ABSTRACT
Stimulation of the ERK/MAPK pathway is required for the exit from pluripotency and onset of differentiation in mouse embryonic stem cells (ESCs). The dynamic behaviour of ERK activity in individual cells during this transition is unclear. Using a FRET-based biosensor, we monitored ERK signalling dynamics of single mouse ESCs during differentiation. ERK activity was highly heterogeneous, with considerable variability in ERK signalling between single cells within ESC colonies. Different triggers of differentiation induced distinct ERK activity profiles. Surprisingly, the dynamic features of ERK signalling were not strongly coupled to loss of pluripotency marker expression, regardless of the differentiation stimulus, suggesting the normal dynamic range of ERK signalling is not rate-limiting in single cells during differentiation. ERK signalling dynamics were sensitive to the degree of cell crowding and were similar in neighbouring cells. Sister cells from a mitotic division also showed more similar ERK activity, an effect that was apparent whether cells remained adjacent or moved apart after division. These data suggest a combination of cell lineage and niche contributes to the absolute level of ERK signalling in mouse ESCs.
KEY WORDS: FRET, ERK, Single cell, Stem cells, Nanog, Epigenetic inheritance
Summary: Live imaging of ERK activity reveals a cell lineage component to signalling strength and suggests the normal variation in ERK activity does not determine the rate of pluripotency exit.
INTRODUCTION
Mouse embryonic stem cells (ESCs) are derived from the inner cell mass of blastocysts, and can give rise to all cells of the developing embryo (Evans and Kaufman, 1981). Under appropriate culture conditions, ESCs can undergo numerous rounds of division whilst maintaining their broad potential. ESCs are commonly cultured in the presence of leukaemia inhibitory factor (LIF), which maintains ESCs in a state of self-renewal through activation of JAK/STAT3 signalling (Niwa et al., 1998; Smith et al., 1988). The broad fate potential of ESCs is maintained through the expression of pluripotency factors such as Oct4 (Pou5f1), Sox2 and Nanog (Avilion et al., 2003; Chambers et al., 2003; Niwa et al., 2000). When cultured in the presence of LIF and serum, pluripotency factor expression can be heterogeneous, with cells in a low expression state exhibiting an increased tendency for differentiation and high-expressing cells displaying an increased propensity for self-renewal (Chambers et al., 2007; Hayashi et al., 2008; Toyooka et al., 2008).
ESCs can also be maintained in a culture condition known as 2i, comprising an inhibitor of the MAPK/ERK pathway and an inhibitor of glycogen synthase kinase-3 (Ying et al., 2008). Heterogeneous expression of pluripotency markers is lost in 2i. Conversely, activation of the ERK pathway, by autocrine FGF4, is required for exit from pluripotency and the onset of differentiation (Kunath et al., 2007; Stavridis et al., 2007). ERK activity has been associated with the up- and downregulation of multiple factors involved in the transition from pluripotency, such as the lineage priming factor Tcf15, and the pluripotency factors Nanog and Klf4 (Davies et al., 2013; Dhaliwal et al., 2018; Hamazaki et al., 2006).
ERK signalling activity in mouse ESCs is usually measured using antibodies against the active phosphorylated form of p44/p42 MAP kinases (also known as Mapk3/Mapk1). This approach only takes snapshots of the status of kinase activity, and does not register the dynamics of the ERK signalling in individual cells. Given the dynamic behaviour of signalling molecules in diverse contexts, the absence of dynamic information at the individual cell level represents a potential knowledge gap in the field. Differences in the dynamic behaviour of signalling molecules can lead to different cellular outcomes (Purvis and Lahav, 2013). A prevalent example of this is the behaviour of ERK in PC12 cells, where sustained activity induces differentiation and transient activity causes proliferation (Cowley et al., 1994; Ryu et al., 2015; Santos et al., 2007). In NRK cells, ERK activity occurs in stochastic pulses following EGF stimulation, with the frequency and duration of these pulses coupled to the rate of proliferation (Albeck et al., 2013; Aoki et al., 2013).
Here, we describe the implementation of an approach for monitoring the ERK activity dynamics of individual ESCs using a FRET (Förster resonance energy transfer)-based biosensor (Harvey et al., 2008; Komatsu et al., 2011). Surprisingly, temporal patterns of ERK activity were only weakly correlated to the rate of exit of cells from pluripotency, suggesting that ERK activity is not normally limiting during this transition. Neighbouring cells showed similar levels of ERK activity and ERK activity was strongly influenced by cell lineage, with related cells showing more similar activity than unrelated cells. These data imply both cell-autonomous and non-autonomous contributions to the absolute strength of ERK signalling in ESCs.
RESULTS
Heterogeneous ERK activity dynamics in mouse ESCs
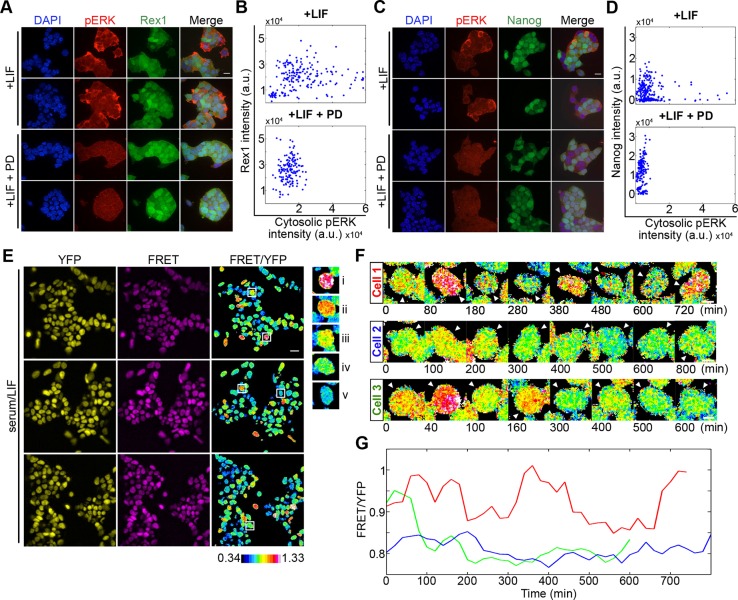
Expression of several regulators of pluripotency is heterogeneous in both standard culture conditions and the mouse embryo. Although ERK pathway activity is required for exit from pluripotency and differentiation of mouse ESCs, the relationship between ERK activity and heterogeneous pluripotency factor expression is unclear. To compare the expression of pluripotency regulators with ERK in single cells, we immunostained GFP knock-in reporter cell lines for Rex1 (Zfp42) (Toyooka et al., 2008) and Nanog (Chambers et al., 2007) with antibodies against phosphorylated forms of p44/42 MAPK (pERK). Over most imaging fields, a small proportion of cells exhibited increased levels of pERK, revealing steady-state ERK activity levels are not uniform in cell populations (Fig. 1A,C, upper panels). High pERK levels were lost following 3 h treatment with PD0325901 (PD), an inhibitor of MEK, confirming the specificity of the immunostaining (Fig. 1A,C, lower panels).
Fig. 1.
Monitoring ERK activity dynamics in single live ESCs. (A) Immunofluorescence of Rex1-GFP reporter ESCs immunostained for pERK (red) and DAPI (blue), cultured in serum+LIF alone or additionally treated with MEK inhibitor PD0325901 (PD; 1 μM) 3 h before fixation. (B) Rex1 reporter intensity plotted against cytosolic pERK levels in serum/LIF (+LIF; r=0.1652, P=0.0220, n=192) and PD (+LIF+PD; r=0.1509, P=0.0681, n=147). (C) Immunofluorescence of Nanog-GFP reporter ESCs immunostained for pERK (red) under the same culture treatments as in A. (D) Nanog reporter intensity plotted against cytosolic pERK level for ESCs in serum/LIF (+LIF; r=−0.089, P=0.1498, n=266 cells) and PD (+LIF+PD; r=0.26, P=0.0005, n=180 cells). Representative experiments from two and three biological replicates for Rex1 and Nanog, respectively, are shown in A-D. (E) ESCs stably expressing the ERK FRET reporter, EKAREV-NLS, cultured in serum/LIF. YFP and FRET channel images shown for three fields of view with the corresponding FRET/YFP ratio image. Examples of cells displaying distinct FRET ratio levels are outlined and shown in the right panel, in order of descending FRET (i-v). Colour key represents the measured FRET/YFP ratios of cells. (F) FRET/YFP ratio filmstrips of individual ESCs in serum/LIF. (G) Changing FRET/YFP levels of the cells shown in F. Red, cell 1; blue, cell 2; green, cell 3. Scale bars: 25 µm (A,C); 20 µm (E); 5 µm (F).
To determine the relationship between pluripotency factor expression and pERK, the Nanog or Rex1 GFP reporter intensity of each cell was plotted against its corresponding cytosolic pERK intensity (Fig. 1B,D). Pearson correlation values over three experimental replicates showed Rex1 reporter expression and pERK levels to be poorly correlated (r=0.14 averaged over two replicates) (Fig. 1B). Nanog reporter expression showed a weak negative correlation (r=−0.097 over three replicates) (Fig. 1D). When a threshold was applied to exclude values below background staining (based upon levels of pERK after PD treatment), the relationships between pERK and pluripotency factor expression remained weak (Rex1: r=−0.095; Nanog: r=−0.14). This absence of a clear relationship between either Nanog or Rex1 and ERK activity is consistent with a recent report describing no correlation between Nanog and levels of Sprouty, a negative regulator of ERK, within individual ESCs (Morgani et al., 2018).
It is increasingly apparent that ERK activity dynamics, as well as absolute levels, are able to direct cell behaviour (Albeck et al., 2013; Aoki et al., 2013; Cowley et al., 1994; Schröter et al., 2015). The absence of a clear relationship between ERK activity and pluripotency factor expression in static images may reflect the importance of signalling dynamics, rather than absolute activity at a single point in time. To study ERK activity dynamics, we employed a FRET-based biosensor, EKAREV (Harvey et al., 2008; Komatsu et al., 2011), which has been used to monitor the spatiotemporal regulation of ERK activity both in vitro (Albeck et al., 2013; Aoki et al., 2013) and in vivo (Hiratsuka et al., 2014). EKAREV is an intramolecular FRET sensor with SECFP as the donor fluorophore and the YFP-like molecule YPet as the acceptor. The fluorophores are separated by a region containing an ERK substrate sequence, followed by a spacer and WW phosphopeptide-binding domain. Active ERK phosphorylates the substrate, permitting substrate association with the WW domain. This interaction ‘closes’ the molecule, bringing the donor and acceptor into close proximity for FRET.
We expressed the EKAREV sensor in E14 mESCs using the PiggyBac transposon system (Ivics et al., 2009), to facilitate more uniform expression. For measuring a wide dynamic range of signal dynamics, whilst maintaining cell health, we used a wide-field system specifically configured for FRET imaging of the donor and acceptor fluorophores (Fig. S1A, Table S1). The EKAREV biosensor contains a nuclear localisation sequence (NLS), resulting in the concentration of signal in nuclei, which facilitated cell tracking and signal quantification using a semi-automated analysis pipeline. To report biosensor activity, we measured the ratio of the sensitised acceptor emission (FRET) to the overall YFP fluorescence (FRET/YFP).
ERK activity levels showed a high level of heterogeneity in ESCs grown under standard (serum/LIF) conditions, as visualised using the EKAREV biosensor (Fig. 1E), in agreement with our immunofluorescence data (Fig. 1A,C). The FRET/YFP ratio was reduced following strong acute inhibition of the MAPK pathway by 3 h treatment with 10 µM PD, indicating FRET ratio levels report on ERK activity (Fig. S1F,G). A strong negative shift in FRET ratio levels was also identified following imaging of ESCs expressing EKAREV with a T/A phospho-site mutation in the substrate domain (EKAREV-TA), demonstrating FRET ratio levels to be dependent on EKAREV phosphorylation (Fig. S1F,G). Longer-term treatment (24 h) with 1 µM PD (the standard concentration used in 2i) resulted in a less substantial negative shift in FRET ratio values (Fig. S1F,G), which may be caused by interactions of EKAREV with other signalling components becoming apparent during adaptation to inhibitor.
FRET time-lapse imaging revealed ESCs display distinct ERK activity patterns in serum/LIF (Fig. 1F,G), with some cells showing small fluctuations over many hours (blue), others showing stronger switching (green) and, more rarely, cells showing oscillations between high and low activity states (red). These traces imply that ERK activity dynamics, as well as activity levels, can be heterogeneous within cell populations.
ERK activity dynamics during differentiation
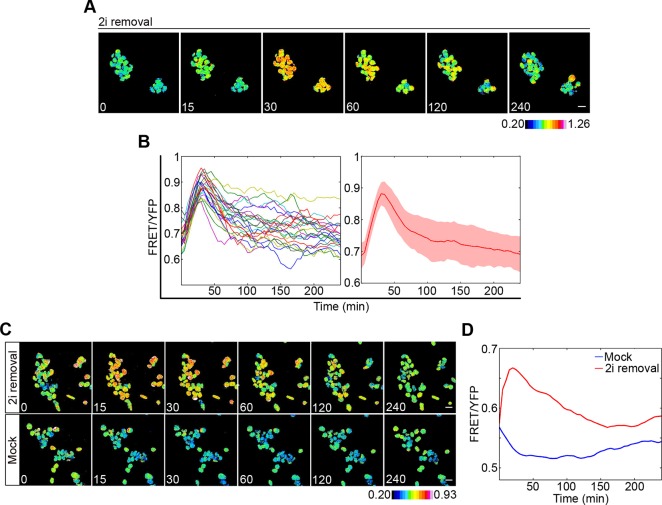
To monitor the single cell dynamics of ERK activity during the exit from pluripotency and the onset of differentiation, we followed the behaviour of the ERK biosensor after removal of 2i from ESC cultures (Ying et al., 2008). ESCs expressing the EKAREV biosensor were cultured in 2i/LIF for a minimum of two passages before media was replaced with non-2i media. FRET time-lapse imaging was carried out following 2i removal over a 4 h period. 2i removal resulted in a sharp increase in ERK activity within minutes, with ERK activity levels peaking around 40 min post 2i removal and then gradually decreasing (Fig. 2A,B). As ERK activity gradually decreased following this initial peak, activity levels became increasingly heterogeneous (Fig. 2B), remaining high in many cells for several hours. To test whether this wave in ERK activation was caused by the removal of 2i and loss of MAPK pathway suppression, cells were cultured in 2i/LIF and media was changed to either media free from 2i (2i removal) or fresh 2i media (Mock). Removal of 2i again resulted in a wave of ERK activity, which could not be detected following treatment with fresh 2i media (Fig. 2C,D). This wave in ERK activation was robust to whether cells were cultured on either gelatin (Fig. 2A,B) or laminin (Fig. S2A,B) and is in agreement with population-based measures of ERK activity (Hamilton et al., 2013; Nett et al., 2018; Yang et al., 2012).
Fig. 2.
Establishing conditions for single cell monitoring of ERK activity dynamics during ESC differentiation. (A) FRET/YFP ratio images of cells at indicated time points (min) following removal of 2i. (B) FRET/YFP ratios of individual cells (left) and the mean for all cells (right) shown in A. Shaded area reflects s.d. (C) FRET/YFP ratio images of ESCs at indicated time points (min) after 2i media was changed to either serum/LIF (2i removal) or serum/LIF/2i (Mock). (D) Mean FRET/YFP levels of all cells shown in E for 2i removal (red) and mock (blue). Colour keys represent the measured FRET/YFP ratios of cells. Scale bars: 20 µm.
Activation kinetics of ERK are weakly correlated with differentiation
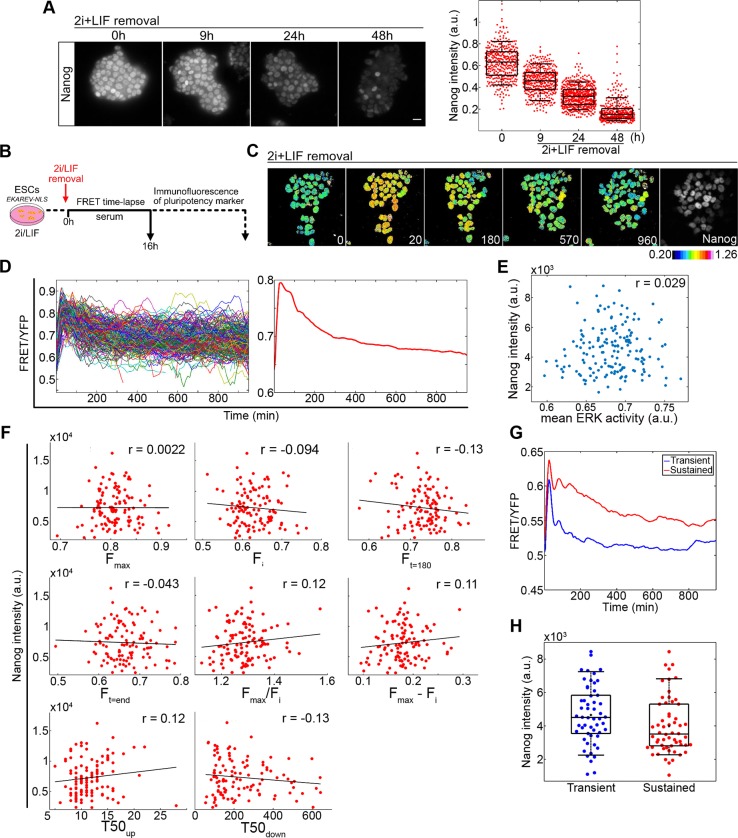
Our data suggests that ERK activation kinetics following the induction of differentiation can be variable across a population. Given the requirement for ERK activity for differentiation, we postulated that these differences in ERK activity profiles between cells could be regulating the asynchronous transition from the pluripotent state (Kalkan et al., 2017; Semrau et al., 2017). To investigate this possibility, we combined monitoring of ERK activity with immunofluorescence against a pluripotency marker. To identify an appropriate marker for loss of pluripotency, EKAREV-NLS-expressing cells were immunostained at specified time points with antibodies against different pluripotency factors following release from 2i/LIF. A clear decrease in the expression of both Nanog and Esrrb was evident 9 h post 2i/LIF withdrawal (Fig. 3A, Fig. S3A,B). In contrast, expression of the core pluripotency factor Sox2 remained largely unchanged (Fig. S3A). Variability in the expression of both Nanog and Esrrb was identified following release from 2i/LIF, with Nanog expression showing more variability at 24 h (CV=0.32) and 48 h (CV=0.55) than Esrrb (CV=0.29 and CV=0.43, respectively), providing a greater range of expression levels for comparison to ERK activity dynamics. We therefore used Nanog expression as an end-point to determine the extent of differentiation.
Fig. 3.
Analysis of ERK activity dynamics and Nanog expression during ESC differentiation. (A) EKAREV-NLS-expressing cells immunostained for Nanog following removal of 2i/LIF at indicated time points. Nanog intensities at each time point from five imaging fields are shown on the right-hand side with box-plots overlaid. Scale bar: 20 µm. (B) Protocol for comparing ERK activity during ESC cell differentiation to expression of pluripotency factors. (C) Ratiometric FRET images of ESCs at indicated time points (min) following release from 2i/LIF followed by immunostaining for Nanog (final panel). (D) FRET/YFP levels of each cell (left) following release from 2i/LIF pooled from five fields of view. Mean FRET/YFP ratio of all cells shown on the right. Data shown are one replicate typical of three biological replicates (879 cells total). See Fig. S3C,D for data from other replicates. (E) Mean ERK activity of each cell plotted against its corresponding Nanog intensity from a typical replicate (r=0.029, P=0.71, n=167 complete tracks). (F) Comparison of final Nanog levels to parameters in the activation kinetics of ERK in response to 2i/LIF withdrawal. Parameters described in Table 1. All measurements were quantified from FRET/YFP values of individual cells following smoothing and interpolation of the data. Scatter plots of Nanog intensity versus parameter descriptor values with Pearson correlation coefficient (r) applied (data from a typical replicate of the three). (G) Average FRET/YFP values of cell tracks grouped by k-mean clustering into transient (blue) or sustained (red) ERK activity. (H) Measured Nanog intensities of individual cells from each cluster shown in G with boxplots overlaid (P=0.013, Kolmogorov–Smirnov test). The slight difference apparent between clusters was not significant over all replicates (P=0.02 and 0.27, Fig. S3G) therefore constitutes a weak effect. In box and whisker plots, whiskers represent percentiles 9 to 91. Box edges are 25th and 75th percentiles.
ERK activity levels were monitored, using 3D time-lapse image acquisition, over 16 h following removal of 2i/LIF. Cells were then fixed and immunostained for Nanog (Fig. 3B,C). Nanog intensities were compared with multiple dynamic features of ERK activity from the same cell. To identify robust features of the data, we carried out three experimental replicates each with multiple imaging fields-of-view, with a total of 879 cells. The wave in ERK activity was observed in all three replicates, with a peak in ERK activity occurring 25±5 min after removal of 2i/LIF. The complete set of ERK activity traces from a single replicate, together with the average ERK activity signal measured over this 16 h image capture period, are shown in Fig. 3D (additional replicates in Fig. S3C).
To determine whether overall ERK activity levels influence the ability of a cell to exit pluripotency, the mean ERK activity of each cell, over the 16 h differentiation time course, was plotted against the corresponding final Nanog intensity (Fig. 3E). Correlations between mean ERK activity and Nanog expression were weak and non-significant (average r=−0.0041 from all experiments; total 563 cells with complete tracks) (Fig. 3E, Fig. S3D). These data indicate that the different ERK activity levels during this differentiation response were not correlated with downregulation of Nanog.
The mean activity level is only one feature of the ERK dynamics. Information influencing the differentiation response may be encoded in other dynamic features of the ERK activity response in individual cells. To test this possibility, we parameterised ERK activity in each cell into a set of descriptors to define response profiles (Cohen-Saidon et al., 2009; Lee et al., 2014) (Table 1). Distinct features of this initial wave associated with ERK activity levels were the maximal (Fmax), initial (Fi) and late (Ft=180 or Ft=end) FRET/YFP values, fold (Fmax/Fi) and amplitude (Fmax−Fi) change, and time to 50% of maximum during the increase (T50up) and decrease (T50down) in activity. Descriptors associated with timing (T50up and T50down) were found to exhibit a greater degree of cell-cell variability compared with descriptors of absolute ERK activity levels (Fig. S3E). In addition, the amplitude of increase in ERK activity (Fmax) was found to be correlated to initial basal levels of ERK activity (Fi) in all three replicates (average r=0.49), suggesting the peak ERK activity level reached is coupled to the initial basal levels in activity (Fig. S3F).
Table 1.
Definition of parameters in the activation kinetics of ERK in response to 2i/LIF withdrawal
To evaluate the possible links between the dynamic features of the ERK response and the rate of passage through early differentiation, we plotted these metrics against the corresponding Nanog intensity of single cells and Pearson correlation coefficients were used to describe the relationship between variables (Fig. 3F). All descriptors of the ERK activity wave were poorly correlated with Nanog expression (Table 1).
To further investigate the dynamic behaviour of ERK activity during the exit from the naïve state, a k-means clustering algorithm was used to group cells based on their ERK activity profile during the 16 h response. Clustering identified two distinct ERK signalling responses, with transient or sustained ERK activity following the initial peak (Fig. 3G). These two distinct patterns of activation were robust between experimental repeats (Fig. S3G). Quantification of Nanog suggested reduced expression in the sustained cluster (Fig. 3H, Fig. S3G). However, this effect was only significant (at P<0.05) in two out of three replicates and differences in Nanog expression between clusters were small, suggesting any relationship between these ERK dynamics and the expression of Nanog is weak.
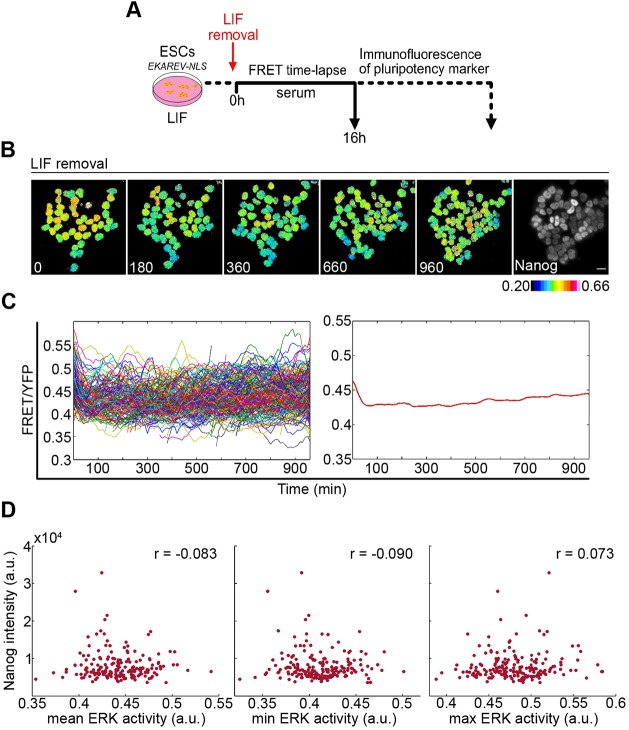
One possibility is that this apparent weak relationship between ERK activity and exit from pluripotency results from the 2i conditions. Under these conditions, expression of several pluripotency markers, including Nanog, is relatively uniform between cells. Could the low heterogeneity in this starting population mask correlations between signalling and rate of exit from pluripotency? To test this possibility, we repeated the differentiation time course using alternative starting conditions of serum/LIF culture, with differentiation triggered by removal of LIF (Fig. 4A). In serum/LIF, pluripotency factor expression is highly heterogeneous (Chambers et al., 2007; Filipczyk et al., 2013; Hayashi et al., 2008; Toyooka et al., 2008), with distinct high and low expression maxima corresponding to cell states with tendencies towards pluripotency and differentiation, respectively.
Fig. 4.
Analysis of ERK activity dynamics and Nanog expression during ESC differentiation after LIF removal. (A) Protocol for analysis of ERK activity dynamics during ESC cell differentiation following LIF removal. (B) Ratiometric FRET images of ESCs at indicated time points (min) following release from LIF followed by immunostaining for Nanog. Scale bar: 20 µm. (C) FRET/YFP levels of each cell (left) after LIF removal, pooled from five fields of view (n=303). Data from a typical experiment from three biological replicates (928 cells total). Mean FRET/YFP ratio of all cells shown on the right. See Fig. S4 for data from other replicates. (D) Mean (P=0.26), minimum (P=0.22) and maximum (P=0.32) ERK activity of each cell plotted against Nanog intensity from a typical replicate (190 cells).
The profile of ERK activity during the 16 h after LIF removal was distinct from the 2i removal profile. Rather than a sharp increase in ERK activity, LIF removal induced a slight dip in ERK activity, persisting over the remainder of the time course (Fig. 4B,C, Fig. S4A). This dip in activity may result from a feeding response, in addition to any loss of stimulatory effects of LIF on ERK (Burdon et al., 1999), as a dip in activation was also observed following treatment with fresh LIF media (Fig. S4B). The LIF removal response explores a different part of the dynamic range of the FRET reporter output compared with 2i removal. The initial FRET levels were slightly higher for serum/LIF compared with 2i/LIF, but the trajectories diverged greatly upon media change (Fig. S4C). The strong surge in FRET following 2i removal may relate to the loss of negative feedback on the ERK pathway following multiple passages with inhibitor.
As with 2i removal, there was no strong positive or negative correlation between ERK activity over the time series and Nanog protein levels at the end of the LIF removal differentiation protocol (Fig. 4D, Fig. S4D), regardless of whether the mean, maximum or minimum ERK activity were considered.
Another potential explanation for the unexpected weak relationship between ERK activity and rate of exit from pluripotency was the dynamic range of the FRET reporter (Gillies et al., 2017). It seemed possible that the strong peak in expression after 2i removal corresponds to some saturating level of activity, with important activity operating above this level. However, the maximal FRET signal (Fmax) after 2i removal showed a similar level of scatter to the initial FRET value (Fi) (Fig. 3F), with no obvious bunching of data at the response peak that would suggest reporter saturation. In addition, a strong relationship between ERK and Nanog was not observed if differentiation was induced by LIF removal (Fig. 4D), where starting conditions show a broad dynamic range of FRET (Fig. 1E, right column), the sharp initial peak did not occur (Fig. 4C), and the cells explored a different range of the FRET reporter output (Fig. S4C). These data imply that the absence of a strong relationship between ERK activity and Nanog can occur below any reporter saturation limits. Following this theme, we then tested to what extent we could identify stronger correlations between Nanog and ERK levels using specific portions of the time series (Fig. S4E). Using both the 2i and LIF removal data, we observed slightly stronger negative correlations between ERK and Nanog for the last 400 min of time series, although the magnitude of these correlations remained weak, and in the case of LIF removal, significant effects were not observed in all replicates.
Overall these data suggest that variability in ERK dynamics during standard differentiation regimes is, at most, a subtle influence on the rate of exit from pluripotency of individual ESCs.
Relationship of ERK activity to local environment
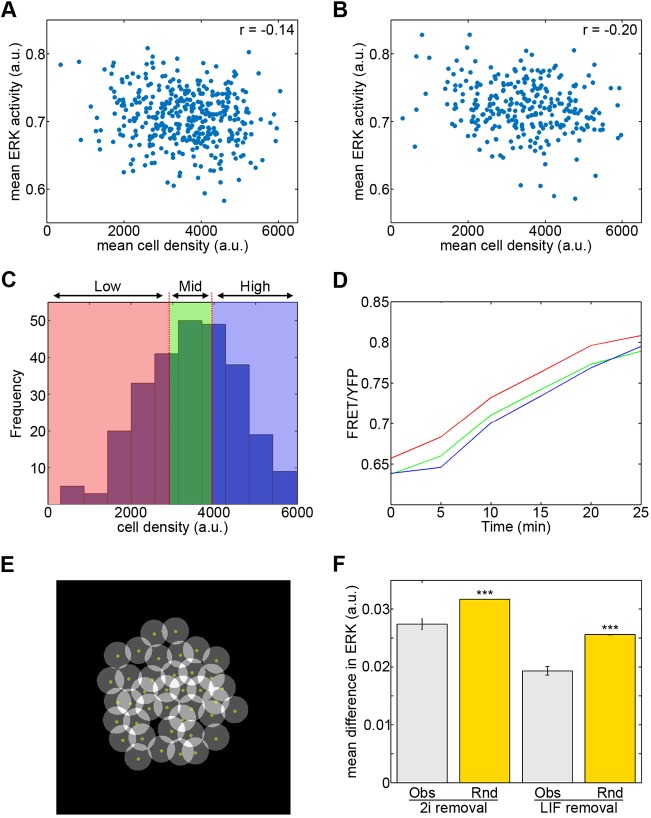
Earlier studies have suggested the culture microenvironment may influence the tendency of a cell towards either pluripotency or differentiation (Cannon et al., 2015) despite the potential for dominant signals in the culture media (such as might be contained in serum) to override local influences. To investigate whether local environmental influences might regulate ERK activity within an ESC colony, we first tested how ERK activity levels of individual cells relate to local cell density. Cell density was determined using a custom-built script to measure the YFP signal surrounding each cell. High density values represent cells in a densely populated area (i.e. at the colony centre) whereas low density values represent cells in more sparsely populated regions (at the colony edge) (Fig. S5A). The mean cell density of each cell over the time series was plotted against its mean ERK activity level, revealing a weak negative correlation between density and ERK activity levels (mean r=−0.18; averaged over three replicates) (Fig. 5A). Comparison of ERK activity and density in the initial peak (first 25 min) of the time series, showed a slightly stronger negative correlation (r=−0.24) (Fig. 5B).
Fig. 5.
Spatial regulation of ERK activity dynamics during differentiation. (A) Mean cell density around each cell over 16 h differentiation (2i removal) plotted against mean ERK (one replicate shown; r=−0.14, P=0.0059, n=388). (B) Mean cell density versus mean ERK activity during the first 25 min of the time series (one replicate; r=−0.20, P=0.0013, n=261). (C) Histogram of average cell density values from the first 25 min for all cells in a single replicate. Colours show binning of cells into three equal groups of low (red), intermediate (green) and high (blue) density. (D) The initial increase in FRET/YFP ratios of cells grouped based on density (colour-coded as in C). Differences in FRET ratio values between high and low cell density group were significant (P=0.0044; see also Fig. S5B). (E) Identification of neighbouring cells. A circle of 25 pixel radius was fixed around the manually selected centroid (yellow dot) of each cell. Cells with overlapping circles were considered neighbours. (F) Mean difference in ERK activity between neighbours (Obs) and randomly assigned neighbours (Rnd). Data shown for differentiation following 2i removal (594 neighbours) and LIF removal (576 neighbours). Error bars represent s.e.m. Differences assessed using Mann–Whitney tests. ***P<0.001. All data are typical of three independent replicates.
To further examine how differences in the activation kinetics of ERK during this initial peak relate to local density, the average density values for each cell were partitioned into three equally sized groups of low, mid and high density cells (Fig. 5C). The mean ERK activity trace of each group was plotted and this analysis demonstrated a significant difference between the ERK response of the high and low density groups (Fig. 5D, Fig. S5B). A higher level of basal ERK activity was consistently seen in the low density cells (Fig. 5D). Although our data did not show a strong relationship between Nanog levels and ERK activity, in most replicates Nanog protein expression was slightly enhanced at higher culture densities (Fig. S5C), in agreement with earlier observations (Cannon et al., 2015).
We next explored whether local differences in cell signalling explain the presence of the two different categories (transient and sustained) of ERK dynamics revealed by k-means clustering. To test this, we developed software to identify the neighbours for each cell in the colony (Fig. 5E). Localisation of cells belonging to each of the k-means clusters showed the proportion of neighbouring cells that were from the same cluster occurred to the extent observed by chance, implying these distinct ERK activity dynamics are randomly spatially distributed within an ESC colony (Fig. S5D,E). To test the sensitivity of our methods, we repeated the analysis on cells manually allocated into clusters based on their location. This test showed our software can detect whether cells of a cluster localise together (Fig. S5F). From these data, it appears that the ERK activity dynamics of a cell are poorly related to the behaviour of its neighbours. However, this approach only incorporates the ERK activity profiles of two groups of cells and, to ensure a significant number of cells could be compared from single colonies, analysis was restricted to a defined portion of the time series.
To overcome these limitations, and further address how the ERK activity of a cell relates to that of its nearest neighbours, we compared the ERK activity of neighbours using a resampling approach. Neighbours were randomly assigned to each cell and the average difference in ERK activity between neighbours was calculated for each randomised data set. A significant difference in the mean difference in ERK activity between real neighbours (Obs) compared with randomly assigned neighbour pairs (Rnd) was identified (Fig. 5F, Fig. S5G) during differentiation after 2i removal. A similar effect was observed in cells differentiating after LIF removal (Fig. 5F). Together, these findings suggest that ESCs in close proximity to one another display more similar levels of ERK activity.
Transmission of ERK activity dynamics along cell lineages
Similar levels of ERK activity in neighbouring cells may result from nearby cells having exposure to similar signalling environments. However, after division, most ESCs remain close to their neighbourhood of origin in a colony, which raises the possibility that local similarities in ERK activity may also result from nearby cells being closely related. Daughter cells from a division will to a certain extent share the contents of their mother cell, and until dilution and turnover of these contents during cell growth and further divisions, these shared contents might harmonise cell behaviour.
To test whether similarities in ERK activity between cells arise as a result of lineage, cell tracks (n=137) for daughter pairs were extracted from each 2i removal experimental replicate. Differences in the mean ERK activity between daughters of a division was calculated (Obs) and this was compared with the expected difference between non-related cells (Rnd) obtained by resampling (10,000 times) the differences in ERK activity between randomised pairs of daughters. The measured difference in ERK activity between daughters was considerably lower than the difference between randomised daughters (Fig. 6A), suggesting that closely related cells display more similar levels of ERK activity. This trend was consistent following analysis on raw and smoothed time series data.
Fig. 6.
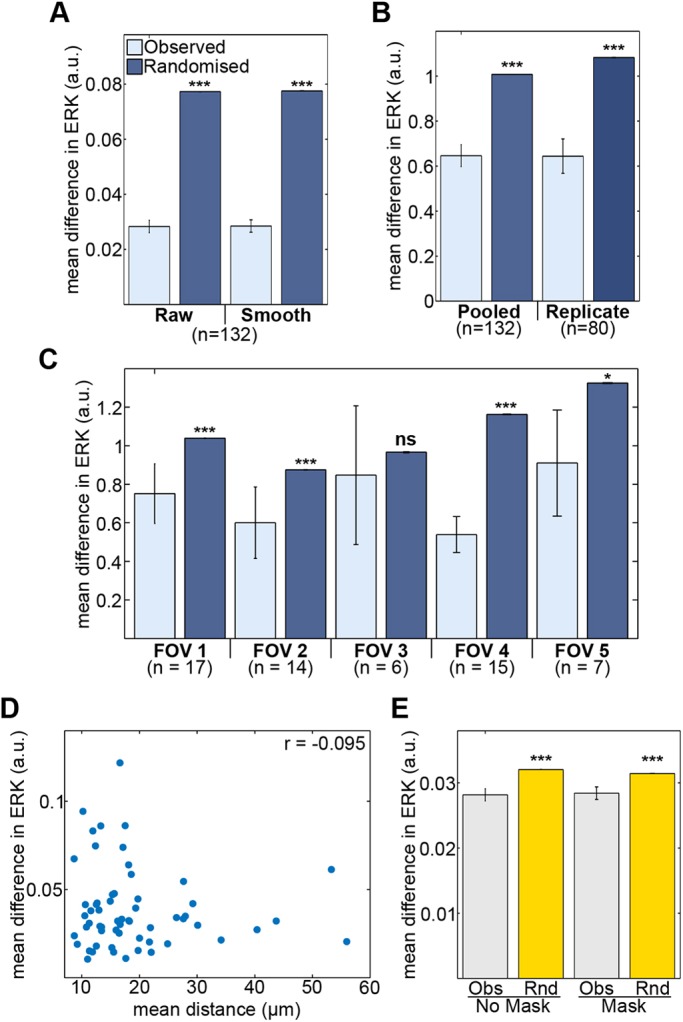
Stability of levels of ERK activity along cell lineages. (A) Daughter cells from a cell division have more similar ERK activity than randomly paired daughters. Analysis was carried out on both raw and filtered (smoothed) FRET traces of daughter cells pooled from three replicates. (B) Differences in mean ERK activity of real and randomised daughter pairs pooled from three replicates and from a single replicate following correction for time-associated differences in ERK activity. (C) Differences in corrected mean ERK activity of real (observed) and randomised daughters from each imaging field of a single replicate. Vertical axes report corrected FRET values from filtered (smoothed) traces. The n-values represent numbers of cell pairs analysed. (D) Scatter plot of mean distance and mean difference in ERK activity between daughter pairs (r=−0.095, P=0.47, n=59). (E) Masking related daughters from neighbour analysis does not alter the increased similarity in ERK activity between neighbouring cells (Obs) compared with non-neighbours (Rnd). Significance testing used Mann–Whitney tests. Error bars represent s.e.m.
One possibility is that the measured similarity of daughters reflects that the random sampling did not take into account the portion of the time series in which a daughter appears. To illustrate, daughters of earlier dividing cells that appear during the initial peak in activation will have a tendency to be different from randomly selected daughters that appear later in the time series when cells exhibit overall lower levels of ERK activity. To address these issues, the mean ERK activity of each daughter was normalised to the average ERK activity of cells from that replicate, for the same segment of time. After this treatment of the randomised data, the ERK activity of related daughters remained more similar than the random expectation (Fig. 6B; Pooled). A further possibility was that the random sampling expands the expected differences between cells because data were pooled from multiple experimental replicates. To test this possibility, the analysis was repeated including the daughter pairs from single replicates, using time-normalised values of mean ERK activity (Fig. 6B; Replicate). The greater similarity of related over randomised cell pairs was robust to this treatment of the data, for all experimental replicates.
Although correlated ERK activity of daughter cells was not affected by experimental replicate, it remained possible that increased differences in ERK activity following randomisation may arise from the comparison of cells in different imaging field of views (FOVs). To test for this possibility, randomisation was carried out on each individual FOV within a single replicate. Again, related cells in a FOV were more similar than the randomised daughter pairs in four out of five FOVs (Fig. 6C). A significant difference could not be identified between the observed and randomised pairs in FOV 3, perhaps due to the low number of divisions; however, the trend was conserved (Fig. 6C). The similarity in ERK activity between sister cells was also a strong feature of the differentiation response to LIF removal (Fig. S6A,B).
Could the similarity in ERK activity of related cells be a result of continued proximity? To evaluate this possibility, the mean difference in ERK activity between daughter pairs was compared with the mean distance between them (Fig. 6D, Fig. S6C). The correlation between inter-daughter distance and difference in ERK activity was weak and non-significant in all replicates. In addition, no correlation between distance and difference in ERK between daughters was identified in the alternative differentiation protocol of LIF removal (Fig. S6C). Considering a much larger effective dataset, which compares the instantaneous relationship between inter-daughter distance and difference in ERK activity for every time point, also showed no strong correlation; indeed, two of the replicates showed a weak negative correlation (Fig. S6D). To give a sense of scale to the plots in Fig. 6D, cells that have 20 µm or less between their centroids tend to be adjacent – they are neighbours. Larger separations indicate sisters separated by one or more intervening cells for all or part of the time trace. These data imply that the similarity in the ERK activity of daughters is not influenced by the degree of cell migration post division. As daughters are in general more similar than neighbours (based on the differences in ERK reporter activity compared with randomised controls), these data suggest the similarity in ERK between progeny of a cell division is not dominated by direct cell contacts.
Is the similarity in the ERK activity of neighbours simply reflecting the proximity of closely related cells? To test this possibility, we repeated the neighbour cell comparisons, this time removing from the analysis sister cells that remained as neighbours. The observed similarity between neighbours was resistant to this treatment of the data, for both 2i removal and LIF removal differentiation (Fig. 6E, Fig. S7A,B). This suggests neighbour similarity emerges from processes other than inheritance of signalling levels from the same parent. Cells will have many neighbours that are not sisters; indeed, masking sisters removed a maximum of 16% of cells from the analysis. Although it remains possible that an inherited effect that persists for multiple cell generations drives similarity between neighbours, we think this is unlikely as more distant relatives in the colony will have had more opportunity for dispersal, and are less likely to remain neighbours.
Other features of the differentiation response could also be extracted from these time series. Nanog protein levels after 16 h differentiation were more similar between real daughter pairs than between randomly selected daughters (Fig. S7C). These findings may relate to previous studies showing expression of pluripotency reporters is more strongly correlated in closely related cells in serum/LIF culture (Cannon et al., 2015; Filipczyk et al., 2015; Singer et al., 2014). Although no strong relationship was identified between time of division and Nanog expression (Fig. S7D), Nanog levels of daughter cells were found to be slightly decreased compared with cells that had not undergone a cell division (Fig. S7E). These data are suggestive of a degree of temporal coupling between cell division and exit from pluripotency.
DISCUSSION
To gain insight into the signalling dynamics underlying stem cell differentiation, we implemented an ERK FRET biosensor to study the changing activity of ERK kinase during the exit from pluripotency of mouse ESCs. ERK activity shows considerable spatial and temporal heterogeneity in ESC cultures undergoing differentiation after removal of 2i, with an initial wave of enhanced activity followed by a long heterogeneous decay in activity over several hours. During differentiation after removal of LIF, a different dynamic behaviour was observed, which lacked the initial increase in ERK activity. ERK dynamics varied considerably within cell populations and despite a strong requirement for the ERK pathway in the differentiation response, the integrated ERK response was only weakly coupled to the rate of differentiation at the single cell level, for both differentiation regimes.
Given the ERK pathway is absolutely essential for differentiation, these findings are unexpected but might be explained if all cells have ‘enough’ ERK activity, so variability in levels of signalling over and above this threshold may have little impact on the rate of differentiation. This interpretation may have parallels with the regulation of cellular metabolism: glycolysis involves around ten enzymatic steps, all of which are crucial, yet few of these steps exert significant control over glycolytic flux (Tanner et al., 2018). An alternative, but compatible, view is that both ERK activity and Nanog regulation are likely to show dense connectivity to multiple other cellular regulators. With such complexity, detecting the portion of signalling specific to exit from pluripotency may require simultaneous study of more than two components.
Understanding the weak coupling between exit from pluripotency and ERK activity is informed by a recent study on RSK, a negative regulator of ERK (Nett et al., 2018). In the absence of RSK, ESCs have elevated levels of ERK phosphorylation and precociously exit pluripotency and differentiate, in population level measurements. Our findings, in single cells, of weak coupling between ERK activity and the rate of differentiation are compatible with the RSK study. Our approach is to monitor the normal dynamic range of ERK signalling, with the regulation governing ERK activity levels, such as by RSK, intact. We propose that the strong effects of RSK depletion on the differentiation response, compared with the weak effects we observe, occur because this perturbation takes cells out of the normal dynamic range. The same study also showed additional peaks in ERK activity in wild-type cells (at the population level) around 15 h of differentiation (Nett et al., 2018), raising the possibility that additional phases of the ERK dynamics beyond the initial wave may be important for developmental progression. In our data, we observed no such later peak, perhaps due to different differentiation conditions; however, we observed a strong reduction in both Nanog and Esrrb expression in the absence of this additional activity, implying later peaks may function beyond the initial phase of differentiation.
A more extreme view is possible, which relates to an earlier study, using inducible ERK depletion, showing that although ERK is required for activation of differentiation genes, it is not required for downregulation of pluripotency genes during differentiation (Chen et al., 2015). This study placed much of the emphasis on the effects usually attributed to ERK on the upstream kinase MEK, which was proposed to operate via other signalling molecules. The emphasis on MEK may be justified; most studies of ERK signalling in ESCs have used MEK inhibition to ablate ERK activity. This approach will also interfere with ERK-independent roles of MEK (Tang et al., 2015), and will not block any MEK-independent regulation of ERK (Chen et al., 2015). It is not clear whether we can extrapolate from the ERK-deficient state, in which the cells are also undergoing telomere shortening, genomic instability, G1 arrest and increased apoptosis, to our more standard culture differentiation conditions. However, our data may be consistent with this earlier study.
ERK dynamics displayed a number of correlates with spatial features of ESC colonies. Firstly, ERK activity was enhanced in regions of the colony with low cell densities, such as the peripheral zones. ERK activity was also correlated between neighbouring cells. Surprisingly for a signalling molecule, the strongest effect on ERK activity detected by the reporter was linked to cell lineage, with closely related cells displaying more similar ERK activity levels than unrelated or more distantly related cells. These observations suggest the magnitudes of the signalling reactions of cells can be cell-autonomously specified, and are not solely a result of extrinsic factors.
Lineage regulation of signalling activity could be accounted for by the expectation that dividing cells transfer their contents to their progeny. No specific ‘epigenetic’ memory mechanism is required for this to allow at least a partial continuation of the cellular phenotype. Protein fluctuation timescales vary from protein to protein and from cell type to cell type, but outlier protein levels could be expected to fluctuate back to the mean within one or two cell cycles (Sigal et al., 2006). It is notable that previous measurements of protein fluctuation time in mouse ESCs suggest more stable protein expression levels down cell lineages (Cannon et al., 2015; Filipczyk et al., 2015; Singer et al., 2014), with fluctuation times of Nanog in pluripotent cell culture of up to six or seven cell cycles. Our observations of stability in the signal strength along cell lineages do not argue against the importance of local signalling; the motility of ESCs is low, and cells that divide often remain adjacent, and those that do separate usually move away by only one or two additional cell diameters over a cell generation. This low motility will favour cells remaining in the same microenvironment, which may contribute to the stability in signalling activity within a lineage. However, we stress that the cell lineage effect is stronger than the neighbour effect, and that removing sister cells from the neighbour analysis has little effect on the magnitude of neighbour correlations. These features of the data argue for distinct effects of both local environment and lineage. Another possibility is that the external signals driving ERK activity may not be strongly limiting under these culture conditions, and so the inherited differences in the levels of signalling components become apparent. This inherited effect may extend to the expression of the signals themselves; for example, a mother cell that has synthesised more Fgf4 mRNA than the population average may pass this on to its progeny, and these would continue to strongly activate ERK in an autocrine manner.
The lineage and niche context of cells have overlapping and non-overlapping effects on ERK activity and pluripotency factor expression. Although we identified that neighbouring cells have more similar ERK activity than more distant cells, this effect does not transfer to Nanog expression. Differences in Nanog levels between cells do not depend on the distance between cells, for either related or unrelated cells in a colony (Cannon et al., 2015). In contrast, both ERK activity (this study) and Nanog levels (Cannon et al., 2015; Filipczyk et al., 2015; Singer et al., 2014; Xenopoulos et al., 2015) show strong persistence along cell lineages despite the weak coupling between the two.
Overall, our observations point to the insights that can be gained by following biochemistry inside living single cells over time during differentiation, and suggest a potential mechanism contributing to cell decision-making based upon cell lineage. It will now be important to test these observations in the early embryo, to evaluate the strengths of the different contributions to ERK dynamics and activity, and the importance of different characteristics of these ERK responses, within a physiologically relevant signalling niche.
MATERIALS AND METHODS
Cell culture
E14TG2a ESCs (obtained from Jenny Nichols, Cambridge Stem Cell Institute, UK) were routinely maintained in serum/LIF culture conditions comprising GMEM (Gibco) supplemented with 10% fetal bovine serum (FBS) and LIF, 1 μM penicillin-streptomycin (Gibco), 2 μM L-glutamine, 1 μM sodium pyruvate, 1 μM non-essential amino acids (Lonza), 7.7 ppm β-mercaptoethanol, plated on gelatin-coated dishes. Cells were also maintained in 2i/LIF conditions (Ying et al., 2008); serum/LIF supplemented with CHIR99021 (3 μM, Axon Medchem) and PD0325901 (1 μM, Axon Medchem). Cells were tested and shown to be clear of mycoplasma. For differentiation out of 2i, ESCs were cultured for a minimum of two passages in 2i/LIF conditions, before being plated at 1.5×104 cells/cm2 in 2i/LIF media on dishes coated with 10 μg/ml laminin (Merck, CC095) or 0.1% gelatin. Medium was then replaced after 20-24 h, or at specified time points, with media free from LIF and 2i. For Nanog and Rex1 reporter cells, the TNGA (Chambers et al., 2007) and OCRG9 (Toyooka et al., 2008) cell lines were used, respectively. A similar protocol was used for differentiation out of LIF except for the starting media being serum/LIF conditions.
Establishment of stable cell lines
The EKAREV-expressing ESC line was established using the piggyBAC transposon system (Ivics et al., 2009). ESCs were co-transformed with EKAREV-NLS plasmid (Aoki et al., 2012) and a pCMV-mPBase transposase expression vector. The E14-EKAREV-TA cell line was generated by transformation of ESCs with EKAREV-TA (from Toru Hiratsuka, King's College London). To generate the SECFP and YPet control cell lines, the SECFP and YPet fluorophore sequences were amplified out of the EKAREV biosensor then cloned back into the expression vector (ppBbsr2-3594nls) plasmid to replace the full EKAREV gene. ESCs were subsequently transformed with either the SECFP or YPet-only plasmids.
Antibodies
For immunofluorescence, cells were washed with PBS, fixed in 4% paraformaldehyde in PBS for 20 min at room temperature and permeabilised with 0.2% Triton X-100 in PBS for 10 min. Following washing in PBS, cells were blocked in 5% bovine serum albumin for 30 min at room temperature and incubated in primary antibody in blocking solution overnight at 4°C. After washing in PBS, cells were incubated in secondary antibody for 1 h at room temperature. Nuclei were stained with 4 μg/ml DAPI where indicated. Primary antibodies were pERK (9101S, Cell Signalling; 1:200); Nanog (14-5761-80, eBiosciences; 1:200); Esrrb (P46705-00, Perseus Proteomics; 1:100); Sox2 (14-9811-82, eBiosciences; 1:100). Species-specific IgG Cy3-conjugated secondary antibodies were used (Jackson ImmunoResearch). For pERK immunostaining, a tyramide amplification reagent (1:200; Alexa Fluor 594, T20950, Life Technologies) was used. Immunostained TNGA and OCRG9 cells were imaged on a Perkin Elmer Ultraview VOX spinning disc confocal microscope with a 60× objective. EKAREV-NLS expressing ESCs stained following the removal of 2i/LIF were imaged using a wide-field system (Muramoto and Chubb, 2008) using a GFP/mCherry filter set (Chroma 59022) and 40×1.30 NA objective. To measure intensities, a point was manually selected within each cell to generate an array of coordinates (Cannon et al., 2015), and intensities were extracted by calculating the median pixel intensity of a 5×5×3 volume centred around each coordinate.
FRET imaging
FRET time-lapse imaging used a wide-field fluorescence system developed for fast, sensitive imaging of mammalian cells. Several confocals were used, but signal was generally too low to be useful (Fig. S1A; Table S1). The system comprised a Zeiss Axiovert 200 stand with an EMCCD camera (C9100-13, Hamamatsu). Illumination was provided by a DG4 light source (Sutter). An xy stage (ASI) with a piezo top was incorporated to facilitate fast capture of 3D stacks at multiple positions in the culture. Images were acquired through a 40× oil objective using filters from Chroma (set 89002). The CFP and YFP exciters were placed in the DG4 light source. The YFP emission filter was in a MAC6000 emission filter wheel (Ludl). The whole system was managed by Volocity Acquisition software. To reduce photodamage, a neutral density filter was used to attenuate illumination to 10% of the lamp intensity. Plating of cells was carried out 20-24 h prior to imaging, onto Ibidi imaging dishes. For short-term 2i removal tests, experiments were carried out over a 4 h period, acquiring a 40-60 µm z-stack, with 1 µm step, of YPF and FRET channel images every 5 min. For more long-term differentiation experiments, a 20-30 µm z-stack, with 2 µm step size, was acquired every 5 min for 3 h, then 10 min thereafter up to 16 h. Four or five imaging fields were collected in parallel. For images from single time frames, ratiometric images between the YFP and FRET channel images were obtained using the RatioPLUS ImageJ plugin, as previously described (Hukasova et al., 2012).
For quantification of FRET from time lapses, we used a custom-built MATLAB graphics interface for cell tracking, incorporating the equation from the RatioPLUS plugin. A cylinder of radius 2 pixels and height of three z-slices was applied to each mouse-clicked coordinate and the average FRET and YFP intensity in each cylinder was measured, and a ratio of the two values (FRET/YFP) was calculated. Fractional intensities were recorded from incomplete pixels on the margin of the cylinder. FRET/YFP intensity time series were smoothed, by applying a moving averaging filter with a span of five frames, to reduce experimental noise. Analysis was carried out using smoothed tracks, unless stated otherwise. Tracking was carried out on YFP channel images because of the strong signal, following application of 25 pixel rolling background subtraction in ImageJ.
To evaluate the contribution of bleed-through donor fluorescence emission to FRET ratio levels, ESCs were transformed with either a YPet or SECFP fluorophore to generate donor and acceptor-only ESCs (Fig. S1B). To determine the FRET signal detected with CFP-only cells, every pixel in the CFP channel image was plotted against every pixel value in the FRET channel for five different imaging fields (Fig. S1C). The gradient of the best-fit line through the data signifies the proportion of FRET signal contributed by bleed-through. The measured FRET ratio was scaled by this gradient (Fig. S1D), ensuring the remaining signal is FRET, not bleed-through. Correction for bleed-through caused a uniform negative shift in quantified FRET ratio values (Fig. S1E). As bleed-through contribution had a negligible effect on the observed relative differences in FRET ratio values between cells in test experiments, bleed-through was not corrected for in the experiments presented here.
Data analysis
For clustering time series data, FRET tracks of single cells of individual experiments were clustered using the k-means MATLAB function. The optimal number of clusters within each data set was assigned using silhouette criteria (Rousseeuw, 1987). Cell tracks being clustered were required to be of equal duration, so incomplete tracks were excluded from this analysis. A MATLAB script was used to localise the cells of each cluster, using the mouse-clicked coordinates recorded during manual tracking. Software is available on request.
For calculation of cell density, a maximum projection of the YFP channel, following background subtraction, was imported into MATLAB and converted into a binary image. Cell position coordinates recorded during tracking were applied to the binary image and a circle of 50 pixel radius was drawn around each coordinate. The sum of binary pixel intensities within each circle was calculated and assigned to its corresponding cell. The greater the number of cells within the circle, the higher the density.
To identify neighbouring cells, cell positions recorded during manual tracking were used to draw a circle of a specified radius around each cell. The radius of the circle was adjusted to incorporate the cell-cell contacts of each cell, with an optimised radius of 25 pixels (9.88 µm). Cells were called neighbours wherever there was overlap between circles (Fig. 5E). This was repeated for all the cells in a population. Unless otherwise stated, neighbours were assigned at the end of the time lapse, which could conceivably result in the exclusion of correlation values between cells that were neighbours at some point. However, ESC motility is generally low, and cells immediately surrounding one another are unlikely to change greatly until mitosis occurs.
For significance testing, tests are described in the relevant figure legend. Significance thresholds: *P<0.05, **P<0.01 and ***P<0.001. Data shown in the main figures are replicates with a representative effect size.
Supplementary Material
Acknowledgements
Imaging was carried out at the MRC LMCB Light Microscopy Facility. We thank Adam Corrigan for assistance with image analysis and Toru Hiratsuka for advice on using the FRET reporter.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.D., M.P., J.R.C.; Methodology: J.D., V.A., M.P., J.R.C.; Software: J.D., V.A.; Validation: J.D.; Formal analysis: J.D., V.A.; Investigation: J.D.; Resources: J.D., J.R.C.; Data curation: J.D., V.A.; Writing - original draft: J.D., J.R.C.; Writing - review & editing: J.D., V.A., M.P., J.R.C.; Visualization: J.D.; Supervision: M.P., J.R.C., V.A.; Project administration: M.P., J.R.C.; Funding acquisition: M.P., J.R.C.
Funding
This work was supported by a Wellcome Trust Senior Fellowship (202867/Z/16/Z to J.R.C.), and Medical Research Council funding (MC_U12266B) to the MRC LMCB University Unit at UCL. J.D. was supported by a London Interdisciplinary Doctoral Training Programme Biotechnology and Biological Sciences Research Council studentship. Deposited in PMC for release after 6 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.172940.supplemental
References
- Albeck J. G., Mills G. B. and Brugge J. S. (2013). Frequency-modulated pulses of ERK activity transmit quantitative proliferation signals. Mol. Cell 49, 249-261. 10.1016/j.molcel.2012.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K., Komatsu N., Hirata E., Kamioka Y. and Matsuda M. (2012). Stable expression of FRET biosensors: a new light in cancer research. Cancer Sci. 103, 614-619. 10.1111/j.1349-7006.2011.02196.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K., Kumagai Y., Sakurai A., Komatsu N., Fujita Y., Shionyu C. and Matsuda M. (2013). Stochastic ERK activation induced by noise and cell-to-cell propagation regulates cell density-dependent proliferation. Mol. Cell 52, 529-540. 10.1016/j.molcel.2013.09.015 [DOI] [PubMed] [Google Scholar]
- Avilion A. A., Nicolis S. K., Pevny L. H., Perez L., Vivian N. and Lovell-Badge R. (2003). Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17, 126-140. 10.1101/gad.224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon T., Stracey C., Chambers I., Nichols J. and Smith A. (1999). Suppression of SHP-2 and ERK signalling promotes self-renewal of mouse embryonic stem cells. Dev. Biol. 210, 30-43. 10.1006/dbio.1999.9265 [DOI] [PubMed] [Google Scholar]
- Cannon D., Corrigan A. M., Miermont A., McDonel P. and Chubb J. R. (2015). Multiple cell and population-level interactions with mouse embryonic stem cell heterogeneity. Development 142, 2840-2849. 10.1242/dev.120741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S. and Smith A. (2003). Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113, 643-655. 10.1016/S0092-8674(03)00392-1 [DOI] [PubMed] [Google Scholar]
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L. and Smith A. (2007). Nanog safeguards pluripotency and mediates germline development. Nature 450, 1230-1234. 10.1038/nature06403 [DOI] [PubMed] [Google Scholar]
- Chen H., Guo R., Zhang Q., Guo H., Yang M., Wu Z., Gao S., Liu L. and Chen L. (2015). Erk signaling is indispensable for genomic stability and self-renewal of mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 112, E5936-E5943. 10.1073/pnas.1516319112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Saidon C., Cohen A. A., Sigal A., Liron Y. and Alon U. (2009). Dynamics and variability of ERK2 response to EGF in individual living cells. Mol. Cell 36, 885-893. 10.1016/j.molcel.2009.11.025 [DOI] [PubMed] [Google Scholar]
- Cowley S., Paterson H., Kemp P. and Marshall C. J. (1994). Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell 77, 841-852. 10.1016/0092-8674(94)90133-3 [DOI] [PubMed] [Google Scholar]
- Davies O. R., Lin C.-Y., Radzisheuskaya A., Zhou X., Taube J., Blin G., Waterhouse A., Smith A. J. H. and Lowell S. (2013). Tcf15 primes pluripotent cells for differentiation. Cell Rep. 3, 472-484. 10.1016/j.celrep.2013.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaliwal N. K., Miri K., Davidson S., Tamim El Jarkass H. and Mitchell J. A. (2018). KLF4 nuclear export requires ERK activation and initiates exit from naive pluripotency. Stem Cell Rep. 10, 1308-1323. 10.1016/j.stemcr.2018.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M. J. and Kaufman M. H. (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154-156. 10.1038/292154a0 [DOI] [PubMed] [Google Scholar]
- Filipczyk A., Gkatzis K., Fu J., Hoppe P. S., Lickert H., Anastassiadis K. and Schroeder T. (2013). Biallelic expression of nanog protein in mouse embryonic stem cells. Cell Stem Cell 13, 12-13. 10.1016/j.stem.2013.04.025 [DOI] [PubMed] [Google Scholar]
- Filipczyk A., Marr C., Hastreiter S., Feigelman J., Schwarzfischer M., Hoppe P. S., Loeffler D., Kokkaliaris K. D., Endele M., Schauberger B. et al. (2015). Network plasticity of pluripotency transcription factors in embryonic stem cells. Nat. Cell Biol. 17, 1235-1246. 10.1038/ncb3237 [DOI] [PubMed] [Google Scholar]
- Gillies T. E., Pargett M., Minguet M., Davies A. E. and Albeck J. G. (2017). Linear integration of ERK activity predominates over persistence detection in Fra-1 regulation. Cell Syst. 5, 549-563.e545. 10.1016/j.cels.2017.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamazaki T., Kehoe S. M., Nakano T. and Terada N. (2006). The Grb2/Mek pathway represses nanog in murine embryonic stem cells. Mol. Cell. Biol. 26, 7539-7549. 10.1128/MCB.00508-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton W. B., Kaji K. and Kunath T. (2013). ERK2 suppresses self-renewal capacity of embryonic stem cells, but is not required for multi-lineage commitment. PLoS ONE 8, e60907 10.1371/journal.pone.0060907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey C. D., Ehrhardt A. G., Cellurale C., Zhong H., Yasuda R., Davis R. J. and Svoboda K. (2008). A genetically encoded fluorescent sensor of ERK activity. Proc. Natl. Acad. Sci. USA 105, 19264-19269. 10.1073/pnas.0804598105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Lopes S. M. C. S., Tang F. and Surani M. A. (2008). Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell 3, 391-401. 10.1016/j.stem.2008.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka T., Fujita Y., Naoki H., Aoki K., Kamioka Y. and Matsuda M. (2014). Intercellular propagation of extracellular signal-regulated kinase activation revealed by in vivo imaging of mouse skin. eLife 4, e05178 10.7554/eLife.05178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hukasova E., Silva Cascales H., Kumar S. R. and Lindqvist A. (2012). Monitoring kinase and phosphatase activities through the cell cycle by ratiometric FRET. J. Vis. Exp., e3410 10.3791/3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics Z., Li M. A., Mátés L., Boeke J. D., Nagy A., Bradley A. and Izsvák Z. (2009). Transposon-mediated genome manipulation in vertebrates. Nat. Methods 6, 415-422. 10.1038/nmeth.1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkan T., Olova N., Roode M., Mulas C., Lee H. J., Nett I., Marks H., Walker R., Stunnenberg H. G., Lilley K. S. et al. (2017). Tracking the embryonic stem cell transition from ground state pluripotency. Development 144, 1221-1234. 10.1242/dev.142711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu N., Aoki K., Yamada M., Yukinaga H., Fujita Y., Kamioka Y. and Matsuda M. (2011). Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol. Biol. Cell 22, 4647-4656. 10.1091/mbc.e11-01-0072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunath T., Saba-El-Leil M. K., Almousailleakh M., Wray J., Meloche S. and Smith A. (2007). FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development 134, 2895-2902. 10.1242/dev.02880 [DOI] [PubMed] [Google Scholar]
- Lee R. E. C., Walker S. R., Savery K., Frank D. A. and Gaudet S. (2014). Fold change of nuclear NF-κB determines TNF-induced transcription in single cells. Mol. Cell 53, 867-879. 10.1016/j.molcel.2014.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgani S. M., Saiz N., Garg V., Raina D., Simon C. S., Kang M., Arias A. M., Nichols J., Schröter C. and Hadjantonakis A.-K. (2018). A Sprouty4 reporter to monitor FGF/ERK signaling activity in ESCs and mice. Dev. Biol. 441, 104-126. 10.1016/j.ydbio.2018.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramoto T. and Chubb J. R. (2008). Live imaging of the Dictyostelium cell cycle reveals widespread S phase during development, a G2 bias in spore differentiation and a premitotic checkpoint. Development 135, 1647-1657. 10.1242/dev.020115 [DOI] [PubMed] [Google Scholar]
- Nett I. R., Mulas C., Gatto L., Lilley K. S. and Smith A. (2018). Negative feedback via RSK modulates Erk-dependent progression from naïve pluripotency. EMBO Rep. 19, e45642 10.15252/embr.201745642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Burdon T., Chambers I. and Smith A. (1998). Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 12, 2048-2060. 10.1101/gad.12.13.2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Miyazaki J.-I. and Smith A. G. (2000). Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24, 372-376. 10.1038/74199 [DOI] [PubMed] [Google Scholar]
- Purvis J. E. and Lahav G. (2013). Encoding and decoding cellular information through signaling dynamics. Cell 152, 945-956. 10.1016/j.cell.2013.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseeuw P. J. (1987). Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 20, 53-65. 10.1016/0377-0427(87)90125-7 [DOI] [Google Scholar]
- Ryu H., Chung M., Dobrzynski M., Fey D., Blum Y., Lee S. S., Peter M., Kholodenko B. N., Jeon N. L. and Pertz O. (2015). Frequency modulation of ERK activation dynamics rewires cell fate. Mol. Syst. Biol. 11, 838-838. 10.15252/msb.20156458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos S. D. M., Verveer P. J. and Bastiaens P. I. H. (2007). Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat. Cell Biol. 9, 324-330. 10.1038/ncb1543 [DOI] [PubMed] [Google Scholar]
- Schröter C., Rué P., Mackenzie J. P. and Martinez Arias A. (2015). FGF/MAPK signaling sets the switching threshold of a bistable circuit controlling cell fate decisions in embryonic stem cells. Development 142, 4205-4216. 10.1242/dev.127530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semrau S., Goldmann J. E., Soumillon M., Mikkelsen T. S., Jaenisch R. and Van Oudenaarden A. (2017). Dynamics of lineage commitment revealed by single-cell transcriptomics of differentiating embryonic stem cells. Nat. Commun. 8, 1096 10.1038/s41467-017-01076-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal A., Milo R., Cohen A., Geva-Zatorsky N., Klein Y., Liron Y., Rosenfeld N., Danon T., Perzov N. and Alon U. (2006). Variability and memory of protein levels in human cells. Nature 444, 643-646. 10.1038/nature05316 [DOI] [PubMed] [Google Scholar]
- Singer Z. S., Yong J., Tischler J., Hackett J. A., Altinok A., Surani M. A., Cai L. and Elowitz M. B. (2014). Dynamic heterogeneity and DNA methylation in embryonic stem cells. Mol. Cell 55, 319-331. 10.1016/j.molcel.2014.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. G., Heath J. K., Donaldson D. D., Wong G. G., Moreau J., Stahl M. and Rogers D. (1988). Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 336, 688-690. 10.1038/336688a0 [DOI] [PubMed] [Google Scholar]
- Stavridis M. P., Lunn J. S., Collins B. J. and Storey K. G. (2007). A discrete period of FGF-induced Erk1/2 signalling is required for vertebrate neural specification. Development 134, 2889-2894. 10.1242/dev.02858 [DOI] [PubMed] [Google Scholar]
- Tang Z., Dai S., He Y., Doty R. A., Shultz L. D., Sampson S. B. and Dai C. (2015). MEK guards proteome stability and inhibits tumor-suppressive amyloidogenesis via HSF1. Cell 160, 729-744. 10.1016/j.cell.2015.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner L. B., Goglia A. G., Wei M. H., Sehgal T., Parsons L. R., Park J. O., White E., Toettcher J. E. and Rabinowitz J. D. (2018). Four key steps control glycolytic flux in mammalian cells. Cell Syst. 7, 49-62.e48. 10.1016/j.cels.2018.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka Y., Shimosato D., Murakami K., Takahashi K. and Niwa H. (2008). Identification and characterization of subpopulations in undifferentiated ES cell culture. Development 135, 909-918. 10.1242/dev.017400 [DOI] [PubMed] [Google Scholar]
- Xenopoulos P., Kang M., Puliafito A., Di Talia S. and Hadjantonakis A. K. (2015). Heterogeneities in Nanog expression drive stable commitment to pluripotency in the mouse blastocyst. Cell Rep. 10, 1508-1520. 10.1016/j.celrep.2015.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.-H., Kalkan T., Morrisroe C., Smith A. and Sharrocks A. D. (2012). A genome-wide RNAi screen reveals MAP kinase phosphatases as key ERK pathway regulators during embryonic stem cell differentiation. PLoS Genet. 8, e1003112 10.1371/journal.pgen.1003112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.-L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P. and Smith A. (2008). The ground state of embryonic stem cell self-renewal. Nature 453, 519-523. 10.1038/nature06968 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.