Summary:
Organisms often respond to changing environments by altering development of particular traits. These plastic traits exhibit genetic variation, i.e., genotypes respond differently to the same environmental cues. Theoretical studies have demonstrated the importance of this variation, which is targeted by natural selection, in adapting plastic responses to maximize fitness [1,2]. However, little is known about the underlying genetic mechanisms. We identify two laterally transferred genes that contribute to variation in a classic example of phenotypic plasticity: the pea aphid’s ability to produce winged offspring in response to crowding. We discovered that aphid genotypes vary extensively for this trait, and that aphid genes of viral origin are upregulated in response to crowding solely in highly-inducible genotypes. We knocked-down expression of these genes to demonstrate their functional role in wing plasticity. Through phylogenetic analysis we found that these genes likely originated from a virus that infects rosy apple aphids and causes their hosts to produce winged offspring [3]. The function of these genes has therefore been retained following transfer to pea aphids. Our results uncover a novel role for co-opted viral genes, demonstrating that they are used to modulate ecologically-relevant, plastic phenotypes. Our findings also address a critical question about the evolution of environmentally-sensitive traits: whether or not the genes that control the expression of plastic traits also underlie variation in plasticity. The genes we identify originated from outside aphids themselves, and thus our work shows that genes formerly unrelated to plasticity can fine-tune the strength of plastic responses to the environment.
Keywords: Phenotypic plasticity, lateral gene transfer, genetic variation, Acyrthosiphon pisum, densovirus, gene expression, polyphenism
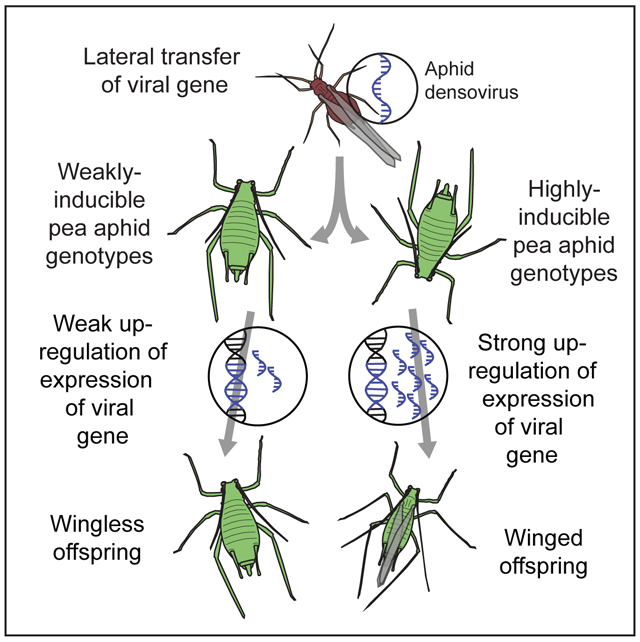
Graphical Abstract
eTOC Blurb
Parker and Brisson study the genetic mechanisms underlying variation in the pea aphid’s plastic production of winged offspring in response to crowding. They identify two functional aphid genes that were laterally transferred from a densovirus. These findings show how co-opted genes can fine-tune the strength of plastic responses to the environment.
Results & Discussion:
Pea aphids (Acyrthosiphon pisum) exhibit a textbook example of phenotypic plasticity, where crowded conditions trigger the production of winged rather than wingless offspring. Both morphs are genetically identical to each other and to their mothers due to parthenogenetic reproduction. As in other wing dimorphic insects, winged aphids can disperse to new environments but produce fewer offspring than their wingless counterparts, which leads to a clear trade-off between reproduction and dispersal [4]. The fitness of a particular clone depends on the ability to appropriately sense environmental conditions and produce wingless offspring with high fecundity at low densities or to produce winged offspring when an environment is deteriorating.
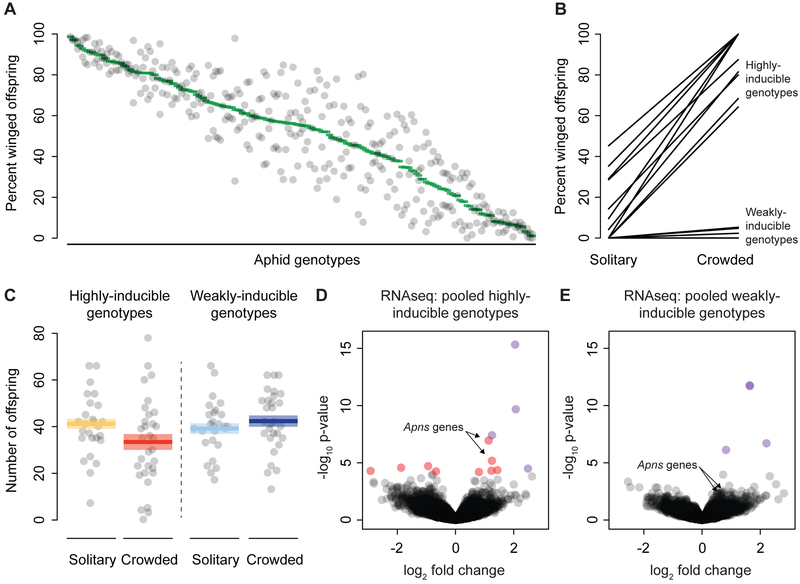
We first characterized the variation in wing plasticity present in an aphid population. We used a panel of 192 aphid lines with unique genotypes collected in Ithaca, NY [5]. We raised aphids from each genotype at low density, subjected them to crowding (12 aphids) in a dish for 12hrs, and then counted the percentage of winged offspring they produced over the 24hrs after crowding. Aphid genotypes exhibited the full range of phenotypic variation, from near-0% to 100% of winged offspring produced in response to the crowding treatment (Figure 1A). This distribution indicates polygenic control of this variation. We then focused on a panel of 10 “highly-inducible” and 10 “weakly-inducible” genotypes, which we confirmed produce high or low levels of winged offspring in response to crowded but not solitary conditions (Figure 1B).
Figure 1: Genetic variation in aphid plasticity.
A: The percentage of winged offspring produced by crowded aphids of 192 genotypes. For each genotype (x-axis), a green bar shows the mean of two experimental replicates shown by the grey points. B: A panel of 10 highly and 10 weakly-inducible genotypes was exposed to solitary or crowded conditions, shown on the x-axis. The y-axis shows the percentage of winged offspring, with each genotype represented by a black line. C: The fecundity of each of the 10 highly and 10 weakly-inducible genotypes in response to solitary and crowded conditions is shown. The number of offspring born to each group of three aphids on a single plant produced in the 48hrs after treatment is shown along the y-axis. The grey points are experimental replicates. The mean of each combination of phenotype and treatment is shown with a colored bar, with standard error represented by the lighter colored boxes. D, E: Volcano plot resulting from RNA sequencing of pooled highly-inducible genotypes (D) or weakly-inducible genotypes (E). Within each plot, the x-axis shows the log2 fold change of each expressed gene in the aphid genome, with higher expression in crowded conditions to the right, and the y-axis shows the negative log10 of the p-value. Four genes that were statistically significantly differentially expressed by both pools of genotypes (FDR<0.1) are shown in purple (note that two points are largely overlapping in the weakly-inducible plot). An additional 9 genes which were significantly differentially expressed only in highly-inducible genotypes are shown in red.
We investigated if a higher degree of plasticity comes with a fecundity cost. Highly- and weakly-inducible genotypes did not differ in their overall fecundity (χ2=0.95, 1DF, p=0.33; Figure 1C), but did differ in how crowded conditions influenced fecundity (treatment * highly- vs. weakly- inducible phenotype; χ2=27, 1DF, p=2.3 × 10−7). These results suggest that the plastic response itself is costly to aphids, since crowding led to a reduction in the number of aphids born to highly-inducible genotypes, but did not have a similar effect on weakly-inducible genotypes. These fecundity costs are in addition to the transgenerational costs of producing low-fecundity winged offspring [6], stressing the importance to an aphid clone of appropriately responding to environmental cues.
To explore the mechanistic basis of variation in plasticity, we sequenced transcriptomes from highly-and weakly-inducible aphid genotypes (aphids from 10 genotypes pooled per phenotype) under both solitary and crowded conditions (Table S1), and we identified genes differentially expressed in response to crowding. Four genes were differentially expressed in both highly- and weakly-inducible genotypes (Figures 1D & 1E; Table S2), indicating that some aspects of the response to crowding are shared among genotypes. More importantly, an additional nine genes were differentially expressed only in highly-inducible genotypes (Table S2), revealing that some effects of the genetic variation for this plasticity can be discerned at the transcription level. Our highly-inducible differentially-expressed gene list was remarkably similar to a previous RNAseq study of a highly-inducible pea aphid genotype, with five genes overlapping despite different experimental conditions [7] (Table S2).
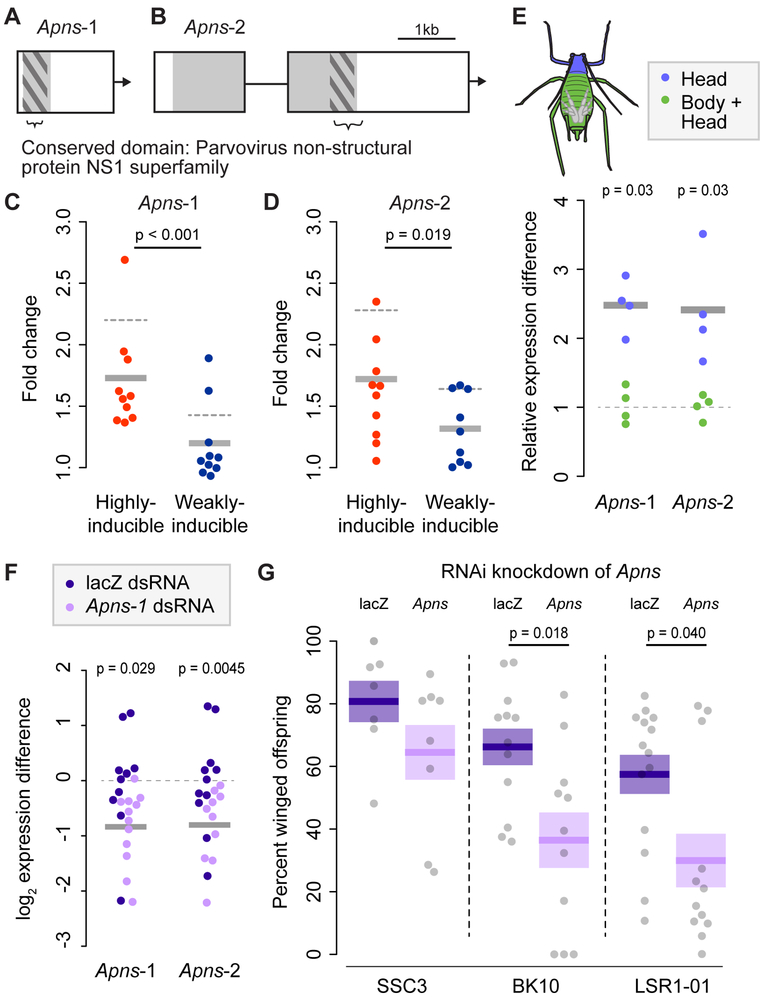
We focused specifically on two of the genes exclusively upregulated by highly-inducible genotypes, which we call Apns-1 (Figure 2A; ACYPI085607) and Apns-2 (Figure 2B; ACYPI36509). The putative proteins of both genes contain a “parvovirus non-structural protein NS1 superfamily” conserved domain (domain E-value; Apns-1: 2.09e-07, Apns-2: 4.42e-14). The presence of this viral domain suggests that these genes could be the result of lateral gene transfers into the aphid genome. Both genes are within pea aphid genomic scaffolds and therefore appear to be true genome integrations. To confirm this finding, we used reads from Nanopore sequencing of a pea aphid genotype different than that used in the aphid genome project. Both Apns genes show contiguity with aphid sequence, with single long reads spanning both the Apns and nearby aphid genes (Figure S1).
Figure 2: Differential expression and function of A. pisum non-structural proteins.
A & B: Models of Apns-1 (ACYPI085607) and Apns-2 (ACYPI36509). The conserved domain is bracketed at the bottom of each figure, and the open reading frame is shown in grey. The region used for phylogenetic alignment is shown by dark grey hashes. A scale bar for both genes (1kb) is shown at the top. C & D: Show the fold change in gene expression of Apns-1 (C) and Apns-2 (D) as measured by qRT-PCR in response to crowding relative to solitary conditions, with each genotype shown by a single point (averaged across biological replicates). The mean difference across the 10 genotypes for each phenotype is shown by a solid grey bar, with the differential expression value from the RNA-seq shown with dotted grey line. E: Shows expression levels of the Apns genes in heads versus whole body samples. F: Shows the results of RNA interference (RNAi) knock-down of the Apns genes as measured by qRT-PCR. The y-axis shows the log2 expression difference of each sample relative to the average expression of control (lacZ injected) aphids. The grey bars show the average expression difference of the Apns-1 dsRNA-injected aphids. G: Shows the results of expression knockdown on the percentage of winged aphids. We conducted three replicates of the experiment each with a different aphid genotype, shown along the x-axis. The percentage of winged offspring born to groups of four aphids is shown on the y-axis. Treatment (injection with dsRNA from lacZ (control) or Apns-1, expected to affect both Apns genes) is shown along the top of the figure. Each grey point represents one biological replicate, and the purple boxes show the mean +/− standard error of each combination of experiment × treatment. See Figures S1 and S2 for more information.
To measure expression of these genes in individual genotypes (rather than in pooled samples as in the RNAseq above), we repeated the crowding assay and used qRT-PCR to measure gene expression in the 10 highly and 10 weakly-inducible genotypes. We found that highly-inducible genotypes upregulate both Apns genes more strongly in response to crowding than do weakly-inducible genotypes (Figures 2C & S2A; LMM on ΔCTvalues; Apns-1: χ2=15.6, 1Df, p<0.001; Apns-2: Figures 2D & S2B, χ2=5.47, 1Df, p=0.019), confirming our RNAseq data. We also found that the expression of both genes is enriched in heads relative to whole-body samples (Figure 2E; Wilcoxon rank-sum tests on ΔCT values; Apns-1: W=16, p=0.029; Apns-2: W=16, p=0.029).
To determine the function of these genes in the aphid plastic wing response, we used RNA interference (RNAi, Figure 2F) to knock down expression. We used three lines (not used above) that were known from previous studies to be highly-inducible (using similar protocols, lines SSC3, BK10 & LSR1-01 produced 92.3%, 82.2%, and 62.0% winged offspring in response to crowding, respectively [8]). We injected dsRNA of Apns-1 into uncrowded aphids, exposed aphids to crowded conditions, and measured the percentage of winged offspring born to aphids 48-72hrs after injection. Because of the similarity of the two Apns genes, dsRNA generated from Apns-1 cDNA sequence led to the knock-down of both viral genes, resulting in a 42% and 43% reduction in expression of Apns-1 and Apns-2, respectively (Figure 2F; Wilcoxon tests on ΔCT values; Apns-1: W=86, p=0.029; Apns-2: W=111, p=0.0045). dsRNA injection significantly reduced the proportion of winged offspring born to aphids from two of the three genotypes tested (Figure 2G; Wilcoxon tests; SSC3: W=15, p=0.15; BK10: W=27, p=0.018; LSR1-01: W=44, p=0.040), demonstrating that this laterally transferred putative viral gene has a functional role in aphid wing plasticity. We repeated the experiment using the same genotypes but instead crowded aphids before dsRNA injection (Figures S2C-S2H). This method produced qualitative similar results but the effects were not statistically significant.
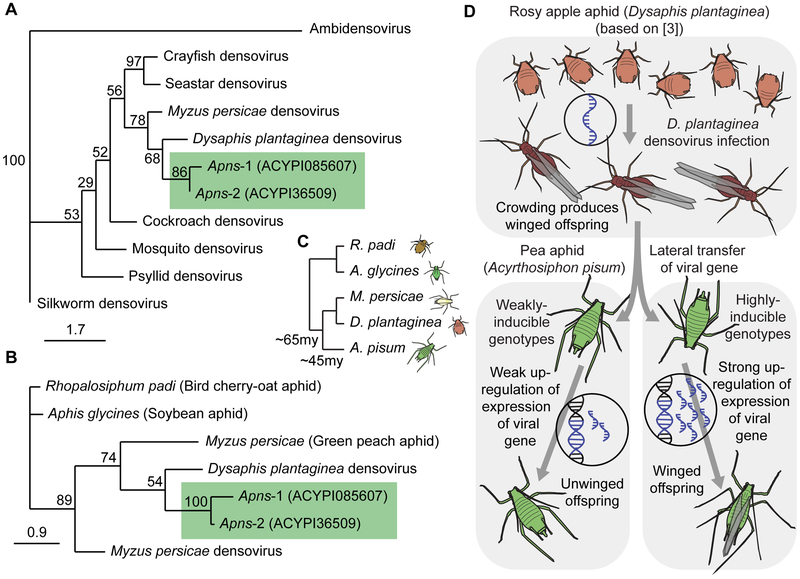
Having established a role for the Apns genes in pea aphid wing plasticity, we next explored the origin of these putative viral genes in the pea aphid genome. Densoviruses are single-stranded DNA viruses with small (4-6kb) genomes that are related to parvoviruses and infect a wide diversity of arthropods [9]. Two examples of densoviruses infecting aphids have been investigated: one from the peach potato aphid, Myzus persicae [10], and one from the rosy apple aphid, Dysaphis plantaginea [3]. We performed a phylogenetic reconstruction of arthropod densoviruses and found that the pea aphid genes Apns-1 and Apns-2 clustered within the densovirus sequences, and cluster most closely with D. plantaginea densovirus (DplDNV) (Figure 3A; Alignment Figure S3A). This phylogenetic placement is consistent with the genes originating via lateral gene transfer.
Figure 3: Origin of pea aphid densovirus genes and their retention of function.
A: Unrooted, bootstrap consensus tree showing the protein phylogenetic relationships among densovirus sequences from different invertebrate hosts, related viruses (parvovirus and ambidensovirus), and pea aphid Apns-1 and Apns-2. B: Unrooted, bootstrap consensus tree created from amino acid directed nucleotide alignments showing the phylogenetic relationships among densovirus sequences (from Myzus persicae and Dysaphis plantaginea) and aphid homologs (Rhopalosiphum padi, Aphis glycines, Myzus persicae, and pea aphid Apns-1 and Apns-2). For both trees, only nucleotide sequence alignable with the shorter pea aphid copy (Apns-1) were used. 1000 bootstrap trees were generated with maximum likelihood. C: Species tree for the aphid species referred to in Figure 3B. The relationships among species are inferred from trees in Kim et al. [32] and Hardy et al. [33]. Divergence times are based on [32]. D: Model illustrating densovirus gene domestication and subsequent retention of function. See Figure S3 and Table S3 for more information.
Densovirus genes, including both structural and nonstructural proteins, have previously been found to be integrated and expressed in the genomes of pea aphids [11] and several other aphid species [12]. We therefore performed a further phylogenetic analysis using publicly-available sequences of annotated aphid genes with homology to densoviruses (Table S3) [13,14]. This analysis again showed that the pea aphid genes grouped with most similarity to DplDNV (Figure 3B, compare to the species tree in Figure 3C), again suggesting that these genes resulted from a lateral gene transfer from a DplDNV-like densovirus. This virus is efficiently transmitted from rosy apple aphids (D. plantaginea) to their offspring (vertical transmission), and is described as a viral mutualist. This is because the production of winged offspring in rosy apple aphids is dependent on infection with DplDNV, and viral infection increases host mobility and promotes dispersal [3]. Densovirus-free rosy apple aphids do not produce winged offspring even in response to crowding and poor host plant conditions. Intriguingly, therefore, the free-living densovirus most closely related to the integrated densovirus genes in the pea aphid causes the production of winged aphids. We note, however, that support for the branch containing the Apns genes and DplDNV was low in both analyses, and an alternative possibility is that the Apns genes originated from a lateral transfer from Myzus persicae densovirus or an uncharacterized aphid virus. We performed an extension of this analysis using aphid sequences from two additional genomes (Diuraphis noxia and M. cerasi) that again supported our finding that the Apns genes originate from LGT of an aphid densovirus, and might further suggest that sequences closely related to DplDNV have repeatedly integrated into multiple aphid genomes (Figure S3; though see below). Future study of the timing and origin of the Apns genes in aphids is needed.
We suggest that the densovirus genes have the same effect on winged morph induction in the pea aphid that densovirus infection has in the D. plantaginea, and these genes likely have retained their function after introduction into the pea aphid genome (Summarized in Figure 3D). It is unclear whether D. plantaginea densovirus actively induces winged forms in its host using this nonstructural protein, or whether D. plantaginea responds to viral infection by producing winged offspring, and future work is needed to uncover the precise mechanism by which these proteins act. However, densovirus nonstructural proteins are generally involved in virus replication and transcriptional activation of capsid genes [15]. Non-structural proteins of vertebrate parvoviruses can activate transcription factors and induce epigenetic modifications in hosts through histone acetylation [16]. Apns-1 and Apns-2 may, therefore, induce transcription of genes related to wing morph determination, potentially acting in the brain given the enriched head gene expression levels.
Lateral gene transfers are an important source of phenotypic change in prokaryotes, but only recently have we begun to appreciate the frequency and importance of lateral transfers from microbes in eukaryotic evolution [17]. Most laterally transferred DNA is not expressed by eukaryotic hosts, and is quickly inactivated or eroded. Examples of functional lateral gene transfer are therefore uncommon, but some prominent examples come from the integration of viruses into host genomes [18] and from the transfer of DNA from vertically-transmitted bacteria to their animal hosts [19]. Our study provides a clear example of a functional lateral gene transfer from a vertically-transmitted viral partner to its host.
Genetic variation in plastic traits has been documented in a diversity of taxa, from natural variation among isolates of the nematode Pristionchus pacificus in the production of dimorphic adult mouth forms [20] to predator-mediated plasticity in the shape of Chthamalus anisopoma barnacles [21]. This variation has been theorized to be important in adapting plastic responses to fitness optima [2]. Our results provide insight into the molecular mechanisms underlying genetic variation in phenotypically plastic traits. This lateral gene transfer event appears to be part of a modulation or “fine-tuning” of the sensitivity of the plastic response to the environment. The densovirus gene insertion event is much more recent than the evolution of the wing plasticity itself, which is ancient to aphids and common to many major groups [22]. This finding therefore sheds light on an important question about how phenotypically plastic traits evolve: whether genes that underlie variation in a plastic trait also control expression of the trait, or whether genes from outside of these developmental pathways are co-opted to modify the strength of a plastic response to environmental cues. The answer in this case is clearly the latter. Not only are the Apns genes from outside the developmental genetic pathway for the aphid wing plasticity, they are from outside aphids themselves.
STAR Methods:
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Benjamin Parker (bjp@utk.edu).
Experimental model and subject details
Pea aphids (Acyrthosiphon pisum).
Aphids were maintained on fava bean plants (Vicia faba, Improved Long Pod, Harris Seeds, Rochester NY) in climate controlled incubators kept at 20°C and a 16L:8D light cycle and a light intensity of ~5500 lux. Aphids reproduce via pathogenesis under these (summer) conditions—genetically-distinct lineages were maintained in separate cages, with several developing nymphs moved to new plants each week prior to use in the experiments. All of the aphids used in this study were adult (~10 days old) pathogenetic females; specific information on the collection location for the genotypes used in each experiment is reported in the Method Details, below. In each experiment individuals reared together in a cage were divided randomly into treatment groups.
Method details
Variation in wing plasticity from a natural aphid population.
The aphid panel used in this study was collected in May of 2015 in two alfalfa (Medicago sativa) fields (each ~0.2 ha, 800 m apart) in Ithaca, NY [5]. Lines were genotyped at 14 microsatellite loci and lines with identical microsatellite repeats were removed from the panel, leaving 192 total unique genotypes included in the experiment. We carried out two replicates of a standardized crowding assay [7,8]. We reared aphids from each genotype on bean plants at low densities (<7 individuals per plant) for three generations. We then placed adult aphids that had begun reproducing in 3.5 cm (diameter) Petri dishes for 24 hrs at a density of 12 aphids per dish as in our previous work [7]. Aphids were then placed on a bean plant for a further 24 hrs and allowed to reproduce, and the percentage of offspring produced during this period that developed wings was recorded.
Wing induction and fecundity assays.
20 aphid genotypes were selected using the data from the phenotypic screen of the genotypes described above. 10 genotypes produced a low percentage of winged-offspring after crowding (weakly-inducible genotypes: genotypes 73, 88, 183, 218, 223, 248, 375, 396, 405, & 495), and 10 genotypes produced a high percentage of winged-offspring (highly-inducible genotypes: genotypes 42, 125, 179, 202, 217, 281, 286, 445, 473, & 500). We specifically chose genotypes that did not harbor the facultative endosymbiont Regiella insecticola as previous work has suggested that infection with this bacterium alters wing induction [23]. We also verified that these genotypes did not harbor DplDNV using established PCR protocols [3]. We verified the phenotypes of these genotypes using the protocol described above, but included a non-crowded (solitary) treatment (one aphid per Petri dish) to verify that solitary conditions do not lead to high levels of wing induction [7].
We determined if the induction of winged offspring from crowded conditions led to measurable effects on aphid fitness, and if these effects differed across the highly and weakly inducing aphid genotypes. We again reared aphids at low densities for three generations (< 7 aphids per plant) and randomly assigned 10 day-old (adult) aphids to either a solitary or crowding treatment. After 24 hrs we moved the aphids on to fresh bean plants in a plastic cage with a mesh top for 48 hrs in groups of three aphids per plant, and then counted the number of offspring in each cage.
Data were analyzed using linear mixed models implemented in R 3.5.0 using the lme4 package [24]. Treatment (solitary vs. crowded) and phenotype (highly vs weakly inducible genotypes) were modeled as fixed effects, with genotype nested within phenotype and modeled as a random effect. Counts of the number of offspring were modeled using a Poisson distribution [25]. Minimal models were derived by first removing the interaction term between treatment and phenotype, then the main effect of phenotype, and then treatment, with model comparisons performed using ANOVA and χ2 tests to determine the statistical significance of each model factor.
Pooled Gene Expression Study using RNAseq.
We reared aphids at low densities as above, and then kept them in Petri dishes in solitary or crowded conditions for 12 hrs. We flash-froze aphids in liquid nitrogen, and later thawed them in RNAlater-ICE (Invitrogen) at room temperature for 30 m, during which we dissected out and discarded embryos (so that embryo transcripts would not be included in the RNAseq libraries) and pooled the adult carcasses into groups of 10 aphids (each aphid from a different genotype). We extracted RNA using Trizol and an isopropanol precipitation with an ethanol wash. We removed genomic DNA from the total RNA samples using Zymo DNAse I, and cleaned the RNA using the Zymo RNA Clean and Concentrator-5 kit under recommended protocols. RNA quality was verified on an Agilent 2100 Bioanalyzer. We used the Illumina TruSeq kit under recommended protocols and 300 ng starting total RNA to construct 16 cDNA libraries (2 ‘phenotypes’ of pooled highly- and weakly-inducible aphid genotypes × 2 treatments (solitary vs crowded) × 4 biological replicates). The libraries were then pooled into two groups of eight samples, and were further purified using AmpureXP beads. Pooled libraries were sequenced across two lanes of an Illumina HiSeq2500v4 sequencer (eight libraries per lane with a target of 250 million 100 bp reads per lane), with two of the biological replicates for each treatment pooled in each lane.
We trimmed the raw reads for the presence of Illumina adapter sequences using Cutadapt v.1.2.1, and quality trimmed using fastq-mcf (ea-utils software package, -q 20). We aligned the reads to the pea aphid reference genome v.2 [26] using tophat2 [27] preserving strand orientation, and calculated read counts using htseq-count [28] and the “union” overlap mode, using a modified version of the ACYPI OGS v.2.1b (http://bipaa.genouest.org/sp/acyrthosiphon_pisum/) genome annotation file. Read counts were analyzed using EdgeR v.3.18.1 in R v.3.4.1. Genes with a minimum threshold of aligned reads, calculated by (counts / library size) × 106 > 0.5 in at least 4 libraries, were retained in the analysis. Genes with an FDR of less than 0.1 were interpreted as statistically significantly differentially expressed.
Quantitative PCR measures of densovirus-derived gene expression.
We measured gene expression of Apns-1 (ACYPI085607) and Apns-2 (ACYPI36509) in the panel of highly- and weakly-inducible aphid genotypes described above. We reared aphids at low densities for three generations and then exposed them to crowded (12 aphids per dish) or solitary conditions as above. After 12 hrs we dissected and discarded embryos and stored adult carcasses in Trizol at −80°C. Each biological replicate contained 4-6 dissected aphids, and we collected 4 replicates per genotype per treatment. We extracted RNA from each sample as above, and made cDNA using the BioRAD iScript cDNA kit.
We designed quantitative PCR primers that amplified short fragments of the expressed region of both viral genes (Apns-1 F: GCAAACGTCGTTTCTGCCTT & R: ACGACTACGAATCTGGCACG; Apns-2 F: AGTATCCTTGTTGTCCGCCC & R: GCGCACCAATTCCTAAGAGC). Reactions were run on a Bio-RAD CFX96 Real-Time System machine, with an initial step of 95°C for 3 m and 40 cycles of 95°C for 10 s and 60°C for 30 s using two endogenous control genes (ACYPI009769: Glyceraldehyde 3-phosphate dehydrogenase; F: CGGGAATTTCATTGAACGAC & R: TCCACAACACGGTTGGAGTA, and ACYPI009382: NADH dehydrogenase; F: CGAGGAGAACATGCTCTTAGAC & R: GATAGCTTGGGCTGGACATATAG, which were similarly expressed in both morphs). Each 20 μL reaction included a 1X PCR buffer, Mg2+ at 2 mM, dNTPs at 0.2 mM, EvaGreen at 1X, 0.025 units/μL of Invitrogen taq, and 75.6 ng cDNA. We optimized the efficiencies of each primer by to 100 +/− 5% using a serial dilution of 1 ng to 100 ng cDNA by altering primer concentrations (g3PDH: 400 nM forward, 350 nM reverse; NADH: 350F, 300R; ACYPI36509: 200 nM; ACYPI085607: 200 nM). We ran three technical replicates for each reaction. We calculated −ΔCT values by CT_Targe – (CT_g3PDH + CT_NADH)/2. These −ΔCT values were analyzed using linear mixed models after testing for model assumptions. Treatment (crowded vs. solitary) and phenotype (highly- vs. weakly-inducible aphid genotypes) were modeled as fixed effects, and aphid genotype (nested within phenotype) was modeled as a random effect. Minimal models were derived as above (first removing the interaction term, then the main effects of treatment and phenotype), and significance was determined using model comparisons via ANOVA and χ2 statistics.
We tested for enrichment of expression of the viral genes in aphid heads relative to the whole body using qPCR. Aphids from a highly-inducible genotype (genotype 445) were crowded in dishes (12 per dish as above), and after 12 hrs embryos were removed as above. We removed heads from dissected carcasses using a razor blade, cutting heads just below the first leg pair (see figure 2E, top). Head samples contained tissue from approximately 15 aphids per biological replicate, and carcass samples contained tissue from 4 aphids per replicate. Four biological replicates were collected per tissue-type. Dissected tissue was stored in Trizol, and RNA extraction and qPCR were carried out as above. −ΔCT values for each gene were analyzed using non-parametric statistics (Wilcoxon rank-sum tests).
Expression knock-down using RNA interference.
We designed primers that amplify 586 bp of Apns-1 (ACYPI085607; F: TCCGTTTCAATAGCTTCCGAA & R: ACTGCTGCACCGATGAAGAA) with T7 promoter (TAATACGACTCACTATAGGG) sequences included on the 5’ end of each primer. We amplified these fragments using standard PCR conditions and Phusion HF PCR Mastermix and primers at 0.5 μM, using cDNA synthesized from RNA extracted from adult pea aphids (BioRad iScript cDNA Synthesis kit using recommended conditions). PCR product was purified and concentrated before dsRNA synthesis using ethanol and NaOAc precipitation, with the concentrated PCR product at approximately 500 ng/uL. Double-stranded RNA (dsRNA) was synthesized using the MEGAscript RNAi Kit using standard protocols but with an overnight transcription incubation, followed by nuclease digestion with DNAse I and RNAse (under recommended protocols) to remove DNA and ssRNA. dsRNA was concentrated as needed using ethanol and LiCl precipitation and suspended in water at a concentration of ~3300 ng/uL. Using the same protocols, we amplified a portion of the lacZ operon from the TOPO TA vector, and made dsRNA from this PCR product to use as a control.
We reared aphids from three different genotypes at low densities (7 aphids per plant), and then injected 60-104 adult aphids from each genotype with 0.3 μL dsRNA from either Apns-1 (ACYPI085607) or lacZ using a glass capillary needle (“Injection-first method”). After a further 24 hrs at low density, we exposed aphids to crowded conditions of 20 adult aphids per plant for 24 hrs. Following crowding, we moved aphids back to 4 adults per plant for 24 hrs. Offspring born during this 48-72 hrs post-injection time point were reared to 4th instar. We assigned each plant a random number so that data collection was blind to treatment, and we screened the offspring for the presence of wing buds to assess wing induction. We choose the 48-72 hour time point to screen for effects of RNAi knock-down because the results of a pilot experiment showed the largest effect of knock-down on the proportion of winged offspring during this period. Data from each of the three genotypes were analyzed separately using rank-sum tests as above, and an adjusted p-value (p < 0.0167) was used to determine statistical significance.
We performed a different version of the experiment where we crowded aphids before dsRNA injection (“Crowding first method”). We reared aphids from the same three genotypes at low densities for several generations, and crowded adult female aphids on small bean plants for 24 hrs at 20 aphids / plant. We then injected 76-120 aphids from each genotype with dsRNA from ACYPI085607 or lacZ as above. Aphids were then housed at low densities, and offspring born during the 48-72 time point were later screened for wing buds. We note that treatment (dsRNA from lacZ vs. Apns-1) had no significant effect on the reproductive output of injected aphids (F = 0.7058, df=1, p = 0.4022).
To validate expression knock-down from RNAi, we collected aphids from plants at 72 hrs after dsRNA injection, removed embryos as above, and stored samples in Trizol at −80°C. We extracted RNA as above, and then selectively enriched for mRNA using the NEBNext Poly(A) mRNA Magnetic Isolation Module under recommended conditions. Each sample was purified three times. We then converted mRNA to cDNA using Oligo(dT)20 primers with the Superscript IV First-Trand Synthesis System (ThermoFisher), followed by treatment with E. coli RNase H under recommended protocols. qPCR was then run on these cDNA samples as above. Expression knock-down was compared separately for each gene with non-parametric rank-sum tests on −ΔCT values as above.
Analysis of Viral Horizontal Gene Transfer.
We used the Dysaphis plantaginea densovirus nonstructural gene sequence (ACG50803.1) to query the pea aphid genome using blastp. Putative homologs (<e-10) were examined for expression in Aphidbase and in data from [29] and unpublished data (see Table S3).
The protein sequence of Apns-1 (ACYPI085607) was used in blastp searches to identify the putative homologs from other aphid species shown in Table S3. We created nucleotide alignments of the Apns-1 (ACYPI085607) homologs, plus sequence from the Myzus persicae densovirus (NP_874376.1) and the D. plantaginea densovirus (ACG50803.1) directed from amino acid sequence alignments using TranslatorX [30] We truncated the alignment to the portion containing the Apns-1 (ACYPI085607) protein. We used maximum likelihood analysis, as implemented in W-IQ-TREE [31], to recreate the phylogenetic relationships among the sequences using “Sequence type = Codon” and “Substitution model = Auto”. The consensus tree was constructed from 1000 bootstrap trees. We conducted an extension of this analysis using blast results from two additional aphid genomes: a blastp search of Myzus cerasi and a tblastn search of Diuraphis noxia (Figure S3B). We added these sequences to the previous alignment, altering the number of “T”s in a homopolymer run to maintain the reading frame for two D. noxia sequences and one M. cerasi sequence.
Quantification and statistical analysis
All statistical analyses were conducted in R v.3.5.0. Figures 1 and 2 were made in R v.3.5.0 and edited for style and layout using Adobe Illustrator CS6. The phylogenies in Figure 3 were made in W-IQ-TREE. The specific details of the types of statistical analyses used can be found in the Method Details section above. Statistical significance of experimental data was determined when p < 0.05, except in one case (Figure 2G) when a more stringent criterion (p < 0.0167) was used to account for multiple statistical tests. All experimental data was collected blinded (e.g. each experimental replicate was assigned a random number and data was collected using this number with no knowledge of treatment) as described in the Method Details above.
Data and software availability
Raw reads from the RNAseq study can be obtained on NCBI’s Sequence Read Archive (SRA) with accession numbers SAMN11482118-SAMN11482133. Gene lists produced from analyses of the RNAseq data and all of the experimental data from this study will be uploaded to the Dryad digital repository upon acceptance.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE |
SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| Fava bean plants (Viciafaba), Improved Long Pod variety | Harris Seeds, Rochester NY | Cat#00096-00-01-330 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| RNAlater-ICE Frozen Tissue Transition Solution | ThermoFisher Scientific | Cat#4427575 |
| Trizol Reagent | ThermoFisher Scientific | Cat#15596026 |
| 2-Propanol | Fisher Chemical | Cat#A416-500 |
| EvaGreen | VWR | Cat#89138-984 |
| Taq DNA polymerase (Invitrogen) | Invitrogen via Fisher Scientific | Cat#10-342-053 |
| Critical Commercial Assays | ||
| RNA Clean & Concentrator-5 (DNAse included) | Zymo Research | Cat#R1013 |
| TruSeq Stranded mRNA Library Prep Kit | Illumina | Cat#RS-122-2101 |
| iScript™ cDNA Synthesis Kit | BioRad USA | Cat#1708890 |
| Phusion® High-Fidelity PCR Master Mix with HF Buffer | New England Biolabs | Cat#M0531S |
| MEGAscript™ RNAi Kit | ThermoFisher Scientific | Cat#AM1626 |
| NEBNext® Poly(A) mRNA Magnetic Isolation Module | New England Biolabs | Cat#E7490S |
| SuperScript™ IV First-Strand Synthesis System | Invitrogen via ThermoFisher Scientific | Cat#18091200 |
| Oligonucleotides | ||
| qPCR Apns-1 F: | GCAAACGTCGTTTCTGCCTT | |
| qPCR Apns-1 R: | ACGACTACGAATCTGGCACG | |
| qPCR Apns-2 F: | AGTATCCTTGTTGTCCGCCC | |
| qPCR Apns-2 R: | GCGCACCAATTCCTAAGAGC | |
| qPCR G3PDH F: | CGGGAATTTCATTGAACGAC | |
| qPCR G3PDH F: | TCCACAACACGGTTGGAGTA | |
| qPCR NADH F: | CGAGGAGAACATGCTCTTAGAC | |
| qPCR NADH R: | GATAGCTTGGGCTGGACATATAG | |
| RNAi Apns F: | TAATACGACTCACTATAGGGTCCGTTTCAATAGCTTCCGAA | |
| RNAi Apns R: | TAATACGACTCACTATAGGGACTGCTGCACCGATGAAGAA | |
| Software and Algorithms | ||
| R | https://www.r-project.org/ | v.3.5.0 |
| Cutadapt | https://cutadapt.readthedocs.io/en/stable/ | v.1.2.1 |
| fastq-mcf (ea-utils) | https://expressionanalysis.github.io/ea-utils/ | v.1 |
| tophat | https://ccb.jhu.edu/software/tophat/index.shtml | v.2 |
| htseq | https://htseq.readthedocs.io/en/release_0.11.1/ | v.1 |
| edgeR | https://bioconductor.org/packages/release/bioc/html/edgeR.html | v.3.18.1 |
| TranslatorX | http://www.translatorx.co.uk/?COLLCC=3253199103& | |
| IQ-TREE | http://iqtree.cibiv.univie.ac.at/ | |
Highlights.
Aphid genotypes vary extensively in the plastic production of winged offspring
Two aphid genes of viral origin are upregulated only in highly-winged genotypes
Knockdown of these genes leads to a lower percentage of winged offspring
The genes likely retained their function after lateral transfer from a densovirus
Acknowledgements:
Angela Douglas and Seung Ho Chung provided the aphid lines used in the plasticity screen, which were invaluable to this work. Binshuang Li and Mary Grantham assisted with the RNAseq and Nanopore sequence analysis. Jennifer Keister, Brandon Courteau, and Omid Saleh Ziabari provided valuable technical assistance. Ryan Bickel, Jack Werren, and Angela Douglas kindly provided feedback on a draft version of the manuscript. Sequencing of transcriptome libraries was carried out by the University of Rochester Genomics Research Center. Rhopalosiphum padi DNA sequence data were downloaded from AphidBase, which acknowledges funding from ERC Starting Grant APHIDHOST-310190 to Jorunn Bos at the James Hutton Institute, UK. This work was funded by NSF IOS 1749514 and NIGMS 5R01GM116867 to JAB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests: The authors declare no competing interests.
References:
- 1.Leimar O, and McNamara JM (2015). The evolution of transgenerational integration of information in heterogeneous environments. The American Naturalist. 185:E55–69. [DOI] [PubMed] [Google Scholar]
- 2.Hazel WN, Smock R, and Johnson MD (1990). A polygenic model for the evolution and maintenance of conditional strategies. Proc. Biol. Sci. 242:181–7. [DOI] [PubMed] [Google Scholar]
- 3.Ryabov EV, Keane G, Naish N, Evered C, and Winstanley D (2009). Densovirus induces winged morphs in asexual clones of the rosy apple aphid, Dysaphis plantaginea. PNAS. 106:8465–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sutherland O (1969). The role of crowding in the production of winged forms by two strains of the pea aphid, Acyrthosiphon pisum. J. Insect Phys 15:1385–410. [Google Scholar]
- 5.Chung SH, Parker BJ, Blow F, Brisson JA, and Douglas AE (In Review). Genetic determinants of nutritional function in an intracellular symbiosis. [DOI] [PubMed] [Google Scholar]
- 6.Zera AJ, and Denno RF (1997). Physiology and ecology of dispersal polymorphism in insects. Annu. Rev. Entomol 42:207–30. [DOI] [PubMed] [Google Scholar]
- 7.Vellichirammal NN, Madayiputhiya N, and Brisson JA (2016). The genomewide transcriptional response underlying the pea aphid wing polyphenism. Mol. Ecol 25:4146–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grantham ME, Antonio CJ, O'Neil BR, Zhan YX, and Brisson JA (2016). A case for a joint strategy of diversified bet hedging and plasticity in the pea aphid wing polyphenism. Biol. Letts 12: 20160654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tijssen P, Pénzes JJ, Yu Q, Pham HT, and Bergoin M (2016). Diversity of small, single-stranded DNA viruses of invertebrates and their chaotic evolutionary past. J. Invert. Pathol 140:83–96. [DOI] [PubMed] [Google Scholar]
- 10.van Munster M, Dullemans A, Verbeek M, van den Heuvel J, Reinbold C, Brault V, Clerivet A, and van der Wilk F (2003). A new virus infecting Myzus persicae has a genome organization similar to the species of the genus Densovirus. J. Gen. Virol 84:165–72. [DOI] [PubMed] [Google Scholar]
- 11.Liu H, Fu Y, Xie J, Cheng J, Ghabrial SA, Li G, Peng Y, Yi X, and Jiang D (2011). Widespread endogenization of densoviruses and parvoviruses in animal and human genomes. J. Virol 85:9863–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clavijo G, van Munster M, Monsion B, Bochet N, and Brault V (2016). Transcription of densovirus endogenous sequences in the Myzus persicae genome. J. Gen. Virol 97:1000–9. [DOI] [PubMed] [Google Scholar]
- 13.Wenger JA, Cassone BJ, Legeai F, Johnston JS, Bansal R, Yates AD, Coates BS, Pavinato VAC, and Michel A (2017). Whole genome sequence of the soybean aphid, Aphis glycines. Insect Biochem. and Mol. Biol S0965-1748(17)30005-X. [DOI] [PubMed] [Google Scholar]
- 14.Mathers TC, Chen Y, Kaithakottil G, Legeai F, Mugford ST, Baa-Puyoulet P, Bretaudeau A, Clavijo B, Colella S, Collin O, et al. (2017). Rapid transcriptional plasticity of duplicated gene clusters enables a clonally reproducing aphid to colonise diverse plant species. Genome Biol. 18:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Legendre D, and Rommelaere J (1992). Terminal regions of the NS-1 protein of the parvovirus minute virus of mice are involved in cytotoxicity and promoter trans inhibition. J. Virol 66:5705–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iseki H, Shimizukawa R, Sugiyama F, Kunita S, Iwama A, Onodera M, Nakauchi H, and Yagami K (2005). Parvovirus nonstructural proteins induce an epigenetic modification through histone acetylation in host genes and revert tumor malignancy to benignancy. J. Virol 79:8886–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunning Hotopp JC, Clark ME, Oliveira DCSG, Foster JM, Fischer P, Muñoz Torres MC, Giebel JD, Kumar N, Ishmael N, Wang S et al. (2007). Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science. 317:1753–6. [DOI] [PubMed] [Google Scholar]
- 18.Aswad A, and Katzourakis A (2012). Paleovirology and virally derived immunity. Trends Ecol. Evol 27:627–36. [DOI] [PubMed] [Google Scholar]
- 19.Husnik F, and McCutcheon JP (2018). Functional horizontal gene transfer from bacteria to eukaryotes. Nature Rev. Microbio 16:67–79. [DOI] [PubMed] [Google Scholar]
- 20.Ragsdale EJ, Müller MR, Rödelsperger C, and Sommer RJ (2013). A developmental switch coupled to the evolution of plasticity acts through a sulfatase. Cell. 155:922–33. [DOI] [PubMed] [Google Scholar]
- 21.Lively C (1999). Developmental strategies in spatially variable environments: barnacle shell dimorphism and strategic models of selection In The Ecology and Evolution of Inducible Defenses, Tollrian R and Harvell CD, eds. (Princeton: Princeton University Press; ), pp. 245–58. [Google Scholar]
- 22.Lambers DHR (1966). Polymorphism in aphididae. Annu. Rev. Entomol 11:47–78. [Google Scholar]
- 23.Reyes ML, Laughton AM, Parker BJ, Wichmann H, Fan MH, Sok D, Hrcek J, Acevedo T, and Gerardo NM (2019). The influence of symbiotic bacteria on reproductive strategies and wing polyphenism in pea aphids responding to stress. J. Animal Ecol 88:601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bates D, Machler M, Bolker B, and Walker S (2015). Fitting linear mixed-effects models using lme4. J. Statistical Software 67:1–48. [Google Scholar]
- 25.O’Hara RB, and Kotze DJ (2010). Do not log-transform count data. Methods in Ecol. and Evol 1:118–22. [Google Scholar]
- 26.International Aphid Genomics Consortium. (2010). Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 8:e1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anders S, Pyl PT, and Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purandare SR, Bickel RD, Jaquiéry J, Rispe C, and Brisson JA (2014). Accelerated evolution of morph-biased genes in pea aphids. Mol. Biol. and Evol 31:2073–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abascal F, Zardoya R, and Telford MJ (2010). TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38:W7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trifinopoulos J, Nguyen L-T, von Haeseler A, and Minh BQ (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44:W232–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim H, Lee S, Jang Y (2011). Macroevolutionary patterns in the Aphidini aphids (Hemiptera: Aphididae): diversification, host association, and biogeographic origins. PLoS One. 6:e24749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hardy NB, Peterson DA, von Dohlen CD (2015). The evolution of life cycle complexity in aphids: Ecological optimization or historical constraint? Evolution. 69:1423–32. [DOI] [PubMed] [Google Scholar]
- 34.Notredame C, Higgins DG, and Heringa J (2000). T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol 302:205–17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw reads from the RNAseq study can be obtained on NCBI’s Sequence Read Archive (SRA) with accession numbers SAMN11482118-SAMN11482133. Gene lists produced from analyses of the RNAseq data and all of the experimental data from this study will be uploaded to the Dryad digital repository upon acceptance.