Abstract
Lipid droplets (LDs) are ubiquitous and highly dynamic subcellular organelles required for the storage of neutral lipids. LD number and size distribution are key parameters affected not only by nutrient supply but also by lipotoxic stress and metabolic regulation. Current methods for LD quantification lack general applicability and are either based on time consuming manual evaluation or show limitations if LDs are high in numbers or closely clustered. Here, we present an ImageJ-based approach for the detection and quantification of LDs stained by neutral lipid dyes in images acquired by conventional wide-field fluorescence microscopy. The method features an adjustable preprocessing procedure that resolves LD clusters. LD identification is based on their circular edges and central fluorescence intensity maxima. Adaptation to different cell types is mediated by a set of interactive parameters. Validation was done for three different cell lines using manual evaluation of LD numbers and volume measurement by 3D rendering of confocal datasets. In an application example, we show that overexpression of the acyl-CoA synthetase, FATP4/ACSVL5, in oleate-treated COS7 cells increased the size of LDs but not their number.
Keywords: endoplasmic reticulum, fatty acid/metabolism, fluorescence microscopy, triglycerides, image quantification, ImageJ/Fiji
Lipid droplets (LDs) are spherical organelles surrounded by a phospholipid monolayer and equipped with a unique set of proteins. After they were portrayed for many years as inert storage organelles for neutral lipids, recent research suggests an additional versatile repertoire of metabolic and regulatory functions in the cellular lipid metabolism. The importance of LDs is further emphasized by their presence in almost every known eukaryotic cell type (1), the abundance of functional contact sites to other organelles [reviewed in (2, 3)], and the involvement in metabolic disorders, such as type II diabetes, hepatic steatosis, and obesity (4–6).
Alterations of lipid metabolism are often apparent through changes of the LD morphology (number and volume of LDs per cell) as well as the capability of cells for de novo LD formation (7–13). Although such changes might be easily qualitatively described using different microscopy techniques, the quantification of LD numbers and size is at least equally important to characterize an identified phenotype.
LD analysis is frequently carried out manually (9, 14, 15) and often involves preprocessing steps (9, 16) to quantify LDs. While manual processing of images may ensure the highest precision, it frequently eliminates the feasibility of high-throughput experiments and large data set analysis.
Existing computational approaches for LD quantification, however, have the problems that: i) methods are either proof-of-principle developments or rely on advanced microscopy techniques (e.g., confocal microscopy, electron microscopy, coherent anti-Stokes Raman scattering microscopy) that are often not available to researchers (17–23); ii) the algorithm needs to be converted for use in common image analysis software (12, 24, 25); or iii) the procedure is optimized for a specific cell line or LD morphology (including threshold-based methods) (26–29).
MATERIALS AND METHODS
Cell lines and cell culture
All cell lines were cultured in DMEM (Thermo Fisher Scientific, Waltham, MA) containing 4.5 g/l glucose supplemented with 10% FBS (Life Technologies, Carlsbad, CA) and 1% penicillin/streptomycin (Life Technologies). Cells were maintained in a humidified atmosphere with 5% CO2 and were split when 80% confluency was reached.
Cells stably expressing the LD marker protein, A3Nt-GFP-FLAG (human ACSL3-N-terminus, M1-L135) (30), the murine fatty acid transport protein 4 (msFATP4-FLAG), and an empty retroviral plasmid (pRVH-1) were generated as described elsewhere (31). Briefly, phoenixGP cells were transfected with A3Nt-GFP-FLAG.pRIJ, msFATP4-FLAG.pRIJ, or pRVH-1.pRIJ, respectively, together with pVSV-G for virus pseudotyping. Viruses were harvested 48 h after transfection by filtering the supernatant through 0.45 μm PVDF membrane pores. COS7 and U2OS were seeded in 6-well plates and incubated with 1 ml (A3Nt-GFP-FLAG.pRIJ, pRVH-1.pRIJ) or 100 μl (msFATP4-FLAG.pRIJ; together with 900 μl normal growth medium) of the virus solution supplemented with 4 μg/ml polybrene for 24 h. After an additional 24 h in normal growth medium, the cells were trypsinized and selected for the stable genomic integration of A3Nt-GFP-FLAG.pRIJ, pRVH-1.pRIJ, or msFATP4-FLAG.pRIJ with puromycin (2 μg/ml for U2OS and 6 μg/ml for COS7). Untransduced cells served as a selection control. The cells were never allowed to reach confluencies above 90% during the selection process. Stably expressing cell lines are identified in this work by outlining the stably expressed construct as a prefix (e.g., FATP4.COS7).
Metabolic states and de novo formation of LDs
For experiments with cells maintained under normal growth conditions (DMEM/FCS), U2OS, COS7, and A431 as well as derivative cell lines were grown on coverslips in 12 wells (30,000/well for COS7 and A431; 50,000/well for U2OS).
To induce LD growth, the normal growth medium was supplemented for 24 h with 600 μM (COS7, U2OS) and 100 μM (A431) oleic acid (OA; Sigma, St. Louis, MO) bound to fatty acid-free BSA (Sigma) in a molar ratio of 6:1.
For starvation, the cells were incubated in serum-free DMEM (1 g/l glucose) containing 300 μM fatty acid-free BSA (COS7, A431) or 200 μM fatty acid-free BSA (U2OS).
For the induction of de novo formed LDs, stably expressing A3Nt.U2OS and A3Nt.COS7 were seeded in 12-well plates (60,000/well) and starved as described above for 24 h. After four washes with PBS, OA:BSA (6:1) was added to the culture medium (FCS-free DMEM, 4.5 g/l glucose) at a final concentration of 600 μM (A3Nt.COS7) and 300 μM OA:BSA (6:1) (A3Nt.U2OS) for 10 min to induce LD formation. Subsequently, the cells were washed three times by dipping the coverslips in beakers filled with PBS and instantly fixed with 4% PFA for 20 min at room temperature.
Microscopy data sets
After the indicated treatment, as described above, the cells were washed three times with PBS and fixed. After three additional washes with PBS, the LDs were either stained with BODIPY493/503 (2 μg/ml in PBS, Invitrogen D3922; Invitrogen, Waltham, MA), LD540 [0.25 μg/ml in PBS, kindly provided by Christoph Thiele (32)], or Nile Red [50 μg/ml in PBS; Sigma (33)] for 15 min at room temperature. The coverslips were rinsed in PBS shortly for three times and were washed additionally three times for 5 min each in PBS before the coverslips were embedded in MOWIOL (Calbiochem, San Diego, CA). Coverslips with cell lines stably expressing A3Nt-GFP were directly embedded in MOWIOL after fixation.
Widefield images were acquired using an Olympus BX41 microscope equipped with a 60× oil immersion PlanApo NA 1.40 objective and an F-view II CCD camera operated by the cell^D software. Emission signals were detected using longpass filters (U-MNG2 for LD540) or bandpass filters (U-M41028 for BODIPY493/503 and Nile Red). The images were saved as a 16-bit TIF image.
For each lipophilic dye and metabolic state, 100 images from two experiments were acquired for COS7, U2OS, and A431 (2,700 images in total; Fig. 2, supplemental Figs. S1, S2). For A3Nt.COS7 and A3Nt.U2OS, 150 images from three experiments per condition (starved, after the 10 min OA pulse) were acquired [600 images in total (Fig. 3)]. Furthermore, 150 images for A3Nt.U2OS under normal growth conditions were acquired (supplemental Fig. S3).
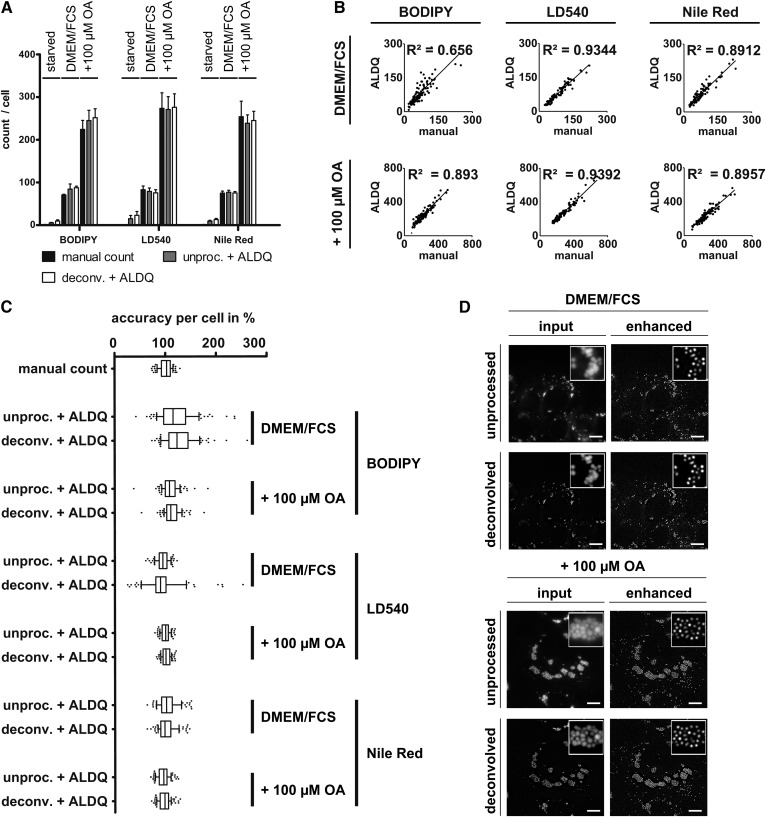
Fig. 2.
Quantification of LDs stained by lipophilic dyes in A431 cells. A431 cells were delipidated (starved) or cultured without (DMEM/FCS) or with (+100 μM OA) oleate supplementation for 24 h. LDs were counterstained with BODIPY493/503, LD540, or Nile Red. To assess the impact of image noise, the images were either deconvolved (deconv.) or left unprocessed (unproc.) and subsequently analyzed by ALDQ. A: Congruency of LD number detection by two independent quantitation techniques in A431 cells. LDs were manually counted (manual count) to which the ALDQ derived number (unproc. + ALDQ) was compared. Automatically determined LD numbers were highly similar to the manual count independent of the metabolic state. Suppression of image noise (deconv. + ALDQ) did not affect the results notably. Bars represent the mean ± SD of n = 2 independent experiments including 100 cells. B: Linear regression. The number of LDs derived by manual counting (x axis) was plotted against the number determined by ALDQ (y axis). Linear regression shows a strong correlation of both counting methods with indicated coefficients of determination (R2). Each analysis included the coordinates of 100 analyzed cells from n = 2 independent experiments. C: ALDQ accuracy is similar to manual counting accuracy. The LD number determined by ALDQ was plotted as a percentage of each cell’s manual LD count. Slim boxes and medians close to 100% indicate a precise LD detection per cell by ALDQ. Remarkably, the deviation per cell by ALDQ is often highly similar to the deviation of two examiners (“manual count”; see also the Materials and Methods). Similar to A, accuracy changes by prior deconvolution (deconv. + ALDQ) could not be observed. Each boxplot includes 100 cells in total from n = 2 independent experiments. D: Heavy clustering of LDs is resolved by preprocessing iterations. Representative images of LD540-stained A431 cells under standard growth condition (DMEM/FCS) and after oleate supplementation (+100 μM OA). A431 cells show extensive clustering of cytosolic LDs. The LDs were enhanced with twelve iterations (enhanced), which resolved the LD clusters. Note that prior deconvolution (deconvolved) can facilitate LD separation by reducing image noise, but does not affect the enhancement result by ALDQ (enhanced) perceptibly compared with raw images (unprocessed). Scale bar 10 μm.
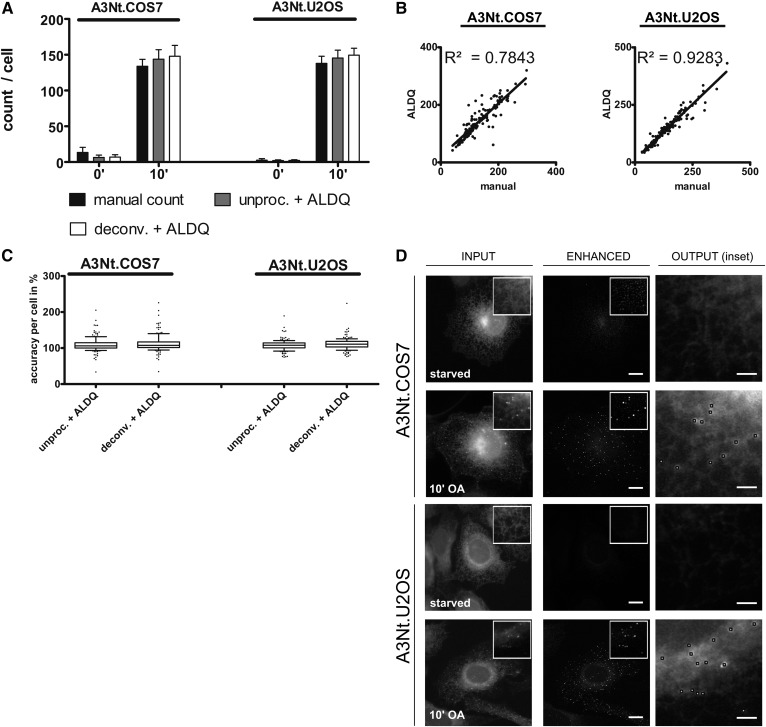
Fig. 3.
Detection of de novo formed LDs stained by the LD protein marker, A3Nt-GFP. The LD marker, A3Nt-GFP, was stably expressed by retroviral delivery in COS7 and U2OS cells. The cells were starved for 24 h (0’) prior to de novo LD formation induced by an oleate pulse for 10 min (10’). After imaging, the images were either deconvolved (deconv.) or left unprocessed (unproc.). A: Comparative analysis of LD detection methods. LD numbers per cell were determined by ALDQ (unproc. + ALDQ) and compared with manual counting (manual count). Automated counting by ALDQ slightly overestimates the LD number in both A3Nt.COS7 and A3Nt.U2OS cells. Prior deconvolution (deconv. + ALDQ) was used to suppress image noise, but did not result in accuracy changes. Bars represent the mean ± SD from n = 3 independent experiments including 150 cells in total. B: Linear regression. For each cell, the number of the manual count (x axis) was plotted against the automated measurement by ALDQ (y axis). Linear regression indicates a good correlation of manual and automated counts indicated by the shown coefficients of determination (R2). C: Single cell accuracy analysis. Manually determined LD numbers per cell were arbitrarily set to 100% and the corresponding ALDQ-derived number (ALDQ) was calculated as a percentage and plotted. Slim boxes, a median close to 100% and only a few outliers indicate high precision of ALDQ independent of prior deconvolution, which suppressed image noise. Each boxplot includes 150 cells in total from n = 3 independent experiments. D: Preprocessing iterations suppress ER-derived background, which enhances LD contrast. Shown are representative images of the indicated cell lines (input) before (starved) and after (10’) the oleate pulse. Cells were processed using 10 or 12 (COS7 10’ OA) preprocessing iterations (enhanced). Note that most fluorescent signal by ER-localized A3Nt-GFP is suppressed while LDs (shown in 10’ OA insets) are enhanced. Scale bar 10 μm and 2 μm (output).
Confocal microscopy was performed using a Nikon Ti Eclipse microscope equipped with an Ultraview VoX confocal spinning disc system (Perkin Elmer, Waltham, MA) and a 100× Apo PLAN VC, NA 1.40 oil objective. For each cell stained with LD540 (50 cells for each cell line and metabolic state; 300 images in total), 50 z-slices (0.2 μm spacing) were acquired using a 514 nm laser for excitation and a 567 nm emission filter and exported by the Volocity software (Perkin Elmer).
For manual assessment of LD numbers, one cell per image was randomly selected and LDs were counted using the multipoint function of ImageJ/Fiji and saved as a region of interest (ROI). In total, 490,627 LDs from 3,450 images were counted manually.
To evaluate a possible LD selection bias, a second examiner counted 180 randomly selected images of lipophilic dye-stained cells (6.66% of all images), which resulted in a deviation per cell from the previous examiner of 99.88 ± 12.64% (mean ± SD). The images covered all cell lines and dyes under normal growth conditions and oleate-treated conditions equally. The boxplots resulting from all cells counted by the two examiners are depicted in Fig. 2 and supplemental Figs. S1 and S2.
3D rendering and LD volume quantification
To validate the volume estimation algorithm, 3D rendering of LD540-positive structures in cells either treated with (600 μM OA) or without (DMEM/FCS) 600 μM OA:BSA (6:1) overnight was used. LD540 staining was selected because of its exceptional signal-to-noise ratio (SNR). The cells were imaged with a spinning disc confocal microscope and exported as described above. The resulting z-stacks were converted to Imaris files (.ims) and blindly deconvolved using the Autoquant software (Media Cybernetics, Rockville, MD). The LDs were segmented using the surface detection of thresholded z-stacks by Imaris (version 9.1; Bitplane, Concord, MA) (23). ROIs were drawn for each cell individually. Falsely segmented LDs were manually fused, whereas surfaces that clearly resulted from bright background structures, such as the ER and mitochondria, were removed manually (occurrence of maximum three particles per image in <10% of the cells without oleate treatment). The analysis settings were kept constant for each cell line and for each condition. Snapshots (Figs. 1A, 4A) were taken using the built-in snapshot function of Imaris. To validate the LD detection algorithm, sum intensity projections (SIPs) of raw z-stacks were created and the resulting pixel intensity values were normalized between 0 and 65,535 (16-bit image). The SIP images were used for the volume estimation by the proposed method [automated LD quantification (ALDQ)].
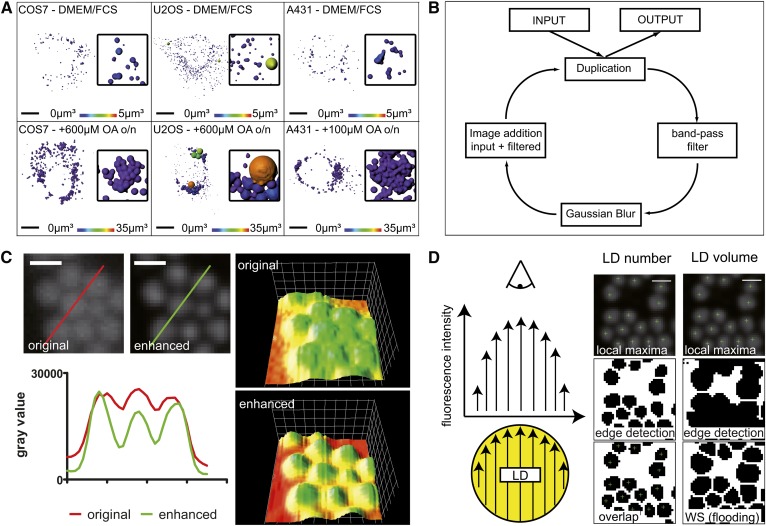
Fig. 1.
LD structure enhancement before detection of their local maxima and edges. A: LD populations in COS7, U2OS, and A431. Cells were cultured in standard growth medium (DMEM/FCS) or with oleate supplementation (+OA) for 24 h on coverslips. LDs were stained by the lipophilic dye, LD540, and imaged under a confocal microscope followed by 3D rendering by the Imaris software. Images color coded for the LD volume. The cell lines are differing in number, size, and clustering of LDs. Scale bar 10 μm, insets enlarged four times. B: LD structure enhancement. The input image is processed using a designed band pass filter. Subsequently, the gray values of the filtered image are added onto the original resulting in a 32-bit image, which is subsequently scaled back to the input image depth. The preprocessing iteration is repeated until LDs are satisfyingly enhanced. C: Contrast enhancement of LDs. LD540-stained LDs of COS7 after 24 h oleate treatment as an unprocessed image (original) and after preprocessing by three preprocessing iterations (enhanced). The corresponding 3D surface plots (right) are color coded for the relative pixel intensity. Gray values along the colored lines were plotted in the adjacent diagram. While the overall intensity is not altered drastically after the preprocessing step, the local maxima are significantly enhanced against the local minima. Scale bar 1 μm. D: LD identification by a central fluorescence maximum and a circular edge (schematic). Left: In widefield microscopy, central fluorescence maxima of LDs stained by lipophilic dyes arise from the observed fluorescence intensity (arrow length) distribution. The fluorescence intensity maximizes in the center due to the largest extension of the sphere. Middle: LD number determination. Local maxima (green crosshair) and edges are detected independently and overlaid. Only if the local maximum shares coordinates with an edge-defined particle, is it considered a LD. Right: LD volume estimation. The binary image of edge-defined particles (middle) represents the boundaries for the following segmentation by watershed flooding (WS; bottom). Each segmented particle is handled as the area of the largest LD cross-section and the volume of the corresponding sphere is calculated accordingly. Scale bar 1 μm.
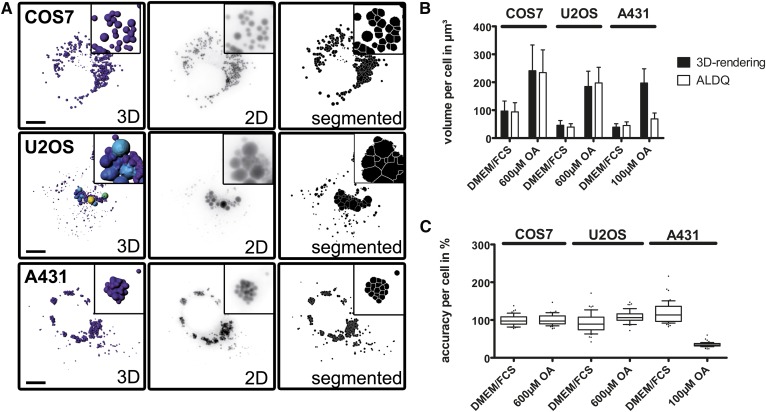
Fig. 4.
Volume estimation by ALDQ. A: Comparison of two volume estimation techniques. The indicated cell lines were treated with oleate for 24 h. Confocal imaging of LD540-stained LDs was followed by 3D rendering (3D) for volume measurement by the Imaris software. The corresponding z-stack was converted to a 2D image (2D, inverted for better visibility) and segmented by a flooding watershed algorithm (segmented) by ALDQ. The same was done for cells cultured without oleate treatment (not shown). Scale bar 10 μm, insets are enlarged 3.5 times. B: Comparison of volume analyses by different quantitation techniques. While the true volume determined by 3D rendering is approximated well in COS7 and U2OS cells by ALDQ, the volume of LDs in A431 supplemented with oleate is heavily underestimated. Bars represent the mean ± SD of 50 analyzed cells. C: Single cell accuracy analysis. The volume determined by ALDQ is plotted as a percentage compared with the volume determined by 3D rendering. Slim boxes representing the 25th to 75th percentile are surrounding the median that is close to 100%, suggesting high accuracy of the volume estimation by ALDQ (with the exception of A431 treated with oleate). Each boxplot corresponds to 50 analyzed cells.
Deconvolution of 2D images
The point spread function was calculated for each LD dye using the BX41 widefield microscope setup and the ImageJ/Fiji plugin “Diffraction PSF 3D” (34). Images were then deconvolved using the “Iterative Deconvolve 3D” plugin (34) with 10 iterations.
LD detection
To ease the localization and separation of LDs in a later step, an iterative operation was applied to increase the signal of LD structures in an image. In each iteration, the difference between the current image and its smoothed version using a Gaussian filter is first calculated. After all negative values are set to zero, the difference is smoothed again by a Gaussian filter. The current image is then updated by adding the smoothed difference and subsequently scaled back to its input image depth.
For LD counting, edges are detected (35) and local intensity maxima are determined in the preprocessed image. Edge-defined particles were obtained using thresholding followed by hole-filling and watershedding. An edge-defined particle sharing coordinates with a local maximum is then counted as a “true” LD.
For the volume estimation, LD edges were detected and processed as described for the number measurement, excluding the watershed step. Edge-defined particles marked with a local maximum are kept while those without a corresponding local maximum are excluded using geodesic reconstruction (36) with the local maxima coordinates as markers. The preprocessed input image is smoothed by Gaussian blurring, and individual LDs are subsequently detected using image flooding (37, 38) where the detected edges served as flooding boundaries. The areas of the segmented particles are measured, and the radius and corresponding volume are calculated assuming that the area describes a circle.
The bioimage analysis workflow described above was developed as an ImageJ/Fiji (39) script using the IJ1 macro language. The script is available via the ImageJ/Fiji update site (http://sites.imagej.net/Fuellekrug-Lab/). The additional plugin used for the image flooding step (http://bigwww.epfl.ch/sage/soft/watershed/) and the MorphoLibJ (36) (ImageJ/Fiji update site “IJPB-plugins,” http://sites.imagej.net/IJPB-plugins/) can be downloaded via the same update site as the ImageJ/Fiji script.
The results output table displays the number of total local maxima detected, the number of edges, the number of true LDs (overlayed), and the approximated volume. The script saves: i) the raw image; ii) the local maxima image shown as an overlay of green and red dots according to true positive or false positive LDs; iii) the input with labeled edges drawn as yellow ROI outlines; and iv) the segmented particles by flooding if the LD volume was computed. Furthermore, ROIs are saved for the local maxima (true LDs and excluded maxima) and edge coordinates are stored in a .zip file. In addition, a .csv file is saved containing each LD’s volume separately allowing a LD volume histogram of each cell (Fig. 5).
Fig. 5.
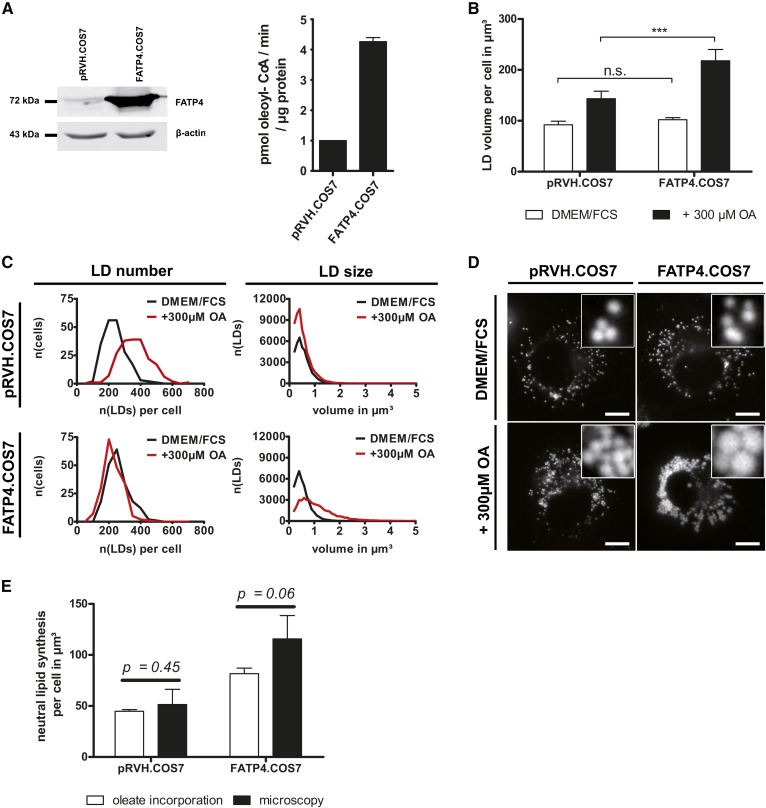
FATP4 overexpression enhances neutral lipid storage and increases LD size after OA incubation. A: FATP4 overexpression in COS7 cells. Left: Representative Western blot signals of FATP4 and β-actin from three independent experiments. Right: Oleoyl-CoA synthetase activity is increased 4.3-fold in FATP4.COS7 compared with control cells. Bars represent the mean ± SD (n = 2). B: Enhanced neutral lipid storage in FATP4.COS7. Total LD volumes of 200 cells stained by LD540 from four independent experiments were quantified by ALDQ. Ectopic FATP4 expression does not increase the LD volume under normal growth conditions (DMEM/FCS). In contrast, neutral lipid levels are strikingly increased after OA treatment in FATP4.COS7 compared with pRVH.COS7 (+300 μM OA). Bars represent the mean ± SD of n = 4 independent experiments. Student’s t-test with unequal variances; ***P < 0.001. C: FATP4 overexpression increases LD size but not LD number after OA treatment. Frequency plots show the amount of LDs per cell (LD number) as well as the volume distribution (LD size) in both FATP4.COS7 and pRVH.COS7 at normal growth conditions (DMEM/FCS) and after 24 h oleate treatment (+300 μM OA). After OA treatment, the number of LDs in pRVH.COS7 increases, while the LD volume distribution shows no difference. In contrast, FATP4.COS7 gains large LDs while maintaining their original LD number after OA treatment. In total, the data from 200 cells from four independent experiments were included. n(x): number of x. D: LD morphology is altered after FATP4 overexpression. Representative images of pRVH.COS7 and FATP4.COS7 with indicated treatments. There is no difference under normal growth conditions. After OA treatment, FATP4.COS7 contains remarkably larger LDs compared with control cells. Scale bar 10 μm. E: Comparison of microscopic and biochemical measurement of neutral lipid synthesis. The indicated cell lines were incubated in the presence of 300 μM [14C]oleate for 24 h. The volume of triolein per cell was calculated by the oleate incorporation into neutral lipids according to the Materials and Methods section. For the microscopy analysis, the difference of the LD volume per cell after and before oleate treatment (see B) was plotted. Bars represent the mean ± SD of n = 4 independent experiments. P values were calculated with Student’s t-test with unequal variances.
A detailed and illustrated protocol describing the installation, workflow, and usage of the script is provided as supplemental File F2.
ACS activity measurement
Measurement of acyl-CoA synthetase (ACS) activity was performed as described recently (40). Briefly, lysates of cells lysed with KTx-buffer [130 mM KCl, 25 mM Tris-HCl (pH 7.4), 1% Triton X-100] were incubated for 10 min at 30°C with the reaction mix {100 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 200 μM DTT, 10 mM ATP, 200 μM CoA, 0.1% Triton X-100, and 20 μM [14C]oleate (10 Ci/mol) bound to 5 μM fatty acid-free BSA}. The reaction was terminated by addition of Dole’s solution [isopropanol:heptane:H2SO4, 40:10:1 (v/v)]. Free oleate was extracted by four washes with heptane. The radioactivity of the aqueous phase, corresponding to the amount of produced oleoyl-CoA, was determined by liquid scintillation counting (LS 6500; Beckman-Coulter, Brea, CA).
Fatty acid incorporation, TLC, and molar quantification of triglyceride levels
pRVH.COS7 and FATP4.COS7 were seeded in triplicates at 50,000 cells/well in 12-well plates and incubated for 24 h in normal growth medium supplemented with 300 μM [14C]oleate (0.3 Ci/mol; Moravek, Brea, CA) bound to 50 μM fatty acid-free BSA. Cells were subsequently lysed for 30 min in the wells by 1% SDS/0.5 M NaOH and protein levels were determined by BCA assay (Pierce, Waltham, MA). To quantify the total amount of incorporated oleate per well, an aliquot of the cell lysate was subjected to scintillation counting.
Cellular lipids from separate wells were extracted as described in detail recently (23) and spotted onto TLC plates (Merck KGaA, Darmstadt, Germany). The lipids were developed in ethanol:chloroform:triethylamine:water [40:35:35:9 (v/v)], and the TLC plates were subsequently exposed to a phosphorimaging plate for 24 h and scanned by the BAS-1500 imaging system (Fuji, Tokyo, Japan). The lipid classes were densitometrically quantified with the AIDA software v.4.27 (Raytest, Straubenhardt, Germany) as a percentage of the total signal in one lane.
Cells on coverslips were seeded in parallel and were cultured in normal growth medium (DMEM/FCS) or in the presence of 300 μM [14C]oleate for 24 h. The cells were fixed, stained with LD540, and embedded as described above and used for the microscopy analysis (Fig. 5B–D).
To quantify the protein amount per cell, 500,000 cells were pelleted by 100 g and resuspended in 1 ml of PBS. The cells in PBS were counted twice again and collected by centrifugation at 200 g in a tabletop centrifuge. The pellet was resuspended in 1% SDS/0.5 M NaOH and incubated for 30 min at room temperature. The lysate was measured in three dilutions (undiluted, 1:2, and 1:4), and the protein amount was quantified by the BCA assay. The average protein content per cell was 452 ± 35 pg (mean ± SD) for pRVH.COS7 and 648 ± 93 pg (mean ± SD) for FATP4.COS7 (n = 4).
The incorporation of oleate into neutral lipids per cell was calculated using the total oleate incorporation per cell and the percentage of neutral lipids determined from the TLC plate (23). For simplification, it was assumed that only triolein (density, 0.915 g/cm3; molecular weight, 885.432 g/mol) was synthesized. Using the density and the molecular weight, the mean volume of triolein per cell was calculated and plotted as shown in Fig. 5E.
Antibodies
Primary antibodies used for Western blot were mouse anti-actin (Sigma; 1:40,000) and rabbit anti-FATP4 [(41); 1:5,000]. Secondary antibodies were goat anti-mouse 680RD and goat anti-rabbit 800CW (Licor, Lincoln, NE).
Statistics and programs
ImageJ/Fiji (1.52b) was used for image analysis and to generate 3D surface plots. Figures were assembled using Adobe Photoshop CS2 and Adobe Illustrator CS2 (Adobe, San José, CA). The boxes of the boxplots in Figs. 2–4 and supplemental Figs. S1 and S2 are enclosed by the 25th and 75th percentile range divided by a line representing the median; whiskers are extending to the 10th and 90th percentile, respectively. Cells outside this range are depicted as outliers. Student’s t-test with unequal variances was used for the calculation of statistical significances. Differences of two groups with P values below 0.05 were considered statistically significant.
RESULTS
The quantification of LD number and size from microscopy images is a vital task in LD research. Here, we propose an easily accessible, interactive, and semi-automated quantification tool for LD quantification using a designed band-pass filter to amplify LD structures while suppressing image noise. The settings can be adjusted to adapt the algorithm to a variety of LD morphologies in different cell lines.
Preprocessing iterations improve contrast of LDs against their surrounding structures
The algorithm was designed to detect the geometric characteristics of LDs given by their spherical nature, which are a central fluorescence intensity maximum and a circular edge, in line with a previous approach (Fig. 1D) (42).
However, the contrast of individual LDs in clusters or in close proximity to the ER was often not sufficient to detect LDs by their local fluorescence maxima (not shown). To overcome this limitation, an adjustable and iterative preprocessing step was developed to enhance the contrast of individual LDs (Fig. 1B, C).
Henceforth, the analysis procedure including the preprocessing step followed by the detection of local maxima and edges and the final evaluation thereof is termed ALDQ. Further image manipulations prior to the analysis are specifically outlined.
ALDQ algorithm detects LDs stained by lipophilic dyes
The algorithm was first evaluated on LDs stained with the common LD staining dyes, BODIPY493/503, LD540, and Nile Red (Fig. 2; supplemental Figs. S1, S2), using manual determination of LD numbers as a validation. Cells were cultured in normal growth medium (DMEM/FCS) or supplemented with oleate (+OA), which led to enlargement and clustering of LDs. Delipidation (starved), in contrast, resulted in the absence of intracellular LDs.
LD populations are highly diverse among different cell types and among cells in a given population. The proposed algorithm was therefore tested on three cell lines that differ extensively regarding their LD morphologies. COS7 contained LDs that are homogenous in size and well distributed throughout the cytoplasm, showing only sparse clustering even after extensive oleate treatment. U2OS contained an unusual abundance of tiny LDs (>300 per cell on average; ∼400–600 nm diameter) under normal growth conditions, whereas after oleate treatment, they showed a heterogeneous population of a few mildly clustered large LDs together with a high number of small LDs. A431 cells are small globular cells with the characteristic of size equivalent, but heavily clustered LDs under normal growth and oleate-induced conditions (Fig. 1A).
Preprocessing of the images by ALDQ resulted in a remarkable contrast enhancement of LDs, even if they were heavily clustered. In addition, fluorescent background signal mainly derived from stained perinuclear ER was suppressed (Fig. 2D; supplemental Figs. S1D, S2D).
Independent of the metabolic state, the lipophilic dye, and the cell line used, the ALDQ-derived LD counts and manually assessed numbers were found to be highly similar, as indicated comprehensively by the mean of two experiments, single cell accuracy analysis, and linear regression (Fig. 2; supplemental Figs. S1, S2).
The impact of image noise on the accuracy of ALDQ was assessed by blindly deconvolving the raw image. This was followed by a second analysis using the same algorithm input parameters for a direct comparison. Intriguingly, no striking differences between raw and deconvolved images were observed, both regarding the LD morphology after preprocessing and the determined LD numbers (Fig. 2; supplemental Figs. S1, S2). However, deconvolution led to partial separation of LDs in clusters prior to the preprocessing step.
LDs stained by the marker protein, A3Nt-GFP, are confidently detected
LD marker proteins are often used in studies focusing on de novo LD formation because of their ability to indicate the presence of tiny LDs more sensitively compared with lipophilic dyes (7, 9). However, LD proteins show secondary localizations either on the ER or in the cytoplasm (43). Although the LD surface is accentuated due to an accumulation of marker protein, an abundance of fluorescent signal is still derived from the ER or cytoplasm, which challenges a specific detection of LD-associated signal.
To assess the capability of ALDQ in quantifying de novo formed LDs stained by the LD marker protein, A3Nt-GFP, the ER/LD fusion protein was stably expressed in COS7 and U2OS by retroviral transduction. The cells were first delipidated for 24 h to deplete preexisting LDs. Subsequently, an oleate pulse was applied for 10 min to induce de novo formation of LDs.
Enhancing the images by ALDQ resulted in a striking suppression of ER-derived fluorescence signal and increased the contrast of LDs even against pronounced ER staining (Fig. 3D). The following detection of local maxima and edges resulted in confident recognition of LDs, as indicated by three analysis methods (Fig. 3A–C). In line with the experiments performed using the lipophilic dyes, prior deconvolution of the images did not result in major changes regarding the accuracy of the ALDQ analysis (Fig. 3A–C).
LD volume is estimated by the area of the largest cross-section
The calculation of the LD volume usually requires a 3D image analysis because volume is a 3D parameter and cannot be measured directly from a 2D image. Nevertheless, because the volume of a sphere is proportional to the area of the largest cross-section, the volume can be estimated if this parameter is determined from a 2D image.
Following this idea, the volume measurement component of ALDQ was developed to detect the largest LD cross-section by image flooding (Figs. 1D, 4A) (37, 38). The area of each flooding-derived particle is measured and the volume of the corresponding sphere is computed accordingly.
To validate the ALDQ volume approximation results, LD540-stained LDs were 3D rendered by the Imaris software as described in the Materials and Methods. The volume measurement by Imaris and the volume estimation by ALDQ were highly consistent for COS7 and U2OS cells (Fig. 4). In contrast, A431 cells loaded with oleate show large globular LD clusters, which are presented as one planar LD layer in the corresponding 2D image (Fig. 4A). Although these layers are segmented convincingly, the volume of the clusters is strongly underestimated (Fig. 4B, C).
FATP4 overexpression changes LD size and increases neutral lipid storage in COS7 cells
To apply the method, the ER-resident ACS, FATP4/ACSVL5 (41), was overexpressed in COS7 by stable retroviral integration (FATP4.COS7), resulting in an approximately 4-fold increase in oleoyl-CoA synthetase activity (Fig. 5A). Cells expressing an empty retroviral vector (pRVH.COS7) served as a control. Both the LD number and the LD volume per cell were determined from cells counterstained with the lipophilic dye, LD540.
Surprisingly, when cultured under normal growth conditions, no differences could be found between FATP4.COS7 and pRVH.COS7 regarding the LD number and LD volume per cell (Fig. 5B–D).
In contrast, after OA supplementation, the storage of neutral lipids was strikingly enhanced in FATP4.COS7 compared with pRVH.COS7, as shown by both LD volume measurement by ALDQ and incorporation of radioactively labeled oleate into neutral lipids (Fig. 5B, E). While OA treatment of pRVH.COS7 induced an increase in LD numbers per cell but no changes regarding the individual LD size, FATP4.COS7 contained notably larger LDs while the LD number per cell was not altered (Fig. 5C, D).
DISCUSSION
ALDQ is a user-friendly algorithm
We aimed for a quantification algorithm that is both simple to setup and easily customized. Consequently, we chose the open source software, ImageJ/Fiji, and limited the changeable analysis parameters to 10, of which only 5 needed frequent adjustments in our hands (supplemental Tables S1–S5).
The iterative preprocessing step involves three parameters: The number of preprocessing iterations (“1”) and the associated Gaussian blur (“blur during preprocessing iterations”; “2”) control the degree of LD enhancement (supplemental Figs. S4, S5). Blurring of the image after the preprocessing procedure (“3”) increases the specificity of maxima detection by image smoothing. The two remaining parameters that needed frequent changes define the noise tolerance for the maximum detection and the local threshold radius for the edge detection. Together, these five parameters mainly determine the sensitivity and specificity of the LD detection.
To allow visualization of the defined parameters at crucial stages of the analysis, the script features an optional “preview mode” that stops at the above-mentioned steps and displays the resulting image. If it is necessary, each time, the user is able to adjust the settings. Once the analysis is complete, the identified LDs are displayed as an overlay on the input image and confirmed by the user.
Over-enhancement of images resulted in multiple artificial fluorescence maxima of a single LD, while under-enhancement decreased the sensitivity of LD detection due to insufficient LD separation. Larger LDs were more sensitive to over-enhancement compared with smaller LDs (supplemental Fig. S4). Over-enhancement of large LDs could be controlled by higher associated Gaussian blurring, which, however, gradually reduced the efficiency of small LD separation (supplemental Fig. S5). Therefore, the usage of higher Gaussian blurring (“blur during preprocessing iterations”) poses only an option for homogenous populations of large LDs. For heterogeneous LD populations, a combination of prior deconvolution, which partially separates LDs by noise cancelling (Fig. 2D; supplemental Figs. S1D, S2D), and reducing the number of preprocessing iterations is the most feasible way to prevent over-separation while maintaining the necessary LD separation.
General recommendations for the setting adjustments are provided in supplemental File F2.
In addition, the script saves the input images marked with the detected maxima, edges, and LD cross-sections in the respective input folder (see also the Materials and Methods and supplemental File F2). Together with the provided troubleshooting guide (Table 2 of supplemental File F2), these images provide assistance on adjusting the settings (Table 1 of supplemental File F2) and allow visualization of the overall accuracy of ALDQ detection.
Broad applicability of ALDQ is ensured by a variety of tested experimental conditions
We show that the algorithm convincingly identifies LDs in 2D images acquired using widefield microscopy, one of the simplest and most broadly available image acquisition techniques for fluorescence detection (Figs. 2, 3; supplemental Figs. S1, S2). In addition, the data also suggest that flattened z-stacks (SIP) derived from confocal microscopy are a possible basis for the analysis (Fig. 4). Although not investigated, we assume that images from any microscopy technique can be analyzed as long as the resulting image shows a central intensity maximum of the LD.
Three lipophilic dyes and one LD protein marker were tested, showing a high accuracy of the analysis method independent of the LD stain. Furthermore, three different cell lines in three metabolic states were included in the experiments to ensure that the algorithm is able to process different LD sizes, intracellular LD distributions, and degrees of LD clustering (Fig. 1A).
To match the requirements given by the different LD morphologies, the number of preprocessing iterations and associated Gaussian blurring can be varied adequately for each cell line, metabolic state, and dye, as described above (supplemental Tables S1–S5). Upon different experimental conditions the LD morphology might change drastically (Fig. 5, supplemental Fig. S2), which may force the user to change the analysis settings. In general, for scientifically accurate analyses, the analysis method should be kept constant when two experimental conditions are compared. However, the algorithm uses a morphologically based segmentation, and it is therefore obvious that the preprocessing procedure needs to be adjusted according to the LD morphology present. To avoid an analysis bias, a close evaluation of the ALDQ output is strictly necessary to control the accuracy of ALDQ.
Deconvolution as a method of denoising images did not result in major changes regarding the accuracy of ALDQ, indicating that the algorithm is robust toward image noise (Figs. 2, 3; supplemental Figs. S1, S2). However, as described above, deconvolution prior to the analysis partially separated LDs by noise reduction and can therefore be used to lower the number of preprocessing iterations, if needed.
A direct comparison of the ALDQ LD volume measurement and labeling of triglycerides by [14C]oleate showed overall consistent absolute neutral lipid volumes per cell (Fig. 5E). Those results indicate that the algorithm is a reliable method to identify and describe changes of the intracellular LD content correctly. However, as demonstrated by the overestimation of LD volumes in FATP4.COS7 by ALDQ, independent methodological approaches should always accompany the microscopic analysis to confirm an identified phenotype.
Optimal experimental setup
For an optimal quantification result, the choice of the LD stain is an important parameter. In our hands, staining of LDs by LD540 and Nile Red resulted in more precise measurements by ALDQ compared with BODIPY493/503 in most of the cases (Fig. 2B, C; supplemental Figs. S1B, C and S2B, C). These results are best explained by the superior SNR of LD540 and Nile Red compared with BODIPY493/503 in our hands. Because the staining conditions were not varied and we only subjectively evaluated the SNRs judged by the contrast of stained LDs and the background staining, these observations cannot be generalized. However, the correlation of the observed SNR and precision of the output values was clearly notable in the conducted experiments and should be considered upon utilization of the algorithm.
Furthermore, we observed that the number of manually counted LDs in U2OS cells under normal growth conditions differ remarkably depending on the lipophilic dye used, despite the fact that the coverslips were located in the same tissue culture well (supplemental Fig. S3). U2OS LDs under normal growth conditions are exceptionally small, which might hamper the detection. Therefore, the mismatch shown in supplemental Fig. S3 might overstate the differences to be expected in other cell lines. However, we think that this experiment underlines the necessity for a deliberate choice of the LD staining.
Limitations of ALDQ
The ALDQ algorithm was developed for the detection of the hydrophobic core of LDs stained by lipophilic dyes. Images where LD surface proteins are labeled are currently not suitable for the analysis, because such staining results in a fluorescent circle enclosing the hydrophobic LD interior without forming specific fluorescence maxima. The only exception is the surface staining of tiny LDs, as they comprise a distinct fluorescence intensity maximum due to their punctuate appearance (Fig. 3).
Volume estimation by ALDQ is dependent on the LD morphology in a certain cell line. As shown for A431 (Fig. 4), 3D LD clusters cannot be depicted equivalently in a 2D image, leading to an information loss that results in an underestimation of the LD volume. A homogenous planar distribution of LDs is therefore a necessity for the precise estimation of the LD volume in a 2D image.
ALDQ is a semi-automated quantification technique that needs user-defined input for cell border definition. This could be avoided by combining ALDQ, for instance, with the Voronoi ImageJ/Fiji plugin to detect cell boundary or by using machine learning algorithms to separate the cell body background from the coverslip background. Indeed, the resulting masks could be used as input ROIs for the ALDQ algorithm. However, this task is beyond the scope of the study.
LD morphology alteration by FATP4 overexpression
We found that overexpression of the ER-localized ACS, FATP4, was sufficient for enhanced storage of neutral lipids (Fig. 5), as suggested also for other ACS enzymes (7, 44, 45). This effect has been explained by enhanced fatty acid uptake and esterification through metabolic trapping rather than fatty acid transport across the plasma membrane (46, 47). In addition, we observed that FATP4 overexpression facilitated the formation of large LDs (Fig. 5). Previous evidence was provided for the hypothesis that LD-localized TG synthesizing enzymes are sufficient for the formation of large LDs (14), although in our setup this would be only indirectly possible. LD fusion events, although they have been rarely observed, would pose another explanation for the described phenotype. Interestingly, the protein responsible for LD fusions, CIDEC/Fsp27, is a target of the fatty acid-activated PPAR-α (48). An enhanced fatty acid uptake by FATP4 overexpression could facilitate increased PPAR activation by fatty acids and, in turn, upregulation of CIDEC/Fsp27, which would mediate the LD fusion events.
In summary, we provide a highly accurate LD quantification approach that can be easily adjusted to a variety of LD morphologies.
Supplementary Material
Acknowledgments
The authors thank Dr. Vibor Laketa and the Infectious Diseases Imaging Platform (IDIP; Center for Integrative Infectious Disease Research, Heidelberg, Germany) for the support with the confocal microscope and 3D rendering, and Simone Sander and Juan Liao for testing early versions of the script.
Footnotes
Abbreviations:
- ACS
- acyl-CoA synthetase
- ALDQ
- automated lipid droplet quantification
- LD
- lipid droplet
- OA
- oleic acid
- ROI
- region of interest
- SIP
- sum intensity projection
- SNR
- signal-to-noise ratio
This work was supported by German Research Foundation Grants FU 340/7-1 (J.F.) and 112927078-TRR 83 (R.B.) and the Stiftung Nephrologie Heidelberg (M.P., J.F.). T.E. was supported by the MD/PhD program of the Medizinischen Fakultät Heidelberg, Universität Heidelberg.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Murphy D. J. 2001. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog. Lipid Res. 40: 325–438. [DOI] [PubMed] [Google Scholar]
- 2.Gao Q., and Goodman J. M.. 2015. The lipid droplet-a well-connected organelle. Front. Cell Dev. Biol. 3: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuldiner M., and Bohnert M.. 2017. A different kind of love - lipid droplet contact sites. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1862: 1188–1196. [DOI] [PubMed] [Google Scholar]
- 4.Mashek D. G., Khan S. A., Sathyanarayan A., Ploeger J. M., and Franklin M. P.. 2015. Hepatic lipid droplet biology: getting to the root of fatty liver. Hepatology. 62: 964–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olofsson S. O., Andersson L., Haversen L., Olsson C., Myhre S., Rutberg M., Mobini R., Li L., Lu E., Boren J., et al. . 2011. The formation of lipid droplets: possible role in the development of insulin resistance/type 2 diabetes. Prostaglandins Leukot. Essent. Fatty Acids. 85: 215–218. [DOI] [PubMed] [Google Scholar]
- 6.Krahmer N., Farese R. V., and Walther T. C.. 2013. Balancing the fat: lipid droplets and human disease. EMBO Mol. Med. 5: 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kassan A., Herms A., Fernández-Vidal A., Bosch M., Schieber N. L., Reddy B. J. N., Fajardo A., Gelabert-Baldrich M., Tebar F., Enrich C., et al. . 2013. Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J. Cell Biol. 203: 985–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salo V. T., Belevich I., Li S., Karhinen L., Vihinen H., Vigouroux C., Magré J., Thiele C., Hölttä-Vuori M., Jokitalo E., et al. . 2016. Seipin regulates ER-lipid droplet contacts and cargo delivery. EMBO J. 35: 2699–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H., Becuwe M., Housden B. E., Chitraju C., Porras A. J., Graham M. M., Liu X. N., Thiam A. R., Savage D. B., Agarwal A. K., et al. . 2016. Seipin is required for converting nascent to mature lipid droplets. eLife. 5: e16582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi Y., Shinoda A., Furuya N., Harada E., Arimura N., Ichi I., Fujiwara Y., Inoue J., and Sato R.. 2013. Perilipin-mediated lipid droplet formation in adipocytes promotes sterol regulatory element-binding protein-1 processing and triacylglyceride accumulation. PLoS One. 8: e64605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mason R. R., Mokhtar R., Matzaris M., Selathurai A., Kowalski G. M., Mokbel N., Meikle P. J., Bruce C. R., and Watt M. J.. 2014. PLIN5 deletion remodels intracellular lipid composition and causes insulin resistance in muscle. Mol. Metab. 3: 652–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDonough P. M., Agustin R. M., Ingermanson R. S., Loy P. A., Buehrer B. M., Nicoll J. B., Prigozhina N. L., Mikic I., and Price J. H.. 2009. Quantification of lipid droplets and associated proteins in cellular models of obesity via high-content/high-throughput microscopy and automated image analysis. Assay Drug Dev. Technol. 7: 440–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris C. A., Haas J. T., Streeper R. S., Stone S. J., Kumari M., Yang K., Han X., Brownell N., Gross R. W., Zechner R., et al. . 2011. DGAT enzymes are required for triacylglycerol synthesis and lipid droplets in adipocytes. J. Lipid Res. 52: 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilfling F. 2013. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell. 24: 384–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kory N., Thiam A. R., Farese R. V. Jr., and Walther T. C.. 2015. Protein crowding is a determinant of lipid droplet protein composition. Dev. Cell. 34: 351–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gluchowski N. L., Chitraju C., Picoraro J. A., Mejhert N., Pinto S., Xin W., Kamin D. S., Winter H. S., Chung W. K., Walther T. C., et al. . 2017. Identification and characterization of a novel DGAT1 missense mutation associated with congenital diarrhea. J. Lipid Res. 58: 1230–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rinia H. A., Burger K. N. J., Bonn M., and Müller M.. 2008. Quantitative label-free imaging of lipid composition and packing of individual cellular lipid droplets using multiplex CARS microscopy. Biophys. J. 95: 4908–4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nunn A. D. G., Scopigno T., Pediconi N., Levrero M., Hagman H., Kiskis J., and Enejder A.. 2016. The histone deacetylase inhibiting drug Entinostat induces lipid accumulation in differentiated HepaRG cells. Sci. Rep. 6: 28025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim K., Lee S., Yoon J., Heo J., Choi C., and Park Y.. 2016. Three-dimensional label-free imaging and quantification of lipid droplets in live hepatocytes. Sci. Rep. 6: 36815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rohwedder A., Zhang Q., Rudge S. A., and Wakelam M. J. O.. 2014. Lipid droplet formation in response to oleic acid in Huh-7 cells is mediated by the fatty acid receptor FFAR4. J. Cell Sci. 127: 3104–3115. [DOI] [PubMed] [Google Scholar]
- 21.Campos V., Rappaz B., Kuttler F., Turcatti G., and Naveiras O.. 2018. High-throughput, nonperturbing quantification of lipid droplets with digital holographic microscopy. J. Lipid Res. 59: 1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schnitzler J. G., Bernelot Moens S. J., Tiessens F., Bakker G. J., Dallinga-Thie G. M., Groen A. K., Nieuwdorp M., Stroes E. S. G., and Kroon J.. 2017. Nile Red Quantifier: a novel and quantitative tool to study lipid accumulation in patient-derived circulating monocytes using confocal microscopy. J. Lipid Res. 58: 2210–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poppelreuther M., Sander S., Minden F., Dietz M. S., Exner T., Du C., Zhang I., Ehehalt F., Knuppel L., Domschke S., et al. . 2018. The metabolic capacity of lipid droplet localized acyl-CoA synthetase 3 is not sufficient to support local triglyceride synthesis independent of the endoplasmic reticulum in A431 cells. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1863: 614–624. [DOI] [PubMed] [Google Scholar]
- 24.Dejgaard S. Y., and Presley J. F.. 2014. New automated single-cell technique for segmentation and quantitation of lipid droplets. J. Histochem. Cytochem. 62: 889–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dejgaard S. Y., and Presley J. F.. 2018. New method for quantitation of lipid droplet volume from light microscopic images with an application to determination of PAT protein density on the droplet surface. J. Histochem. Cytochem. 66: 447–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deutsch M. J., Schriever S. C., Roscher A. A., and Ensenauer R.. 2014. Digital image analysis approach for lipid droplet size quantitation of Oil Red O-stained cultured cells. Anal. Biochem. 445: 87–89. [DOI] [PubMed] [Google Scholar]
- 27.Moessinger C., Klizaite K., Steinhagen A., Philippou-Massier J., Shevchenko A., Hoch M., Ejsing C. S., and Thiele C.. 2014. Two different pathways of phosphatidylcholine synthesis, the Kennedy Pathway and the Lands Cycle, differentially regulate cellular triacylglycerol storage. BMC Cell Biol. 15: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rambold A. S., Cohen S., and Lippincott-Schwartz J.. 2015. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy and mitochondrial fusion dynamics. Dev. Cell. 32: 678–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Markgraf D. F., Klemm R. W., Junker M., Hannibal-Bach H. K., Ejsing C. S., and Rapoport T. A.. 2014. An ER protein functionally couples neutral lipid metabolism on lipid droplets to membrane lipid synthesis in the ER. Cell Reports. 6: 44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poppelreuther M., Rudolph B., Du C., Grossmann R., Becker M., Thiele C., Ehehalt R., and Fullekrug J.. 2012. The N-terminal region of acyl-CoA synthetase 3 is essential for both the localization on lipid droplets and the function in fatty acid uptake. J. Lipid Res. 53: 888–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuck S., Manninen A., Honsho M., Fullekrug J., and Simons K.. 2004. Generation of single and double knockdowns in polarized epithelial cells by retrovirus-mediated RNA interference. Proc. Natl. Acad. Sci. USA. 101: 4912–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spandl J., White D. J., Peychl J., and Thiele C.. 2009. Live cell multicolor imaging of lipid droplets with a new dye, LD540. Traffic. 10: 1579–1584. [DOI] [PubMed] [Google Scholar]
- 33.Greenspan P., Mayer E. P., and Fowler S. D.. 1985. Nile red: a selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 100: 965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dougherty R. 2005. Extensions of DAMAS and benefits and limitations of deconvolution in beamforming (AIAA Paper 2005-2961. doi:10.2514/6.2005-2961. 11th AIAA/CEAS Aeroacoustics Conference. Monterey, CA, May 23–25, 2005).
- 35.Phansalkar N., More S., Sabale A., and Joshi M.. 2011. Adaptive local thresholding for detection of nuclei in diversity stained cytology images (2011 International Conference on Communications and Signal Processing. 218–220. doi:10.1109/ICCSP.2011.5739305).
- 36.Legland D., Arganda-Carreras I., and Andrey P.. 2016. MorphoLibJ: integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics. 32: 3532–3534. [DOI] [PubMed] [Google Scholar]
- 37.Sage D., and Unser M.. 2003. Teaching image-processing programming in Java. IEEE Signal Process. Mag. 20: 43–52. [Google Scholar]
- 38.Tsukahara M., Mitrovic S., Gajdosik V., Margaritondo G., Pournin L., Ramaioli M., Sage D., Hwu Y., Unser M., and Liebling T. M.. 2008. Coupled tomography and distinct-element-method approach to exploring the granular media microstructure in a jamming hourglass. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 77: 061306. [DOI] [PubMed] [Google Scholar]
- 39.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. . 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Füllekrug J., and Poppelreuther M.. 2016. Measurement of long-chain fatty acyl-CoA synthetase activity. Methods Mol. Biol. 1376: 43–53. [DOI] [PubMed] [Google Scholar]
- 41.Milger K., Herrmann T., Becker C., Gotthardt D., Zickwolf J., Ehehalt R., Watkins P. A., Stremmel W., and Füllekrug J.. 2006. Cellular uptake of fatty acids driven by the ER-localized acyl-CoA synthetase FATP4. J. Cell Sci. 119: 4678–4688. [DOI] [PubMed] [Google Scholar]
- 42.Kumagai S., and Hotta K.. 2012. Counting and radius estimation of lipid droplet in intracellular images (Conference Proceedings - IEEE International Conference on Systems, Man, and Cybernetics. 67–71. doi:10.1109/ICSMC.2012.6377678).
- 43.Kory N., Farese R. V. Jr., and Walther T. C.. 2016. Targeting fat: mechanisms of protein localization to lipid droplets. Trends Cell Biol. 26: 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mashek D. G., McKenzie M. A., Van Horn C. G., and Coleman R. A.. 2006. Rat long chain acyl-CoA synthetase 5 increases fatty acid uptake and partitioning to cellular triacylglycerol in McArdle-RH7777 cells. J. Biol. Chem. 281: 945–950. [DOI] [PubMed] [Google Scholar]
- 45.Hatch G. M., Smith A. J., Xu F. Y., Hall A. M., and Bernlohr D. A.. 2002. FATP1 channels exogenous FA into 1,2,3-triacyl-sn-glycerol and down-regulates sphingomyelin and cholesterol metabolism in growing 293 cells. J. Lipid Res. 43: 1380–1389. [DOI] [PubMed] [Google Scholar]
- 46.Digel M., Ehehalt R., Stremmel W., and Füllekrug J.. 2009. Acyl-CoA synthetases: fatty acid uptake and metabolic channeling. Mol. Cell. Biochem. 326: 23–28. [DOI] [PubMed] [Google Scholar]
- 47.Füllekrug J., Ehehalt R., and Poppelreuther M.. 2012. Outlook: membrane junctions enable the metabolic trapping of fatty acids by intracellular acyl-CoA synthetases. Front. Physiol. 3: 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langhi C., and Baldan A.. 2015. CIDEC/FSP27 is regulated by peroxisome proliferator-activated receptor alpha and plays a critical role in fasting- and diet-induced hepatosteatosis. Hepatology. 61: 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.