donkey serum (017-000-121, Jackson ImmunoResearch Laboratories,) dissolved in PBS containing 0.1% Tween 20 and 0.1% Triton X-100. Without washing, blocking solution was removed from slides and combinations of primary antibodies were added to PBS containing 0.1% Tween 20 and 0.1% Triton X-100. Primary antibodies to detect IFNg+ T cells include goat anti-CD3 epsilon (clone M-20, Santa Cruz Biotechnology), mouse anti-human CD8 (clone 144B, GeneTex), and rabbit anti-human IFNg (Biorbyt, orb214082). To visualize ABC, we incubated slides with goat anti-human CD20 (LifeSpan Biosciences, LS-B11144), rabbit anti-Tbet (H-210, Santa Cruz Biotechnology) and biotinylated mouse anti-human CD11c (clone 118/A5, Thermo Fisher Scientific). To identify IL1B+ monocytes, we used a mixture of goat anti human CD14 (119–13402, RayBiotech) biotinylated rabbit anti-human IL1b (OABF00305-Biotin, Aviva Systems Biology) and mouse anti-human CD16 (clone DJ130c, LifeSpan Biosciences). Finally, slides were probed with rabbit monoclonal anti-human CD90 (2694–1, Epitomics), rat anti-human HLADR (cloneYE2/36 HLK, LifeSpan Biosciences) and mouse anti-human CD45 (clone F10-89-4, abcam) to detect fibroblasts, Class II expressing cells and hematopoietic cells, respectively. Slides with primary antibodies were incubated in a humid chamber at room temperature, overnight. Next morning, primary antibodies for triple T cell stain and for detecting ABC’s were revealed with Alexa Fluor 568 donkey anti-goat IgG (A-11057, Thermo Fisher Scientific), Alexa Fluor 488 donkey anti-rabbit (771-546-152, Jackson ImmunoResearch Laboratories) and Alexa fluor 647 donkey anti-mouse (715-606-151, Jackson ImmunoResearch Laboratories). Primary antibodies in the stain for monocytes were revealed with Alexa Fluor 568 donkey anti-goat Ig G, Alexa fluor 488 streptavidin (S11223, Thermo Fisher Scientific) and Alexa Fluor 647 donkey anti-mouse Ig G. Primary antibodies in the stain for fibroblasts and hematopoietic cells were detected with Cy3 donkey anti-rabbit (711-166-152, Jackson ImmunoResearch Laboratories), Alexa Fluor 488 donkey anti-rat Ig G (A-21208, Thermo Fisher Scientific) and Alexa Fluor 647 donkey anti-mouse Ig G. After 2 hours of incubation, slides were washed and mounted with Vectashield mounting media with DAPI (H-1200, Vector Laboratories). Pictures were taken with an Axioplan Zeiss microscope and recorded with a Hamamatsu camera. Double immunofluorescence pictures were obtained by merging individual channels in NIH Image J software.
Estimation of number of cells by counting nuclei
To estimate number of cells, we counted number of nuclei in 5 random 200× fields that show synovial lining with Image J NIH software. Briefly, original color TIFF files were first transformed into 8-bit grayscale images. We use similar settings to adjust threshold in 8-bit images (Lower threshold level: 0, Upper threshold level: 60). Next, we used process: binary: watershed to separate nuclei. In the analyze icon, we select analyze particles and we use equal settings to count particles in our images (Size (pixel2): 50-infinity, circularity 0.00–1.00, Show: outlines) and we selected to display results. We visually confirmed that individual nuclei were outlined in the final image and calculate the average number of cells/200× field in individual samples.
Tissue samples classification based on leukocyte infiltration
We classified RA tissue samples into leukocyte-poor RA and leukocyte-rich RA based on Mahalanobis distance from OA samples computed on leukocyte abundance measured by flow cytometry. We first took OA samples as a reference, and calculated a multivariate normal distribution of the percentages of live T cells, B cells, and monocytes. Here we used the mahalanobis function in R: data x = a matrix of all 51 samples by flow gates of T cells, B cells, and monocytes; center = mean of T cells, B cells, and monocytes for all OA samples; covariance = covariance of T cells, B cells, and monocytes for all OA samples. We calculated the square root to get Mahalanobis distance for each sample,
We then defined the maximum value of all OA samples (4.5) as a threshold to define 19 leukocyte-rich RA (>4.5) and 17 leukocyte-poor RA (<4.5) samples in our cohort (Supplementary Fig. 1d).
Bulk RNA-seq gene expression quantification
We sorted cells into the major immune and stromal cell populations: T cells, B cells, monocytes and synovial fibroblasts. We then performed RNA sequencing. Full-length cDNA and sequencing libraries were performed using Illumina Smart-eq2 protocol51. Libraries were sequenced on MiSeq from Illumina to generate 35 base paired-end reads. Reads were mapped to Ensembl version 83 transcripts using kallisto 0.42.4 and summed expression of all transcripts for each gene to get transcripts per million (TPM) for each gene.
Bulk RNA-seq quality control
For quality control of bulk RNA-seq data, we began by defining common genes as the set of genes detected with at least 1 mapped fragment in 95% of the samples. Then, for each sample, we computed the percent of common genes detected in that sample. Low quality samples are those that have less than 99% of common genes detected, and these were discarded. We found that the low-quality samples also had low cell counts (Supplementary Fig. 11a). After discarding 25 low quality samples, we used 167 good quality samples, including 45 fibroblast samples, 46 monocyte samples, 47 T cell samples, and 29 B cell samples in all bulk RNA-seq analyses. Cell lineage markers, PDGFRA, C1QA, CD3D, and CD19, are expressed selectively by fibroblasts, monocytes, T cells, and B cells, respectively (Supplementary Fig. 11c).
Single-cell RNA-seq gene expression quantification
Single-cell RNA-seq was performed using the CEL-Seq2 method47 with the following modifications. Single cells were sorted into 384-well plates containing 0.6 μL 1% NP-40 buffer in each well. Then, 0.6 μL dNTPs (10mM each; NEB) and 5 nl of barcoded reverse transcription primer (1 μg/μL) were added to each well along with 20 nL of ERCC spike-in (diluted 1:800,000). Reactions were incubated at 65°C for 5 min, and then moved immediately to ice. Reverse transcription reactions were carried out, as previously described (Hashimshony et al., 2016), and cDNA was purified using 0.8× volumes of Agencourt RNAClean XP beads (Beckman Coulter). In vitro transcription reactions (IVT) were performed, as described followed by EXO-SAP treatment. Amplified RNA (aRNA) was fragmented at 80°C for 3 min and purified using Agencourt RNAClean XP beads (Beckman Coulter). The purified aRNA was converted to cDNA using an anchored random primer and Illumina adaptor sequences were added by PCR. The final cDNA library was purified using Agencourt RNAClean XP beads (Beckman Coulter). Paired-end sequencing was performed on the HiSeq 2500 in High Output Run Mode with a 5% PhiX spike-in using 15 bases for Read 1, 6 bases for the Illumina barcode and 36 bases for Read 2. We mapped Read2 to human reference genome hg19 using STAR 2.5.2b, and removed samples with outlier performance using Picard. We quantified gene levels by counting UMIs (Unique Molecular Identifiers) and transforming the counts to Log2(CPM+1) (Counts Per Million).
Single-cell RNA-seq quality control
For quality control of single-cell RNA-seq data, we filtered out molecules that are likely to be contamination between cells, and we used several metrics to exclude poor quality cells. We identified molecules that are likely to represent cell-to-cell cross-contamination as follows. Many single-cell RNA-seq library preparation protocols include pooling and amplification of cDNA molecules from a large number of cells. This can introduce cell-to-cell contamination. We found that molecules represented by a small number of reads are more likely to be contaminant molecules derived from other cells. We developed a simple algorithm to set a threshold for the minimum number of reads per molecule, and we ran it separately for each quadrant of 96 wells in each 384-well plate. We used 2 marker genes expected to be exclusively expressed in each of the 4 cell types: PDGFRA and ISLR for fibroblasts, CD2 and CD3D for T cells, CD79A and RALGPS2 for B cells, and CD14 and C1QA for monocytes. We counted nonzero expression of these genes in the correct cell type as a true positive and nonzero expression in the incorrect cell type as a false positive. Then we tried each threshold for reads per molecule from 1–20 and chose the threshold that maximizes the ratio of true positive to false positive (Supplementary Fig. 14). This left us with 7,127 cells and 32,391 genes. Next, we discarded cells with fewer than 1,000 genes detected with at least one fragment. We also discarded cells that had more than 25% of molecules coming from mitochondrial genes. This left us with 5,265 cells. We discarded genes that had nonzero expression in fewer than 10 cells. We show all post-QC single cells based on the number of genes detected and percent of molecules from mitochondrial genes for each identified cluster (Supplementary Fig. 15).
Mass cytometry sample processing and quality control
We collected 6 leukocyte-rich, 9 leukocyte-poor RA, and 11 OA samples for mass cytometry analysis, and processed the samples, as described previously22. Briefly, we analyzed samples on a Helios instrument (Fluidigm) after antibody staining and fixation (Supplementary Table 2). Mass cytometry data were normalized using EQ™ Four Element Calibration Beads (Fluidigm), as previously described52. Cells were first gated to live DNA+ cells prior to gating for specific cell populations using the following scheme: B cells (CD3−CD14−CD19+), fibroblasts (CD45−PDPN+), monocytes (CD3−CD14+), and T cells (CD3+CD14−). All biaxial gating was performed using FlowJo 10.0.7.
Integrative computational pipeline for scRNA-seq clustering
We developed a graph-based unbiased clustering pipeline based on canonical correlation analysis to take advantage of the shared variation between single-cell RNA-seq and bulk RNA-seq. We used this computational pipeline to analyze single cells from each cell type. The overall flowchart is shown in Supplementary Fig. 2. We describe the details of each step as follows:
We first selected the highly variable genes such that the mean and standard deviation are in the top 80% of the density distributions from the single-cell RNA-seq matrix C (g genes by m cells, c1,…,m) and bulk RNA-seq matrix (g genes by n samples, s1,…,n), respectively. We focused on the highly variable genes detected in both scRNA-seq and bulk RNA-seq datasets.
Based on the shared highly variable genes, we integrated single-cell RNA-seq with bulk RNA-seq by finding a linear projection of bulk samples and single cells such that the correlation between the genes are maximized using the CCA method53. CCA finds two vectors a and b that maximize the linear correlations cor(CVs1,CVc1) where and . Each bulk sample si gets a coefficient ai and each cell ci gets a coefficient bi. The linear combination of all samples s1,…,n arranges bulk genes along the canonical variate CVs1 and the linear combination of all cells c1,…,m arranges single-cell genes along CVc1 CCA defines the coefficients a1,…,n and b1,…,n that arrange the genes from the two datasets in such a way that the correlation between CVs1 and CVc1 is maximized. After CCA finds the first pair of canonical variates, the next pair is computed on the residuals, and so on.
We calculated the cell-to-cell similarity matrix using Euclidean distance on the top ten CCA canonical variates.
We built up a K-nearest neighbors (KNN) graph based on the cell-to-cell similarity matrix (Euclidean distance) based on local ordinal embedding (LOE), a graph embedding method. We then converted the KNN neighbor relation matrix into an adjacency matrix using the graph.adjacency function from igraph R package;
We clustered the cells using the Infomap algorithm for community detection by applying a cluster_infomap function from igraph R package to decompose the cell-to-cell adjacency matrix into major modules by minimizing a description of the information flow;
We then constructed a low dimensional embedding using tSNE based on the cell-to-cell distance matrix using the following parameters: perplexity = 50 and theta = 0.5;
We identified and prioritized significantly differentially expressed genes for each distinct cluster based on percent of non-zero expressing cells, AUC score54, and fold-change;
For pathway analysis, we downloaded gene sets from Gene Ontology (GO) terms on April 201755,56. This included 9,797 GO terms and 15,693 genes. We also used the immunological signatures from 4872 hallmark gene sets from MSigDB57 to test enrichment of all the tested genes sorted by decreased AUC scores for each cluster by 105 permutation tests55. We used the liger R package (https://github.com/JEFworks/liger) to do gene set enrichment analysis (GSEA).
To identify the most reasonable and stable clusters, we ran this pipeline repeatedly while tuning the number of top canonical variates (4, 8, 12, 16, and 20) that were incorporated for the cell-to-cell similarity matrix, and the number of k (50, 100, 150, 200, 250, and 300) to build up the K-nearest neighbors’ graph. We chose the clusters that yielded the greatest number of differentially expressed genes. We used Silhouette analysis58,59 on the cell-to-cell Euclidean distance matrix to evaluate our clustering results (Supplementary Fig. 2b). For each cell, the silhouette width s(i) is defined as follows:
where a(i) is the average dissimilarity between a cell and all the other cells in the same cluster and b(i) is the average distance between a cell and all cells in the nearest cluster to which the cell does not belong. The measure range is [−1, 1], where a value near 1 indicates a cell is far from neighboring clusters, a value near 0 indicates a cell is near a decision boundary, and a value near −1 indicates the cell is closer to a neighboring cluster than its own cluster.
Thus, for each pair of single-cell RNA-seq and bulk RNA-seq, we ran our pipeline on the shared samples that have both datasets for each cell type (Figure 1b). For integrating fibroblast data, we used 45 bulk RNA-seq samples, 1,844 single cells and 7,016 shared highly variable genes; for integrating monocyte data, we used 47 bulk RNA-seq samples, 750 single cells and 7,016 shared highly variable genes; for integrating T cell data, we used 47 bulk RNA-seq samples, 1,716 single cells and 7,003 shared highly variable genes; for integrating B cell data, we used 29 bulk RNA-seq samples, 1,142 single cells and 7,023 shared highly variable genes.
Mass cytometry clustering
We created mass cytometry datasets for analysis by concatenating cells from all individuals for each cell type. For donors with more than 1,000 cells, we randomly selected 1,000 cells to ensure that samples were equally represented. In this way, we created downsampled datasets of 25,161 fibroblasts from 23 patients, 15,298 monocytes from 26 patients, 19,985 T cells from 26 patients, and 8,179 B cells from 23 patients for analysis. We then applied the tSNE algorithm (Barnes-Hut implementation) to each dataset using the following parameters: perplexity = 30 and theta = 0.5. We used all markers except those used to gate each population in the SNE clustering. To identify high-dimensional populations, we used a modified version of DensVM23. DensVM performs kernel density estimation across the dimensionally reduced SNE map to build a training set, then assigns cells to clusters by their expression of all markers using an SVM classifier. We modified the DensVM code to increase the range of potential bandwidths searched during the density estimation step and to return the SVM model generated from the tSNE projection. We summarized the details of the clusters with proportion of cells from each disease cohort in Supplementary Table 3.
Disease association test of cell populations
We tested whether abundances of individual populations were altered in RA case samples compared to OA controls using two ways. First, we assessed whether marker genes (AUC > 0.7, 20 < n < 100) characteristic of each scRNA-seq cluster were differentially expressed in the same direction in scRNA-seq and bulk RNA-seq datasets. Second, we applied MASC19, a single cell association method for testing whether case-control status influences the membership of single cells in any of multiple cellular subsets while accounting for technical confounds and biological variation. We specified donor identity and batch as random-effect covariates.
Integration of bulk RNA-seq with mass cytometry
We used CCA to associate the abundances of mass cytometry clusters with gene expression in bulk RNA-seq. We started by selecting the samples that had both data types. The mass cytometry data matrix has samples and clusters, where the values represent proportions of cells from each sample in each cluster. The bulk RNA-seq data matrix has samples and genes, where the values represent proportions of gene abundance from each sample in each gene. CCA identifies canonical variates (a linear combination of bulk RNA-seq genes and a linear combination of mass cytometry cluster proportions) that maximize correlation of samples along each canonical variate. In other words, it tries to arrange samples from each dataset in a similar order along each canonical variate. We ran CCA separately for fibroblasts, monocytes, T cells, and B cells. For fibroblasts, we associated 2,299 genes with 8 mass cytometry clusters on 22 samples. For monocytes, we associated 2,161 genes with 5 mass cytometry clusters on 25 samples. For T cells, we associated 2,255 genes with 9 mass cytometry clusters on 26 samples. For B cells, we associated 22,95 genes with 10 mass cytometry clusters on 17 samples.
Finding correspondence between scRNA-seq clusters and mass cytometry clusters
1) For each cell type, we ran CCA with mass cytometry clusters with bulk RNA-seq. Each gene is correlated with each canonical variate (CV). Also, each mass cytometry cluster is correlated with each CV. By visualizing these correlations, we can see the positions of bulk RNA-seq genes and mass cytometry clusters in the same space (Figure 4h).
2) We then associated single-cell RNA-seq clusters with mass cytometry clusters by projecting cluster markers (AUC > 0.7) for each single-cell RNA-seq cluster in the CCA space acquired from step 1).
3) We took the average across the cluster marker genes for each single-cell RNA-seq cluster for each CV and obtained an “average CV” matrix.
4) Based on the “average CV” matrix, we computed Spearman correlation between the scRNA-seq average CV and the CV for mass cytometry clusters.
5) Next, we generated a null distribution for the Spearman correlations by shuffling the scRNA-seq gene names and then repeating steps 2–4 10,000 times.
6) For the 10,000 replicates of CCA matrix, we repeated from step 2 to step 5. Then, we counted how many times the correlation of each pair was greater than the observed value from step 4).
7) Finally, we converted the to a permutation p to a z − score.
Differential expression analysis with bulk RNA-seq
We classified all the samples into OA, leukocyte-poor RA, and leukocyte-rich RA synovial tissues based on the quantitative analysis of T cells, B cells, and monocytes by flow cytometry. PCA on bulk RNA-seq samples showed separation of leukocyte-rich and leukocyte-poor RA on the first or second principal components. For differential analysis, we used the limma R package to identify significantly differentially expressed genes. We used the Benjamini-Hochberg method to estimate false discovery rate (FDR).
Identification of markers for distinct scRNA-seq clusters
Based on the single-cell RNA-seq clusters, we identified cluster marker genes by comparing the cells in one cluster with all other clusters from the same cell type, based on Log2(CPM+1). We prioritized cluster marker genes using three criteria: 1) percent of non-zero expressing cells > 60%; 2) are under the receiver-operator curve (AUC)54 > 0.7; and 3) fold-change (FC) > 2.
Intracellular flow cytometry of synovial tissue T cell stimulation
Disaggregated synovial tissue cells were incubated with Fixable Viability Dye (eBioscience) and Fc blocking antibodies (eBioscience) followed by staining for surface markers in Brilliant Stain Buffer (BD Bioscience). Cell were then fixed and permeabilized using an intracellular staining kit (eBioscience), followed by intracellular staining for granzymes or cytokines. Antibodies used in this study include anti-CD45 (clone HI30) from BD Biosciences; anti-CD3 (clone UCHT1), anti-CD8 (clone SK1), anti-CD14 (clone M5E2), anti-CD4 (clone RPA-T4), anti-HLA-DR (clone L243), anti-granzyme B (clone GB11), and anti-granzyme K (clone GM26E7) from Biolegend; and anti-IFNG (clone 4S.B3) and anti-TNF (clone MAb11) from eBioscience. Data were collected on a BD Fortessa flow cytometer and analyzed using FlowJo 10.5 software. Disaggregated synovial tissue cells were incubated with a cell stimulation cocktail containing PMA and ionomycin (eBioscience) in RPMI with 10% fetal calf serum (Gemini). After 15 minutes, brefeldin A (eBioscience) was added. The cells were incubated at 37C 5% CO2 for an additional 2 hours. The cells were then collected and stained for intracellular cytokines following the protocol above and the data was shown in Supplementary Fig. 10.
Statistics
Results are shown as mean with 95% confidence intervals. The statistics tests used were t-test and Kolmogorov-Smirnov test, unless otherwise stated, as described with one-sided or two-sided in the figure legends. Benjamini-Hochberg FDR < 0.01 and Fold-change > 2 were considered to be statistically significant when appropriate.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Accelerating Medicines Partnership (AMP) in Rheumatoid Arthritis and Lupus Network. AMP is a public-private partnership (AbbVie Inc., Arthritis Foundation, Bristol-Myers Squibb Company, Lupus Foundation of America, Lupus Research Alliance, Merck Sharp & Dohme Corp., National Institute of Allergy and Infectious Diseases, National Institute of Arthritis and Musculoskeletal and Skin Diseases, Pfizer Inc., Rheumatology Research Foundation, Sanofi and Takeda Pharmaceuticals International, Inc.) created to develop new ways of identifying and validating promising biological targets for diagnostics and drug development Funding was provided through grants from the National Institutes of Health (UH2-AR067676, UH2-AR067677, UH2-AR067679, UH2-AR067681, UH2-AR067685, UH2-AR067688, UH2-AR067689, UH2-AR067690, UH2-AR067691, UH2-AR067694, and UM2-AR067678). This work is also supported in part by funding from the Ruth L. Kirschstein National Research Service Award (F31AR070582) from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (K.S.). K.W. is supported by a Rheumatology Research Foundation Scientist Development Award, and KL2/Catalyst Medical Research Investigator Training award (an appointed KL2 award) from Harvard Catalyst | The Harvard Clinical and Translational Science Center (National Center for Advancing Translational Sciences, National Institutes of Health Award KL2 TR002542). D.A.R. is supported by NIAMS K08 AR072791–01. L.T.D. is supported by NIAMS K01 AR066063. J.H.A. is supported by R21 AR071670, and the Bertha and Louis Weinstein research fund. K.S. is supported by NIAMS F31-AR070582. S.R. is supported by 1R01AR063759-01A1 and Doris Duke Charitable Foundation Grant #2013097. A.H.J. is supported by an Arthritis National Research Foundation Grant. A.F., C.D.B. and J.D.T. were supported by the Arthritis Research UK Rheumatoid Arthritis (#20298), and by the National Institute for Health Research (NIHR)’s Birmingham Biomedical Research Centre program, supported by the National Institute for Health Research/Wellcome Trust Clinical Research Facility at University Hospitals Birmingham NHS Foundation Trust.
Footnotes
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data Availability
The single-cell RNA-seq data, bulk RNA-seq data, mass cytometry data, flow cytometry data, and the clinical and histological data this study are available at ImmPort (https://www.immport.org/shared/study/SDY998 and https://www.immport.org/shared/study/SDY999, study accession codes SDY998 and SDY999). The raw single-cell RNA-seq and mass cytometry data are deposited in dbGAP (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001457.v1.p1. The source code repository of the computational and statistical analysis is located athttps://github.com/immunogenomics/amp_phase1_ra. Data can also be viewed on 3 different websites at https://immunogenomics.io/ampra, https://immunogenomics.io/cellbrowser/, and https://portals.broadinstitute.org/single_cell/study/amp-phase-1.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Gibofsky A Epidemiology, pathophysiology, and diagnosis of rheumatoid arthritis: A Synopsis. Am. J. Manag. Care 20, S128–35 (2014). [PubMed] [Google Scholar]
- 2.McInnes IB & Schett G The pathogenesis of rheumatoid arthritis. N. Engl. J. Med 365, 2205–2219 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Orr C et al. Synovial tissue research: a state-of-the-art review. Nat. Rev. Rheumatol 13, 463–475 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Wolfe F et al. The mortality of rheumatoid arthritis. Arthritis Rheum. 37, 481–494 (1994). [DOI] [PubMed] [Google Scholar]
- 5.Namekawa T, Wagner UG, Goronzy JJ & Weyand CM Functional subsets of CD4 T cells in rheumatoid synovitis. Arthritis Rheum. 41, 2108–2116 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Gizinski AM & Fox DA T cell subsets and their role in the pathogenesis of rheumatic disease. Curr. Opin. Rheumatol 26, 204–210 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Reparon-Schuijt CC et al. Secretion of anti-citrulline-containing peptide antibody by B lymphocytes in rheumatoid arthritis. Arthritis Rheum. 44, 41–47 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Mulherin D, Fitzgerald O & Bresnihan B Synovial tissue macrophage populations and articular damage in rheumatoid arthritis. Arthritis Rheum. 39, 115–124 (1996). [DOI] [PubMed] [Google Scholar]
- 9.Kinne RW, Bräuer R, Stuhlmüller B, Palombo-Kinne E & Burmester GR Macrophages in rheumatoid arthritis. Arthritis Res. 2, 189–202 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Müller-Ladner U et al. Synovial fibroblasts of patients with rheumatoid arthritis attach to and invade normal human cartilage when engrafted into SCID mice. Am. J. Pathol 149, 1607–1615 (1996). [PMC free article] [PubMed] [Google Scholar]
- 11.Pap T, Müller-Ladner U, Gay RE & Gay S Fibroblast biology. Role of synovial fibroblasts in the pathogenesis of rheumatoid arthritis. Arthritis Res. 2, 361–367 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennis G Jr et al. Synovial phenotypes in rheumatoid arthritis correlate with response to biologic therapeutics. Arthritis Res. Ther 16, R90 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orange DE et al. Machine learning integration of rheumatoid arthritis synovial histology and RNAseq data identifies three disease subtypes. Arthritis Rheumatol (2018). doi: 10.1002/art.40428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindberg J et al. Variability in synovial inflammation in rheumatoid arthritis investigated by microarray technology. Arthritis Res. Ther 8, R47 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephenson W et al. Single-cell RNA-seq of rheumatoid arthritis synovial tissue using low-cost microfluidic instrumentation. Nat. Commun 9, 791 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papalexi E & Satija R Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol 18, 35–45 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Schelker M et al. Estimation of immune cell content in tumour tissue using single-cell RNA-seq data. Nat. Commun 8, 2032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rao DA et al. Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature 542, 110–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fonseka CY et al. Mixed-effects association of single cells identifies an expanded effector CD4+ T cell subset in rheumatoid arthritis. Sci. Transl. Med 10, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villani A-C et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizoguchi F et al. Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis. Nat. Commun 9, 789 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donlin LT et al. Methods for high-dimensonal analysis of cells dissociated from cyropreserved synovial tissue. Arthritis Res. Ther 20, 139 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becher B et al. High-dimensional analysis of the murine myeloid cell system. Nat. Immunol 15, 1181–1189 (2014). [DOI] [PubMed] [Google Scholar]
- 24.De Maesschalck R, Jouan-Rimbaud D & Massart DL The Mahalanobis distance. Chemometrics Intellig. Lab. Syst 50, 1–18 (2000). [Google Scholar]
- 25.Krenn V et al. Grading of Chronic Synovitis - A Histopathological Grading System for Molecular and Diagnostic Pathology. Pathology - Research and Practice 198, 317–325 (2002). [DOI] [PubMed] [Google Scholar]
- 26.van der Maaten L & Hinton G. Visualizing Data using t-SNE. J. Mach. Learn. Res 9, 2579–2605 (2008). [Google Scholar]
- 27.Todd DJ et al. XBP1 governs late events in plasma cell differentiation and is not required for antigen-specific memory B cell development. J. Exp. Med 206, 2151–2159 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubtsov AV et al. CD11c-Expressing B Cells Are Located at the T Cell/B Cell Border in Spleen and Are Potent APCs. J. Immunol 195, 71–79 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pillai S Now you know your ABCs. Blood 118, 1187–1188 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Ellebedy AH et al. Defining antigen-specific plasmablast and memory B cell subsets in human blood after viral infection or vaccination. Nat. Immunol 17, 1226–1234 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S et al. IL-21 drives expansion and plasma cell differentiation of autoreactive CD11c hi T-bet+ B cells in SLE. Nat. Commun 9, 1758 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pitzalis C, Kelly S & Humby F New learnings on the pathophysiology of RA from synovial biopsies. Curr. Opin. Rheumatol 25, 334–344 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Filer A The fibroblast as a therapeutic target in rheumatoid arthritis. Curr. Opin. Pharmacol 13, 413–419 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Westra H-J et al. Fine-mapping and functional studies highlight potential causal variants for rheumatoid arthritis and type 1 diabetes. Nat. Genet 50, 1366–1374 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Al-Mayouf SM et al. Loss-of-function variant in DNASE1L3 causes a familial form of systemic lupus erythematosus. Nat. Genet 43, 1186–1188 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Snelling SJB et al. Dickkopf-3 is upregulated in osteoarthritis and has a chondroprotective role. Osteoarthritis Cartilage 24, 883–891 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee EB et al. Tofacitinib versus methotrexate in rheumatoid arthritis. N. Engl. J. Med 370, 2377–2386 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Zizzo G, Hilliard BA, Monestier M & Cohen PL Efficient clearance of early apoptotic cells by human macrophages requires M2c polarization and MerTK induction. J. Immunol 189, 3508–3520 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frara N et al. Transgenic Expression of Osteoactivin/gpnmb Enhances Bone Formation In Vivo and Osteoprogenitor Differentiation Ex Vivo. J. Cell. Physiol 231, 72–83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jenks SA et al. Distinct Effector B Cells Induced by Unregulated Toll-like Receptor 7 Contribute to Pathogenic Responses in Systemic Lupus Erythematosus. Immunity 49, 725–739.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smolen JS How well can we compare different biologic agents for RA? Nat. Rev. Rheumatol 6, 247–248 (2010). [DOI] [PubMed] [Google Scholar]
- 42.McInnes IB et al. The role of interleukin-15 in T-cell migration and activation in rheumatoid arthritis. Nat. Med 2, 175 (1996). [DOI] [PubMed] [Google Scholar]
- 43.McInnes IB & Liew FY Cytokine networks-towards new therapies for rheumatoid arthritis. Nat. Clin. Pract. Rheumatol 1, 31 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Hicks SC, Townes FW, Teng M & Irizarry RA Missing data and technical variability in single-cell RNA-sequencing experiments. Biostatistics (2017). doi: 10.1093/biostatistics/kxx053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parkhomenko E, Tritchler D & Beyene J Sparse canonical correlation analysis with application to genomic data integration. Stat. Appl. Genet. Mol. Biol 8, Article 1 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Witten DM, Tibshirani R & Hastie T A penalized matrix decomposition, with applications to sparse principal components and canonical correlation analysis. Biostatistics 10, 515–534 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hashimshony T et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 17, 77 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bendall SC et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bjornson ZB, Nolan GP & Fantl WJ Single-cell mass cytometry for analysis of immune system functional states. Curr. Opin. Immunol 25, 484–494 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peterson VM et al. Multiplexed quantification of proteins and transcripts in single cells. Nat. Biotechnol 35, 936–939 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Picelli S et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Finck R et al. Normalization of mass cytometry data with bead standards. Cytometry A 83, 483–494 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.González I, Déjean S, Martin P & Baccini A CCA: An R Package to Extend Canonical Correlation Analysis. Journal of Statistical Software, Articles 23, 1–14 (2008). [Google Scholar]
- 54.Sing T, Sander O, Beerenwinkel N & Lengauer T ROCR: visualizing classifier performance in R. Bioinformatics 21, 3940–3941 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Subramanian A et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ashburner M et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liberzon A et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reynolds AP, Richards G, de la Iglesia B & Rayward-Smith VJ Clustering Rules: A Comparison of Partitioning and Hierarchical Clustering Algorithms. J. Math. Model. Algorithms 5, 475–504 (2006). [Google Scholar]
- 59.Rousseeuw PJ Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math 20, 53–65 (1987). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.