Background
Oxycodone is a semi-synthetic opioid derived from thebaine and is an opioid-receptor agonist used to provide analgesia to patients [1]. It has been in clinical use since 1917 [2] and is recommended for the treatment of moderate to severe pain [3], although it may not be effective in the treatment of some painful conditions, such as diabetic neuropathy [4]. The molecular structure of oxycodone contains two planar rings, two aliphatic rings and four chiral centers. Oxycodone molecules can form up to 16 stereoisomers and are not highly lipophilic [1, 5].
Oxycodone provides a similar level of analgesia as morphine, but has an increased bioavailability and half-life, prolonging the analgesic effect [6]. Compared to morphine, patients taking oxycodone experience a reduced frequency of side effects, particularly delirium [3, 7, 8]. This is thought to be due to the fact that accumulation of morphine metabolites in patients can lead to toxicity [9], while this is not the case for oxycodone. These characteristics can make oxycodone a preferable option for analgesia in patients. However, like other opioids, oxycodone and its active metabolite oxymorphone are liable to abuse and can lead to opioid dependence [10]. Indeed, oxycodone has been found to have an abuse liability similar to that of heroin [11].
Pharmacokinetics
Oxycodone can be administered in a number of different forms, including orally, rectally, intravenously and as an epidural. Some studies have observed differences in oxycodone pharmacokinetics between different administration forms [12, 13], including that intravenous oxycodone has a greater area under the curve (AUC) than oral or rectal administration, while others have found that the pharmacokinetics or pharmacodynamics of oral, rectal and intravenous oxycodone are similar [14, 15]. Regardless of the method of administration, the pharmacokinetics of oxycodone are dose-independent [16].
Delivering oxycodone as an epidural has been shown to be especially effective in providing analgesia. Cerebrospinal fluid (CSF) samples from patients receiving an oxycodone epidural show that the AUC of oxycodone was significantly higher than in CSF samples from patients receiving intravenous oxycodone [17]. Oxycodone concentrations in the CSF of epidural patients were 1000–10000 times higher than that of oxymorphone, suggesting that oxycodone provides the majority of the analgesic effect in these cases. Patients receiving an oxycodone epidural also required less rescue analgesia than those receiving intravenous oxycodone.
To reduce the abuse potential of immediate-release tablets, oxycodone is available in a controlled-release tablet form. Controlled-release oxycodone has a similar AUC to the immediate-release form, but a smaller Cmax and later Tmax [18, 19]. However, it should be noted that controlled-release oxycodone can be tampered with, so does not completely eliminate the risk of abuse [20]. Both immediate-release and controlled-release oxycodone reach steady-state concentrations after 24 hours of dosing [19] and taking oral oxycodone with or without food has little impact on the drug’s pharmacokinetics or pharmacodynamics [18, 21].
The age of a patient taking oxycodone can have a substantial impact on their exposure to the drug. The clearance of oxycodone decreases as patients age, prolonging the half-life [22]. Pediatric patients exhibit a 20–40% increase in oxycodone clearance compared to adults [23, 24], while studies in elderly patients have found that plasma concentrations, AUC and half-life of oxycodone are increased compared to younger patients [25–28]. Exposure to the inactive metabolite noroxycodone is also increased in elderly patients [25, 28].
Neonates and infants are an exception to this general trend, as oxycodone clearance increases with age in the first six months following birth [29–31]. In addition, in vitro studies of hepatocytes from neonate, infant, pediatric and adult populations found that fewer metabolites of oxycodone are seen in samples from patients younger than six months old [24], indicating that metabolism of oxycodone is not fully matured within this age group. This reduced clearance puts young infants at an increased risk of adverse side effects [31].
A study of oxycodone pharmacokinetics in women in labor and neonates found that pregnant women had greater clearance, a lower volume of distribution and a shorter elimination half-life compared to women who were not pregnant. Oxycodone concentrations in neonates were similar to maternal concentrations, indicating that maternal plasma concentrations of oxycodone can be used to predict fetal exposure. Neonates had only minor exposure to oxycodone metabolites, suggesting that the parent drug can cross the placenta more efficiently than its metabolites [32].
Clearance of oxycodone is severely decreased in patients with impaired liver function, with patients awaiting a liver transplant excreting more unchanged oxycodone than after they had received a transplant. Following liver transplantation, oxycodone pharmacokinetics were similar to values seen in healthy adults [33]. Cancer patients with cachexia also have an increased exposure to both oxycodone and its active metabolite oxymorphone [34]. This is thought to be due to a reduction in CYP3A4/5 enzyme activity, discussed in greater detail in the ‘Metabolism’ section below.
Absorption and distribution
Oral oxycodone has a bioavailability of 60–70%, with little difference in bioavailability between different forms of orally administered oxycodone (e.g. tablet, capsule, liquid) [27, 35]. Assessment of intranasal and sublingual absorption of oxycodone has found that bioavailability of these forms is low, potentially due to the low lipophilicity of the oxycodone molecule (Papp = 0.7) [5, 36, 37]. Absorption of oxycodone is higher in females than in males and increases with age [38].
Once absorbed into the blood, oxycodone is between 38–45% protein-bound [5, 39]. The vast majority of this binding is to albumin, with some additional binding to α1-acid glycoprotein (5–10% of the total protein-bound fraction) [39].
Metabolism
Figure 1 depicts the metabolism of oxycodone in hepatocytes. Oxycodone is primarily metabolized in the liver by CYP3A4/5, which mediates the N-demethylation of oxycodone to noroxycodone. Around 45% of a total dose of oxycodone is metabolized through this pathway [13, 16, 28, 40, 41]. Some noroxycodone is also produced as a result of first-pass metabolism by CYP3A4/5 in the intestine [13, 35]. Noroxycodone is considered to be inactive; when noroxycodone formation is blocked by the inhibition of CYP3A4/5, the analgesic effect of oxycodone is increased [42, 43].
Figure 1.
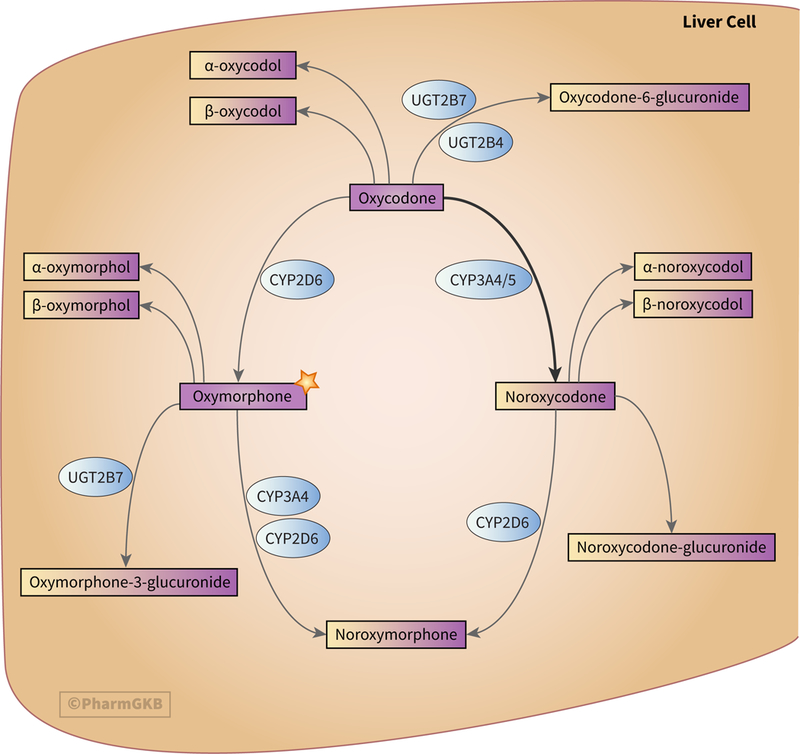
Stylized diagram showing oxycodone metabolism in the liver. The star denotes the significance of oxymorphone as the only active metabolite of oxycodone and that it is itself used clinically. A fully clickable version of this figure can be found at https://www.pharmgkb.org/pathway/PA166170927.
A further 19% of the total dose of oxycodone undergoes O-demethylation in the liver to form the active metabolite oxymorphone, a reaction catalyzed by CYP2D6 [16, 40, 41, 44]. Like its parent drug, oxymorphone also exerts an analgesic effect on the body [45–48] and is considered to be more potent than morphine while causing fewer side effects [49, 50]. However, other work has found it to be a weaker analgesic than oxycodone [51]. Oxymorphone can be directly administered to patients to provide analgesia and has been shown not to inhibit or induce CYP2C9 or CYP3A4 activity. This makes oxymorphone an attractive option for analgesia in patients taking multiple medications as the number of potential drug-drug interactions is reduced [52].
Both of oxycodone’s primary oxidative metabolites, oxymorphone and noroxycodone, are further metabolized to form the secondary metabolite noroxymorphone. The O-demethylation of noroxycodone to noroxymorphone is mediated by CYP2D6, while both CYP3A4 and CYP2D6 are able to catalyze the N-demethylation of oxymorphone to noroxymorphone [40].
A number of studies have investigated the effects of inhibition or induction of CYP2D6 and CYP3A4/5 on oxycodone metabolism. Inhibition of CYP2D6 by quinidine in healthy subjects blocked production of the active metabolite oxymorphone and increased the AUC of oxycodone [53]. Interestingly, these pharmacokinetic changes did not appear to have an impact on the results of psychomotor tests or subjective symptoms reported by the subjects, although potential impacts of CYP2D6 inhibition on the analgesic effects of oxycodone and oxymorphone were not investigated [53]. A case report of a patient prescribed oxycodone along with the CYP2D6 inhibitor fluoxetine described how the patient’s oxycodone dosage increased to overcome the inhibition effects of fluoxetine [44].
Some studies of the effects of CYP2D6 inhibition by paroxetine did not find any changes in the pharmacokinetics or clinical effects between oxycodone alone and co-administration of oxycodone and paroxetine [54, 55], while other studies have found that co-administration of paroxetine increased the AUC of noroxycodone and reduced the formation of oxymorphone and noroxymorphone, although it is unclear whether this had any impact on oxycodone-induced analgesia [25, 56].
Inhibition of CYP3A4/5 by numerous drugs, including antifungal agents and some antibiotics, reduces the formation of noroxycodone, increasing oxycodone exposure and causing the metabolism of oxycodone to shift towards the CYP2D6-mediated formation of oxymorphone [25, 42, 55, 57–59]. This increased exposure to oxycodone and its active metabolite can lead to an increase in clinical effects of the drug, including increased analgesia [42, 43]. Conversely, CYP3A4/5 induction by drugs such as rifampin and carbamazepine decreased the AUC and bioavailability of oxycodone and oxymorphone and, in most studies, also decreased drug effect, including analgesia [25, 35, 60, 61]. However, another study found no change in the effects of oxycodone when CYP3A4/5 is induced [62].
Inhibition of both CYP2D6 and CYP3A4/5, for example by concomitant use of paroxetine and itraconazole, increases exposure to oxycodone in subjects while reducing the concentrations of the oxidative metabolites noroxycodone, oxymorphone and noroxymorphone. However, no significant change in the pharmacological response was observed, suggesting that oxycodone provides the majority of the drug activity [54, 56, 63, 64].
Oxycodone, oxymorphone and noroxycodone can all undergo reduction or glucuronidation to produce other metabolites. 6-keto reduction of oxycodone, oxymorphone and noroxycodone forms oxycodol, oxymorphol and noroxycodol respectively. Each of these reductive metabolites has two stereoisomers, labelled α- and β- [16, 65]. The majority of oxycodone glucuronidation is carried out by UGT2B7 and UGT2B4 [41, 65], while the majority of oxymorphone molecules are glucuronidated by UGT2B7 to form oxymorphone-3-glucuronide [16, 66–68]. It is currently unclear how glucuronidation of noroxycodone is catalyzed.
The pharmacokinetics of oxycodone metabolism differ between males and females. Females have been found to have lower serum concentrations of the active molecules oxycodone and oxymorphone and higher concentrations of CYP3A4/5-derived metabolites than males [28, 38, 69]. It has been suggested that this is due to CYP3A4/5 having a higher activity in females than males [69], however no work has been carried out to determine whether this change in pharmacokinetics affects the achievement of analgesia in females compared to males.
Excretion
Approximately 72% of an oxycodone dose is excreted in the urine; 8% as oxycodone, 47% as oxidative metabolites and 18% as reduced metabolites [13, 16]. At the time of writing, it is not known how the remaining 28% of the original dose is excreted. 97% of excreted oxycodone is unconjugated [70]. Of the oxidative metabolites, 93% of oxymorphone is excreted in its glucuronidated form, while noroxycodone is mostly unconjugated when excreted [13, 70].
When oxymorphone is administered directly to a subject, only 50% of the dose is excreted in the urine. 44% of the original dose is excreted as oxymorphone-3-glucuronide with the remaining 6% excreted as unchanged oxymorphone or as reduced metabolites [70, 71].
The elimination of oxycodone and its metabolites is impaired in cancer patients with kidney dysfunction [72], potentially increasing oxycodone exposure [1]. However, a study of oxycodone pharmacokinetics in premature babies with immature renal function found that clearance of oxycodone was not affected [29].
Pharmacodynamics
Oxycodone and oxymorphone act on opioid receptors in the dorsal horn of the spinal cord [73]. Despite its low lipophilicity, current evidence from work in rats and sheep suggests that oxycodone is capable of crossing the blood-brain barrier without the need for an active transporter, such as p-glycoprotein [74]. Indeed, inhibition of p-glycoprotein in rats had no discernable effect on oxycodone’s ability to cross the blood-brain barrier and provide analgesia [75].
There are suggestions that an active transporter may be involved in the ability of oxymorphone to cross the blood-brain barrier in rats [76]. This transporter is unlikely to be p-glycoprotein (coded for by the gene ABCB1), as in vitro work has shown that oxymorphone is neither a substrate nor inhibitor of p-glycoprotein [77]. In the cases of both oxycodone and oxymorphone, further work is needed to establish whether these observations made in animal models are applicable to humans.
Once inside the brain, oxycodone binds the μ-opioid receptor (MOR) with 95.7% relative affinity [16]. Oxycodone exhibits only minimal binding at the κ- and δ-opioid receptors (KOR and DOR) [16]. Oxymorphone has a greater binding affinity to MOR than oxycodone, while noroxycodone and the reduced metabolites α- and β-oxycodol have reduced binding affinities to MOR compared to oxycodone [16].
Research investigating the ability of oxycodone and its metabolites to cause activation of the G-proteins coupled to MOR in vitro has found that oxycodone is a partial agonist, with oxymorphone showing 30- to 40-fold greater potency than the parent drug [16, 78]. Noroxycodone, which is considered an inactive metabolite, is less potent than oxycodone [78]. The fact that oxycodone is capable of G-protein activation supports the idea that oxycodone does not need to be bioactivated to be an effective analgesic [78].
Despite the fact that oxymorphone is an active analgesic, the results of some studies suggest that the analgesic effect of oxycodone is almost completely derived from the parent drug. As mentioned previously, inhibition of both CYP2D6 and CYP3A4/5 did not significantly affect the clinical effects of oxycodone [54, 56, 63, 64], while Klimas et al have calculated that, following oxycodone administration, oxymorphone concentrations at MOR are too low to provide a substantial analgesic effect and that oxycodone provides 83–95% of analgesia [79]. However, this is contradicted by reports of patients with the CYP2D6 poor metabolizer (PM) phenotype, who were unable to achieve sufficient analgesia using oxycodone [7, 80, 81], discussed in further detail in the Pharmacogenomics section. These reports imply that the CYP2D6-mediated metabolism of oxycodone to oxymorphone is involved in the analgesic effect of oxycodone.
Andreassen et al found that increased oxymorphone concentrations in patients were correlated in an increase in pain intensity [82]. This is thought to be due to patients with more severe pain taking an increased dose of oxycodone, thereby raising the concentrations of oxycodone metabolites, including oxymorphone, rather than oxymorphone having a noxious effect on patients. Further study is merited to resolve this discrepancy about the underlying biochemistry of oxycodone analgesia in the published literature.
Pharmacogenomics
CYP2D6 has been the focus for much of the pharmacogenetic work on oxycodone. In contrast to the well-defined impact of CYP2D6 variants on the metabolism of the opioids codeine and tramadol [83, 84], the effects of CYP2D6 phenotypes on oxycodone are less clear and the evidence is contradictory. It has been suggested that patients who are CYP2D6 PMs and are also taking a drug which inhibits the activity of CYP3A4/5 may have a substantially increased exposure to oxycodone and risk of oxycodone toxicity as they are less able to metabolize oxycodone through the CYP2D6 route [43].
There is a correlation of ethnicity to allelic variation of Cytochrome P450 genes, including CYP2D6. Although race is a social construct, some African ethnic groups appear to have a higher frequency of CYP2D6 decreased-function or non-functional alleles, which can contribute to a PM phenotype, as compared to Asian and Caucasian groups. As examples, the decreased function CYP2D6*17 and *29 alleles have been reported at increased frequencies in certain African populations. CYP2D6*17 has been found at a frequency of 34% in a Zimbabwean (Shona) population compared to frequencies of 0–0.9% in Asian and Caucasian populations, while CYP2D6*29 has been reported at a frequency of 20% in a Tanzanian population compared with a low frequency of 0% in Asian and Caucasian populations [85].
Additionally, the non-functional CYP2D6*3 allele has been reported at frequencies of 0–9% in African populations (9% in a Zimbabwean San population) compared with frequencies of 0–3.5% in Asian and Caucasian populations [85]. Lower CYP2D6 activity has also been observed in US populations of predominantly African descent [86], in part attributable to the previously described variant alleles occurring at a higher frequency in this population compared to US populations of predominantly European descent [86].
A number of case studies and observations made in publications suggest that a patient’s CYP2D6 metabolizer status may affect their response to oxycodone. There are reports of CYP2D6 PMs who were not able to sufficiently control their pain using oxycodone or required an increased dose of oxycodone compared to other metabolizer phenotypes [7, 80, 81]. Furthermore, patients identified as CYP2D6 UMs appear to experience a greater analgesic effect from oxycodone than CYP2D6 PMs [87, 88]. CYP2D6 UMs have also been found to have increased exposure to oxymorphone compared to other metabolizer phenotypes [26, 59] and there are multiple reports of CYP2D6 UMs experiencing adverse drug effects as a result of taking oxycodone [87, 89].
As discussed previously, these observations appear to contradict the assumption that oxymorphone does not make a significant contribution to oxycodone-induced analgesia. However, it should be emphasized that the majority of this evidence is based on observations made in either case reports on during the course of research studies rather than conclusions from fully analyzed datasets. It is also important to note that other studies, including large, multicenter studies, have found no difference in oxycodone exposure or response to oxycodone between CYP2D6 metabolizer phenotypes in both adult and pediatric patients [90–93].
Based on this evidence, the Royal Dutch Pharmacists Association - Pharmacogenetics Working Group (DPWG) have issued guidelines for prescribing oxycodone with regards to a patient’s CYP2D6 metabolizer phenotype, in which they recommend that CYP2D6 UMs, intermediate metabolizers (IMs) and PMs are prescribed an alternative analgesic [94, 95]. There is currently insufficient data for the group to recommend adjusting dosage of oxycodone for different CYP2D6 phenotypes. An annotated version of these guidelines can be found on the PharmGKB website at https://www.pharmgkb.org/guideline/PA166104973.
A clinical study by Jannetto and Bratanow attempted to use pharmacogenetics to inform prescription of opioids, including oxycodone. Although they saw no correlation between the steady-state concentration of oxycodone and CYP2D6 genotype, they subsequently discovered that the two CYP2D6 PMs in their study cohort had not adhered to the dosing schedule, which would have affected the results [96].
A study of opioid addiction, including patients using oxycodone, in different CYP2D6 metabolizer phenotypes has indicated that CYP2D6 PMs may have a degree of protection against opioid addiction [97]. In the case of oxycodone, it is suggested that CYP2D6 PMs are less able to produce oxymorphone and experience a reduced effect of oxycodone [97]. This would appear to contradict the notion that the majority of the effects of oxycodone are mediated by oxycodone itself rather than oxymorphone.
The authors also suggest that CYP2D6 UMs may also be partially protected from oxycodone dependence as they metabolize oxycodone too quickly to experience a positive reinforcing effect [97]. This hypothesis is partially supported by a mathematical model, which shows that CYP2D6 PMs have a reduced risk of becoming dependent on oxycodone [98].
As with CYP2D6, allelic variation in CYP3A4/5 in also correlated with ethnicity. Studies have shown a significantly higher incidence of CYP3A4*1B allelic frequency in populations of African descent (African 66–86%; European 2–4%, Asian 0%) [99, 100]. This allele is thought to increase expression of the CYP3A4 enzyme, leading to increased enzyme activity [101]. In the case of oxycodone, CYP3A4 UMs may have higher rates of poor analgesia due to the increased formation of the inactive metabolite noroxycodone. More study is needed to correlate CYP2D6 and CYP3A4/5 activity and clinical effects of oxycodone in sub-populations with increased frequencies of alleles which confer decreased or increased function.
Research investigating the effect of the non-functional CYP3A5*3 allele on oxycodone pharmacokinetics has found that the *3 allele has no effect on concentrations of oxycodone compared to patients who carried at least one CYP3A5*1 allele [90, 102]. However, there is conflicting information about whether the *3 allele affects noroxycodone plasma concentrations following administration of oxycodone [90, 102]. One study also found that the *3/*3 genotype was associated with increased oxycodone dose escalation [102], the reasons for which are not known at this time.
A study in Han Chinese patients with cancer pain observed a slight association between the TT genotype at rs7439366 in UGT2B7 and a reduced analgesic response to oxycodone compared to patients with the CC genotype (p=0.047). However, this genotype had no impact on the dosing needs of patients. Additionally, the CYP3A4*1G allele was found to have no effect on oxycodone dosage or analgesic response in these patients [103].
Other pharmacogenetics work has investigated the impact of variants in the transporter ABCB1 and the μ-, κ- and δ-opioid receptors, coded for by the genes OPRM1, OPRK1 and OPRD1 respectively. The variant rs1045642 in ABCB1 has been found to have no effect on oxycodone pharmacokinetics or analgesia [102, 104, 105], although one study found that carriers of the C allele at rs1045642 experienced more pronounced side effects than subjects with the T allele [104]. The T allele of rs2032582 in ABCB1 is associated with increased CNS depression in mothers of neonates [106] and an increased frequency of adverse drug reactions in healthy subjects [104]. However, conflicting evidence is given for its association with response to oxycodone in terms of pharmacokinetics and analgesia [104, 105].
There is evidence that the G allele at the SNP rs1799971 in OPRM1 is associated with lower pain tolerance thresholds and an increase in oxycodone dosage required to achieve analgesia compared to the AA genotype [104, 107, 108], while other work has found no link between rs1799971 and response to oxycodone [105]. rs589046, rs9479757, rs533586 and rs563649 in OPRM1 in addition to rs419335 and rs2234918 in OPRD1 are also associated with response to oxycodone in healthy volunteers under experimental pain conditions [108].
It should be noted that there is conflicting evidence regarding the association between rs1799971 and response to opioids more generally and it is possible that the nature of this association is dependent on both the specific opioid being used and the clinical context in which it is being used. While one meta-analysis looking at consumption of postsurgical opioids found that, similar to the oxycodone studies, the GG genotype is associated with increased opioid dosage [109], a meta-analysis of response to fentanyl in women in labor found that the G allele was associated with improved response to fentanyl [110].
Conclusion
There is still a great deal of uncertainty about many aspects of the pharmacokinetics and pharmacodynamics of oxycodone, chief among them being the relative contributions of oxycodone and oxymorphone to the overall analgesic effect and how the effects of oxycodone are modulated by polymorphisms in pharmacogenes. Many of the pharmacogenomic studies discussed here were carried out in Caucasian populations or in cohorts where Caucasian patients formed the overwhelming majority. There is therefore a pressing need for large-scale clinical studies to be carried out in ethnically diverse populations to clarify how CYP2D6 and CYP3A4/5 genotypes impact the effectiveness of oxycodone as an analgesic, particularly within the current context of the opioid crisis. The results of these studies could better inform analgesic selection, as in the DPWG guideline, or oxycodone dosing for patients.
Acknowledgements
The authors thank Li Gong for critical reading of the manuscript. This work is supported by the NIH/NIGMS grant GM61374.
Footnotes
Conflict of interest: RBA is a stockholder in Personalis Inc. and 23andMe, and a paid advisor for Karius
References
- 1.Olkkola KT, et al. , Does the pharmacology of oxycodone justify its increasing use as an analgesic? Trends Pharmacol Sci, 2013. 34(4): p. 206–14. [DOI] [PubMed] [Google Scholar]
- 2.Falk E, Eukodal, ein neues Narkotikum. Munchener Medizinische Wochenschrift, 1917. 20: p. 381–384. [Google Scholar]
- 3.Riley J, et al. , Oxycodone: a review of its use in the management of pain. Curr Med Res Opin, 2008. 24(1): p. 175–92. [DOI] [PubMed] [Google Scholar]
- 4.Gaskell H, et al. , Oxycodone for neuropathic pain in adults. Cochrane Database Syst Rev, 2016. 7: p. Cd010692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poyhia R and Seppala T, Liposolubility and protein binding of oxycodone in vitro. Pharmacol Toxicol, 1994. 74(1): p. 23–7. [DOI] [PubMed] [Google Scholar]
- 6.Poyhia R, et al. , The pharmacokinetics of oxycodone after intravenous injection in adults. Br J Clin Pharmacol, 1991. 32(4): p. 516–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maddocks I, et al. , Attenuation of morphine-induced delirium in palliative care by substitution with infusion of oxycodone. J Pain Symptom Manage, 1996. 12(3): p. 182–9. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt-Hansen M, et al. , Oxycodone for cancer-related pain. Cochrane Database Syst Rev, 2017. 8: p. Cd003870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gretton SK, et al. , Plasma morphine and metabolite concentrations are associated with clinical effects of morphine in cancer patients. J Pain Symptom Manage, 2013. 45(4): p. 670–80. [DOI] [PubMed] [Google Scholar]
- 10.Dart RC, et al. , Trends in opioid analgesic abuse and mortality in the United States. N Engl J Med, 2015. 372(3): p. 241–8. [DOI] [PubMed] [Google Scholar]
- 11.Comer SD, et al. , Abuse liability of prescription opioids compared to heroin in morphine-maintained heroin abusers. Neuropsychopharmacology, 2008. 33(5): p. 1179–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leow KP, et al. , Comparative oxycodone pharmacokinetics in humans after intravenous, oral, and rectal administration. Ther Drug Monit, 1992. 14(6): p. 479–84. [DOI] [PubMed] [Google Scholar]
- 13.Poyhia R, et al. , The pharmacokinetics and metabolism of oxycodone after intramuscular and oral administration to healthy subjects. Br J Clin Pharmacol, 1992. 33(6): p. 617–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leow KP, et al. , Single-dose and steady-state pharmacokinetics and pharmacodynamics of oxycodone in patients with cancer. Clin Pharmacol Ther, 1992. 52(5): p. 487–95. [DOI] [PubMed] [Google Scholar]
- 15.Leow KP, Cramond T, and Smith MT, Pharmacokinetics and pharmacodynamics of oxycodone when given intravenously and rectally to adult patients with cancer pain. Anesth Analg, 1995. 80(2): p. 296–302. [DOI] [PubMed] [Google Scholar]
- 16.Lalovic B, et al. , Pharmacokinetics and pharmacodynamics of oral oxycodone in healthy human subjects: role of circulating active metabolites. Clin Pharmacol Ther, 2006. 79(5): p. 461–79. [DOI] [PubMed] [Google Scholar]
- 17.Kokki M, et al. , Central nervous system penetration of oxycodone after intravenous and epidural administration. Br J Anaesth, 2014. 112(1): p. 133–40. [DOI] [PubMed] [Google Scholar]
- 18.Benziger DP, et al. , Differential effects of food on the bioavailability of controlled-release oxycodone tablets and immediate-release oxycodone solution. J Pharm Sci, 1996. 85(4): p. 407–10. [DOI] [PubMed] [Google Scholar]
- 19.Reder RF, et al. , Steady-state bioavailability of controlled-release oxycodone in normal subjects. Clin Ther, 1996. 18(1): p. 95–105. [DOI] [PubMed] [Google Scholar]
- 20.Peacock A, et al. , Methods and predictors of tampering with a tamper-resistant controlled-release oxycodone formulation. Int J Drug Policy, 2015. 26(12): p. 1265–72. [DOI] [PubMed] [Google Scholar]
- 21.Scheidel B, et al. , Bioavailability of oxycodone after administration of a new prolonged-release once-daily tablet formulation in healthy subjects, in comparison to an established twice-daily tablet. Int J Clin Pharmacol Ther, 2017. 55(11): p. 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saari TI, et al. , Oxycodone clearance is markedly reduced with advancing age: a population pharmacokinetic study. Br J Anaesth, 2012. 108(3): p. 491–8. [DOI] [PubMed] [Google Scholar]
- 23.Olkkola KT, et al. , Pharmacokinetics and ventilatory effects of intravenous oxycodone in postoperative children. Br J Clin Pharmacol, 1994. 38(1): p. 71–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korjamo T, et al. , Metabolism of oxycodone in human hepatocytes from different age groups and prediction of hepatic plasma clearance. Front Pharmacol, 2011. 2: p. 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liukas A, et al. , Plasma concentrations of oral oxycodone are greatly increased in the elderly. Clin Pharmacol Ther, 2008. 84(4): p. 462–7. [DOI] [PubMed] [Google Scholar]
- 26.Liukas A, et al. , Elimination of intravenous oxycodone in the elderly: a pharmacokinetic study in postoperative orthopaedic patients of different age groups. Drugs Aging, 2011. 28(1): p. 41–50. [DOI] [PubMed] [Google Scholar]
- 27.Kokki M, et al. , Absorption of different oral dosage forms of oxycodone in the elderly: a cross-over clinical trial in patients undergoing cystoscopy. Eur J Clin Pharmacol, 2012. 68(10): p. 1357–63. [DOI] [PubMed] [Google Scholar]
- 28.Elder NM, et al. , Observations of urinary oxycodone and metabolite distributions in pain patients. J Anal Toxicol, 2014. 38(3): p. 129–34. [DOI] [PubMed] [Google Scholar]
- 29.Kokki M, et al. , Maturation of oxycodone pharmacokinetics in neonates and infants: Oxycodone and its metabolites in plasma and urine. Br J Clin Pharmacol, 2017. 83(4): p. 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valitalo P, et al. , Maturation of Oxycodone Pharmacokinetics in Neonates and Infants: a Population Pharmacokinetic Model of Three Clinical Trials. Pharm Res, 2017. 34(5): p. 1125–1133. [DOI] [PubMed] [Google Scholar]
- 31.Pokela ML, et al. , Marked variation in oxycodone pharmacokinetics in infants. Paediatr Anaesth, 2005. 15(7): p. 560–5. [DOI] [PubMed] [Google Scholar]
- 32.Kokki M, et al. , Intravenous oxycodone for pain relief in the first stage of labour--maternal pharmacokinetics and neonatal exposure. Basic Clin Pharmacol Toxicol, 2012. 111(3): p. 182–8. [DOI] [PubMed] [Google Scholar]
- 33.Tallgren M, et al. , Pharmacokinetics and ventilatory effects of oxycodone before and after liver transplantation. Clin Pharmacol Ther, 1997. 61(6): p. 655–61. [DOI] [PubMed] [Google Scholar]
- 34.Naito T, et al. , Cancer cachexia raises the plasma concentration of oxymorphone through the reduction of CYP3A but not CYP2D6 in oxycodone-treated patients. J Clin Pharmacol, 2013. 53(8): p. 812–8. [DOI] [PubMed] [Google Scholar]
- 35.Nieminen TH, et al. , Rifampin greatly reduces the plasma concentrations of intravenous and oral oxycodone. Anesthesiology, 2009. 110(6): p. 1371–8. [DOI] [PubMed] [Google Scholar]
- 36.Weinberg DS, et al. , Sublingual absorption of selected opioid analgesics. Clin Pharmacol Ther, 1988. 44(3): p. 335–42. [DOI] [PubMed] [Google Scholar]
- 37.Takala A, et al. , Pharmacokinetic comparison of intravenous and intranasal administration of oxycodone. Acta Anaesthesiol Scand, 1997. 41(2): p. 309–12. [DOI] [PubMed] [Google Scholar]
- 38.Kaiko RF, et al. , Pharmacokinetic-pharmacodynamic relationships of controlled-release oxycodone. Clin Pharmacol Ther, 1996. 59(1): p. 52–61. [DOI] [PubMed] [Google Scholar]
- 39.Leow KP, et al. , Determination of the serum protein binding of oxycodone and morphine using ultrafiltration. Ther Drug Monit, 1993. 15(5): p. 440–7. [DOI] [PubMed] [Google Scholar]
- 40.Lalovic B, et al. , Quantitative contribution of CYP2D6 and CYP3A to oxycodone metabolism in human liver and intestinal microsomes. Drug Metab Dispos, 2004. 32(4): p. 447–54. [DOI] [PubMed] [Google Scholar]
- 41.Romand S, et al. , Characterization of oxycodone in vitro metabolism by human cytochromes P450 and UDP-glucuronosyltransferases. J Pharm Biomed Anal, 2017. 144: p. 129–137. [DOI] [PubMed] [Google Scholar]
- 42.Hagelberg NM, et al. , Voriconazole drastically increases exposure to oral oxycodone. Eur J Clin Pharmacol, 2009. 65(3): p. 263–71. [DOI] [PubMed] [Google Scholar]
- 43.Saari TI, et al. , Effects of itraconazole on the pharmacokinetics and pharmacodynamics of intravenously and orally administered oxycodone. Eur J Clin Pharmacol, 2010. 66(4): p. 387–97. [DOI] [PubMed] [Google Scholar]
- 44.Otton SV, et al. , Inhibition by fluoxetine of cytochrome P450 2D6 activity. Clin Pharmacol Ther, 1993. 53(4): p. 401–9. [DOI] [PubMed] [Google Scholar]
- 45.Gimbel JS, et al. , Efficacy and safety of oxymorphone immediate release for the treatment of mild to moderate pain after ambulatory orthopedic surgery: results of a randomized, double-blind, placebo-controlled trial. Arch Phys Med Rehabil, 2005. 86(12): p. 2284–9. [DOI] [PubMed] [Google Scholar]
- 46.Aqua K, et al. , Efficacy and tolerability of oxymorphone immediate release for acute postoperative pain after abdominal surgery: a randomized, double-blind, active- and placebo-controlled, parallel-group trial. Clin Ther, 2007. 29(6): p. 1000–12. [DOI] [PubMed] [Google Scholar]
- 47.Gimbel J and Ahdieh H, The efficacy and safety of oral immediate-release oxymorphone for postsurgical pain. Anesth Analg, 2004. 99(5): p. 1472–7; table of contents. [DOI] [PubMed] [Google Scholar]
- 48.Hale ME, Dvergsten C, and Gimbel J, Efficacy and safety of oxymorphone extended release in chronic low back pain: results of a randomized, double-blind, placebo- and active-controlled phase III study. J Pain, 2005. 6(1): p. 21–8. [DOI] [PubMed] [Google Scholar]
- 49.Beaver WT, et al. , Comparisons of the analgesic effects of oral and intramuscular oxymorphone and of intramuscular oxymorphone and morphine in patients with cancer. J Clin Pharmacol, 1977. 17(4): p. 186–98. [DOI] [PubMed] [Google Scholar]
- 50.Eddy NB and Lee LE Jr., The analgesic equivalence to morphine and relative side action liability of oxymorphone (14-hydroxydihydro morphinone). J Pharmacol Exp Ther, 1959. 125(2): p. 116–21. [PubMed] [Google Scholar]
- 51.Babalonis S, et al. , Pharmacodynamic effects of oral oxymorphone: abuse liability, analgesic profile and direct physiologic effects in humans. Addict Biol, 2016. 21(1): p. 146–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams M, et al. , Oxymorphone extended release does not affect CYP2C9 or CYP3A4 metabolic pathways. J Clin Pharmacol, 2005. 45(3): p. 337–45. [DOI] [PubMed] [Google Scholar]
- 53.Heiskanen T, Olkkola KT, and Kalso E, Effects of blocking CYP2D6 on the pharmacokinetics and pharmacodynamics of oxycodone. Clin Pharmacol Ther, 1998. 64(6): p. 603–11. [DOI] [PubMed] [Google Scholar]
- 54.Gronlund J, et al. , Exposure to oral oxycodone is increased by concomitant inhibition of CYP2D6 and 3A4 pathways, but not by inhibition of CYP2D6 alone. Br J Clin Pharmacol, 2010. 70(1): p. 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kummer O, et al. , Effect of the inhibition of CYP3A4 or CYP2D6 on the pharmacokinetics and pharmacodynamics of oxycodone. Eur J Clin Pharmacol, 2011. 67(1): p. 63–71. [DOI] [PubMed] [Google Scholar]
- 56.Gronlund J, et al. , Effect of inhibition of cytochrome P450 enzymes 2D6 and 3A4 on the pharmacokinetics of intravenous oxycodone: a randomized, three-phase, crossover, placebo-controlled study. Clin Drug Investig, 2011. 31(3): p. 143–53. [DOI] [PubMed] [Google Scholar]
- 57.Gronlund J, et al. , Effect of telithromycin on the pharmacokinetics and pharmacodynamics of oral oxycodone. J Clin Pharmacol, 2010. 50(1): p. 101–8. [DOI] [PubMed] [Google Scholar]
- 58.Liukas A, et al. , Inhibition of cytochrome P450 3A by clarithromycin uniformly affects the pharmacokinetics and pharmacodynamics of oxycodone in young and elderly volunteers. J Clin Psychopharmacol, 2011. 31(3): p. 302–8. [DOI] [PubMed] [Google Scholar]
- 59.Nieminen TH, et al. , Grapefruit juice enhances the exposure to oral oxycodone. Basic Clin Pharmacol Toxicol, 2010. 107(4): p. 782–8. [DOI] [PubMed] [Google Scholar]
- 60.Sakamoto A, et al. , Oxycodone Resistance Due to Rifampin Use in an Osteosarcoma Patient with Tuberculosis. Am J Case Rep, 2017. 18: p. 1130–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee HK, et al. , Negative urine opioid screening caused by rifampin-mediated induction of oxycodone hepatic metabolism. Clin Chim Acta, 2006. 367(1–2): p. 196–200. [DOI] [PubMed] [Google Scholar]
- 62.Nieminen TH, et al. , St John’s wort greatly reduces the concentrations of oral oxycodone. Eur J Pain, 2010. 14(8): p. 854–9. [DOI] [PubMed] [Google Scholar]
- 63.Gronlund J, et al. , Miconazole oral gel increases exposure to oral oxycodone by inhibition of CYP2D6 and CYP3A4. Antimicrob Agents Chemother, 2011. 55(3): p. 1063–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nieminen TH, et al. , Oxycodone concentrations are greatly increased by the concomitant use of ritonavir or lopinavir/ritonavir. Eur J Clin Pharmacol, 2010. 66(10): p. 977–85. [DOI] [PubMed] [Google Scholar]
- 65.Moore KA, et al. , Tentative identification of novel oxycodone metabolites in human urine. J Anal Toxicol, 2003. 27(6): p. 346–52. [DOI] [PubMed] [Google Scholar]
- 66.Adams MP and Ahdieh H, Single- and multiple-dose pharmacokinetic and dose-proportionality study of oxymorphone immediate-release tablets. Drugs R D, 2005. 6(2): p. 91–9. [DOI] [PubMed] [Google Scholar]
- 67.Foulds JA, et al. , OPRM1 genotype and naltrexone response in depressed alcohol-dependent patients. Pharmacogenet Genomics, 2015. 25(5): p. 270–3. [DOI] [PubMed] [Google Scholar]
- 68.Coffman BL, et al. , The glucuronidation of opioids, other xenobiotics, and androgens by human UGT2B7Y(268) and UGT2B7H(268). Drug Metab Dispos, 1998. 26(1): p. 73–7. [PubMed] [Google Scholar]
- 69.Andreassen TN, et al. , Influences on the pharmacokinetics of oxycodone: a multicentre cross-sectional study in 439 adult cancer patients. Eur J Clin Pharmacol, 2011. 67(5): p. 493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yee DA, et al. , Observations on the urine metabolic ratio of oxymorphone to oxycodone in pain patients. J Anal Toxicol, 2012. 36(4): p. 232–8. [DOI] [PubMed] [Google Scholar]
- 71.Cone EJ, et al. , Oxymorphone metabolism and urinary excretion in human, rat, guinea pig, rabbit, and dog. Drug Metab Dispos, 1983. 11(5): p. 446–50. [PubMed] [Google Scholar]
- 72.Kirvela M, et al. , The pharmacokinetics of oxycodone in uremic patients undergoing renal transplantation. J Clin Anesth, 1996. 8(1): p. 13–8. [DOI] [PubMed] [Google Scholar]
- 73.Kokki HK,M, Central Nervous System Penetration of the Opioid Oxycodone, in Neuropathology of Drug Addictions and Substance Misuse, Preedy VP, Editor. 2016, Elsevier; p. 457–466. [Google Scholar]
- 74.Villesen HH, et al. , Cerebral kinetics of oxycodone in conscious sheep. J Pharm Sci, 2006. 95(8): p. 1666–76. [DOI] [PubMed] [Google Scholar]
- 75.Bostrom E, Simonsson US, and Hammarlund-Udenaes M, Oxycodone pharmacokinetics and pharmacodynamics in the rat in the presence of the P-glycoprotein inhibitor PSC833. J Pharm Sci, 2005. 94(5): p. 1060–6. [DOI] [PubMed] [Google Scholar]
- 76.Sadiq MW, et al. , Oxymorphone active uptake at the blood-brain barrier and population modeling of its pharmacokinetic-pharmacodynamic relationship. J Pharm Sci, 2013. 102(9): p. 3320–31. [DOI] [PubMed] [Google Scholar]
- 77.Metcalf MD, et al. , Opioids and efflux transporters. Part 4: influence of N-substitution on P-glycoprotein substrate activity of noroxymorphone analogues. Bioorg Med Chem Lett, 2014. 24(15): p. 3592–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thompson CM, et al. , Activation of G-proteins by morphine and codeine congeners: insights to the relevance of O- and N-demethylated metabolites at mu- and delta-opioid receptors. J Pharmacol Exp Ther, 2004. 308(2): p. 547–54. [DOI] [PubMed] [Google Scholar]
- 79.Klimas R, et al. , Contribution of oxycodone and its metabolites to the overall analgesic effect after oxycodone administration. Expert Opin Drug Metab Toxicol, 2013. 9(5): p. 517–28. [DOI] [PubMed] [Google Scholar]
- 80.Susce MT, Murray-Carmichael E, and de Leon J, Response to hydrocodone, codeine and oxycodone in a CYP2D6 poor metabolizer. Prog Neuropsychopharmacol Biol Psychiatry, 2006. 30(7): p. 1356–8. [DOI] [PubMed] [Google Scholar]
- 81.Foster A, Mobley E, and Wang Z, Complicated pain management in a CYP450 2D6 poor metabolizer. Pain Pract, 2007. 7(4): p. 352–6. [DOI] [PubMed] [Google Scholar]
- 82.Andreassen TN, et al. , Is oxycodone efficacy reflected in serum concentrations? A multicenter, cross-sectional study in 456 adult cancer patients. J Pain Symptom Manage, 2012. 43(4): p. 694–705. [DOI] [PubMed] [Google Scholar]
- 83.Crews KR, et al. , Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin Pharmacol Ther, 2014. 95(4): p. 376–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gong L, et al. , PharmGKB summary: tramadol pathway. Pharmacogenet Genomics, 2014. 24(7): p. 374–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rajman I, et al. , African Genetic Diversity: Implications for Cytochrome P450-mediated Drug Metabolism and Drug Development. EBioMedicine, 2017. 17: p. 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gaedigk A, et al. , Unique CYP2D6 activity distribution and genotype-phenotype discordance in black Americans. Clin Pharmacol Ther, 2002. 72(1): p. 76–89. [DOI] [PubMed] [Google Scholar]
- 87.Samer CF, et al. , Genetic polymorphisms and drug interactions modulating CYP2D6 and CYP3A activities have a major effect on oxycodone analgesic efficacy and safety. Br J Pharmacol, 2010. 160(4): p. 919–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zwisler ST, et al. , The hypoalgesic effect of oxycodone in human experimental pain models in relation to the CYP2D6 oxidation polymorphism. Basic Clin Pharmacol Toxicol, 2009. 104(4): p. 335–44. [DOI] [PubMed] [Google Scholar]
- 89.de Leon J, Dinsmore L, and Wedlund P, Adverse drug reactions to oxycodone and hydrocodone in CYP2D6 ultrarapid metabolizers. J Clin Psychopharmacol, 2003. 23(4): p. 420–1. [DOI] [PubMed] [Google Scholar]
- 90.Stamer UM, et al. , CYP2D6 genotype dependent oxycodone metabolism in postoperative patients. PLoS One, 2013. 8(3): p. e60239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Balyan R, et al. , CYP2D6 pharmacogenetic and oxycodone pharmacokinetic association study in pediatric surgical patients. Pharmacogenomics, 2017. 18(4): p. 337–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zwisler ST, et al. , Impact of the CYP2D6 genotype on post-operative intravenous oxycodone analgesia. Acta Anaesthesiol Scand, 2010. 54(2): p. 232–40. [DOI] [PubMed] [Google Scholar]
- 93.Andreassen TN, et al. , Do CYP2D6 genotypes reflect oxycodone requirements for cancer patients treated for cancer pain? A cross-sectional multicentre study. Eur J Clin Pharmacol, 2012. 68(1): p. 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Swen JJ, et al. , Pharmacogenetics: from bench to byte. Clin Pharmacol Ther, 2008. 83(5): p. 781–7. [DOI] [PubMed] [Google Scholar]
- 95.Swen JJ, et al. , Pharmacogenetics: from bench to byte--an update of guidelines. Clin Pharmacol Ther, 2011. 89(5): p. 662–73. [DOI] [PubMed] [Google Scholar]
- 96.Jannetto PJ and Bratanow NC, Utilization of pharmacogenomics and therapeutic drug monitoring for opioid pain management. Pharmacogenomics, 2009. 10(7): p. 1157–67. [DOI] [PubMed] [Google Scholar]
- 97.Tyndale RF, Droll KP, and Sellers EM, Genetically deficient CYP2D6 metabolism provides protection against oral opiate dependence. Pharmacogenetics, 1997. 7(5): p. 375–9. [DOI] [PubMed] [Google Scholar]
- 98.Linares OA, et al. , The CYP2D6 gene determines oxycodone’s phenotype-specific addictive potential: implications for addiction prevention and treatment. Med Hypotheses, 2014. 82(3): p. 390–4. [DOI] [PubMed] [Google Scholar]
- 99.Bains RK, African variation at Cytochrome P450 genes: Evolutionary aspects and the implications for the treatment of infectious diseases. Evol Med Public Health, 2013. 2013(1): p. 118–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Marwa KJ, et al. , Cytochrome P450 single nucleotide polymorphisms in an indigenous Tanzanian population: a concern about the metabolism of artemisinin-based combinations. Malar J, 2014. 13: p. 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Amirimani B, et al. , Increased transcriptional activity of the CYP3A4*1B promoter variant. Environ Mol Mutagen, 2003. 42(4): p. 299–305. [DOI] [PubMed] [Google Scholar]
- 102.Naito T, et al. , CYP3A5*3 affects plasma disposition of noroxycodone and dose escalation in cancer patients receiving oxycodone. J Clin Pharmacol, 2011. 51(11): p. 1529–38. [DOI] [PubMed] [Google Scholar]
- 103.Li J, et al. , The impact of UGT2B7 C802T and CYP3A4*1G polymorphisms on pain relief in cancer patients receiving oxycontin. Support Care Cancer, 2018. [DOI] [PubMed]
- 104.Zwisler ST, et al. , The antinociceptive effect and adverse drug reactions of oxycodone in human experimental pain in relation to genetic variations in the OPRM1 and ABCB1 genes. Fundam Clin Pharmacol, 2010. 24(4): p. 517–24. [DOI] [PubMed] [Google Scholar]
- 105.Zwisler ST, et al. , Lack of association of OPRM1 and ABCB1 single-nucleotide polymorphisms to oxycodone response in postoperative pain. J Clin Pharmacol, 2012. 52(2): p. 234–42. [DOI] [PubMed] [Google Scholar]
- 106.Lam J, et al. , Putative association of ABCB1 2677G>T/A with oxycodone-induced central nervous system depression in breastfeeding mothers. Ther Drug Monit, 2013. 35(4): p. 466–72. [DOI] [PubMed] [Google Scholar]
- 107.Cajanus K, et al. , How much oxycodone is needed for adequate analgesia after breast cancer surgery: effect of the OPRM1 118A>G polymorphism. J Pain, 2014. 15(12): p. 1248–56. [DOI] [PubMed] [Google Scholar]
- 108.Olesen AE, et al. , The genetic influences on oxycodone response characteristics in human experimental pain. Fundam Clin Pharmacol, 2015. 29(4): p. 417–25. [DOI] [PubMed] [Google Scholar]
- 109.Choi SW, et al. , Effects of Single Nucleotide Polymorphisms on Surgical and Post-surgical Opioid Requirements - A Systematic Review and Meta-analysis. Clin J Pain, 2017. [DOI] [PubMed]
- 110.Song Z, et al. , Effects of OPRM1 A118G polymorphism on epidural analgesia with fentanyl during labor: a meta-analysis. Genet Test Mol Biomarkers, 2013. 17(10): p. 743–9. [DOI] [PubMed] [Google Scholar]


