Abstract
Coenzyme Q (CoQ or ubiquinone) serves as an essential redox-active lipid in respiratory electron and proton transport during cellular energy metabolism. CoQ also functions as a membrane-localized antioxidant protecting cells against lipid peroxidation. CoQ deficiency is associated with multiple human diseases; CoQ10 supplementation in particular has noted cardioprotective benefits. In Saccharomyces cerevisiae, Coq10, a putative START domain protein, is believed to chaperone CoQ to sites where it functions. Yeast coq10 deletion mutants (coq10Δ) synthesize CoQ inefficiently during log phase growth and are respiratory defective and sensitive to oxidative stress. Humans have two orthologs of yeast COQ10, COQ10A and COQ10B. Here, we tested the human co-orthologs for their ability to rescue the yeast mutant. We showed that expression of either human ortholog, COQ10A or COQ10B, rescues yeast coq10Δ mutant phenotypes, restoring the function of respiratory-dependent growth on a nonfermentable carbon source and sensitivity to oxidative stress induced by treatment with PUFAs. These effects indicate a strong functional conservation of Coq10 across different organisms. However, neither COQ10A nor COQ10B restored CoQ biosynthesis when expressed in the yeast coq10Δ mutant. The involvement of yeast Coq10 in CoQ biosynthesis may rely on its interactions with another protein, possibly Coq11, which is not found in humans. Coexpression analyses of yeast COQ10 and human COQ10A and COQ10B provide additional insights to functions of these START domain proteins and their potential roles in other biologic pathways.
Keywords: antioxidants, lipids/chemistry, lipids/peroxidation, mass spectrometry, mitochondria, Saccharomyces cerevisiae, ubiquinone, steroidogenic acute regulatory protein-related lipid transfer
Coenzyme Q (CoQ) is a lipid composed of a fully substituted redox-active benzoquinone ring attached to a long polyisoprenoid chain. The polyisoprenoid chain of CoQn, with n ≥ 6 isoprene units, anchors CoQ at the mid-plane of the membrane phospholipid bilayers. The reversible reduction and oxidation of CoQ and CoQH2 enables the transport of electrons and protons necessary for cellular respiration. CoQ also serves as an important electron acceptor for enzymes involved in fatty acid β-oxidation, oxidation of proline and sulfide, and pyrimidine biosynthesis (1–3). The reduced or hydroquinone form of CoQH2 serves as a chain-terminating antioxidant that slows lipid peroxidation (2).
Although CoQ exists in most biological membranes, its synthesis occurs exclusively inside the mitochondria in eukaryotes, or in the cytosol in Escherichia coli, catalyzed by a cohort of enzymes, many of which are organized in a complex known as the CoQ synthome (also known as complex Q) in eukaryotes, or the Ubi metabolon in E. coli (4–6). In Saccharomyces cerevisiae, currently known members of the CoQ synthome consist of Coq3–Coq9 and Coq11 (4, 5). Together, they modify the quinone head group through a series of methylation (Coq3 and Coq5), deamination (Coq6, Coq9), and hydroxylation (Coq6, Coq7, Coq9) reactions (4, 5). The definitive functions of the remaining members of the CoQ synthome (Coq4, Coq8, and Coq11) are yet to be fully characterized. Attachment of the polyisoprenoid chain to the aromatic ring precursor precedes the ring modification steps. Coq1, a hexaprenyl pyrophosphate synthetase, condenses farnesyl pyrophosphate with three molecules of isopentenyl pyrophosphate to form hexaprenyldiphosphate, which is transferred to the 4-hydroxybenzoic acid (4HB) or para-aminobenzoic acid (pABA) ring at the C3 position by Coq2 (4, 5). The number of isoprene units (n) in the polyisoprenoid chain of CoQn varies between organisms, as determined by the specific polyprenyl diphosphate synthase (7), and consists of six isoprene units in S. cerevisiae (CoQ6), eight isoprene units in E. coli (CoQ8), and predominantly ten isoprene units in Schizosaccharomyces pombe and humans (CoQ10) (8). Each of the yeast coq1Δ–coq9Δ mutants shows complete abolishment of CoQ6 biosynthesis and fails to respire (5). Their defects in respiration can be readily restored by exogenous supplementation with CoQ6 (5).
The coq10Δ mutant is unusual among the yeast coq mutants because it produces wild-type content of CoQ6 at stationary phase, yet its de novo synthesis of CoQ6 during log phase is inefficient (9, 10). Despite having normal or nearly normal steady state levels of CoQ6, the coq10Δ mutant displays a respiratory-deficient phenotype shown by anemic growth on medium containing a nonfermentable carbon source and decreased NADH and succinate oxidase activities (10). In addition, the coq10Δ mutant is sensitive to lipid peroxidation induced by exogenously added PUFAs (9). Thus, the CoQ6 present in the coq10Δ mutant is not utilized efficiently for either respiration or for its function as an antioxidant.
The NMR structure of CC1736, a Coq10 ortholog in Caulobacter crescentus, identified it as a member of the steroidogenic acute regulatory protein-related lipid transfer (START) domain superfamily (11). This family includes proteins that bind polycyclic compounds, such as cholesterol and polyketides, in a signature hydrophobic cavity (11). The START domain typically spans ∼210 residues (12) and folds into a helix-grip structure consisting of antiparallel β-sheets flanked by one α-helix on each side (13). START domain-containing proteins are primarily involved in nonvesicular transport of lipids between membranes (14). For instance, STARD4 is a START domain protein that binds and transports cholesterol from the plasma membrane to mitochondria, the ER, and the endocytic recycling compartment, equilibrating cholesterol content among cellular membranes to fit their biophysical properties and physiological needs (15). Purified CC1736 binds to CoQn with variable polyisoprenoid chain lengths and to the farnesylated analog of a late-stage CoQ intermediate, demethoxy-CoQ3 (DMQ3) (9). Coq10 polypeptides isolated from S. cerevisiae and S. pombe copurify with CoQ6 and CoQ10, respectively (10, 16). Similarly, CoQ8 copurifies with the S. pombe Coq10 polypeptide expressed in E. coli (16). These observations have led to the current hypothesis that the Coq10 polypeptide is a putative CoQn chaperone, necessary for delivering CoQ from its site of synthesis and/or the pool of free CoQ to sites of function.
Complex III inhibitors, antimycin A and myxothiazol, enhance reactive oxygen species (ROS) formation by blocking oxidation of cytochrome bH at the N-site or inhibiting reduction of cytochrome bL at the P-site, respectively (17–19). Thereby, antimycin A induces ROS through reverse electron flow from cytochrome bL to CoQ to form the semiquinone radical (20), whereas myxothiazol-dependent ROS production results from incomplete CoQH2 oxidation by slow reduction of the Rieske iron-sulfur protein (18, 19). Mitochondria isolated from yeast coq10Δ produce significantly elevated ROS in the presence of antimycin A, but not myxothiazol, suggesting that in the absence of the Coq10 polypeptide, electron transfer from CoQH2 to the Rieske iron-sulfur protein is defective (20). This specific requirement for the presence of the Coq10 START domain polypeptide for functional electron transfer by complex III is further substantiated by the binding of both oxidized and reduced forms of a photo-reactive azido-quinone probe to the Coq10 polypeptide (21).
CoQ deficiencies are associated with human disease and the beneficial effects of CoQ10 supplementation in therapeutic regimens are increasingly appreciated (1, 4). Mutations in several genes encoding CoQ biosynthetic enzymes result in primary CoQ deficiency and cause encephalopathy, cerebellar ataxia, cardiomyopathy, nephrotic syndrome, and myopathy (1, 4). CoQ deficiency can also occur secondary to mutations in aprataxin, electron transfer flavoprotein dehydrogenase, or serine/threonine-protein kinase B-Raf (3). CoQ10 supplementation rescues the proteinuria in patients with nephrotic syndrome, provided that therapy is initiated early (22). Patients who develop myalgia under statin administration are often prompted to take CoQ10 supplements to mitigate adverse symptoms (23). Long term CoQ10 treatment has also been shown to improve symptoms and reduce major adverse cardiovascular events when it is used as adjunctive treatment in patients with chronic heart failure (24, 25).
Yeast is a superb model organism in which to study CoQ biosynthesis because many of the enzymes involved in CoQ biosynthesis are functionally conserved from yeast to humans (4, 5). In this work, we test the human co-orthologs of yeast Coq10, COQ10A and COQ10B, for their ability to complement the yeast coq10Δ mutant. We show that expression of human COQ10A or COQ10B rescues yeast coq10Δ-defective respiration and its sensitivity to oxidative stress, and restores steady-state levels of Coq polypeptides. However, neither COQ10A nor COQ10B expression is able to stabilize the yeast CoQ synthome or rescue the partial defect in de novo CoQ6 biosynthesis characteristic of the yeast coq10Δ mutant.
MATERIALS AND METHODS
Yeast strains and growth media
S. cerevisiae strains used in this study are described in Table 1. Growth media for yeast included YPD (1% Bacto yeast extract, 2% Bacto peptone, 2% dextrose), YPG (1% Bacto yeast extract, 2% Bacto peptone, 3% glycerol), and YPGal (1% Bacto yeast extract, 2% Bacto peptone, 2% galactose, 0.1% dextrose) (26). Synthetic dextrose/minimal-complete (SD-Complete) and synthetic dextrose/minimal minus uracil (SD-Ura) [0.18% Difco yeast nitrogen base without amino acids and ammonium sulfate, 0.5% (NH4)2SO4, 0.14% NaH2PO4, 2% dextrose, complete amino acid supplement, or amino acid supplement lacking uracil] were prepared as described (27). Solid media contained an additional 2% Bacto agar.
TABLE 1.
Genotype and source of yeast strains
| Strain | Genotypea | Source |
| W303 1B | MAT α, ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | R. Rothsteinb |
| W303 coq1Δ | MAT α, ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 coq1::LEU2 | (99) |
| CC303 | MAT α, ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 coq3::LEU2 | (100) |
| W303 coq4Δ | MAT a, ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 coq4::TRP1 | (101) |
| W303 coq5Δ | MAT α, ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 coq5::HIS3 | (27) |
| W303 coq6Δ | MAT a, ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 coq6::LEU2 | (102) |
| W303 coq7Δ | MAT α, ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 coq7::LEU2 | (103) |
| W303 coq8Δ | MAT a, ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 coq8::HIS3 | (101) |
| W303 coq9Δ | MAT α, ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 coq9::URA3 | (104) |
| W303 coq10Δ | MAT a, ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 coq10::HIS3 | (10) |
| W303 coq11Δ | MAT a, ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 coq11::HIS3 | This study |
Mating type a (MAT a) is in bold to distinguish it from mating type α (MAT α).
Dr. Rodney Rothstein, Department of Human Genetics, Columbia University, New York, NY.
Construction of single- and multi-copy yeast expression vectors of human COQ10A and COQ10B
Plasmids used in this study are listed in Table 2. Generation of single-copy (pQM) and multi-copy (pRCM) yeast expression vectors was previously described (9, 28). Both pQM and pRCM contain the yeast CYC1 promoter and the first 35 residues of the yeast COQ3 ORF, corresponding to the proposed Coq3 mitochondrial leader sequence to direct import of human proteins into yeast mitochondria. To generate the single- and multi-copy yeast expression vectors of human COQ10A, the human COQ10A ORF (mRNA #1, Fig. 1A), encoding residues 44-247, was PCR amplified from pHCOQ10/ST1 (10) with primers 5′-ggccATCGATATGAGGTTTCTGACCTCCTGC-3′ and 5′-ggccGGTACCTCAAGTCTGGTGCACCTC-3′, and cloned into pQM and pRCM vectors using the restriction enzymes, ClaI and KpnI (New England BioLabs), to generate pQM COQ10A and pRCM COQ10A, respectively. Similarly, full-length human COQ10B ORF (mRNA #1, Fig. 1A), encoding residues 1-238, was PCR amplified from COQ10B cDNA clone (GeneCopoeia) with primers 5′-ggccATCGATATGGCAGCTCGGACTGGTCAT-3′ and 5′-ggccGGTACCTTATGTGTGATGGACTTCATGAAGCATTAACTCC-3′ to generate pQM COQ10B and pRCM COQ10B.
TABLE 2.
Yeast expression vectors
| Plasmid | Relevant Genes/Markers | Source |
| pQM | pAH01 with COQ3 mito leader, single-copy | (28) |
| pQM COQ10A | pQM with human COQ10A; single-copy | This work |
| pQM COQ10B | pQM with human COQ10B; single-copy | This work |
| pRCM | pCH1 with COQ3 mito leader; multi-copy | (9) |
| pRCM COQ10A | pRCM with human COQ10A; multi-copy | This work |
| pRCM COQ10B | pRCM with human COQ10B; multi-copy | This work |
Fig. 1.
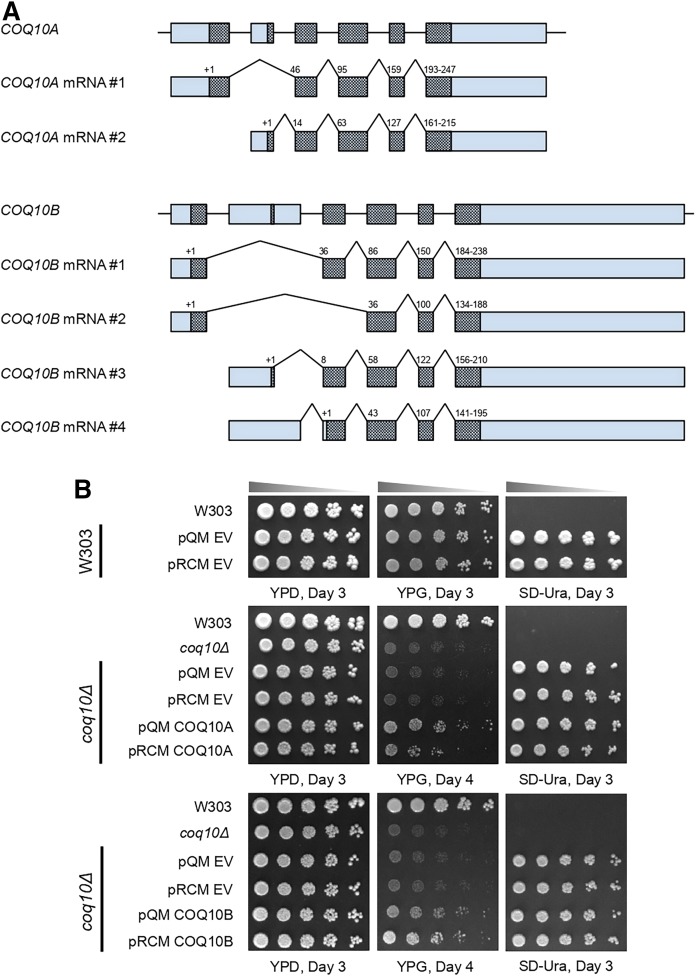
Expression of either human COQ10A or COQ10B restores respiratory growth of the yeast coq10Δ mutant. A: A schematic representation of alternative splicing of human COQ10A and COQ10B mRNA. Alternative splicing and different translation initiation sites result in two isoforms of COQ10A and four isoforms of COQ10B. Boxes represent the exons, and the protein coding sequences are shaded within the exons. Both exons and the protein coding sequences were drawn relative to their corresponding number of base pairs. The introns are represented by horizontal lines and are not drawn to scale. The translation initiation sites were marked as “+1” on top of the protein coding sequence, and the numbers denote the amino acid residue numbers at the beginning of the coding sequence within each exon. The N-terminal sequences of all isoforms of COQ10A and COQ10B are listed in supplemental Table S1, with predicted mitochondria targeting sequence highlighted in bold. B: Wild-type W303, coq10Δ mutant, coq10Δ expressing single-copy (pQM), multi-copy (pRCM) human COQ10A, COQ10B, or their respective empty vectors were inoculated in SD-Complete or SD-Ura liquid medium overnight, from which a series of 5-fold dilutions of the overnight culture were prepared and plated on to YPD, YPG, and SD-Ura plate medium. Aliquots of 2 μl of samples from serial dilutions were plated in each spot, starting at 0.2 OD600/ml in the first spot to the left. Pictures were taken 3–4 days after incubation at 30°C.
Complementation of yeast coq10Δ by human COQ10A and COQ10B
Each of the following plasmids, pQM (empty vector), pQM COQ10A, pQM COQ10B, pRCM (empty vector), pRCM COQ10A, and pRCM COQ10B, was transformed into wild-type W303 or coq10Δ, and the transformed cells were selected on SD-Ura plates as described (29). A single colony from each SD-Ura plate was inoculated in SD-Ura liquid medium. Wild-type W303 and coq10Δ were each inoculated in SD-Complete liquid medium. Cultures were incubated overnight at 30°C 250 rpm. The overnight cultures were diluted to 0.2 OD600/ml with sterile water, from which a series of 5-fold dilutions were prepared. An aliquot of 2 μl of sample from the dilution series was plated onto YPD and YPG plate medium and incubated at 30°C. Pictures were taken after 3–4 days.
Fatty acid sensitivity assay
A fatty acid sensitivity assay was performed as described (30, 31) with slight modifications. Briefly, yeast W303 wild-type, cor1Δ, coq9Δ, and coq10Δ were inoculated in SD-Complete liquid medium, and coq10Δs harboring the designated plasmids were inoculated in SD-Ura liquid medium and incubated overnight at 30°C 250 rpm. Overnight cultures were back diluted to 0.2 OD600/ml with fresh SD-Complete or SD-Ura liquid medium and incubated for 6 h at 30°C 250 rpm to logarithmic phase. The cells were harvested, washed twice with sterile water, and suspended in 0.1 M phosphate buffer with 0.2% dextrose (pH 6.2) to a cell density of 0.2 OD600/ml. To test yeast sensitivity to PUFA-induced oxidative stress, ethanol-diluted oleic acid (Nu-Chek Prep) or α-linolenic acid (Nu-Check Prep) was added to aliquots of 5 ml cell suspension in phosphate buffer with 0.2% dextrose to a final concentration of 200 μM. Identical 5 ml cell suspensions were prepared with 0.1% (v/v) ethanol as a vehicle control. After a 4 h incubation at 30°C 250 rpm, cell viability was assessed with plate dilution assay by spotting 2 μl of sample from a series of 5-fold dilution onto YPD plates. Cell viability was also ascertained before addition of fatty acids, and labeled as 0 h.
Mitochondria isolation from yeast coq10Δ expressing human COQ10A or COQ10B
Precultures of yeast coq10Δ transformed with pQM COQ10A, pQM COQ10B, pRCM COQ10A, or pRCM COQ10B in YPD were back diluted with YPGal and grown overnight at 30°C 250 rpm until the cell density had reached ∼3.0 OD600/ml. Preparation of spheroplasts with Zymolyase-20T (MP Biomedicals) and extraction of mitochondria in the presence of Complete EDTA-free protease inhibitor mixture (Roche), phosphatase inhibitor cocktail set II (EMD Millipore), and phosphatase inhibitor cocktail set 3 (Sigma-Aldrich) over Nycodenz (Sigma-Aldrich) density gradient were previously described (32). Purified mitochondria were flash-frozen in liquid nitrogen and stored at −80°C until use.
Immunoblot analysis of steady state Coq polypeptide levels
Protein concentration in gradient-purified mitochondria was measured by the BCA assay (Thermo Fisher Scientific). Purified mitochondria were resuspended in SDS sample buffer [50 mM Tris (pH 6.8), 10% glycerol, 2% SDS, 0.1% bromophenol blue, and 1.33% β-mercaptoethanol], and an aliquot of 25 μg of mitochondrial protein from each sample was loaded in individual lanes and separated by SDS gel electrophoresis on 12% Tris-glycine polyacrylamide gels. Proteins were subsequently transferred to 0.45 μm nitrocellulose membrane (Bio-Rad) and blocked with blocking buffer (0.5% BSA, 0.1% Tween 20, 0.02% SDS in phosphate-buffered saline). Representative Coq polypeptides and mitochondrial malate dehydrogenase Mdh1 (loading control) were detected with rabbit polyclonal antibodies prepared in blocking buffer at the dilutions listed in Table 3. Polyclonal antibodies against human COQ10A (Proteintech) and COQ10B (Abcam) were commercially obtained and used at dilutions recommended by the companies. The secondary IRDye 680LT goat anti-rabbit IgG antibody (LiCOR) was used at 1:10,000 dilution in the same blocking buffer. Immunoblot images were visualized with a LiCOR Odyssey infrared scanner (LiCOR), and relative protein levels were quantified by band densitometry using ImageJ software (https://imagej.nih.gov/ij/).
TABLE 3.
Description and source of antibodies
| Antibody | Working Dilution | Source |
| Coq1 | 1:10,000 | (99) |
| Coq3 | 1:200 | (105) |
| Coq4 | 1:2,000 | (106) |
| Coq5 | 1:5,000 | (107) |
| Coq6 | 1:200 | (102) |
| Coq7 | 1:1,000 | (108) |
| Coq8 | Affinity purified, 1:30 | (63) |
| Coq9 | 1:1,000 | (109) |
| Coq10 | Affinity purified, 1:400 | This work |
| Coq11 | 1:500 | This work |
| Mdh1 | 1:10,000 | L. McAlister-Henna |
| COQ10A | 1:500 | Proteintech |
| COQ10B | 1 μg/ml | Abcam |
Dr. Lee McAlister-Henn, Department of Molecular Biophysics and Biochemistry, University of Texas Health Sciences Center, San Antonio, TX.
Analysis of high molecular weight complexes with two-dimensional Blue Native/SDS-PAGE
Two-dimensional Blue Native/SDS-PAGE (BN/SDS-PAGE) was performed as described (33–35). Purified mitochondria at 4 mg/ml were solubilized for 1 h in ice-cold solubilization buffer [11 mM HEPES (pH 7.4), 0.33 M sorbitol, 1× NativePAGE sample buffer (Thermo Fisher Scientific), 16 mg/ml digitonin (Biosynth)] in the presence of the previously described mixtures of protease and phosphatase inhibitors. After centrifugation (100,000 g, 10 min), the protein concentration in the soluble fraction was measured by BCA assay, and NativePAGE 5% G-250 sample additive (Thermo Fisher Scientific) was added to the soluble fraction to a final concentration of 0.25%. Soluble protein from each sample (80 μg) was separated on NativePAGE 4–16% Bis-Tris gel (Thermo Fisher Scientific) in the first dimension, followed by separation on 12% Tris-glycine polyacrylamide gel in the second dimension. The molecular weight standards for Blue Native gel electrophoresis and SDS gel electrophoresis were obtained from GE Healthcare (Sigma-Aldrich) and Bio-Rad, respectively. Immunoblot analysis of the CoQ synthome was performed as described above using antibodies against Coq4 and Coq9. A separate COQ10A- or COQ10B-containing complex was detected using commercial antibodies against COQ10A and COQ10B, respectively.
Analyses of de novo and steady state levels of CoQ6 and CoQ6-intermediates
Metabolic labeling with 13C6-labeled ring precursors and subsequent analyses of labeled and unlabeled CoQ6 and CoQ6-intermediates in yeast whole-cell lipid extract by RP-HPLC MS/MS were previously described (9, 36). Briefly, overnight cultures of yeast wild-type, coq10Δ in SD-Complete medium, and coq10Δ expressing pQM, pQM COQ10A, pQM COQ10B, pRCM, pRCM COQ10A, or pRCM COQ10B in SD-Ura medium were back diluted with fresh SD-Complete or SD-Ura medium to 0.1 OD600/ml and grown to 0.5 OD600/ml (early-log phase) at 30°C 250 rpm. The 13C6-labeled precursors, pABA or 4HB, were dissolved in ethanol and added to yeast cultures at a final concentration of 5 μg/ml (equivalent to 34.9 μM 13C6-pABA and 34.7 μM 13C6-4HB). Vehicle control samples contained a final concentration of 0.1% (v/v) ethanol. The cultures were incubated with the labeled precursors or ethanol for 5 h at 30°C 250 rpm before triplicates of 5 ml culture were harvested for lipid extraction. The cell density measured by OD600 at the time of harvest was recorded.
For lipid extraction, collected yeast cell pellets were dissolved in 2 ml of methanol, and lipids were extracted twice in the presence of internal standard CoQ4, each time with 2 ml of petroleum ether followed by vigorous vortex. The organic phase from two extractions was combined and dried under a stream of N2 gas. A series of CoQ6 standards (Avanti) containing CoQ4 were prepared and lipid extracted concurrently with yeast samples to construct a CoQ6 standard curve. The dried lipids were reconstituted in 200 μl of 0.5 mg/ml benzoquinone prepared in ethanol to oxidize all lipid species for MS analysis with a 4000 QTRAP linear MS/MS spectrometer (Applied Biosystems). Aliquots (20 μl) of each reconstituted lipid extract were injected into a Luna phenyl-hexyl column (100 × 4.6 mm, 5 μm; Phenomenex). The HPLC mobile phase consisted of solvent A (95:5 methanol/isopropanol, 2.5 mM ammonium formate) and solvent B (isopropanol, 2.5 mM ammonium formate). As the percentage of solution B was increased linearly from 0% to 10%, representative CoQ6 and CoQ6-intermediates eluted off the column at distinct retention times and were monitored under multiple reaction monitoring mode scanning the precursor to product ion transitions listed in Table 4. For each analyte, the precursor to product ion transitions of both protonated ion species and its ammonium adduct ion species were tracked. The ammonium adducts provide much stronger ion signals for detection of isoprenoids by positive-ion electrospray ionization MS. Analyst 1.4.2 software (Applied Biosystems) was used for data acquisition and processing. In each sample, the amount of CoQ6 and CoQ6-intermediates was calculated from the sum of peak areas of each analyte and its corresponding ammonium adduct at a specific retention time, corrected for the recovery of internal standard CoQ4. Statistical analyses were performed using GraphPad Prism with two-way ANOVA multiple comparisons comparing the mean of each sample to the mean of its corresponding empty vector control, and comparing the mean of wild-type to the mean of coq10Δ.
TABLE 4.
Precursor-to-product ion transitions
| m/z [M+H]+ | m/z [M+NH4]+ | |
| HAB | 546.4/150.0 | 563.0/150.0 |
| 13C6-HAB | 552.4/156.0 | 569.0/156.0 |
| HHB | 547.4/151.0 | 564.0/151.0 |
| 13C6-HHB | 553.4/157.0 | 570.4/157.0 |
| DMQ6 | 561.4/167.0 | 578.0/167.0 |
| 13C6-DMQ6 | 567.4/173.0 | 584.0/173.0 |
| IDMQ6 | 560.6/166.0 | 577.0/166.0 |
| 13C6-IDMQ6 | 566.6/172.0 | 583.0/172.0 |
| CoQ4 | 455.4/197.0 | 472.0/197.0 |
| CoQ6 | 591.4/197.0 | 608.0/197.0 |
| 13C6-CoQ6 | 597.4/203.0 | 614.0/203.0 |
Homology modeling of human COQ10A and COQ10B
Secondary structural elements and disordered regions within the COQ10A and COQ10B ORFs were predicted using PsiPred (37) and DisoPred (38). Secondary structural alignments and initial model generation were performed using the SwissModel server (39). Using COQ10A as the query sequence, 50 homologous protein templates were identified with sequence identities ranging from 6.94% to 28.68%, and sequence similarity between 25% and 30%. Nine templates with the best alignment of the secondary structural elements were selected for model building in SwissModel. Of the nine models generated, four models with either low QMEAN scores or high identity to the template were selected for further improvement. As the sequence coverage of the models differs from each other, a combination of all four models was used to generate the final model that covers residues 87-228 of COQ10A. The final steps of model refinement, rebuilding strange loops, and improvement of side chain packing and backbone distortion were completed using custom script in PyRosetta (40). A total of 16,000 decoys were generated and the convergence of the refinement was assessed by checking RMSD of all decoys to the lowest energy decoy versus Rosetta energy. The top five decoys with the lowest Rosetta energy were selected and model quality was assessed via Qmean (41), Verify3D (42), Errat (43), and MolProbity (44). The decoy that scored equally well in all four metrics was chosen as the final model of COQ10A. A similar approach was used to create a homology model of COQ10B from a combination of six models that covered residues 79-219 of COQ10B.
Molecular docking of CC1736
Molecular docking was completed using Autodock vina (45), and substrate molecules were produced using phenix.ELBOW (46) and verified with phenix.REEL (46). Docking of each substrate was executed with a 20 × 20 × 20 Å grid box that encompassed the entirety of the hydrophobic pocket of CC1736. Docking was performed with an exhaustiveness of 18 and nine docking solutions were produced per run.
Yeast Coq10 ortholog similarity clustering and Coq10 coexpression analysis
The protein similarity network was constructed using the EFI-EST tool (http://efi.igb.illinois.edu/efi-est/) (47) with an alignment score of 30, with human COQ10A as the BLASTp seed sequence, retrieving 8,095 hits. Protein nodes were collapsed at >75% identity. The network was visualized with the yFiles organic layout provided with the Cytoscape software (48). The nodes in the network were colored by taxonomy as provided by the UniProt database (49). Information associated with proteins included in this analysis can be found in supplemental Table S2. Gene neighborhoods of bacterial homologs were retrieved with the EFI-GNT tool (https://efi.igb.illinois.edu/efi-gnt/). The phylogenetic analysis was performed using MAFFT (50) for sequence alignment and IQ Tree (51) as implemented on the CIPRES (52, 53) web portal with 1,000 bootstrap replicates (54). Before tree reconstruction, the multiple sequence alignment (MSA) was trimmed to remove poorly aligned sequence at the N terminus, and the edited MSA can be found in supplemental Table S2.
RESULTS
Expression of either COQ10A or COQ10B from humans restores respiratory growth of the yeast coq10Δ mutant
Human COQ10A and COQ10B are co-orthologs of yeast COQ10 located on human chromosome 12 and chromosome 2, respectively (55). The polymorphisms in COQ10A (P79H, P231S) and COQ10B (L48F) are thought to be one of the genetic factors predisposing patients to statin-associated myopathy (56, 57). The underlying molecular mechanisms of statin-associated myopathy are proposed to be isoprenoid depletion, inhibition of CoQ biosynthesis, disruption of cholesterol homeostasis, or disturbance of calcium metabolism (58). COQ10A and COQ10B each contain six exons, which give rise to two isoforms of COQ10A and four isoforms of COQ10B as a result of alternative splicing and translation initiation (Fig. 1A). Each of the two COQ10A mRNA transcripts contains a unique 5′ UTR and translation initiation site (Fig. 1A, supplemental Table S1). According to the UniProt database (49), COQ10A mRNA #1 encodes the longer isoform of COQ10A, with the first 15 residues predicted to be the mitochondrial targeting sequence (supplemental Table S1). COQ10A mRNA #2 encodes COQ10A isoform 2 with a unique N terminus. Two of the COQ10B mRNA transcripts share an identical 5′ UTR and translation initiation site, whereas the other two COQ10B mRNA transcripts each have a unique 5′ UTR and translation initiation site (Fig. 1A, supplemental Table S1). Based on the UniProt database, COQ10B mRNA #1 encodes the longest isoform, and its mitochondrial targeting sequence consists of the first 37 residues (supplemental Table S1). COQ10B isoform 2 encoded by mRNA #2 lacks one of the exons present in mRNA #1 (Fig. 1A). COQ10B isoforms 3 and 4 contain distinct N-terminal sequences and their subcellular localization is unknown (Fig. 1A, supplemental Table S1). Although RNA processing predicts several isoforms of COQ10A and COQ10B, both isoforms of COQ10A and all four isoforms of COQ10B retain amino acid residues important for START domain formation.
Expression of human COQ10A from a multi-copy vector was previously shown to complement respiratory growth of the yeast coq10Δ mutant on a nonfermentable carbon source (10, 16). Here, we examined the functional complementation of the yeast coq10Δ by both single-copy (pQM) and multi-copy (pRCM) expression of human COQ10A or COQ10B. The cDNA expressed for COQ10A corresponded to residues 44-247 of isoform 1 (COQ10A mRNA #1), and the cDNA expressed for COQ10B corresponded to residues 1-238 of isoform 1 (COQ10B mRNA #1). Yeast coq10Δ and coq10Δ with empty vectors pQM or pRCM show slow growth on nonfermentable glycerol-containing medium (Fig. 1B). Expression of either COQ10A or COQ10B in single-copy or multi-copy rescued the glycerol growth of the coq10Δ mutant (Fig. 1B). Interestingly, single-copy COQ10A and both single- and multi-copy COQ10B seem to complement the defective growth of the coq10Δ mutant better as compared with multi-copy COQ10A (Fig. 1B). These results identify human COQ10A and COQ10B as functional co-orthologs of yeast Coq10.
Human COQ10A and COQ10B share low sequence identity but high structural similarity with other Coq10 orthologs
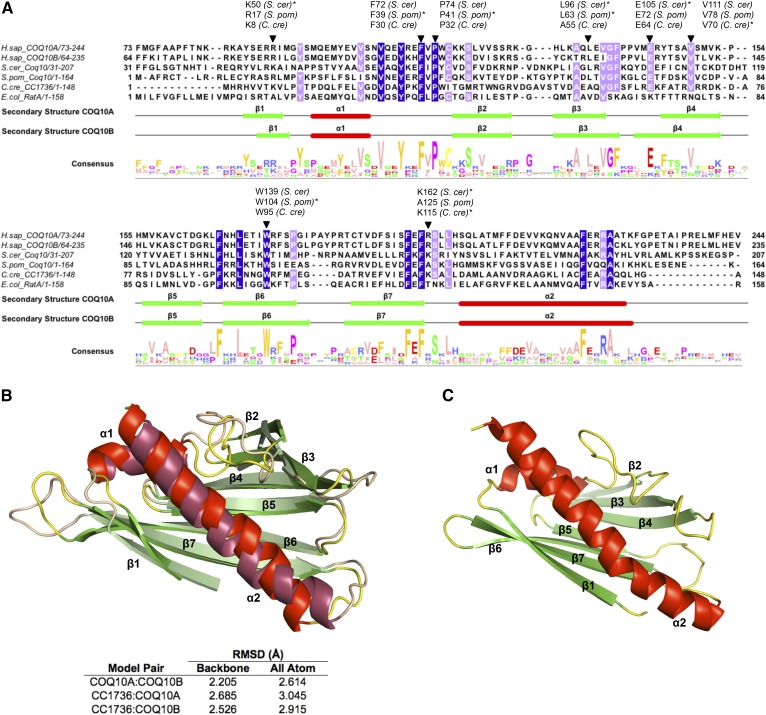
In vitro lipid binding assays have shown that the C. crescentus Coq10 ortholog, CC1736, binds to isoforms of CoQn with varying polyisoprenoid chain length (n = 2, 3, 6, or 10) and to a farnesylated analog (n = 3) of a late-stage CoQ intermediate, DMQ3 (9). CC1736 does not bind to a farnesylated analog of an early-stage CoQ intermediate, farnesyl-hydroxybenzoate (9). This observation strongly implies that the quinone moiety, but not the polyisoprenoid chain, is the structural determinant of Coq10 ligand interaction. Several amino acid residues have been identified through site-directed mutagenesis, and a substitution of these residues on Coq10 orthologs results in altered ligand-binding affinity and respiratory defects (9, 16, 21, 59). Among these residues, a combination of positive charge provided by residue K8, paired hydrophobic residues A55 and V70, and a salt bridge formed by E64 and K115 are thought to confer the ligand specificity of CC1736 (11). Mutations in the equivalent residues in S. cerevisiae Coq10 result in defects in respiration as measured by oxygen consumption, H2O2 release, or NADH-cytochrome c reductase activity (9, 59). Here, we constructed sequence alignment of four Coq10 orthologs and mapped these residues relative to positions in isoform 1 of the human COQ10A and COQ10B amino acid sequence (Fig. 2A). From the MSA, we noticed several additional conserved residues (shaded blue and lilac in Fig. 2A) besides those previously identified. Most noticeably, aromatic amino acids (tyrosine, phenylalanine, and tryptophan) and nonpolar amino acids (valine, leucine, and alanine) are highly enriched among these conserved residues (Fig. 2A). Residues F39 and P41 of S. pombe Coq10 are believed to be within the region for CoQ binding, and mutation of F39A and P41A reduces the photo-labeling yield with azido-quinone to about 50% of the wild-type control (21). Replacement of L63 or W104 with alanine on S. pombe Coq10 reduces CoQ binding affinity to about 50%, and a double point mutant further reduces the binding affinity to only 25% (16). Based on homology modeling, the N-terminal regions of human COQ10A and COQ10B do not seem to form defined secondary structure and were excluded from the predicted secondary structures shown in Fig. 2A. Thus, exon skipping in COQ10B isoform 2 (Fig. 1A) as well as variable N-terminal sequences on all COQ10A and COQ10B isoforms that result from alternative splicing and translation initiation (Fig. 1A) are not expected to change the secondary structural elements of the START domain of COQ10A and COQ10B.
Fig. 2.
Human COQ10A and COQ10B share low sequence identity with Coq10 orthologs, but are predicted to contain conserved START domain structures for lipid binding. A: Sequence alignment of human COQ10A (residues 73-244) and COQ10B (residues 64-235) with Coq10 orthologs in S. cerevisiae, S. pombe, C. crescentus, and E. coli. MSA was constructed using Tcoffee, with identical residues shaded in blue and highly conserved residues shaded in lilac. Residues that have been previously tested and deemed critical for ligand binding are indicated with an inverted triangle, and the asterisk indicates the organism in which the site-directed mutagenesis study was performed. The secondary structures of COQ10A and COQ10B were predicted using JPred and adjusted based on their refined models in B. Alpha-helices are shown in red and labeled α1-α2 and β-sheets are shown in green and labeled β1–β7. The figure was assembled in Jalview. B: Overlay of predicted homologous models of human COQ10A and COQ10B were generated using the structures of MSMEG_0129 from Mycobacterium smegmatis [Protein Data Bank identification (PDB ID): 5Z8O] as a template. The predicted structures of COQ10A (dark shade) and COQ10B (bright shade) were colored respectively, based on their predicted secondary structures. The COQ10A and COQ10B share a similar START domain, a hydrophobic cavity consists of α-helix (red) and anti-parallel β-sheets (green). C: NMR structure of CC1736 (PDB ID: 1T17), a Coq10 ortholog in C. crescentus (11). Structures in B and C were generated using PyMOL.
Based on the MSA, we also noticed that although human COQ10A and COQ10B exhibit relatively low sequence identity with other Coq10 orthologs (∼26–31%), they are predicted to share similar helix-grip structures (Fig. 2B). The predicted structures of COQ10A and COQ10B are almost identical to each other (Fig. 2B) and similar to the solved structure of CC1736 (Fig. 2C). The core of both COQ10A and COQ10B consists of two α-helices and seven antiparallel β-sheets, and is predicted to form a hydrophobic cavity shielding its CoQ lipid ligand from the aqueous environment (Fig. 2B). The structural feature of a helix-grip fold identifies both COQ10A and COQ10B as distinctive members of the START domain protein family.
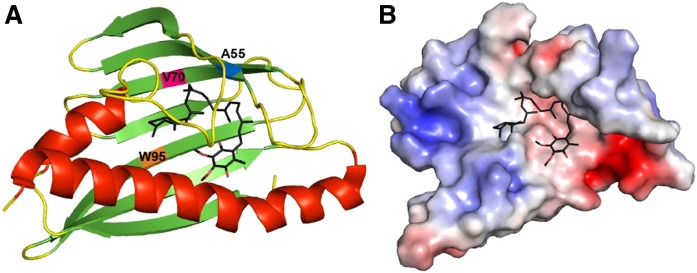
In order to reveal the most likely binding site for the CoQ lipid ligand and to identify additional residues that may confer CoQ lipid ligand binding specificity in COQ10A and COQ10B, we attempted to dock CoQ6 to the known structure of CC1736. Consistent with previous in vitro binding assay results, we observed that docking of farnesyl-hydroxybenzoate to CC1736 occurred with free energy values significantly higher (more positive and less favorable) than CoQ6 docked at the central cavity of CC1736 (Fig. 3A, B). The docking solutions of CoQ6 to CC1736 consistently showed that the CoQ6 folds into a boomerang-like structure, with its hexaprenyl tail making contact with residues A55, V70, and W95 lining the surface inside the cavity (Fig. 3A). However, the orientation of the benzoquinone head group was slightly more variable between docking solutions. Several START domain proteins have been reported to undergo ligand-induced conformational change such that the readily accessible entryway to the central cavity becomes partially constricted, shielding the ligand from the aqueous environment (60–62). Thus, it is highly likely that the flexible orientation of the quinone head group may be a result of a less compact conformation of the central cavity in the structure of a ligand-free CC1736. Further studies that would allow a more in-depth characterization of the conformational change of Coq10 or its orthologs are needed to decipher the CoQ ligand binding interaction.
Fig. 3.
CoQ6 docks in the hydrophobic cavity of CC1736. A: CoQ6 (colored in black) was docked to the NMR structure of CC1736 using AutoDock. The α-helices are shown in red and β-sheets are shown in green. Docking structures were produced using Autodock vina (45). B: Electrostatic surface of CC1736 showing the cavity docked with CoQ6 with some residues hidden for clarity (red, negative; blue, positive). The polyisoprenoid chain is threaded through the hydrophobic cleft created by residues Ala55, Val70, and Trp95.
Expression of human COQ10A or COQ10B restores steady state levels of Coq polypeptides
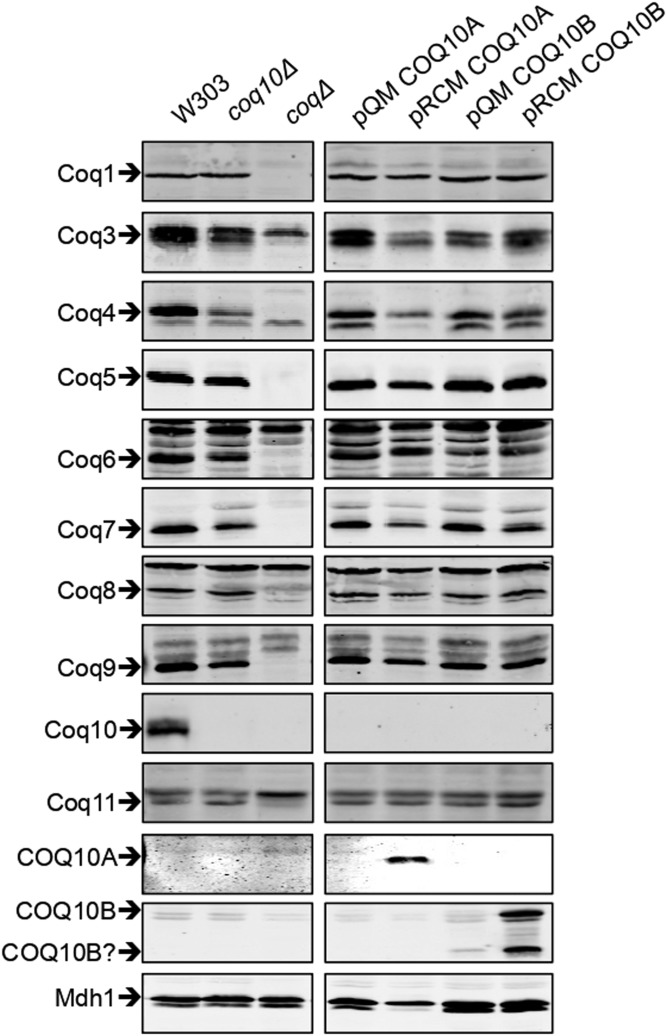
Coq polypeptide components of the CoQ synthome serve as enzymes required for CoQ biosynthesis and/or play structural roles necessary for formation or stability of the CoQ synthome (5, 34, 63). Yeast Coq10 has not been detected as part of the CoQ synthome, but its absence causes destabilization of Coq3, Coq4, Coq6, Coq7, and Coq9, as well as the overall CoQ synthome (34, 63). We tested the ability of human COQ10A or COQ10B to restore steady state levels of Coq polypeptides when expressed in the yeast coq10Δ mutant. We noticed that the steady state level of Coq5 is slightly reduced and the level of Coq8 is slightly increased in the coq10Δ mutant, in addition to other affected Coq polypeptide levels shown previously (63). The presence of single-copy pQM COQ10A fully restores steady state levels of Coq5 and Coq6 (Fig. 4, supplemental Fig. S1) and partially restores steady state levels of Coq3, Coq4, Coq7, and Coq9 (Fig. 4, supplemental Fig. S1). The single-copy pQM COQ10B fully restores the steady state level of Coq5 (Fig. 4, supplemental Fig. S1) and restores steady-state levels of Coq4, Coq7, and Coq9 to a minimal degree (Fig. 4, supplemental Fig. S1), but seems to have a negative effect on the levels of Coq3, Coq6, and Coq8 (Fig. 4, supplemental Fig. S1). Neither COQ10A nor COQ10B expressed on a multi-copy pRCM vector functions as well as the corresponding single-copy pQM vector, despite the fact that COQ10A and COQ10B are barely detected when expressed from a single-copy vector (Fig. 4). The multi-copy pRCM COQ10A restores the steady state level of Coq6 and partially restores the levels of Coq5 and Coq9 (Fig. 4, supplemental Fig. S1), while the steady state levels of Coq3, Coq4, and Coq7 remain similar to the coq10Δ mutant, if not lower (Fig. 4, supplemental Fig. S1). Yeast cells expressing COQ10A from a multi-copy vector show a nearly 50% reduction of Coq8 content compared with the wild-type control (Fig. 4, supplemental Fig. S1). Overexpression of Coq8 promotes assembly of subcomplexes of the CoQ synthome (34), and Coq8 deficit may explain the ineffective rescue of coq10Δ by the multi-copy COQ10A and its corresponding poor growth phenotype on YPG plate medium (Fig. 1B). The multi-copy pRCM COQ10B restores Coq5 and Coq9 (Fig. 4, supplemental Fig. S1), but has no effect on the Coq3, Coq4, Coq6, and Coq7 levels (Fig. 4, supplemental Fig. S1). Opposite from the effect of expressing multi-copy pRMC COQ10A, the presence of multi-copy pRCM COQ10B leads to a 50% increase of Coq8 compared with the wild-type cells (Fig. 4, supplemental Fig. S1). It is quite intriguing that pRCM COQ10B results in a slight reduction of the Coq1 level, while the level of Coq11 is nearly 2.5-fold more compared with the wild-type cells (Fig. 4, supplemental Fig. S1). The anti-COQ10A antibody specifically recognizes COQ10A and does not cross-react with human COQ10B (Fig. 4). Anti-COQ10B antibody is also antigen-specific, but it gives two intense bands, migrating at ∼30 kDa and ∼17 kDa (Fig. 4). In human cells, only the higher molecular mass band is detected, and the lower molecular mass band may correspond to a processed form of COQ10B unique to the yeast cells.
Fig. 4.
Expression of human COQ10A or COQ10B restores steady state levels of Coq polypeptides. An aliquot of 25 μg of purified mitochondrial protein from wild-type, coq10Δ, or coq10Δ expressing single- or multi-copy COQ10A or COQ10B was applied to each lane and separated on 12% Tris-glycine SDS-PAGE gels. Purified mitochondria from coq1Δ and coq3Δ–coq11Δ mutants were included as negative controls for Western blotting against each of the Coq polypeptides. Purified mitochondria from coq10Δ were used as coqΔ control for blotting against human COQ10A and COQ10B. Yeast mitochondrial malate dehydrogenase (Mdh1) was included as loading control. Two panels (left and right) are immunoblots derived from the same nitrocellulose membrane. Relative protein levels were quantified by band densitometry in supplemental Fig. S1 using ImageJ software.
Expression of human COQ10A or COQ10B fails to restore the CoQ synthome in the yeast coq10Δ mutant
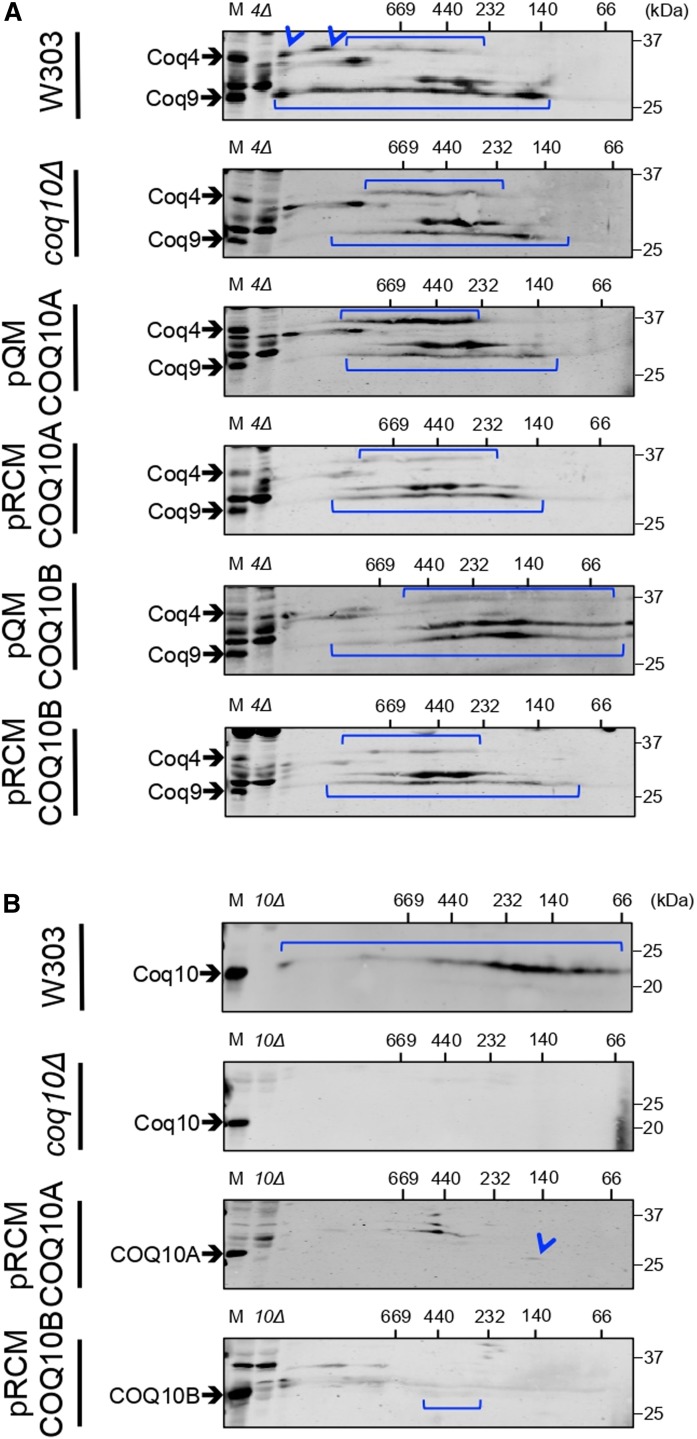
The decreased steady state levels of component Coq polypeptides in the yeast coq10Δ mutant are directly related to the destabilization of the CoQ synthome (34). Here, we assessed the stability of the CoQ synthome in coq10Δ expressing single- or multi-copy COQ10A or COQ10B by two-dimensional BN/SDS-PAGE. In the first dimension, multi-subunit protein complexes are resolved under nondenaturing native conditions, followed by separation into individual polypeptide constituents in the second dimension by traditional SDS-PAGE under denaturing conditions. Proteins that represent subunits from the same multi-subunit complex are found in one vertical line, and a designated polypeptide component that is present in several distinct complexes or subcomplexes is indicated by a horizontal signal. Thus, the CoQ synthome that is represented by its component Coq4 or Coq9 polypeptide is shown by two horizontal lines that align with the Coq4 and Coq9 bands present in the sample of intact mitochondria from each sample in the lane labeled as “M” (Fig. 5). In wild-type yeast cells, the CoQ synthome is represented by a complex array of high molecular mass signals, spanning a range of about 400 kDa to >1 MDa for Coq4 and from ∼140 kDa to >1 MDa for Coq9 (Fig. 5A). In contrast, the CoQ synthome in the coq10Δ mutant appears destabilized, indicated by the disappearance of complexes much larger than 669 kDa and an appearance of complexes limited to a distribution between 140 kDa and slightly greater than 669 kDa for both Coq4 and Coq9 signals (Fig. 5A). Although expression of single-copy pQM COQ10A restored the steady state polypeptide levels of both Coq4 and Coq9 (Fig. 4), the distribution of high molecular mass signals for Coq4 and Coq9 remains limited to a range similar to that of the coq10Δ mutant (Fig. 5A). Single-copy pQM COQ10B reinforced the CoQ synthome assemblies below 440 kDa, as shown by intense Coq9 signal (Fig. 5A), but it also failed to restore the CoQ synthome at a much greater molecular mass (Fig. 5A). Neither multi-copy COQ10A nor COQ10B expression appears to confer a stabilization effect on the CoQ synthome (Fig. 5A). Thus, the expression of either COQ10A or COQ10B, while having a rather dramatic effect on steady state Coq polypeptide levels, exerts a negligible effect on the high molecular mass signals that characterize efficient CoQ synthesis and the presence of the CoQ synthome.
Fig. 5.
Expression of either human COQ10A or COQ10B partially restores the yeast CoQ synthome. An aliquot of 80 μg of purified mitochondrial protein from wild-type, coq10Δ, or coq10Δ expressing single- or multi-copy COQ10A or COQ10B was resolved on a two-dimensional BN/SDS-PAGE and blotted against Coq4 and Coq9 (A) or against Coq10, COQ10A, or COQ10B (B). An aliquot of 25 μg of unsolubilized intact mitochondria from each designated sample was loaded in the lane labeled “M”, and the same amount of intact coq4Δ or coq10Δ mitochondria were included as negative controls. The yeast coq4Δ mutant lacks both Coq4 and Coq9 polypeptides (63) and was used as a negative control for blotting against both proteins on the same membrane.
Coq10 comigrates with Coq2 and Coq8 on two-dimensional BN/SDS-PAGE (64), but so far there is no direct evidence showing Coq10 interaction with other known Coq polypeptides or with the CoQ synthome. On a sucrose gradient, native Coq10 from yeast mitochondrial extract sediments at a fraction that corresponds to a molecular mass of approximately 140 kDa (10). Given that the monomeric molecular mass of mature Coq10 is 20 kDa, Coq10 must be present in a complex that consists of an oligomeric form of Coq10 and/or with other partner proteins (10). We tested to determine whether human COQ10A or COQ10B might also assemble into complexes. Because the signal intensities of both COQ10A and COQ10B expressed from single-copy vectors are quite weak, we decided to examine their complex formation when expressed from the multi-copy vector. On the two-dimensional BN/SDS-PAGE, wild-type yeast Coq10-containing complex is distributed across the entire range of high molecular mass standards, but predominantly concentrated between 66 and 232 kDa (Fig. 5B), and the Coq10-containing complex is absent in the coq10Δ mutant (Fig. 5B). Although the signal intensities of COQ10A and COQ10B are low, it appears that COQ10A forms a discrete complex at ∼140 kDa (Fig. 5B), and COQ10B is dispersed across between 232 and 440 kDa (Fig. 5B). Knowing the total amount of protein subjected to analysis by BN/SDS-PAGE, it is possible that a significant amount of COQ10A and COQ10B may migrate at a much smaller size corresponding to its monomeric molecular mass, not observed by the first dimension gel matrix.
Expression of human COQ10A or COQ10B fails to restore CoQ biosynthesis in the yeast coq10Δ mutant
In yeast, CoQ is produced from two distinct quinone ring precursors, pABA or 4HB (5). Early-stage intermediates, 4-amino-3-hexaprenylbenzoic acid (HAB) and 4-hydroxy-3-hexaprenylbenzoic acid (HHB), are the first polyisoprenylated intermediates emerging from pABA and 4HB, respectively (5). Subsequent modifications of the ring of HAB or HHB give rise to late-stage intermediates, 4-imino-demethoxy-Q6H2 (IDMQ6H2) or demethoxy-Q6H2 (DMQ6H2). It is believed that only DMQ6H2 gets directly converted to CoQ6H2, whereas IDMQ6H2 represents a dead-end late-stage product; the deamination of HAB is mediated by the Coq6/Coq9 step in the pABA pathway (5). To examine how well single- and multi-copy COQ10A or COQ10B rescue yeast coq10Δ de novo CoQ biosynthesis, we labeled the cells with 13C6-pABA or 13C6-4HB in order to measure the efficiency of CoQ production from both pathways.
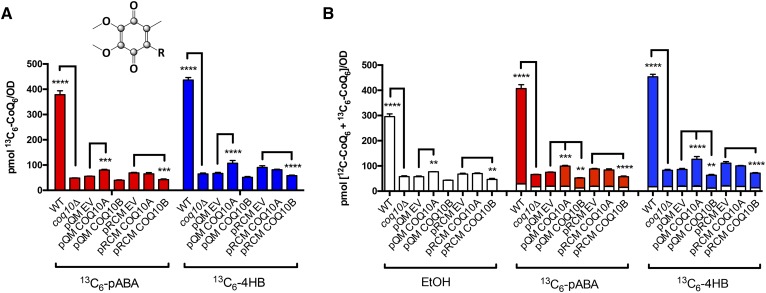
Consistent with previously published results (9), we observed that the yeast coq10Δ mutant produced less 13C6-CoQ6 (Fig. 6A) but significantly higher amounts of the early intermediates, 13C6-HAB (supplemental Fig. S2A) and 13C6-HHB (supplemental Fig. S2B) during log phase growth. The yeast coq10Δ mutant also makes less late-stage intermediates, 13C6-DMQ6 (supplemental Fig. S2C) and 13C6-IDMQ6 (supplemental Fig. S2D), de novo compared with the wild-type cells. One caveat to this result is that when the yeast coq10Δ mutant is transformed with empty vector pQM or pRCM, these empty vectors seem to make a significant difference on the levels of all representative CoQ6-intermediates as well as in CoQ6 when compared with the coq10Δ mutant (Fig. 6, supplemental Fig. S2). We suspected that this might be a result of different medium used to culture the coq10Δ mutant (SD-Complete) and coq10Δ mutant with empty vectors (SD-Ura). Therefore, we compared the yeast coq10Δ expressing single- or multi-copy COQ10A or COQ10B to their respective empty vector controls for the statistical analyses. If COQ10A or COQ10B were to restore efficient de novo CoQ biosynthesis, we would expect lower amounts of 13C6-HAB and 13C6-HHB and a higher amount of 13C6-CoQ6. However, expression of either COQ10A or COQ10B has only a minimal effect on the de novo biosynthesis of 13C6-CoQ6. Single-copy COQ10A seems to make slightly more 13C6-CoQ6 from labeled pABA and 4HB (Fig. 6A), and has slightly higher levels of total CoQ6 (Fig. 6B) when compared with the empty vector control. In contrast, expression of COQ10B, particularly in multi-copy, decreases de novo 13C6-CoQ6 as well as the total CoQ6 content (Fig. 6A, B). While single- and multi-copy COQ10A and multi-copy COQ10B synthesize 13C6-DMQ6 (supplemental Fig. S2C) and 13C6-IDMQ6 (supplemental Fig. S2D), the conversion from early-stage intermediates to late-stage intermediates is slow, as indicated by a buildup of the labeled 13C6-HAB (supplemental Fig. S2A) and 13C6-HHB (supplemental Fig. S2B).
Fig. 6.
Expression of human COQ10A or COQ10B has a minimal effect on de novo CoQ6 biosynthesis and total CoQ6 content. The de novo production of CoQ6 was measured from yeast whole-cell lipid extracts from wild-type, coq10Δ, coq10Δ expressing single- or multi-copy COQ10A or COQ10B, or their respective empty vector labeled with 13C6-pABA (red) or 13C6-4HB (blue) for 5 h. Expression of single- or multi-copy COQ10A or COQ10B has almost negligible effect on both de novo 13C6-CoQ6 (A) and total CoQ6 (B) when compared with their respective empty vector controls. The statistical analyses were performed using two-way ANOVA multiple comparisons from three biological replicates, comparing yeast coq10Δ expressing single- or multi-copy COQ10A or COQ10B to their respective empty vector controls, and comparing yeast coq10Δ mutant to the wild-type control. The error bar indicates mean ± SD, and the statistical significance is represented by *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
In addition to 13C6-labeled CoQ6 and CoQ6-intermediates, we also quantified the unlabeled CoQ6 and CoQ6-intermediates (Fig. 6B, supplemental Fig. S2E–H). Similar to what we observed with the 13C6-labeled CoQ6 and CoQ6-intermediates, the yeast coq10Δ mutant expressing single- or multi-copy COQ10A or COQ10B has only a negligible effect on the steady state levels of CoQ6 and CoQ6-intermediates. To summarize, neither single- nor multi-copy expression of COQ10A or COQ10B functionally restores de novo CoQ biosynthesis to the wild-type level, and has negligible effects on the synthesis of early stage CoQ6-intermediates relative to the empty vector control. Given that the high molecular weight CoQ synthome is known to be necessary for efficient CoQ6 biosynthesis in yeast, it is not surprising that each of the human COQ10 orthologs fails to restore the yeast de novo CoQ6 production.
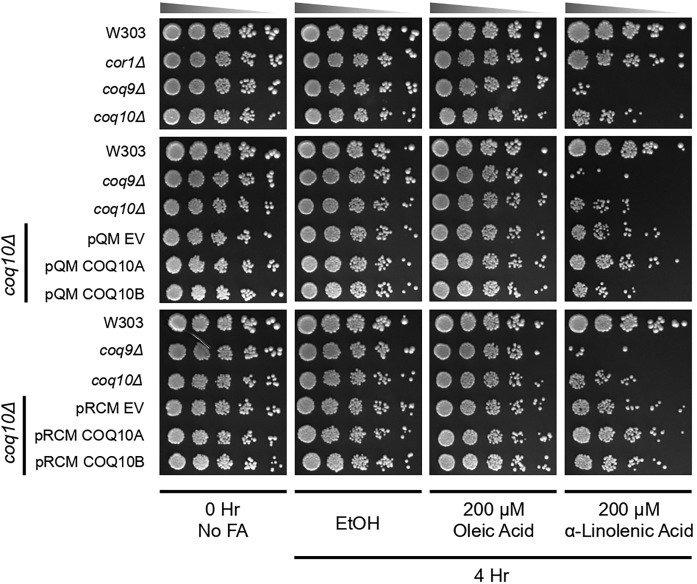
Human COQ10A rescues PUFA sensitivity of the yeast coq10Δ mutant
PUFAs are particularly susceptible to oxidative damage caused by ROS-dependent abstraction of hydrogen atoms at bis-allylic positions, generating carbon-centered free radicals (65). In the presence of oxygen, the resulting peroxyl radicals trigger a chain reaction of lipid peroxidation and propagate oxidative damage to other macromolecules. Collectively, this oxidative damage results in DNA mutations, protein fragmentation, and formation of protein-protein cross-links (66). As shown in Fig. 7, the yeast CoQ-less mutant (coq9Δ) is sensitive to α-linolenic acid due to the absence of antioxidant protection offered by CoQ (30, 31). The yeast coq10Δ mutant is also sensitive to treatment with α-linolenic acid, presumably because the chaperone function of the Coq10 polypeptide is necessary for the ability of CoQ to function as an antioxidant (9). This chaperone function of Coq10 is independent of its role in respiration per se, because a CoQ-replete respiratory-deficient mutant lacking a subunit of complex III (cor1Δ) is resistant to α-linolenic acid (Fig. 7). Thus, we assessed whether the sensitivity to PUFA treatment of yeast coq10Δ was rescued by the expression of single- or multi-copy human COQ10A or COQ10B. Expression of either single- or multi-copy COQ10A rescued yeast coq10Δ sensitivity to treatment with α-linolenic acid (Fig. 7). Multi-copy COQ10B partially rescued yeast coq10Δ sensitivity to α-linolenic acid, while single-copy COQ10B did not have a significant effect (Fig. 7). As expected, yeast strains tested were resistant to monounsaturated oleic acid (Fig. 7). Thus, our data suggest that COQ10A and COQ10B are only partially able to complement the yeast coq10Δ mutant; both human co-orthologs are capable of rescuing defective respiration and PUFA sensitivity in the yeast coq10Δ mutant, but fail to rescue the defect in CoQ6 biosynthesis.
Fig. 7.
Human COQ10A rescues PUFA sensitivity of the yeast coq10Δ mutant. Wild-type W303, respiratory-deficient mutant cor1Δ, CoQ-less mutant coq9Δ, coq10Δ, and coq10Δ expressing single- or multi-copy COQ10A or COQ10B, or their respective empty vector were grown in SD-Complete or SD-Ura liquid medium to log phase. Harvested cells were washed twice with sterile water and resuspended in phosphate buffer with 0.2% dextrose (pH 6.2) to 0.2 OD600/ml. The resuspended cells were incubated with oleic acid or α-linolenic acid prepared in ethanol at a final concentration of 200 μM for 4 h. Cells were also incubated with only ethanol as vehicle control. Aliquots of cell suspension from each sample were removed before addition of fatty acids [0 h (0 Hr), no fatty acids], and 4 h (4 Hr) post fatty acid incubation to assess cell viability by plate dilution assay. An aliquot of 2 μl of a series of 5-fold diluted samples was plated in each spot starting at 0.2 OD600/ml in the first spot on YPD plate medium. Pictures were taken 2 days after incubation at 30°C.
COQ10 family analysis
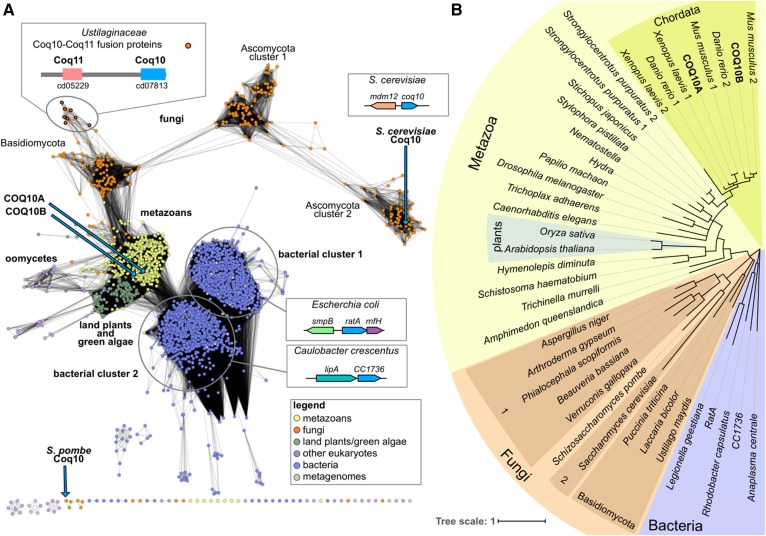
A protein sequence similarity network analysis reveals that the COQ10-like family can be divided into six main similarity clusters that are generally grouped by taxonomy (Fig. 8A). The COQ10-like proteins from land plants and green algae (with the exception of prasinophyte homologs) are closely related to animal homologs and, in the phylogenetic tree reconstruction, are nestled within the metazoan clade (Fig. 8B). Two groups of bacterial proteins are found in the COQ10-like family. Proteins encoded by genes (referred to as yfjG/ratA in E. coli) in the highly conserved smpB-ratA-yfjF gene neighborhood (67, 68) dominate bacterial cluster 1. RatA from E. coli (see sequence alignment in Fig. 2A) has a proposed role in cell cycle arrest as a response to stresses such as nutrient starvation (69). Upon induction, RatA blocks 70S ribosome association and inhibits the translation initiation step (69). The first gene in the neighborhood, smpB, also encodes a protein that interacts with the ribosome through a complex formed with tmRNA that is essential for rescuing stalled ribosomes (70, 71). The third gene in the conserved operon, yfjF, encodes a protein of unknown function, which was renamed RatB, an assumed antitoxin of RatA, but evidence suggests that RatB does not function as an antitoxin to RatA (69). YfjF is homologous to RnfH, a Rhodobacter capsulatus homolog encoded by a gene at the end of the rnf operon (72). Rnf is an enzyme complex homologous to but distinct from the bacterial respiratory complex, Na+-dependent NADH:ubiquinone oxidoreductase (Na+-NQR) (73). However, a role for RnfH in the Rnf complex is unsubstantiated and many rnf operons do not encode an RnfH orthologs (74).
Fig. 8.
The COQ10 family of proteins. A: A protein similarity network of proteins similar to COQ10 is shown. Each node (circle) represents one or more protein sequences, and each edge (solid line) represents similarity between two proteins (threshold set at an alignment score of 30). Nodes are colored by taxonomy as indicated in the legend. The locations of nodes representing human COQ10A and COQ10B, as well as previously characterized Coq10 orthologs from S. cerevisiae and S. pombe, are indicated with blue arrows. The disconnection of S. pombe Coq10 from the rest of the network is due to the low similarity (as measured by the BLASTp Evalue) between S. pombe Coq10 and other COQ10 homologs (with the exception of Coq10 from other Schizosaccharomyces species). The predominant operons observed for the two distinct bacterial clusters represented by E. coli and C. crescentus are shown as cartoons. A schematic of the Coq11-Coq10 protein fusion observed in genomes from Ustilaginaceae is also shown, and corresponding nodes are indicated with a thick border. Coq11 contains the cd05229 domain, an atypical SDR domain, and Coq10 contains the cd07813 domain, a SRPBCC (START/RHO_α_C/PITP/Bet_v1/CoxG/CalC) domain with a deep hydrophobic ligand-binding pocket. Protein and organism information for each node is available in supplemental Table S2. B: A phylogenetic tree of selected COQ10 homologs from each of the sequence similarity network protein clusters. Background shading corresponds to taxonomy as indicated. Branches with less than 50% bootstrap support were deleted.
Proteins encoded by genes that are often physically clustered with lipA orthologs, which encode lipoyl synthases, dominate bacterial cluster 2. Mining coexpression databases revealed that the characterized COQ10 from S. pombe is coexpressed with AIM22 (second-ranked coexpressed gene) (75), encoding a putative lipoate-protein ligase A. These two functional inferences (conserved gene clusters in bacteria and coexpression in S. pombe) may point to a yet uncharacterized role of COQ10 in lipoic acid biosynthesis or regulation; lipoic acid is a prosthetic group covalently attached to several dehydrogenases within the mitochondria. We also note that the top-ranked coexpressed gene in S. cerevisiae, MDM12, is positioned head-to-head with COQ10 (75), suggesting that the two genes share a bi-directional promoter. The mitochondrial distribution and morphology protein (Mdm12) is the cytosolic subunit of the ER-mitochondria encounter structure (ERMES) for establishing the ER-mitochondria contact sites, the absence of which causes severe mitochondrial morphological defects, defects in respiration, and rapid loss of mitochondrial DNA (76, 77).
The Coq10-Coq11 fusion proteins found in genomes from Ustilaginaceae (78) are also shown in Fig. 8. Additional domains found in the present analysis fused to the COQ10 domain (PF03364) (encoded by at least two different species) include PF00098 (zinc-finger domain), PF00227/PF10584 (proteasome subunits), and PF00378 (enoyl-CoA hydratase/isomerase family). Additionally, both COQ10A and COQ10B were previously found to interact with the enoyl-CoA hydratase, ECH1, which in turn has been observed to interact with COQ2, COQ3, COQ4, COQ6, COQ7, and COQ8A (79, 80). Mining coexpression databases, both COQ3 and COQ8A also coexpress with COQ10A (81). The human ECH1 localizes to the matrix of mitochondria and participates in β-oxidation of unsaturated fatty acids (82), and the interaction of ECH1 with both COQ10A and COQ10B aligns well with their roles as chaperones to deliver CoQ as a cofactor for the reaction.
DISCUSSION
This study provides another piece of evidence supporting the functional conservation of yeast and human proteins involved in CoQ biosynthesis. Unlike yeast, humans have two isoforms of COQ10, which may have evolved by a duplication event during chordate evolution (Fig. 8B). RNA-seq analysis of different human tissues suggests that although both COQ10A and COQ10B are universally expressed, COQ10A seems predominantly expressed in heart and skeletal muscle cells (83, 84). Knowing that COQ10A and COQ10B share 66% sequence identity (84% similarity), the enrichment of COQ10A in human heart and skeletal muscle implies a functional specialization of the protein. From our experimental results, it is curious to note that single-copy COQ10A performs the best in terms of restoring steady state Coq polypeptide levels (Fig. 4), stabilizing the CoQ synthome (Fig. 5), and restoring resistance against PUFAs (Fig. 7), despite the finding that single-copy COQ10A seems to restore respiratory growth phenotype just as well as both single- and multi-copy COQ10B on YPG plate medium (Fig. 1B). In contrast, neither COQ10A nor COQ10B expression restores de novo CoQ synthesis in yeast when compared with the coq10Δ empty vector control (Fig. 6, supplemental Fig. S2). Efficient CoQ biosynthesis requires a properly assembled CoQ synthome, but neither COQ10A nor COQ10B expression fully restores the CoQ synthome assembly (Fig. 5). Importantly, neither COQ10A nor COQ10B expression restores the steady state levels of Coq4, which is known to be the central organizer protein of the CoQ synthome (34). Additionally, the assembly and stability of the CoQ synthome relies on the presence of CoQ6 and CoQ6-intermediates. Studies have shown that reestablishment of de novo CoQ6 biosynthesis and the levels of certain CoQ6-intermediates restores the CoQ synthome assembly and CoQ domain formation (34, 85). The CoQ synthome is responsible for efficient de novo CoQ biosynthesis, and the resulting CoQ and CoQ-intermediates are in turn necessary to stabilize the CoQ synthome. Because neither COQ10A nor COQ10B rescues the defect in de novo CoQ6 production, the amounts of CoQ6 or CoQ6-intermediates may not be sufficient to restore and stabilize the CoQ synthome. However, expression of either of the human orthologs, COQ10A or COQ10B, complemented the coq10Δ glycerol growth (Fig. 1B). Thus, human COQ10A and COQ10B fulfill two independent functions in yeast: 1) both facilitate the function of CoQ in respiration; and 2) both enable CoQ to function as a chain-terminating antioxidant. However, neither COQ10A nor COQ10B function to restore the efficient CoQ biosynthesis in yeast.
One possibility is that a coordinated action of both yeast Coq10 and Coq11 is necessary to restore efficient CoQ biosynthesis in the yeast coq10Δ mutant. In mammalian cells, it is common that the START domain may be coupled with other motifs/domains on the same protein, offering additional functions, such as localization, enzymatic activity, or signaling (12, 13, 86). For instance, the metastatic lymph node 64 (MLN64) is a cholesterol-specific START protein, and it contains a conserved membrane-spanning (MLN64 N terminal) MENTAL domain in addition to the START domain (87). The NMENTAL domain anchors MLN64 to the late endosome membranes, from which the MENTAL domain can capture cholesterol and subsequently transfer it to the cytoplasmic START domain (14, 87, 88). Yeast Coq11 belongs to the short-chain dehydrogenase/reductase (SDR) superfamily, which contains a conserved Rossmann fold with an N-terminal binding site for NAD(H) or NADP(H) (89). The S. cerevisiae coq11Δ does not exhibit an apparent growth defect on a nonfermentable carbon source, but its de novo 13C6-CoQ6 production is significantly lower compared with the wild-type cells (78). S. cerevisiae coq10Δ shares a very similar phenotype except that its growth on nonfermentable carbon sources is impaired but not completely abolished. Thus, it may be possible that, in yeast, the Coq10 and Coq11 functions need to be coordinated in order to achieve wild-type level efficiency for de novo CoQ6 biosynthesis. The presence of Coq10-Coq11 fusion protein in several Ustilaginaceae species (Fig. 8) further consolidates a potential functional link conferred by their physical interaction and/or function in the same biological pathway. In mammalian cells, the closest but distinct homolog to yeast Coq11 is NDUFA9, a NADH dehydrogenase and a complex I ubiquinone reduction-module (Q-module) subunit (78, 90). However, because humans do not have Coq11, a similar interaction between Coq10 and Coq11 either never evolved or was lost during evolution. Therefore, we postulate that neither human COQ10A nor COQ10B was able to interact with yeast Coq11, hence failing to restore de novo CoQ biosynthesis (Fig. 6, supplemental Fig. S2). In contrast, plants and algae do have orthologs of yeast Coq11 and have evolved to use Coq11 or Coq11-like proteins (78). The Arabidopsis thaliana genome encodes four Coq11 orthologs (At1g32220, At5g15910, At5g15480, and At5g10730), and the chloroplast-localized flavin reductase-related protein, At1g32220, is thought to be involved in plastoquinone biosynthesis (49, 78). In COXPRESdb, COQ10A is coexpressed with SDR39U1, which, like Coq11, belongs to the SDR superfamily (75), and SDR39U1 was found to coelute with human COQ9 from a hydroxyapatite column in mitochondria solubilized with Triton X-100 (91). These functional inferences may supply a link between human COQ10 and Coq11-like proteins in CoQ production.
So far, most of the publications on yeast Coq10 are focused on its putative function as a CoQ chaperone for its function in respiration and as a lipophilic antioxidant. The Coq10 similarity network analysis (Fig. 8A) suggests that Coq10 homologs may be functionally linked to lipoic acid synthesis and/or regulation. Lipoic acid is a sulfur-containing cofactor and is essential for enzymatic functions of pyruvate dehydrogenase and α-ketoglutarate dehydrogenase, as well as the glycine cleavage system (92). Lipoic acid is assembled on its cognate proteins from precursor octanoic acid from the fatty acid biosynthesis pathway to a specific lysine residue of the cognate protein by octanoyl transferase (LipB), followed by insertion of sulfur by lipoyl synthetase (LipA) (92). Eukaryotes contain a conserved mitochondrial fatty acid synthetic pathway, independent from cytosolic fatty acid biosynthesis machinery (93). The respiratory competence in yeast is dependent on the ability of mitochondria to synthesize fatty acids, and yeast deletion mutants of enzymes involved in mitochondrial fatty acid synthesis exhibit a respiratory-deficient phenotype and small mitochondria, possibly mediated by inefficient tRNA processing by RNase P cleavage (93). A role for Coq10 or Coq10 homologs in fatty acid synthesis has not been substantiated, but because yeast coq10Δ is respiratory-defective, it would be interesting to examine the mitochondrial morphology in this particular mutant and assess the lipoic acid-dependent enzyme activities.
Another possible functional role of Coq10 relates to the coexpression of COQ10 and MDM12 from a shared promoter in S. cerevisiae (Fig. 8A) (94). The arrangement and coregulation of different genes via promoter sharing is indicative of functional connection and/or physical interactions between the two gene products. In yeast, Mdm12 is positioned on the mitochondrial outer membrane as part of the ERMES complex subunit; Mdm12 bridges Mmm1 localized at the ER membrane with Mdm10 and Mdm34 at the cytosolic site (76). The ERMES complex mediates ER-mitochondrial contacts, essential for lipid exchange between the two organelles (76). Recently, polypeptide members of the CoQ synthome (Coq3–Coq7, Coq9, and Coq11) have been shown to localize selectively to multiple domains (CoQ domains) (85, 95). These CoQ domains are marked by ER-mitochondria contact sites and are established by a START-like domain containing ER membrane sterol transporter protein, Ltc1, in complex with the mitochondrial outer membrane protein, Tom71, as well as by the ERMES complex (85, 95). Presence of the CoQ domain relies on the cooperative assembly of the CoQ synthome components (85). Absence of any one subunit of the ERMES complex also elicits a dramatic effect on the overall stability of the CoQ synthome and CoQ domain formation, but only a negligible effect on the steady state levels of the constituent Coq polypeptides of the CoQ synthome (85, 95). Deletion of LTC1 in the mmm1Δ mutant further enhanced the defect of CoQ domain formation, suggesting a redundant functional role of these two ER-mitochondria tethers for domain positioning in the mitochondria (85). The ERMESΔ mutants accumulate steady state and 13C6-labeled CoQ6-intermediates as a result of inefficient CoQ6 biosynthesis from destabilized CoQ synthome (95). Interestingly, these ERMESΔ mutants retain more 13C6-labeled CoQ6 even though their steady state CoQ6 levels inside the mitochondria are lower compared with the wild-type cells (95). The disruption of CoQ6 homeostasis presumably results from the reduced sequestration of CoQ6 within the mitochondria and/or compromised degradation of CoQ6 at the peroxisomes, which have been reported to colocalize with the ERMES complex at specific mitochondrial subdomains (95, 96). Inside the yeast mitochondria, two separate studies have observed a reduced number of CoQ synthome domain formations in the yeast coq10Δ mutant, which is likely contributed by the partial destabilization of the CoQ synthome as well as component Coq polypeptides in the absence of Coq10 (63, 85, 95). The establishment of CoQ domains within mitochondria relies on CoQ-intermediates and substrate flux (85), which seems to correlate well with the role of Coq10 in mediating efficient CoQ production. However, the exact mechanism underlying the role of Coq10 in coordination of the ER-mitochondria contact sites and CoQ biosynthesis remains to be elucidated.
Similar CoQ domains are also observed in human cells (85). However, mammalian cells do not have orthologs of ERMES polypeptides, and the physical contact between ER and mitochondria is established by homo- or heterodimerization of ER-localized mitofusin (MFN)2, with MFN1 or MFN2 on the mitochondrial outer membrane (97). Loss of MFN2 impairs respiration capacity, originating from a depletion of mitochondrial CoQ content and reduced CoQ-dependent NADH-cytochrome c reductase and succinate-cytochrome c reductase activities (98). Proteomic and metabolic analyses suggest that loss of MFN2 likely affects the isoprene biosynthesis pathway that is upstream of both CoQ and cholesterol biosynthesis, but the effect is only observed as decreased CoQ but not cholesterol levels (98). Similar to the yeast system, the enzymes involved in the isoprene biosynthetic pathway exist in different organelles, including mitochondria, ER, and peroxisomes (98); however, the functional implications of the involvement of human COQ10A and COQ10B in physical coordination of these organelles remain to be uncovered.
In summary, this study indicates that although expression of the human COQ10A or COQ10B orthologs failed to restore efficient de novo synthesis of CoQ6, they nonetheless rescued the respiratory deficiency and the sensitivity to oxidative stress of the yeast coq10Δ mutant. These results indicate that the Coq10 START domain functions as a CoQ chaperone, necessary for respiration-dependent cellular bioenergetics and defense mechanisms against oxidative stress. The COQ10 family protein analyses provide additional insights into the possible roles of Coq10-dependent transport of CoQ, necessary for its function as a cofactor in biological pathways and trafficking between organelles.
Supplementary Material
Acknowledgments
The authors thank Dr. Carol Dieckmann from the University of Arizona for her comments and suggestions on this work.
Footnotes
Abbreviations:
- BN/SDS-PAGE
- Blue Native/SDS-PAGE
- CoQ
- coenzyme Q or ubiquinone
- CoQH2
- reduced coenzyme QH2 hydroquinone or ubiquinol
- CoQn
- coenzyme Qn or ubiquinone-n (refers to a specific isoform of CoQn, where n designates the number of isoprene units in the tail of CoQn, e.g., CoQ6 in Saccharomyces cerevisiae or CoQ10 in humans)
- DMQ6H2
- demethoxy-Q6H2
- DMQ3
- demethoxy-coenzyme Q3
- ERMES
- ER-mitochondria encounter structure
- HAB
- 4-amino-3-hexaprenylbenzoic acid
- 4HB
- 4-hydroxybenzoic acid
- HHB
- 4-hydroxy-3-hexaprenylbenzoic acid
- IDMQ6H2
- 4-imino-demethoxy-Q6H2
- MFN
- mitofusin
- MLN64
- metastatic lymph node 64
- MSA
- multiple sequence alignment
- pABA
- para-aminobenzoic acid
- ROS
- reactive oxygen species
- SD-Complete
- synthetic dextrose/minimal-complete
- SDR
- short-chain dehydrogenase/reductase
- SD-Ura
- synthetic dextrose/minimal minus uracil
- START
- steroidogenic acute regulatory protein-related lipid transfer
This work was supported by National Science Foundation Grant MCB-1330803 (to C.F.C.) and National Institutes of Health Grant T32 GM 008496 (to H.S.T. and M.C.B.). C.E.B-H. is supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research. M.G-J. is supported by the Quantitative and Computational Biosciences Collaboratory Postdoctoral Fellowship. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Alcázar-Fabra M., Trevisson E., and Brea-Calvo G.. 2018. Clinical syndromes associated with coenzyme Q10 deficiency. Essays Biochem. 62: 377–398. [DOI] [PubMed] [Google Scholar]
- 2.Turunen M., Olsson J., and Dallner G.. 2004. Metabolism and function of coenzyme Q. Biochim. Biophys. Acta. 1660: 171–199. [DOI] [PubMed] [Google Scholar]
- 3.Desbats M. A., Lunardi G., Doimo M., Trevisson E., and Salviati L.. 2015. Genetic bases and clinical manifestations of coenzyme Q10 (CoQ10) deficiency. J. Inherit. Metab. Dis. 38: 145–156. [DOI] [PubMed] [Google Scholar]
- 4.Stefely J. A., and Pagliarini D. J.. 2017. Biochemistry of mitochondrial coenzyme Q biosynthesis. Trends Biochem. Sci. 42: 824–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Awad A. M., Bradley M. C., Fernández-del-Río L., Nag A., Tsui H. S., and Clarke C. F.. 2018. Coenzyme Q10 deficiencies: pathways in yeast and humans. Essays Biochem. 62: 361–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hajj Chehade M., Pelosi L., Fyfe C. D., Loiseau L., Rascalou B., Brugière S., Kazemzadeh K., Vo C-D-T., Ciccone L., Aussel L., et al. 2019. A soluble metabolon synthesizes the isoprenoid lipid ubiquinone. Cell Chem. Biol. 26: 482–492.e7. [DOI] [PubMed] [Google Scholar]
- 7.Okada K., Suzuki K., Kamiya Y., Zhu X., Fujisaki S., Nishimura Y., Nishino T., Nakagawa T., Kawamukai M., and Matsuda H.. 1996. Polyprenyl diphosphate synthase essentially defines the length of the side chain of ubiquinone. Biochim. Biophys. Acta. 1302: 217–223. [DOI] [PubMed] [Google Scholar]
- 8.Kawamukai M. 2016. Biosynthesis of coenzyme Q in eukaryotes. Biosci. Biotechnol. Biochem. 80: 23–33. [DOI] [PubMed] [Google Scholar]
- 9.Allan C. M., Hill S., Morvaridi S., Saiki R., Johnson J. S., Liau W. S., Hirano K., Kawashima T., Ji Z., Loo J. A., et al. 2013. A conserved START domain coenzyme Q-binding polypeptide is required for efficient Q biosynthesis, respiratory electron transport, and antioxidant function in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1831: 776–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barros M. H., Johnson A., Gin P., Marbois B. N., Clarke C. F., and Tzagoloff A.. 2005. The Saccharomyces cerevisiae COQ10 gene encodes a START domain protein required for function of coenzyme Q in respiration. J. Biol. Chem. 280: 42627–42635. [DOI] [PubMed] [Google Scholar]
- 11.Shen Y., Goldsmith-Fischman S., Atreya H. S., Acton T., Ma L., Xiao R., Honig B., Montelione G. T., and Szyperski T.. 2005. NMR structure of the 18 kDa protein CC1736 from Caulobacter crescentus identifies a member of the START domain superfamily and suggests residues mediating substrate specificity. Proteins. 58: 747–750. [DOI] [PubMed] [Google Scholar]
- 12.Ponting C. P., and Aravind L.. 1999. START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem. Sci. 24: 130–132. [DOI] [PubMed] [Google Scholar]
- 13.Iyer L. M., Koonin E. V., and Aravind L.. 2001. Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins. 43: 134–144. [DOI] [PubMed] [Google Scholar]
- 14.Alpy F., and Tomasetto C.. 2014. START ships lipids across interorganelle space. Biochimie. 96: 85–95. [DOI] [PubMed] [Google Scholar]
- 15.Martin L. A., Kennedy B. E., and Karten B.. 2016. Mitochondrial cholesterol: mechanisms of import and effects on mitochondrial function. J. Bioenerg. Biomembr. 48: 137–151. [DOI] [PubMed] [Google Scholar]
- 16.Cui T. Z., and Kawamukai M.. 2009. Coq10, a mitochondrial coenzyme Q binding protein, is required for proper respiration in Schizosaccharomyces pombe. FEBS J. 276: 748–759. [DOI] [PubMed] [Google Scholar]
- 17.Wikström M. K., and Berden J. A.. 1972. Oxidoreduction of cytochrome b in the presence of antimycin. Biochim. Biophys. Acta. 283: 403–420. [DOI] [PubMed] [Google Scholar]
- 18.Starkov A. A., and Fiskum G.. 2001. Myxothiazol induces H2O2 production from mitochondrial respiratory chain. Biochem. Biophys. Res. Commun. 281: 645–650. [DOI] [PubMed] [Google Scholar]
- 19.Dröse S., and Brandt U.. 2008. The mechanism of mitochondrial superoxide production by the cytochrome bc1 complex. J. Biol. Chem. 283: 21649–21654. [DOI] [PubMed] [Google Scholar]
- 20.Busso C., Tahara E. B., Ogusucu R., Augusto O., Ferreira-Junior J. R., Tzagoloff A., Kowaltowski A. J., and Barros M. H.. 2010. Saccharomyces cerevisiae coq10 null mutants are responsive to antimycin A. FEBS J. 277: 4530–4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murai M., Matsunobu K., Kudo S., Ifuku K., Kawamukai M., and Miyoshi H.. 2014. Identification of the binding site of the quinone-head group in mitochondrial Coq10 by photoaffinity labeling. Biochemistry. 53: 3995–4003. [DOI] [PubMed] [Google Scholar]
- 22.Montini G., Malaventura C., and Salviati L.. 2008. Early coenzyme Q10 supplementation in primary coenzyme Q10 deficiency. N. Engl. J. Med. 358: 2849–2850. [DOI] [PubMed] [Google Scholar]
- 23.Marcoff L., and Thompson P. D.. 2007. The role of coenzyme Q10 in statin-associated myopathy: a systematic review. J. Am. Coll. Cardiol. 49: 2231–2237. [DOI] [PubMed] [Google Scholar]
- 24.Ayer A., Macdonald P., and Stocker R.. 2015. CoQ10 function and role in heart failure and ischemic heart disease. Annu. Rev. Nutr. 35: 175–213. [DOI] [PubMed] [Google Scholar]
- 25.Mortensen S. A., Rosenfeldt F., Kumar A., Dolliner P., Filipiak K. J., Pella D., Alehagen U., Steurer G., Littarru G. P., and Investigators Q. S. S.. 2014. The effect of coenzyme Q10 on morbidity and mortality in chronic heart failure: results from Q-SYMBIO: a randomized double-blind trial. JACC Heart Fail. 2: 641–649. [DOI] [PubMed] [Google Scholar]
- 26.Burke D., Dawson D., and Stearns T.. 2000. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- 27.Barkovich R. J., Shtanko A., Shepherd J. A., Lee P. T., Myles D. C., Tzagoloff A., and Clarke C. F.. 1997. Characterization of the COQ5 gene from Saccharomyces cerevisiae. Evidence for a C-methyltransferase in ubiquinone biosynthesis. J. Biol. Chem. 272: 9182–9188. [DOI] [PubMed] [Google Scholar]
- 28.Hsu A. Y., Poon W. W., Shepherd J. A., Myles D. C., and Clarke C. F.. 1996. Complementation of coq3 mutant yeast by mitochondrial targeting of the Escherichia coli UbiG polypeptide: evidence that UbiG catalyzes both O-methylation steps in ubiquinone biosynthesis. Biochemistry. 35: 9797–9806. [DOI] [PubMed] [Google Scholar]
- 29.Elble R. 1992. A simple and efficient procedure for transformation of yeasts. Biotechniques. 13: 18–20. [PubMed] [Google Scholar]
- 30.Hill S., Hirano K., Shmanai V. V., Marbois B. N., Vidovic D., Bekish A. V., Kay B., Tse V., Fine J., Clarke C. F., et al. 2011. Isotope-reinforced polyunsaturated fatty acids protect yeast cells from oxidative stress. Free Radic. Biol. Med. 50: 130–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill S., Lamberson C. R., Xu L., To R., Tsui H. S., Shmanai V. V., Bekish A. V., Awad A. M., Marbois B. N., Cantor C. R., et al. 2012. Small amounts of isotope-reinforced polyunsaturated fatty acids suppress lipid autoxidation. Free Radic. Biol. Med. 53: 893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glick B. S., and Pon L. A.. 1995. Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 260: 213–223. [DOI] [PubMed] [Google Scholar]
- 33.Schägger H., Cramer W. A., and von Jagow G.. 1994. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217: 220–230. [DOI] [PubMed] [Google Scholar]
- 34.He C. H., Xie L. X., Allan C. M., Tran U. C., and Clarke C. F.. 2014. Coenzyme Q supplementation or over-expression of the yeast Coq8 putative kinase stabilizes multi-subunit Coq polypeptide complexes in yeast coq null mutants. Biochim. Biophys. Acta. 1841: 630–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wittig I., Braun H. P., and Schagger H.. 2006. Blue native PAGE. Nat. Protoc. 1: 418–428. [DOI] [PubMed] [Google Scholar]
- 36.Marbois B., Xie L. X., Choi S., Hirano K., Hyman K., and Clarke C. F.. 2010. para-Aminobenzoic acid is a precursor in coenzyme Q6 biosynthesis in Saccharomyces cerevisiae. J. Biol. Chem. 285: 27827–27838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buchan D. W., Minneci F., Nugent T. C., Bryson K., and Jones D. T.. 2013. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 41: W349–W357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones D. T., and Cozzetto D.. 2015. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics. 31: 857–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F. T., de Beer T. A. P., Rempfer C., Bordoli L., et al. 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46: W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chaudhury S., Lyskov S., and Gray J. J.. 2010. PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics. 26: 689–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benkert P., Tosatto S. C., and Schomburg D.. 2008. QMEAN: a comprehensive scoring function for model quality assessment. Proteins. 71: 261–277. [DOI] [PubMed] [Google Scholar]
- 42.Lüthy R., Bowie J. U., and Eisenberg D.. 1992. Assessment of protein models with three-dimensional profiles. Nature. 356: 83–85. [DOI] [PubMed] [Google Scholar]
- 43.Colovos C., and Yeates T. O.. 1993. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 2: 1511–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C.. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66: 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trott O., and Olson A. J.. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31: 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams P. D., Afonine P. V., Bunkoczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gerlt J. A., Bouvier J. T., Davidson D. B., Imker H. J., Sadkhin B., Slater D. R., and Whalen K. L.. 2015. Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): a web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta. 1854: 1019–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T.. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.UniProt Consortium T. 2018. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 46: 2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katoh K., and Standley D. M.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nguyen L. T., Schmidt H. A., von Haeseler A., and Minh B. Q.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller M. A., Schwartz T., Pickett B. E., He S., Klem E. B., Scheuermann R. H., Passarotti M., Kaufman S., and O’Leary M. A.. 2015. A RESTful API for access to phylogenetic tools via the CIPRES science gateway. Evol. Bioinform. Online. 11: 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller M. A., Pfeiffer W., and Schwartz T.. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE) in New Orleans, LA. November 14, 2010. 1–8. [Google Scholar]
- 54.Hoang D. T., Chernomor O., von Haeseler A., Minh B. Q., and Vinh L. S.. 2018. UFBoot2: improving the Ultrafast Bootstrap approximation. Mol. Biol. Evol. 35: 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.NCBI Resource Coordinators. 2018. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 46: D8–D13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vrablik M., Zlatohlavek L., Stulc T., Adamkova V., Prusikova M., Schwarzova L., Hubacek J. A., and Ceska R.. 2014. Statin-associated myopathy: from genetic predisposition to clinical management. Physiol. Res. 63(Suppl 3): S327–S334. [DOI] [PubMed] [Google Scholar]
- 57.Peterson T. A., Adadey A., Santana-Cruz I., Sun Y., Winder A., and Kann M. G.. 2010. DMDM: domain mapping of disease mutations. Bioinformatics. 26: 2458–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abd T. T., and Jacobson T. A.. 2011. Statin-induced myopathy: a review and update. Expert Opin. Drug Saf. 10: 373–387. [DOI] [PubMed] [Google Scholar]
- 59.Busso C., Bleicher L., Ferreira-Junior J. R., and Barros M. H.. 2010. Site-directed mutagenesis and structural modeling of Coq10p indicate the presence of a tunnel for coenzyme Q6 binding. FEBS Lett. 584: 1609–1614. [DOI] [PubMed] [Google Scholar]
- 60.Gatta A. T., Sauerwein A. C., Zhuravleva A., Levine T. P., and Matthews S.. 2018. Structural insights into a StART-like domain in Lam4 and its interaction with sterol ligands. Biochem. Biophys. Res. Commun. 495: 2270–2274. [DOI] [PubMed] [Google Scholar]
- 61.Horenkamp F. A., Valverde D. P., Nunnari J., and Reinisch K. M.. 2018. Molecular basis for sterol transport by StART-like lipid transfer domains. EMBO J. 37: e98002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jentsch J. A., Kiburu I., Pandey K., Timme M., Ramlall T., Levkau B., Wu J., Eliezer D., Boudker O., and Menon A. K.. 2018. Structural basis of sterol binding and transport by a yeast StARkin domain. J. Biol. Chem. 293: 5522–5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hsieh E. J., Gin P., Gulmezian M., Tran U. C., Saiki R., Marbois B. N., and Clarke C. F.. 2007. Saccharomyces cerevisiae Coq9 polypeptide is a subunit of the mitochondrial coenzyme Q biosynthetic complex. Arch. Biochem. Biophys. 463: 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tauche A., Krause-Buchholz U., and Rodel G.. 2008. Ubiquinone biosynthesis in Saccharomyces cerevisiae: the molecular organization of O-methylase Coq3p depends on Abc1p/Coq8p. FEMS Yeast Res. 8: 1263–1275. [DOI] [PubMed] [Google Scholar]
- 65.Yin H., Xu L., and Porter N. A.. 2011. Free radical lipid peroxidation: mechanisms and analysis. Chem. Rev. 111: 5944–5972. [DOI] [PubMed] [Google Scholar]
- 66.Porter N. A. 1986. Mechanisms for the autoxidation of polyunsaturated lipids. Acc. Chem. Res. 19: 262–268. [Google Scholar]
- 67.Iyer L. M., Burroughs A. M., and Aravind L.. 2006. The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like beta-grasp domains. Genome Biol. 7: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hudson C. M., Lau B. Y., and Williams K. P.. 2014. Ends of the line for tmRNA-SmpB. Front. Microbiol. 5: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y., and Inouye M.. 2011. RatA (YfjG), an Escherichia coli toxin, inhibits 70S ribosome association to block translation initiation. Mol. Microbiol. 79: 1418–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karzai A. W., Susskind M. M., and Sauer R. T.. 1999. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). EMBO J. 18: 3793–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Karzai A. W., Roche E. D., and Sauer R. T.. 2000. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat. Struct. Biol. 7: 449–455. [DOI] [PubMed] [Google Scholar]
- 72.Jouanneau Y., Jeong H. S., Hugo N., Meyer C., and Willison J. C.. 1998. Overexpression in Escherichia coli of the rnf genes from Rhodobacter capsulatus–characterization of two membrane-bound iron-sulfur proteins. Eur. J. Biochem. 251: 54–64. [DOI] [PubMed] [Google Scholar]
- 73.Reyes-Prieto A., Barquera B., and Juarez O.. 2014. Origin and evolution of the sodium-pumping NADH: ubiquinone oxidoreductase. PLoS One. 9: e96696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Biegel E., Schmidt S., Gonzalez J. M., and Muller V.. 2011. Biochemistry, evolution and physiological function of the Rnf complex, a novel ion-motive electron transport complex in prokaryotes. Cell. Mol. Life Sci. 68: 613–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Okamura Y., Aoki Y., Obayashi T., Tadaka S., Ito S., Narise T., and Kinoshita K.. 2015. COXPRESdb in 2015: coexpression database for animal species by DNA-microarray and RNAseq-based expression data with multiple quality assessment systems. Nucleic Acids Res. 43: D82–D86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Murley A., and Nunnari J.. 2016. The emerging network of mitochondria-organelle contacts. Mol. Cell. 61: 648–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kornmann B., Currie E., Collins S. R., Schuldiner M., Nunnari J., Weissman J. S., and Walter P.. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 325: 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Allan C. M., Awad A. M., Johnson J. S., Shirasaki D. I., Wang C., Blaby-Haas C. E., Merchant S. S., Loo J. A., and Clarke C. F.. 2015. Identification of Coq11, a new coenzyme Q biosynthetic protein in the CoQ-synthome in Saccharomyces cerevisiae. J. Biol. Chem. 290: 7517–7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Floyd B. J., Wilkerson E. M., Veling M. T., Minogue C. E., Xia C., Beebe E. T., Wrobel R. L., Cho H., Kremer L. S., Alston C. L., et al. 2016. Mitochondrial protein interaction mapping identifies regulators of respiratory chain function. Mol. Cell. 63: 621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chatr-Aryamontri A., Oughtred R., Boucher L., Rust J., Chang C., Kolas N. K., O’Donnell L., Oster S., Theesfeld C., Sellam A., et al. 2017. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 45: D369–D379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhu Q., Wong A. K., Krishnan A., Aure M. R., Tadych A., Zhang R., Corney D. C., Greene C. S., Bongo L. A., Kristensen V. N., et al. 2015. Targeted exploration and analysis of large cross-platform human transcriptomic compendia. Nat. Methods. 12: 211–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kovalyov L. I., Kovalyova M. A., Kovalyov P. L., Serebryakova M. V., Moshkovskii S. A., and Shishkin S. S.. 2006. Polymorphism of delta3,5-delta2,4-dienoyl-coenzyme A isomerase (the ECH1 gene product protein) in human striated muscle tissue. Biochemistry (Mosc.). 71: 448–453. [DOI] [PubMed] [Google Scholar]
- 83.Uhlén M., Fagerberg L., Hallström B. M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjöstedt E., Asplund A., et al. 2015. Proteomics. Tissue-based map of the human proteome. Science. 347: 1260419. [DOI] [PubMed] [Google Scholar]
- 84.Fagerberg L., Hallstrom B. M., Oksvold P., Kampf C., Djureinovic D., Odeberg J., Habuka M., Tahmasebpoor S., Danielsson A., Edlund K., et al. 2014. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics. 13: 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Subramanian K., Jochem A., Le Vasseur M., Lewis S., Paulson B. R., Reddy T. R., Russell J. D., Coon J. J., Pagliarini D. J., and Nunnari J.. 2019. Coenzyme Q biosynthetic proteins assemble in a substrate-dependent manner into domains at ER-mitochondria contacts. J. Cell Biol. 218: 1353–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Clark B. J. 2012. The mammalian START domain protein family in lipid transport in health and disease. J. Endocrinol. 212: 257–275. [DOI] [PubMed] [Google Scholar]
- 87.Alpy F., Wendling C., Rio M. C., and Tomasetto C.. 2002. MENTHO, a MLN64 homologue devoid of the START domain. J. Biol. Chem. 277: 50780–50787. [DOI] [PubMed] [Google Scholar]
- 88.Alpy F., and Tomasetto C.. 2006. MLN64 and MENTHO, two mediators of endosomal cholesterol transport. Biochem. Soc. Trans. 34: 343–345. [DOI] [PubMed] [Google Scholar]
- 89.Persson B., and Kallberg Y.. 2013. Classification and nomenclature of the superfamily of short-chain dehydrogenases/reductases (SDRs). Chem. Biol. Interact. 202: 111–115. [DOI] [PubMed] [Google Scholar]
- 90.Baertling F., Sanchez-Caballero L., van den Brand M. A. M., Fung C. W., Chan S. H., Wong V. C., Hellebrekers D. M. E., de Coo I. F. M., Smeitink J. A. M., Rodenburg R. J. T., et al. 2018. NDUFA9 point mutations cause a variable mitochondrial complex I assembly defect. Clin. Genet. 93: 111–118. [DOI] [PubMed] [Google Scholar]
- 91.Yamamoto T., Tamaki H., Katsuda C., Nakatani K., Terauchi S., Terada H., and Shinohara Y.. 2013. Molecular basis of interactions between mitochondrial proteins and hydroxyapatite in the presence of Triton X-100, as revealed by proteomic and recombinant techniques. J. Chromatogr. A. 1301: 169–178. [DOI] [PubMed] [Google Scholar]
- 92.Cronan J. E. 2014. Biotin and lipoic acid: synthesis, attachment, and regulation. Ecosal Plus. 6: doi:10.1128/ecosalplus.ESP-0001-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hiltunen J. K., Schonauer M. S., Autio K. J., Mittelmeier T. M., Kastaniotis A. J., and Dieckmann C. L.. 2009. Mitochondrial fatty acid synthesis type II: more than just fatty acids. J. Biol. Chem. 284: 9011–9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hibbs M. A., Hess D. C., Myers C. L., Huttenhower C., Li K., and Troyanskaya O. G.. 2007. Exploring the functional landscape of gene expression: directed search of large microarray compendia. Bioinformatics. 23: 2692–2699. [DOI] [PubMed] [Google Scholar]
- 95.Eisenberg-Bord M., Tsui H. S., Antunes D., Fernández-del-Río L., Bradley M. C., Dunn C. D., Nguyen T. P. T., Rapaport D., Clarke C. F., and Schuldiner M.. 2019. The endoplasmic reticulum-mitochondria encounter structure complex coordinates coenzyme Q biosynthesis. Contact (Thousand Oaks). 2: 2515256418825409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cohen Y., Klug Y. A., Dimitrov L., Erez Z., Chuartzman S. G., Elinger D., Yofe I., Soliman K., Gartner J., Thoms S., et al. 2014. Peroxisomes are juxtaposed to strategic sites on mitochondria. Mol. Biosyst. 10: 1742–1748. [DOI] [PubMed] [Google Scholar]
- 97.Merkwirth C., and Langer T.. 2008. Mitofusin 2 builds a bridge between ER and mitochondria. Cell. 135: 1165–1167. [DOI] [PubMed] [Google Scholar]
- 98.Mourier A., Motori E., Brandt T., Lagouge M., Atanassov I., Galinier A., Rappl G., Brodesser S., Hultenby K., Dieterich C., et al. 2015. Mitofusin 2 is required to maintain mitochondrial coenzyme Q levels. J. Cell Biol. 208: 429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gin P., and Clarke C. F.. 2005. Genetic evidence for a multi-subunit complex in coenzyme Q biosynthesis in yeast and the role of the Coq1 hexaprenyl diphosphate synthase. J. Biol. Chem. 280: 2676–2681. [DOI] [PubMed] [Google Scholar]
- 100.Do T. Q., Schultz J. R., and Clarke C. F.. 1996. Enhanced sensitivity of ubiquinone-deficient mutants of Saccharomyces cerevisiae to products of autoxidized polyunsaturated fatty acids. Proc. Natl. Acad. Sci. USA. 93: 7534–7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hsu A. Y., Do T. Q., Lee P. T., and Clarke C. F.. 2000. Genetic evidence for a multi-subunit complex in the O-methyltransferase steps of coenzyme Q biosynthesis. Biochim. Biophys. Acta. 1484: 287–297. [DOI] [PubMed] [Google Scholar]
- 102.Gin P., Hsu A. Y., Rothman S. C., Jonassen T., Lee P. T., Tzagoloff A., and Clarke C. F.. 2003. The Saccharomyces cerevisiae COQ6 gene encodes a mitochondrial flavin-dependent monooxygenase required for coenzyme Q biosynthesis. J. Biol. Chem. 278: 25308–25316. [DOI] [PubMed] [Google Scholar]
- 103.Marbois B. N., and Clarke C. F.. 1996. The COQ7 gene encodes a protein in Saccharomyces cerevisiae necessary for ubiquinone biosynthesis. J. Biol. Chem. 271: 2995–3004. [DOI] [PubMed] [Google Scholar]
- 104.Johnson A., Gin P., Marbois B. N., Hsieh E. J., Wu M., Barros M. H., Clarke C. F., and Tzagoloff A.. 2005. COQ9, a new gene required for the biosynthesis of coenzyme Q in Saccharomyces cerevisiae. J. Biol. Chem. 280: 31397–31404. [DOI] [PubMed] [Google Scholar]
- 105.Poon W. W., Barkovich R. J., Hsu A. Y., Frankel A., Lee P. T., Shepherd J. N., Myles D. C., and Clarke C. F.. 1999. Yeast and rat Coq3 and Escherichia coli UbiG polypeptides catalyze both O-methyltransferase steps in coenzyme Q biosynthesis. J. Biol. Chem. 274: 21665–21672. [DOI] [PubMed] [Google Scholar]
- 106.Belogrudov G. I., Lee P. T., Jonassen T., Hsu A. Y., Gin P., and Clarke C. F.. 2001. Yeast COQ4 encodes a mitochondrial protein required for coenzyme Q synthesis. Arch. Biochem. Biophys. 392: 48–58. [DOI] [PubMed] [Google Scholar]
- 107.Baba S. W., Belogrudov G. I., Lee J. C., Lee P. T., Strahan J., Shepherd J. N., and Clarke C. F.. 2004. Yeast Coq5 C-methyltransferase is required for stability of other polypeptides involved in coenzyme Q biosynthesis. J. Biol. Chem. 279: 10052–10059. [DOI] [PubMed] [Google Scholar]
- 108.Tran U. C., Marbois B., Gin P., Gulmezian M., Jonassen T., and Clarke C. F.. 2006. Complementation of Saccharomyces cerevisiae coq7 mutants by mitochondrial targeting of the Escherichia coli UbiF polypeptide: two functions of yeast Coq7 polypeptide in coenzyme Q biosynthesis. J. Biol. Chem. 281: 16401–16409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hsieh E. J., Dinoso J. B., and Clarke C. F.. 2004. A tRNATRP gene mediates the suppression of cbs2–223 previously attributed to ABC1/COQ8. Biochem. Biophys. Res. Commun. 317: 648–653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.