ABSTRACT
Dedicated stem cells ensure postnatal growth, repair and homeostasis of skeletal muscle. Following injury, muscle stem cells (MuSCs) exit from quiescence and divide to reconstitute the stem cell pool and give rise to muscle progenitors. The transcriptomes of pooled MuSCs have provided a rich source of information for describing the genetic programs of distinct static cell states; however, bulk microarray and RNA sequencing provide only averaged gene expression profiles, blurring the heterogeneity and developmental dynamics of asynchronous MuSC populations. Instead, the granularity required to identify distinct cell types, states, and their dynamics can be afforded by single cell analysis. We were able to compare the transcriptomes of thousands of MuSCs and primary myoblasts isolated from homeostatic or regenerating muscles by single cell RNA sequencing. Using computational approaches, we could reconstruct dynamic trajectories and place, in a pseudotemporal manner, the transcriptomes of individual MuSC within these trajectories. This approach allowed for the identification of distinct clusters of MuSCs and primary myoblasts with partially overlapping but distinct transcriptional signatures, as well as the description of metabolic pathways associated with defined MuSC states.
KEY WORDS: Muscle stem cells, Satellite cells, Single cell RNA-seq, Muscle regeneration, Mouse
Summary: Single cell RNA sequencing of adult muscle stem cells and primary myoblasts from homeostatic or regenerating muscles identifies distinct clusters of cell types with partially overlapping but distinct transcriptional signatures.
INTRODUCTION
Growth and homeostasis of the skeletal muscle system, a tissue/organ constituting approximately 35% of the human total body mass (Janssen et al., 2000), rely on tissue-resident skeletal muscle stem cells (MuSCs). Following a period of postnatal proliferation leading to a muscle mass increase of several-fold (White et al., 2010), adult MuSCs enter quiescence (Chargé and Rudnicki, 2004). In response to acute or chronic muscle injury, MuSCs are activated, proliferate and differentiate into either newly formed or pre-existing muscle fibers to ensure compensatory muscle regeneration and repair (Brack and Rando, 2012; Comai and Tajbakhsh, 2014). Whereas quiescent and activated MuSCs express the transcription factor Pax7 and accumulate its cognate protein (Seale et al., 2000; Oustanina et al., 2004), transcripts of the myogenic regulators Myf5 and MyoD are present in quiescent MuSCs (Beauchamp et al., 2000; Crist et al., 2012; Liu et al., 2013; Ryall et al., 2015b; de Morree et al., 2017) but their corresponding proteins are detected only after MuSCs become activated (Zammit et al., 2002). MuSCs retain regenerative capabilities when isolated from donors and transplanted into recipient hosts (Sherwood et al., 2004; Collins et al., 2005; Montarras et al., 2005; Kuang et al., 2007). In one study, a single MuSC transplanted into the muscles of mice extensively proliferated giving rise to tens of thousands of cells contributing to muscle fibers. In contrast, transplanted primary myoblasts were ineffective at engrafting and proliferating (Sacco et al., 2008). Failure of MuSCs to efficiently undergo cell division and regenerate is observed during aging (Brack and Rando, 2007; Brack et al., 2007; Kuang et al., 2007; Chakkalakal et al., 2012; Sousa-Victor et al., 2014; Bernet et al., 2014; Cosgrove et al., 2014; Price et al., 2014; Tierney et al., 2014; Sacco and Puri, 2015; García-Prat et al., 2016; Lukjanenko et al., 2016). Moreover, dystrophin deficiency compromises the ability of MuSCs to undergo asymmetric division resulting in inefficient generation of myogenic precursors (Dumont et al., 2015; Feige et al., 2018). The transitions experienced by MuSCs during development and regeneration are regulated by partially overlapping and dynamic transcriptional programs (Buckingham and Relaix, 2007; Sincennes et al., 2016; Chang et al., 2018). Bulk transcriptome analyses of MuSCs have contributed important mechanistic insights by providing static snapshots of averaged gene expression (Pallafacchina et al., 2010; Liu et al., 2013; Sousa-Victor et al., 2014; Yennek et al., 2014; Ryall et al., 2015b; Alonso-Martin et al., 2016; Aguilar et al., 2016; Machado et al., 2017; van Velthoven et al., 2017; Pala et al., 2018). The granularity required to identify distinct cell types, states, and their transcriptional dynamics can, however, be provided by single cell analysis.
Here, we use droplet-based single cell RNA-seq (scRNA-seq) to capture the transcriptional state of thousands of individual MuSCs and primary myoblasts. Using computational approaches, we infer developmental dynamic trajectories of homeostatic and regenerative adult MuSCs and of primary myoblasts, and describe the relative transcriptional changes, including those relative to metabolic pathways, associated with discrete cell state transitions.
RESULTS
scRNA-seq permits the identification of cell populations of the mouse adult hindlimb skeletal muscles
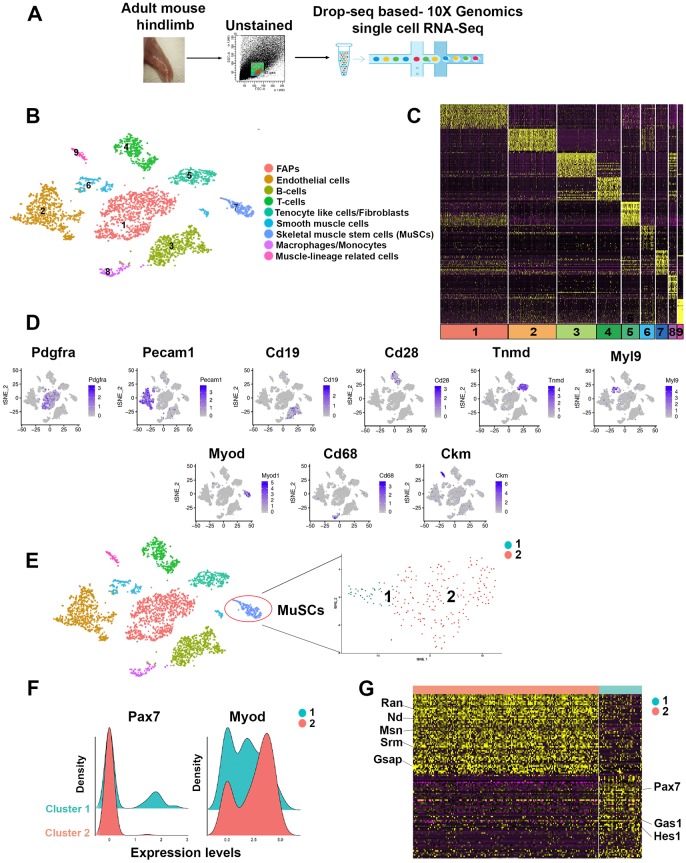
We performed scRNA-seq to identify and enumerate cell populations of mouse adult hindlimb skeletal muscles. Muscles of 3-month-old C56BL/6J mice were dissociated into single cells by enzymatic digestion and live cells were isolated by fluorescence-activated cell sorting (FACS) (Fig. 1A). Cells obtained from two mice were sequenced separately. The two samples were tested for similarity and merged for further analysis (Fig. S1A). After quality control, we retained 4414 cells for downstream analysis. On average, we detected 1221 gene transcripts in each individual cell (156,277 mean reads per cell) (Fig. S1B). These cell and transcript numbers are comparable to those recently reported by the Tabula Muris Consortium for droplet scRNA-seq in limb muscle (Tabula Muris Consortium et al., 2018). The sequencing data were analyzed by t-distributed stochastic neighbor embedding (t-SNE) (Satija et al., 2015). Unsupervised clustering of the quality-controlled cells of the two combined mouse samples returned nine cell clusters, transcriptomes of which were related to fibroadipogenic precursors (FAPs), tenocyte-like cells, smooth muscle cells, endothelial cells, adaptive and innate immunity cells (B- and T-lymphocytes, macrophages, monocytes), and a small cluster expressing genes present in mature skeletal muscle (e.g. creatine kinase muscle, Ckm) (Fig. 1B). These findings are similar to those reported in other studies (Verma et al., 2018; Giordani et al., 2018 preprint).
Fig. 1.
Identification of distinct cell populations in mouse adult skeletal muscle. (A) Scheme of muscle dissection, single cell FACS isolation and scRNA-seq. (B) Graph-based clustering of FACS-isolated single cells identifies distinct clusters corresponding to different cell populations. (C) Heatmap representing the top 20 most variably expressed genes between cell clusters identified. Colors and numbers correspond to the cell clusters shown in B. (D) Expression of representative genes in distinct cell clusters. (E) Clustering and t-SNE plot of freshly FACS-isolated single cells with MuSC characteristics. (F) Kernel density estimation of Pax7 and Myod transcripts in MuSC cluster 1 and cluster 2. (G) Heatmap representing the top 50 most variably expressed genes between the two MuSC clusters.
Visualization of the top 20 most variably expressed genes between cell clusters documented distinct transcriptional programs of the nine clusters (Fig. 1C) and expression of known cell lineage-enriched or -specific transcripts was observed in each of the cell clusters (Fig. 1D). Of the total 4414 cells, 217 (5%) revealed a gene expression pattern that could be assigned to MuSCs. Within this cluster, cells were observed to express variable levels of the MuSC markers vascular cell adhesion molecule 1 (Vcam1), Myod (Myod1), Myf5 and desmin (Des) (Chargé and Rudnicki, 2004) (Fig. S1C). Skeletal muscle dissociation and FACS isolation affect a subset of transcripts in a subpopulation of MuSCs (van den Brink et al., 2017). These manipulations, necessary to obtain MuSCs that can be employed in transplantation protocols (Montarras et al., 2005; Sacco et al., 2008), decrease expression of transcripts represented in niche-associated quiescent MuSCs, such as that of the paired box/homeodomain Pax7 (Seale et al., 2000), and promote accumulation of transcripts enriched in activated MuSCs, such as that encoding MyoD (Machado et al., 2017; van Velthoven et al., 2017). Consistent with these observations, t-SNE analysis of the MuSC cluster revealed the presence of two juxtaposed clusters (clusters 1 and 2; Fig. 1E). Using non-parametric kernel density estimation (KDE), we determined the probability density function of cells expressing Pax7 and Myod in MuSC cluster 1 and cluster 2. In cluster 1, KDE identified two cell populations: one with low, the other with high Pax7. Cluster 2 contained only low-expressing Pax7 cells. KDE for MyoD transcripts identified two cell populations in cluster 2: one with lower, the other with higher Myod. Cluster 1 was enriched for low-expressing Myod cells (Fig. 1F). A heatmap illustrating expression of the top 50 most variable genes in the two MuSC subpopulations is shown in Fig. 1G and the corresponding data are reported in Table S1. Overall, scRNA-seq of mononucleated cells obtained from dissociated hindlimb skeletal muscles permitted the identification of distinct cell lineages, including MuSCs.
scRNA-seq of FACS-purified muscle stem cells
MuSCs derived from hindlimb muscles of two 3-month-old C56BL/6J mice were prospectively FACS-purified as described (Liu et al., 2015) [VCAM1+/CD31 (PECAM1)−/CD45 (PTPRC)−/Sca1 (Ly6a)−] and immediately sequenced (Fig. 2A, Table S1). The two samples were tested for similarity and merged for further analysis (Table S1, MuSCs1 versus MuSCs2 expression sheet; Fig. S2A,B). After quality control, we retained 3081 MuSCs for downstream scRNA-seq analysis. On average, we detected 994 expressed genes in each individual MuSC (Fig. S2C). Using the Chromium platform (10x Genomics), 50,000-70,000 mean reads per cell are generally sufficient to approach saturation, and primary cells with low RNA content and complexity, such as MuSCs, may require less sequencing to achieve saturation reads of 80-90% (https://kb.10xgenomics.com/hc/en-us/articles/115002474263-How-much-sequencing-saturation-should-I-aim-for-; Zhang et al. 2019).
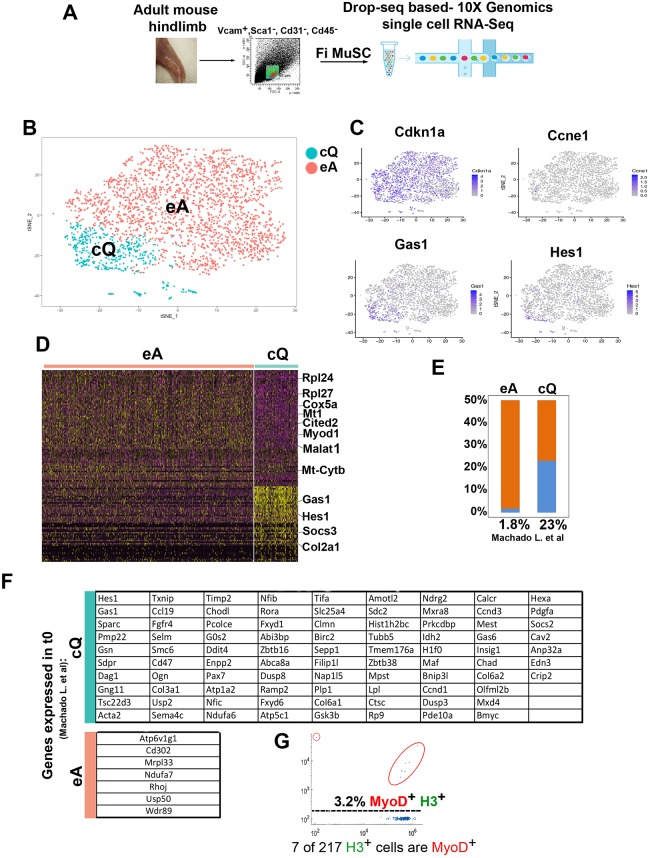
Fig. 2.
Transcriptional characterization of FACS-isolated MuSCs. (A) Scheme of MuSC FACS isolation and scRNA-seq. (B) Graph-based clustering of FACS-isolated MuSCs (VCAM1+/CD31−/CD45−/Sca1−) identifies two clusters: MuSCs close-to-quiescence (cQ) and MuSCs early activation (eA). (C) Expression pattern of the cell cycle inhibitor genes Cdkn1a, cyclin E (Ccne1), growth-arrest 1 (Gas1) and the Notch target Hes1. (D) Heatmap of the top 50 most variably expressed genes between MuSC cQ and MuSC eA. (E) Percentage of transcripts shared between MuSC eA, MuSC cQ and PFA-fixed t0 MuSCs (Machado et al., 2017). (F) Gene list of transcripts shared between MuSC eA, MuSC cQ and PFA-fixed t0 MuSCs (Machado et al., 2017). Prkcdbp, Cavin3; Sdpr, Cavin2; Selm, Selenom; Sepp1, Selenop. (G) Single cell western blot analysis of 217 freshly FACS-isolated MuSCs with histone H3 and MyoD antibodies. MyoD+ cells are encircled in red.
To approximate sequencing saturation, we obtained 236,841 mean reads per cell (Fig. S2C). t-SNE analysis identified two MuSC clusters (Fig. 2B). Transcripts of the cell cycle inhibitor Cdkn1a were present in both clusters whereas those of cyclin E were not detected suggesting that all FACS-isolated MuSCs remained G0-arrested (Fig. 2C). Differentially expressed genes in the two MuSC clusters included growth arrest-specific 1 (Gas1), the Notch target Hes1, Pax7 and the calcitonin receptor (Calcr) in one cluster (MuSCs close-to-quiescence, MuSC cQ) (Fig. 2C,D, Table S1). Genes associated with early activation, such as Myod, Myf5 and ribosomal genes, were instead enriched in the other MuSC cluster (MuSCs early-activation, MuSC eA) (Fig. 2D, Table S1). The MuSC cQ cluster comprised 975 cells (975/3081; 32% of total MuSCs) and the MuSC eA cluster 2108 cells (2108/3081; 68% of total MuSCs). Gene ontology (GO) analysis confirmed that the two MuSC clusters are transcriptionally distinct (Fig. S2D, Table S1). GO terms related to resistance to stress and response to unfolded protein, cell cycle arrest and circadian rhythm were enriched in the MuSC cQ cluster whereas terms indicating activation of ribosome biogenesis, mRNA processing, translation, and protein stabilization were enriched in the MuSC eA cluster (Fig. 2D, Fig. S2D, Table S1). Transcriptome comparison between FACS-isolated MuSCs (ex vivo MuSCs) and in vivo quiescent MuSCs indicated that the MuSC ex vivo transcriptome remains largely reflective of the in vivo transcriptome (van Velthoven et al., 2017). However, another study has reported marked transcriptional differences between MuSCs FACS-isolated from unfixed or paraformaldehyde (PFA)-fixed muscles (Machado et al., 2017). To evaluate the transcriptional state of MuSC cQ and MuSC eA clusters, we compared their respective transcriptomes with that of MuSCs isolated from PFA-fixed muscles to remove genes for which transcription is affected by muscle dissection and FACS isolation (Machado et al., 2017). This analysis revealed that although MuSC eA and MuSCs derived from PFA-fixed muscles shared only 1.8% of transcripts, the percentage increased to 23% in MuSC cQ (Fig. 2E,F).
Myod transcripts are present in quiescent MuSCs but their corresponding protein can be detected only in activated MuSCs. To determine whether FACS-isolated MuSCs cells translated Myod mRNA into the corresponding protein, we captured 217 MuSCs for single cell western blot analysis. Every MuSC was positive for the histone H3 protein. However, MyoD could be detected in only seven out of 217 MuSCs (3.2%) (Fig. 2G). Thus, although tissue and cell manipulations induce transcriptional modifications, these are not immediately followed by MuSC activation as indicated by the paucity of freshly isolated MuSCs expressing detectable levels of the MyoD protein.
scRNA-seq of freshly isolated MuSCs and primary myoblasts identifies discrete transcriptional programs
We wished to compare scRNA-seq profiles of freshly isolated MuSCs, which have not fully committed to the muscle cell lineage (Chargé and Rudnicki, 2004), with those of committed primary myoblasts (PMs). To this end, PMs isolated from the hindlimb muscles of 3-month-old C56BL/6J mice were cultured in growth medium before scRNA-seq. After quality control, we retained 4429 PMs for downstream analysis. On average, we detected 4933 expressed genes in each individual PM (Fig. S3A). To compare MuSCs and PMs directly, we performed t-SNE analysis and computationally merged their corresponding scRNA-seq datasets (Fig. 3A, Fig. S2E,F). t-SNE analysis segregated the transcriptomes corresponding to MuSCs and PMs into five clusters: two belonging to MuSCs and three to PMs (Fig. 3A, Fig. S2G,H). Analysis of the top 20 genes differentially expressed in each of the five clusters is shown in Fig. 3B. Cells present in both PM clusters (PM1 and PM2) expressed transcripts of cyclins Ccnd1 and Ccnd2 (Fig. 3C). Cells of the PM1 cluster were enriched for cell cycle regulators of the G1/S phase (Table S2). A cluster of PMs containing cells that had activated transcription reflective of differentiation (differentiating myocytes, DM) was characterized by myogenin expression (Fig. 3C). In addition to being negative for Ccnd1 and Ccnd2, the cluster corresponding to MuSC cQ expressed growth arrest-specific 1 (Gas1) and the cell cycle inhibitors Cdkn1a and Cdkn2d, whereas Myf5 was preferentially detected in the cluster corresponding to MuSC eA (Table S2). In addition to terms related to the cell cycle, GO analysis of the PM clusters revealed an enrichment for terms related to metabolic pathways (oxidative and glycolytic enzymes, serine biosynthesis enzymes, enzymes of the pentose phosphate pathway), proteostasis (proteome quality and homeostasis), and activation of translation (Fig. 3D, Table S2).
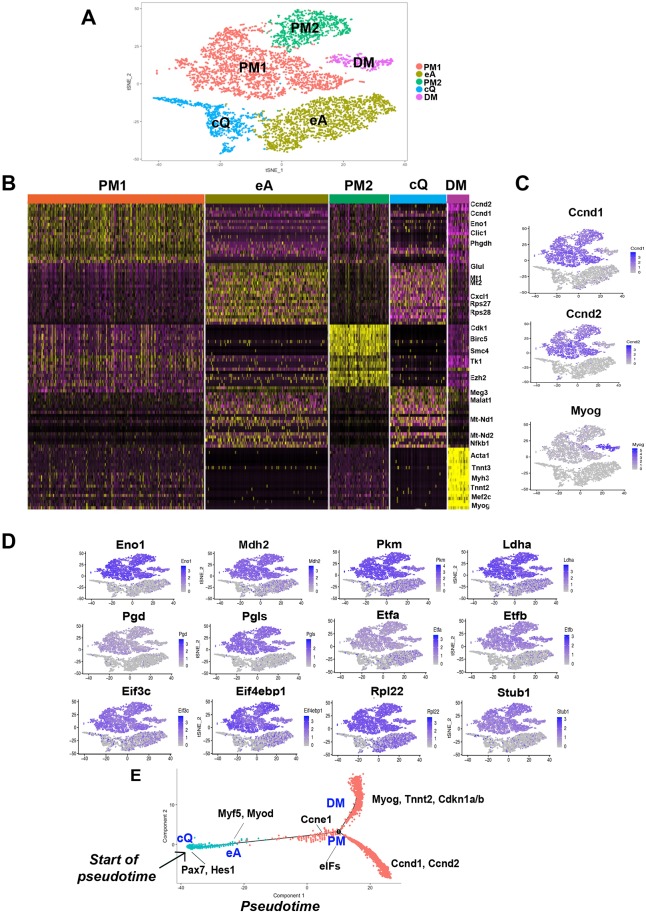
Fig. 3.
Clustering and pseudotemporal trajectories identify transcriptional dynamics of MuSCs and PMs. (A) Graph-based clustering of FACS-isolated MuSCs and PMs showing MuSCs close-to-quiescence (cQ), MuSCs early activation (eA), primary myoblasts cluster 1 (PM1) and cluster 2 (PM2), and differentiating myocytes (DM). (B) Heatmap of the top 20 most variably expressed genes between MuSC cQ, MuSC eA, PM1, PM2 and DM clusters. (C,D) Expression patterns of the cyclin genes Ccnd1, Ccnd2, and myogenin (C) and of glycolytic (Eno1, Pkm, Ldha), mitochondrial (Mdh2, Etfa, Etfb), pentose phosphate (Pgd, Pgls), translation initiation factors (Eif3c, Eif4ebp1), ribosomal (Rpl22) and ubiquitin-ligase (Stub1) genes (D). (E) Pseudotime single cell trajectory reconstructed by Monocle2 for MuSC cQ, MuSC eA and PMs. Pax7 and Hes1 are enriched in MuSC cQ and Myf5 and Myod in MuSC eA. PMs branch off in differentiating myocytes (DM) expressing myogenin (Myog), troponin (Tnnt2) and the cell cycle inhibitors Cdkn1a/b and in PMs that fail to enter differentiation and maintain expression of the cyclins Ccnd1 and Ccdn2.
To obtain temporal resolution of MuSCs and PMs, we employed the pseudotemporal ordering algorithm Monocle2 (Qiu et al., 2017). This algorithm permits single cells to be ordered informatically along trajectories reflecting their progression towards differentiation (Trapnell et al., 2014). Cells located in the initial phase of the trajectory (start of pseudotime) correspond to MuSC cQ, as indicated by expression of Pax7 and the Notch target Hes1 (Fig. 3E). Without an apparent hiatus separating them from the initial phase, cells located further on the trajectory initiate Myf5 transcription and upregulate Myod (MuSC eA). PMs were identified as a distinct group of cells expressing transcripts involved in cell cycle progression (cyclin E and G1/S checkpoint), chromatin remodeling complexes, and regulators of translational initiations (eukaryotic initiation factors, eIFs) (Fig. 3E, Table S2). PMs branched off in two distinct trajectories. One trajectory identified cells progressing towards differentiation, as indicated by activation of myogenin and the muscle structural transcript Tnnt2 (troponin T2) (Fig. 3E). Genes encoding cyclins were downregulated and cell cycle inhibitors Cdkn1a/b upregulated in cells located within this branch (Fig. 3E, DM). Another trajectory stemming out of the PM node identified cells that retained Ccnd1 and Ccnd2 without detectable myogenin expression (Fig. 3E). This branch corresponds to proliferating myoblasts that have not entered the differentiation program and may contain a self-renewal subpopulation of described ‘reserve cells’ (Qiu et al., 2017; Cacchiarelli et al., 2018).
Single cell trajectories of homeostatic, injured MuSCs, and PMs
To evaluate how the MuSC transcriptome is impacted by muscle injury, we generated and compared scRNA-seq datasets of homeostatic (uninjured) and regenerating MuSCs. Tibialis anterior (TA) and extensor digitorum longus (EDL) muscles of two 3-month-old C56BL/6J mice were injured with the myotoxic agent notexin and MuSCs were FACS-isolated 60 h after injury (Fig. 4A, Fig. S3B). After quality control, we retained 3550 MuSCs for downstream scRNA-seq analysis. On average, we detected 1336 expressed genes in each individual MuSC (248,914 mean reads per cell; Fig. S3C). The two samples were tested for similarity and merged for further analysis (Fig. S3D,E, Table S2 60 h Inj1 versus 60 Inj2 sheet). t-SNE analysis of injured MuSCs identified three clusters, MuSCs 60 h Cl1, Cl2 and Cl3, distinct from those of homeostatic MuSCs (Fig. 4A, Fig. S3G,H).
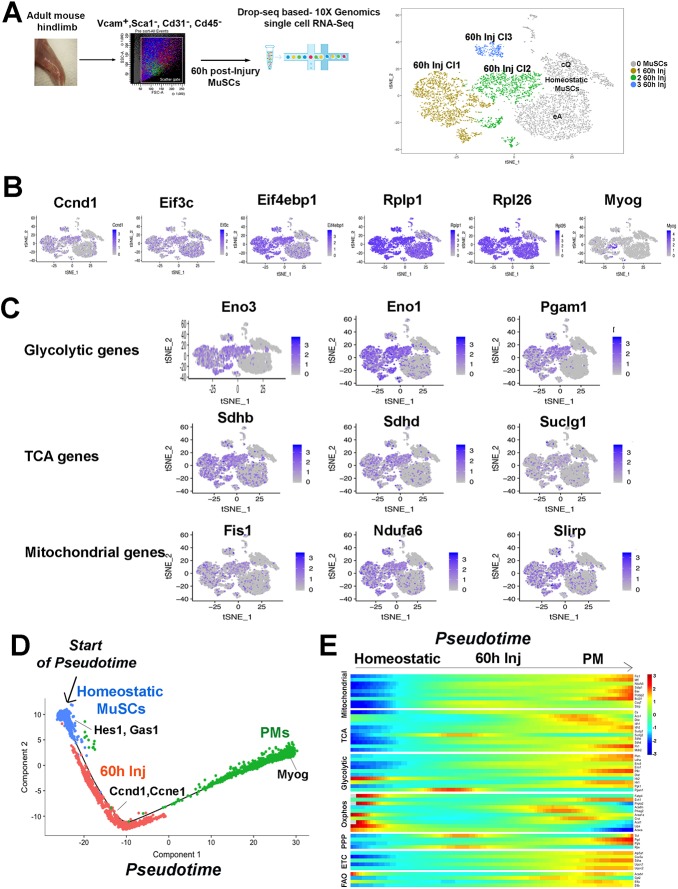
Fig. 4.
Clustering and single cell trajectories of homeostatic MuSCs, injured MuSCs, and PMs. (A) Scheme of MuSC FACS isolation 60 h after muscle injury, scRNA-seq, and graph-based clustering of FACS-isolated homeostatic and MuSCs 60 h after injury. Homeostatic MuSCs (in gray) are composed of two clusters, cQ and eA, and MuSCs 60 h of three clusters, 60 h Cl1, Cl2 and Cl3. (B) Expression pattern for Ccnd1, translation initiation factors (Eif3c, Eif4ebp1), ribosomal genes (Rplp1, Rpl26), and myogenin (Myog). (C) Expression pattern of glycolytic, TCA and mitochondrial genes. (D) Pseudotime single cell trajectory reconstructed by Monocle2 for homeostatic MuSCs, injured MuSCs (60 h Inj) and PMs. Transcripts enriched in distinct cell states are indicated. (E) Pseudotemporal heatmap showing gene expression dynamics for metabolic genes. Genes (row) are clustered and cells (column) are ordered according to pseudotime.
GO analysis revealed activation of common transcriptional programs in the three injured MuSC clusters (Fig. S3F, Table S2). Translational regulators were enriched in all injured MuSCs (Eif3c, Eif4ebp1), which had also activated Ccnd1 transcription (Fig. 4B, Fig. S3F, Table S2). Cells located in one of the injured MuSC clusters expressed myogenin and were enriched for the terms ‘muscle contraction’ and ‘myotube differentiation’, indicating that they had entered differentiation (Fig. 4B, Table S2). As noted in bulk transcriptome analyses of injured or culture-activated MuSCs (Pallafacchina et al., 2010; Liu et al., 2013; Ryall et al., 2015b; Pala et al., 2018), expression of genes involved in oxidation-reduction processes, glycolysis, tricarboxylic acid (TCA) cycle, mitochondrial genes, serine biosynthesis and pentose phosphate pathways was also enriched in injured MuSCs (Fig. 4C, Fig. S3F, Table S2). Among them, we found profilin 1 (Pfn1), deletion of which induces a switch from glycolysis to mitochondrial respiration in hematopoietic stem cells (Zheng et al., 2014), and Park7, encoding a redox-sensitive chaperone protecting neurons against oxidative stress and involved in early-onset Parkinson disease, deletion of which promotes glycolysis in skeletal muscle (Shi et al., 2015). Expression of genes known to regulate metabolic pathways and RNA metabolism (Rplp1, Rpl26) (Fig. 4B, Fig. S3F) and expression of nucleophosmin 1 (Npm1) (Fig. S3I), which promotes RNA metabolism and is essential for embryonic development (Colombo et al., 2005) (Fig. S3F), were also increased.
Pseudotemporal ordering aligned individual homeostatic, injured MuSCs, and PMs, along a linear trajectory. Homeostatic MuSCs were located at one end and PMs were found at the opposite end of this trajectory. Injured MuScs were temporally distinct from homeostatic MuSCs and extended towards PMs (Fig. 4D, Fig. S4A). Correct cell ordering was confirmed by plotting genes known to be expressed in the three different cell states (Fig. 4D). Next, we generated heatmaps of pseudotemporal expression patterns to visualize metabolic pathways comprehensively in homeostatic MuScs, injured MuSCs, and PMs (Fig. 4E, Fig. S4B). Expression of mitochondrial genes progressively increased from homeostatic through injured MuSCs to PMs, serving as a molecular correlate of the increased mitochondria observed in activated and cultured MuSCs (Tang and Rando, 2014; Ryall et al., 2015b). Concordantly, genes of the TCA cycle and the electron transport chain were increased in injured MuSCs and in PMs. With the exception of Hk2 (Pala et al., 2018), glycolysis and pentose phosphate pathways were also activated in injured MuSCs and in PMs (Fig. 4E, Table S2). As previously observed, expression of a subset of oxidative genes (Fabp4, Pnpla2, Acaca, Lipa, Acsl1) was elevated in homeostatic MuSCs and decreased in injured MuSCs and in PMs (Ryall et al., 2015b) (Fig. 4E).
Dynamics of metabolic pathway connectivity in homeostatic MuSCs, injured MuSCs and proliferating myoblasts
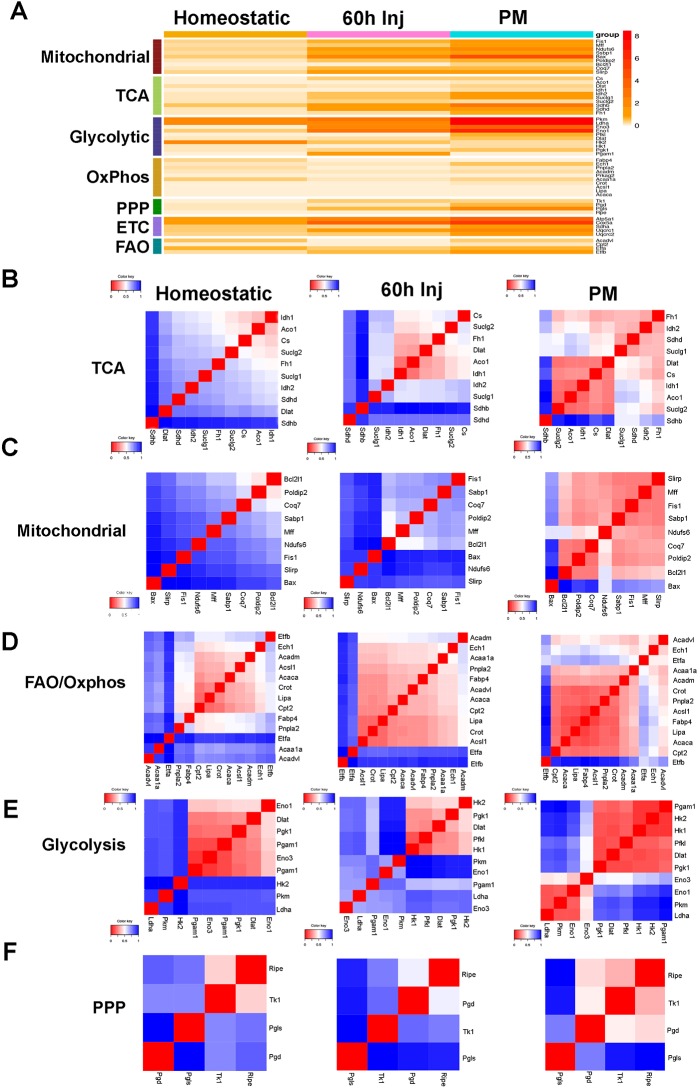
Analysis of average gene expression confirmed pseudotemporal expression patterns indicating that transcripts of metabolic enzymes were increased in both injured MuSCs and proliferating PMs compared with homeostatic MuSCs (Fig. 5A). To evaluate the transcriptional dynamics of different metabolic pathways in homeostatic MuSCs, injured MuSCs and PMs, we generated heatmaps of genetic interactions based on the use of pairwise matrices with Euclidean distance. In such matrices, gene proximity indicates co-expression whereas gene distance indicates absence of co-expression. Grouping genes into clusters based on the similarity between their expression profiles allows evaluation of the presence of functionally connected and related modules (Stuart et al., 2003; Nguyen and Lió, 2009). Connectivity mapping indicated that genes of the TCA cycle established an interacting module in homeostatic MuSCs. This module was stabilized, as revealed by increased gene interactions in injured MuSCs, and extended to encompass the majority of TCA genes in PMs (Fig. 5B). Mitochondrial, oxidative phosphorylation and fatty acid oxidation genes steadily increased their connectivity, both in strength and number, from homeostatic MuSCs to PMs (Fig. 5C,D). In a similar manner, electron transport chain (ETC) genes progressively increased their interaction (Fig. S4C). Glycolytic genes established a well-coordinated network in homeostatic MuSCs (Fig. 5E, left). In injured MuSCs, the connectivity of the glycolytic interaction map transitioned to a different conformation characterized by newly acquired intermediate gene interactions (∼0.75 on the color key) of Ldha with all the glycolytic genes and strengthened connectivity of Pkm (Fig. 5E, middle). Gene connectivity further increased in PMs (Fig. 5E, right). Interactions among members of the pentose phosphate pathway also progressively expanded and stabilized in injured MuSCs and PMs (Fig. 5F). Connectivity of genes involved in ribosomal biogenesis steadily increased from MuSCs to PMs and that of protein-folding genes underwent dynamic changes as well. In contrast, the network of genes for structural proteins remained relatively constant (Fig. S4C).
Fig. 5.
Metabolic pathways gene connectivity in homeostatic MuSCs, injured MuSCs, and proliferating myoblasts. (A) Heatmap of average expression for metabolic genes in homeostatic and injured MuSCs (60 h Inj), and in PMs. (B-F) Distance matrices indicating gene connectivity (highest connectivity, red; lowest connectivity, blue) of metabolic genes in homeostatic and injured MuSCs and in PMs.
This analysis revealed that although expression of metabolic pathway genes is concordantly orchestrated resulting in a progressive enrichment of all the energy-producing pathways from homeostatic and injured MuSCs to PMs, the dynamics and connectivity of genes within the individual metabolic pathways are distinct and identify discrete cell states.
DISCUSSION
We have employed scRNA-seq to identify different cell populations of adult mouse skeletal muscle with a special emphasis on MuSCs. FACS-isolated MuSCs could be identified with distinct transcriptional transition states. This phenomenon has been previously documented using bulk RNA-seq approaches (Machado et al., 2017; van Velthoven et al., 2017) and scRNA-seq of 21 and 235 MuSCs, respectively (van den Brink et al., 2017; Cho and Doles, 2017). Here, we extend those findings by analyzing thousands of MuSCs obtained from homeostatic and notexin-injured muscles and comparing the scRNA-seq datasets with those obtained from proliferating committed PMs. t-SNE analysis of FACS-isolated cells identified two MuSC clusters with overlapping but distinct transcriptomes. Indicating MuSC activation, one of the clusters contained cells with higher Myod and lower Pax7 transcript levels whereas the other cluster was composed of cells retaining a transcriptome closer to that of PFA-fixed MuSCs (Machado et al., 2017). Although freshly isolated MuSCs express Myod and Myf5 transcripts, their cognate proteins are not detected until cell activation (Zammit et al., 2002). This regulatory step is controlled by microRNA 489 (Cheung et al., 2012) and microRNA 31 (Crist et al., 2012), and by the RNA-binding proteins of the tristetraprolins (TTP) family, which destabilize Myod mRNA (Hausburg et al., 2015), and Staufen1, which binds to and prevents translation of Myod mRNA in quiescent MuSCs (de Morree et al., 2017). Our single cell western blot analysis of freshly FACS-isolated MuSCs indicated that the MyoD protein could be detected in only a small percentage of MuSCs (∼3%), suggesting that although Myod transcripts are increased during FACS isolation of MuSCs, they are not immediately translated.
By aligning the individual transcriptomes of MuSCs derived from homeostatic and injured muscles, and primary committed myoblasts along a pseudotemporal single trajectory, we could order them, in an unsupervised fashion, and obtain a dynamic description of the transcriptional events characterizing the progression of the individual cells towards differentiation. As expected, the transcriptomes of close-to-quiescence and early-activation clusters present in freshly isolated MuSCs partially overlapped but the existing differences were bioinformatically recognized, thus allowing the positioning of the two cell clusters in close yet distinct space-time positions along the trajectory. Sharp transcriptome differences between homeostatic and injured MuSCs resulted in injured MuSCs occupying a pseudotemporal position intermediate between homeostatic MuSCs and PMs. Alternate trajectories identified PMs that successfully entered terminal differentiation, as indicated by expression of myogenin, muscle structural genes, and cell cycle inhibitors, or that retained a transcriptome indicative of a committed yet undifferentiated state.
Several chaperones were enriched in the MuSC cQ cluster. Carbon dating experiments indicate an average age of 15.9 years for cells residing in human skeletal muscle (Spalding et al., 2005). Thus, chaperones may afford a quality control mechanism by maintaining appropriate protein folding (Hartl et al., 2011). A concomitant enriched expression of genes involved in ‘response to unfolded protein’ suggests tight proteostatic regulation in the MuSC cQ cluster (García-Prat et al., 2017). The MuSC eA cluster was enriched for ribosome biogenesis, RNA splicing, and translational initiation factors. These GO terms were also present in regenerative MuSCs and PMs. Thus, in addition to their post-transcriptional modifications (Zismanov et al., 2016), expression of translational initiation factors was increased in MuSCs and proliferating PMs. Besides serving as building blocks for muscle growth and repair, MuSCs cross-talk with their immediate cellular environment via extracellular vescicles containing RNA and proteins, mediating intercellular communication. During this process, MuSCs secrete exosomes containing miRNAs regulating extracellular matrix (ECM) remodeling (Fry et al., 2017). Coordinated activation of an extensive network of genes controlling extracellular exosomes biogenesis was observed in MuSC eA, regenerative MuSCs, and PMs supporting a potential role for MuSC-secreted vesicles in ECM remodeling that occurs during muscle repair. Interestingly, transcripts of Ccnd1 were detected in MuSCs cQ (Figs 2 and 3), suggesting a role unrelated to cell cycle regulation for cyclin D1.
Metabolic reprogramming is associated with distinct states of embryonic and somatic cells (Ryall et al., 2015a). In MuSCs, transcriptional activation of both glycolytic and mitochondrial genes occurs during cell activation and proliferation (Pallafacchina et al., 2010; Rocheteau et al., 2012; Liu et al., 2013; Rodgers et al., 2014; Ryall et al., 2015b; Pala et al., 2018) and interfering with either glycolysis or peroxisomal fatty acid oxidation bears functional consequences on cell activation and differentiation (Ryall et al., 2015b; Pala et al., 2018). We took advantage of scRNA-seq granularity to investigate metabolic reprogramming in MuSCs and PMs. Regenerating MuSCs are enriched for all the main energy-producing pathways, including glycolysis (Liu et al., 2013; Pala et al., 2018). Bioenergetics profiles of MuSCs freshly isolated from adult mice revealed a positive basal oxygen consumption rate (OCR)/extracellular acidification rate (ECAR) ratio (indicative of oxidative phosphorylation) (Pala et al., 2018), and cultured MuSCs displayed increased ECAR (indicative of glycolysis) compared with freshly isolated MuSCs (Ryall et al., 2015b). In addition, comparison of transcripts present in both FACS-isolated MuSCS and in vivo 4-thiouracil-labeled MuSC transcriptomes revealed an enrichment of transcripts for mitochondrial-associated functions (TCA, electron transport chain, oxidative reduction) (van Velthoven et al., 2017), suggesting oxidative phosphorylation in quiescent MuSCs (Rodgers et al., 2014; Ryall et al., 2015b). By analyzing gene networks based on pairwise matrices of Euclidean distance, we detected overall increase of all metabolic pathways investigated from homeostatic through injured MuSCs to PMs. In injured muscles, increased glycolysis may generate the glycolytic intermediates necessary for the production of new biomass and metabolites utilized by histone- and DNA-modifying enzymes whereas oxidative phosphorylation would generate the ATP required for MuSC proliferation (Ryall et al., 2015a; Pala et al., 2018). Indeed, metabolites generated by different metabolic pathways regulate the activity of chromatin-modifying enzymes known to control different aspects of MuSC biology (McKinnell et al., 2008; Juan et al., 2011; Kawabe et al., 2012; Liu et al., 2013; Ryall et al., 2015b; Boonsanay et al., 2016; Faralli et al., 2016; Scionti et al., 2017; Das et al., 2017; Tosic et al., 2018; Puri and Mercola, 2012). For instance, the oscillation of intermediary metabolites of the NAD biosynthetic pathways may be relevant for circadian gene expression in MuSCs and skeletal muscle (Nakahata et al., 2009; Ryall et al., 2015b; Solanas et al., 2017; Andrews et al., 2010; Lowe et al., 2018). Lastly, perturbations of specific metabolic pathways induce state changes in MuSCs (Ryall et al., 2015b; Pala et al., 2018), revealing a central role of metabolism in regulating cell state transitions. scRNA-seq has permitted the identification of cellular states and a granular definition of the individual cell transcriptional developmental trajectories accounting for the transition of MuSCs from quiescence to proliferation and differentiation. Additional studies will advance our understanding of the regulatory mechanisms governing these transitions at the individual cell level and provide further insights into how metabolic pathways are transcriptionally tuned.
MATERIALS AND METHODS
Mice and animal care
Mice were housed in a pathogen-free facility and all experiments were performed according to the National Institutes of Health (NIH) Animal Care and Use regulations.
Mononuclear cells and MuSC isolation by FACS from mouse tibialis anterior muscle
Hindlimb muscles from 3-month-old C56BL/6J wild-type mice were dissociated into single cells by enzymatic digestion and live cells were isolated by FACS. Skeletal MuSCs were sorted following described methods (Liu et al., 2015). Briefly, hindlimb muscles from 3-month-old adult wild-type mice were minced and digested with collagenase for 1 h and MuSCs were released from muscle fibers by further digesting the muscle slurry with collagenase/dispase for an additional 30 min. After filtering out the debris, cells were incubated with the following primary antibodies: biotin anti-mouse CD106 (anti-VCAM1, BioLegend 105704; 1:75), PE/Cy7 Streptavidin (BioLegend 405206; 1:75), Pacific Blue anti-mouse Ly-6A/E (anti- Sca1, BioLegend 108120; 1:75), APC anti-mouse CD31 (BioLegend 102510; 1:75) and APC anti-mouse CD45 (BioLegend 103112; 1:75). Satellite cells were sorted by gating VCAM1-positive, Pacific Blue-labeled Sca1-negative, and APC-labeled CD31/CD45-negative cells. SYTOX Green (ThermoFisher Scientific S7020; 1:30,000) was used as a counterstain.
PM culture conditions
PMs derived from the hindlimbs of 3-month-old C56BL/6J wild-type mice were cultured for four passages in HAM's F10 (Invitrogen) supplemented with 2.5 ng/µl of bFGF (Invitrogen), 10% horse serum (Invitrogen), and penicillin and streptomycin (Invitrogen).
Muscle injury
C57/BL6 mice (3-6 months old) were anesthetized using isoflurane (1.5-2.5%) via a vaporizer with anesthetic gas-scavenging equipment. A small incision (3-5 mm) was made on the right-side leg to expose the TA muscle. A volume of 100 µl notexin (2 µM, Accurate Chemical & Scientific) was evenly delivered into TA and EDL muscles using a 29 gauge insulin needle. The incision was closed by wound clips. Buprenorphine (100 µl 0.1 mg/kg diluted in saline) was delivered subcutaneously at the completion of surgery to alleviate the pain. The mice were put on a separate clean bedding cage and were monitored until they became fully awake, then moved back to their original cages.
Single cell sequencing library construction using 10x Genomics Chromium Controller
Cells were collected, washed once with 1×PBS and re-suspended in PBS with 0.04% bovine serum albumin. Cellular suspensions were loaded on a Chromium Instrument (10x Genomics) to generate single cell GEMs. Single cell RNA-seq libraries were prepared using a Chromium Single Cell 3′ Library & Gel Bead Kit v2 (P/N 120737, 10x Genomics). GEM-RT was performed in a C1000 Touch Thermal cycler with 96-Deep Well Reaction Module (Bio-Rad; P/N 1851197): 53°C for 45 min, 85°C for 5 min; held at 4°C. Following retrotranscription, GEMs were broken and the single-strand cDNA was purified with DynaBeads MyOne Silane Beads (Thermo Fisher Scientific; P/N 37002D). cDNA was amplified using the C1000 Touch Thermal cycler with 96-Deep Well Reaction Module: 98°C for 3 min; cycled 12 times: 98°C for 15 s, 67°C for 20 s, and 72°C for 1 min; 72°C for 1 min; held at 4°C. Amplified cDNA product was purified with the SPRIselect Reagent Kit (0.6× SPRI). Indexed sequencing libraries were constructed using the reagents in the Chromium Single Cell 3′ Library & Gel Bead Kit v2, following these steps: (1) end repair and A-tailing; (2) adaptor ligation; (3) post-fragmentation, end repair and A-tailing double size selection cleanup with SPRI-select beads; (4) sample index PCR and cleanup. The barcode sequencing libraries were diluted at 3 nM and sequenced on an Illumina HiSeq3000 using the following read length: 26bp for Read1, 8 bp for I7 Index and 98 bp for Read2.
Data processing and clustering
Sequences from the Chromium platform were de-multiplexed and aligned using CellRanger ver. 2.2.0 from 10x Genomics with default parameters (https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger). For clustering, filtering, variable gene selection and dimensionality reduction were performed using Seurat ver.2.3.4 (Butler et al., 2018) according to the following workflow: (1) Cells with fewer than 200 detected genes or in which a high percentage of UMIs mapped to mitochondrial genes were excluded from further analysis. (2) Cells with a lower percentage (0.06-0.1%) of UMIs mapped to mitochondrial genes were retained for further analysis. (3) The UMI counts per million were log-normalized for each cell using the natural logarithm (Butler et al., 2018). (4) Variable genes were selected using thresholds (0.05-0.1) after calculating the binned values from log average expression and dispersion for each gene (Butler et al., 2018). (5) The expression level of highly variable genes was scaled along each gene, and we regressed out cell-cell variation in gene expression by the number of detected molecules (5000-8000), and mitochondrial gene expression. (6) The scaled data were projected onto a low-dimensional subspace of principal component analysis (PCA) using dimensional reduction. The number of PCA were decided through the assessment of statistical plots (Butler et al., 2018). (7) Cells were clustered using a graph-based clustering approach optimized by the Louvain algorithm with resolution parameters and visualized using two-dimensional tSNE. (8) To define the clusters’ characteristics, we identified markers for all clusters with low minimum percentage of genes detected and reassigned the names of the clusters on the basis of the distribution of the specific markers. In addition, using the top n markers based on log-fold changing values, we generated an expression heatmap for given cells and genes in the clusters.
Single cell trajectory analysis
Trajectory analysis of scRNA-seq was performed using Monocle v.2.8.0 (Qiu et al., 2017). First, datasets containing information of cell differentiation were created using cell IDs selected through Seurat package. Then, we performed dimensional reduction using the DDRTree method to visualize the dataset, ordered the cells by global gene expression levels, and visualized the trajectory of the dataset. To illustrate the trends of gene expression across pseudotime, we generated heatmaps to represent smooth expression curves of the genes.
Patterns of gene interactions during cell differentiation
To study gene interactions during cell differentiation, we used distance matrices of gene expressions. After selecting genes related to metabolic pathways, we collected the values of their gene expressions in each state and calculated the similarities of the gene expressions using Pearson’s correlation coefficient to generate the corresponding heatmaps (Stuart et al., 2003).
Single cell western blot
Freshly FACS-isolated MuSCs (1×105) were resuspended in 1 ml of loading buffer (ProteinSimple) and loaded on small scWest chip run (Milo System, ProteinSimple). MuSCs were settled for 15 min, followed by 5 s lysis and 1 min separation (Milo System, ProteinSimple). scWest chip was incubated with sheep anti-histone H3 (Novus Biologicals, NB100-747; 1:40) and donkey anti-rabbit IgG Alexa 555 (Invitrogen, A-31527; 1:40). Mouse anti-MyoD (Novus Biologicals, NBP2-32883) (100 µg/ml) was revealed by donkey anti-mouse IgG Alexa 647 (Invitrogen, A-31571; 1:20). scWest chip was scanned using a microarray scanner and analysis was performed with Scout Software (Milo System, ProteinSimple).
Supplementary Material
Acknowledgements
We thank Berendia Jackson, Dr Khooshbu Shah and the ProteinSimple team for assistance with single cell western blot analysis and Jim Simone from the Flow Cytometry Section, NIAMS, for assistance with FACS. This study utilized the high-performance computational capabilities of the Helix System at the NIH, Bethesda, MD, USA (https://hpc.nih.gov/systems/helix.html/).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.D., A.H.J., V.S.; Methodology: S.D., A.H.J., K.-D.K., F.N., G.G.-C., X.F., V.S.; Software: K.-D.K.; Validation: A.H.J., K.-D.K., F.N., J.P., V.S.; Formal analysis: S.D., A.H.J., K.-D.K., F.N., V.S.; Investigation: S.D., A.H.J., F.N., G.G.-C., X.F.; Resources: V.S.; Data curation: S.D., A.H.J., K.-D.K., X.F., V.S.; Writing - original draft: S.D., V.S.; Visualization: S.D., K.-D.K.; Supervision: V.S.; Project administration: V.S.; Funding acquisition: V.S.
Funding
This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases Intramural Research Program (AR041126 and AR041164). Deposited in PMC for release after 12 months.
Data availability
Datasets generated in this study are available in the Gene Expression Omnibus repository under accession number GSE126834.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.174177.supplemental
References
- Aguilar C. A., Pop R., Shcherbina A., Watts A., Matheny R. W. Jr, Cacchiarelli D., Han W. M., Shin E., Nakhai S. A., Jang Y. C. et al. (2016). Transcriptional and chromatin dynamics of muscle regeneration after severe trauma. Stem Cell Rep. 7, 983-997. 10.1016/j.stemcr.2016.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Martin S., Rochat A., Mademtzoglou D., Morais J., de Reyniès A., Auradé F., Chang T. H.-T., Zammit P. S. and Relaix F. (2016). Gene expression profiling of muscle stem cells identifies novel regulators of postnatal myogenesis. Front. Cell Dev. Biol. 4, 58 10.3389/fcell.2016.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews J. L., Zhang X., McCarthy J. J., McDearmon E. L., Hornberger T. A., Russell B., Campbell K. S., Arbogast S., Reid M. B., Walker J. R. et al. (2010). CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc. Natl. Acad. Sci. USA 107, 19090-19095. 10.1073/pnas.1014523107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp J. R., Heslop L., Yu D. S. W., Tajbakhsh S., Kelly R. G., Wernig A., Buckingham M. E., Partridge T. A. and Zammit P. S. (2000). Expression of CD34 and Myf5 defines the majority of quiescent adult skeletal muscle satellite cells. J. Cell Biol. 151, 1221-1234. 10.1083/jcb.151.6.1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernet J. D., Doles J. D., Hall J. K., Kelly Tanaka K., Carter T. A. and Olwin B. B. (2014). p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nat. Med. 20, 265-271. 10.1038/nm.3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonsanay V., Zhang T., Georgieva A., Kostin S., Qi H., Yuan X., Zhou Y. and Braun T. (2016). Regulation of skeletal muscle stem cell quiescence by Suv4-20h1-Dependent facultative heterochromatin formation. Cell Stem Cell 18, 229-242. 10.1016/j.stem.2015.11.002 [DOI] [PubMed] [Google Scholar]
- Brack A. S. and Rando T. A. (2007). Intrinsic changes and extrinsic influences of myogenic stem cell function during aging. Stem Cell Rev. 3, 226-237. 10.1007/s12015-007-9000-2 [DOI] [PubMed] [Google Scholar]
- Brack A. S. and Rando T. A. (2012). Tissue-specific stem cells: lessons from the skeletal muscle satellite cell. Cell Stem Cell 10, 504-514. 10.1016/j.stem.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brack A. S., Conboy M. J., Roy S., Lee M., Kuo C. J., Keller C. and Rando T. A. (2007). Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science 317, 807-810. 10.1126/science.1144090 [DOI] [PubMed] [Google Scholar]
- Buckingham M. and Relaix F. (2007). The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu. Rev. Cell Dev. Biol. 23, 645-673. 10.1146/annurev.cellbio.23.090506.123438 [DOI] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E. and Satija R. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411-420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacchiarelli D., Qiu X., Srivatsan S., Manfredi A., Ziller M., Overbey E., Grimaldi A., Grimsby J., Pokharel P., Livak K. J. et al. (2018). Aligning single-cell developmental and reprogramming trajectories identifies molecular determinants of myogenic reprogramming outcome. Cell Syst. 7, 258-268.e253. 10.1016/j.cels.2018.07.006 [DOI] [PubMed] [Google Scholar]
- Chakkalakal J. V., Jones K. M., Basson M. A. and Brack A. S. (2012). The aged niche disrupts muscle stem cell quiescence. Nature 490, 355-360. 10.1038/nature11438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang N. C., Sincennes M. C., Chevalier F. P., Brun C. E., Lacaria M., Segales J., Munoz-Canoves P., Ming H. and Rudnicki M. A. (2018). The dystrophin glycoprotein complex regulates the epigenetic activation of muscle stem cell commitment. Cell Stem Cell 22, 755-768.e756. 10.1016/j.stem.2018.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chargé S. B. and Rudnicki M. A. (2004). Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 84, 209-238. 10.1152/physrev.00019.2003 [DOI] [PubMed] [Google Scholar]
- Cheung T. H., Quach N. L., Charville G. W., Liu L., Park L., Edalati A., Yoo B., Hoang P. and Rando T. A. (2012). Maintenance of muscle stem-cell quiescence by microRNA-489. Nature 482, 524-528. 10.1038/nature10834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho D. S. and Doles J. D. (2017). Single cell transcriptome analysis of muscle satellite cells reveals widespread transcriptional heterogeneity. Gene 636, 54-63. 10.1016/j.gene.2017.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C. A., Olsen I., Zammit P. S., Heslop L., Petrie A., Partridge T. A. and Morgan J. E. (2005). Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell 122, 289-301. 10.1016/j.cell.2005.05.010 [DOI] [PubMed] [Google Scholar]
- Colombo E., Bonetti P., Lazzerini Denchi E., Martinelli P., Zamponi R., Marine J.-C., Helin K., Falini B. and Pelicci P. G. (2005). Nucleophosmin is required for DNA integrity and p19Arf protein stability. Mol. Cell. Biol. 25, 8874-8886. 10.1128/MCB.25.20.8874-8886.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai G. and Tajbakhsh S. (2014). Molecular and cellular regulation of skeletal myogenesis. Curr. Top. Dev. Biol. 110, 1-73. 10.1016/B978-0-12-405943-6.00001-4 [DOI] [PubMed] [Google Scholar]
- Cosgrove B. D., Gilbert P. M., Porpiglia E., Mourkioti F., Lee S. P., Corbel S. Y., Llewellyn M. E., Delp S. L. and Blau H. M. (2014). Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nat. Med. 20, 255-264. 10.1038/nm.3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crist C. G., Montarras D. and Buckingham M. (2012). Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell 11, 118-126. 10.1016/j.stem.2012.03.011 [DOI] [PubMed] [Google Scholar]
- Das S., Morvan F., Morozzi G., Jourde B., Minetti G. C., Kahle P., Rivet H., Brebbia P., Toussaint G., Glass D. J. et al. (2017). ATP citrate lyase regulates myofiber differentiation and increases regeneration by altering histone acetylation. Cell Rep. 21, 3003-3011. 10.1016/j.celrep.2017.11.038 [DOI] [PubMed] [Google Scholar]
- de Morree A., van Velthoven C. T. J., Gan Q., Salvi J. S., Klein J. D. D., Akimenko I., Quarta M., Biressi S. and Rando T. A. (2017). Staufen1 inhibits MyoD translation to actively maintain muscle stem cell quiescence. Proc. Natl. Acad. Sci. USA 114, E8996-E9005. 10.1073/pnas.1708725114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont N. A., Wang Y. X., von Maltzahn J., Pasut A., Bentzinger C. F., Brun C. E. and Rudnicki M. A. (2015). Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat. Med. 21, 1455-1463. 10.1038/nm.3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faralli H., Wang C., Nakka K., Benyoucef A., Sebastian S., Zhuang L., Chu A., Palii C. G., Liu C., Camellato B. et al. (2016). UTX demethylase activity is required for satellite cell-mediated muscle regeneration. J. Clin. Invest. 126, 1555-1565. 10.1172/JCI83239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige P., Brun C. E., Ritso M. and Rudnicki M. A. (2018). Orienting muscle stem cells for regeneration in homeostasis, aging, and disease. Cell Stem Cell 23, 653-664. 10.1016/j.stem.2018.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry C. S., Kirby T. J., Kosmac K., McCarthy J. J. and Peterson C. A. (2017). Myogenic progenitor cells control extracellular matrix production by fibroblasts during skeletal muscle hypertrophy. Cell Stem Cell 20, 56-69. 10.1016/j.stem.2016.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Prat L., Martínez-Vicente M., Perdiguero E., Ortet L., Rodríguez-Ubreva J., Rebollo E., Ruiz-Bonilla V., Gutarra S., Ballestar E., Serrano A. L. et al. (2016). Autophagy maintains stemness by preventing senescence. Nature 529, 37-42. 10.1038/nature16187 [DOI] [PubMed] [Google Scholar]
- García-Prat L., Sousa-Victor P. and Muñoz-Cánoves P. (2017). Proteostatic and metabolic control of stemness. Cell Stem Cell 20, 593-608. 10.1016/j.stem.2017.04.011 [DOI] [PubMed] [Google Scholar]
- Giordani L., He G. J., Negroni E., Sakai H., Law J. Y. C., Siu M. M., Wan R., Tajbakhsh S., Cheung T. H. and Le Grand F (2018). High-dimensional single-cell cartography reveals novel skeletal muscle resident cell populations. bioRxiv doi:10.1101/304683 10.1101/304683 [DOI] [PubMed] [Google Scholar]
- Hartl F. U., Bracher A. and Hayer-Hartl M. (2011). Molecular chaperones in protein folding and proteostasis. Nature 475, 324-332. 10.1038/nature10317 [DOI] [PubMed] [Google Scholar]
- Hausburg M. A., Doles J. D., Clement S. L., Cadwallader A. B., Hall M. N., Blackshear P. J., Lykke-Andersen J. and Olwin B. B. (2015). Post-transcriptional regulation of satellite cell quiescence by TTP-mediated mRNA decay. eLife 4, e03390 10.7554/eLife.03390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen I., Heymsfield S. B., Wang Z. M. and Ross R. (2000). Skeletal muscle mass and distribution in 468 men and women aged 18-88 yr. J. Appl. Physiol. 89, 81-88. 10.1152/jappl.2000.89.1.81 [DOI] [PubMed] [Google Scholar]
- Juan A. H., Derfoul A., Feng X., Ryall J. G., Dell'Orso S., Pasut A., Zare H., Simone J. M., Rudnicki M. A. and Sartorelli V. (2011). Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev. 25, 789-794. 10.1101/gad.2027911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe Y., Wang Y. X., McKinnell I. W., Bedford M. T. and Rudnicki M. A. (2012). Carm1 regulates Pax7 transcriptional activity through MLL1/2 recruitment during asymmetric satellite stem cell divisions. Cell Stem Cell 11, 333-345. 10.1016/j.stem.2012.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S., Kuroda K., Le Grand F. and Rudnicki M. A. (2007). Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell 129, 999-1010. 10.1016/j.cell.2007.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Cheung T. H., Charville G. W., Hurgo B. M. C., Leavitt T., Shih J., Brunet A. and Rando T. A. (2013). Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 4, 189-204. 10.1016/j.celrep.2013.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Cheung T. H., Charville G. W. and Rando T. A. (2015). Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting. Nat. Protoc. 10, 1612-1624. 10.1038/nprot.2015.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe M., Lage J., Paatela E., Munson D., Hostager R., Yuan C., Katoku-Kikyo N., Ruiz-Estevez M., Asakura Y., Staats J. et al. (2018). Cry2 is critical for circadian regulation of myogenic differentiation by Bclaf1-mediated mRNA stabilization of Cyclin D1 and Tmem176b. Cell Rep. 22, 2118-2132. 10.1016/j.celrep.2018.01.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukjanenko L., Jung M. J., Hegde N., Perruisseau-Carrier C., Migliavacca E., Rozo M., Karaz S., Jacot G., Schmidt M., Li L. et al. (2016). Loss of fibronectin from the aged stem cell niche affects the regenerative capacity of skeletal muscle in mice. Nat. Med. 22, 897-905. 10.1038/nm.4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado L., Esteves de Lima J., Fabre O., Proux C., Legendre R., Szegedi A., Varet H., Ingerslev L. R., Barres R., Relaix F. et al. (2017). In situ fixation redefines quiescence and early activation of skeletal muscle stem cells. Cell Rep. 21, 1982-1993. 10.1016/j.celrep.2017.10.080 [DOI] [PubMed] [Google Scholar]
- McKinnell I. W., Ishibashi J., Le Grand F., Punch V. G., Addicks G. C., Greenblatt J. F., Dilworth F. J. and Rudnicki M. A. (2008). Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat. Cell Biol. 10, 77-84. 10.1038/ncb1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montarras D., Morgan J., Collins C., Relaix F., Zaffran S., Cumano A., Partridge T. and Buckingham M. (2005). Direct isolation of satellite cells for skeletal muscle regeneration. Science 309, 2064-2067. 10.1126/science.1114758 [DOI] [PubMed] [Google Scholar]
- Nakahata Y., Sahar S., Astarita G., Kaluzova M. and Sassone-Corsi P. (2009). Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324, 654-657. 10.1126/science.1170803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V.-A. and Lió P. (2009). Measuring similarity between gene expression profiles: a Bayesian approach. BMC Genomics 10 Suppl. 3, S14 10.1186/1471-2164-10-S3-S14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oustanina S., Hause G. and Braun T. (2004). Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J. 23, 3430-3439. 10.1038/sj.emboj.7600346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pala F., Di Girolamo D., Mella S., Yennek S., Chatre L., Ricchetti M. and Tajbakhsh S. (2018). Distinct metabolic states govern skeletal muscle stem cell fates during prenatal and postnatal myogenesis. J. Cell Sci. 131, jcs212977 10.1242/jcs.212977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallafacchina G., François S., Regnault B., Czarny B., Dive V., Cumano A., Montarras D. and Buckingham M. (2010). An adult tissue-specific stem cell in its niche: a gene profiling analysis of in vivo quiescent and activated muscle satellite cells. Stem Cell Res. 4, 77-91. 10.1016/j.scr.2009.10.003 [DOI] [PubMed] [Google Scholar]
- Price F. D., von Maltzahn J., Bentzinger C. F., Dumont N. A., Yin H., Chang N. C., Wilson D. H., Frenette J. and Rudnicki M. A. (2014). Inhibition of JAK-STAT signaling stimulates adult satellite cell function. Nat. Med. 20, 1174-1181. 10.1038/nm.3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri P. L. and Mercola M. (2012). BAF60 A, B, and Cs of muscle determination and renewal. Genes Dev. 26, 2673-2683. 10.1101/gad.207415.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H. A. and Trapnell C. (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979-982. 10.1038/nmeth.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocheteau P., Gayraud-Morel B., Siegl-Cachedenier I., Blasco M. A. and Tajbakhsh S. (2012). A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell 148, 112-125. 10.1016/j.cell.2011.11.049 [DOI] [PubMed] [Google Scholar]
- Rodgers J. T., King K. Y., Brett J. O., Cromie M. J., Charville G. W., Maguire K. K., Brunson C., Mastey N., Liu L., Tsai C. R. et al. (2014). mTORC1 controls the adaptive transition of quiescent stem cells from G0 to G(Alert). Nature 510, 393-396. 10.1038/nature13255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryall J. G., Cliff T., Dalton S. and Sartorelli V. (2015a). Metabolic reprogramming of stem cell epigenetics. Cell Stem Cell 17, 651-662. 10.1016/j.stem.2015.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryall J. G., Dell'Orso S., Derfoul A., Juan A., Zare H., Feng X., Clermont D., Koulnis M., Gutierrez-Cruz G., Fulco M. et al. (2015b). The NAD(+)-dependent SIRT1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells. Cell Stem Cell 16, 171-183. 10.1016/j.stem.2014.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco A. and Puri P. L. (2015). Regulation of muscle satellite cell function in tissue homeostasis and aging. Cell Stem Cell 16, 585-587. 10.1016/j.stem.2015.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco A., Doyonnas R., Kraft P., Vitorovic S. and Blau H. M. (2008). Self-renewal and expansion of single transplanted muscle stem cells. Nature 456, 502-506. 10.1038/nature07384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R., Farrell J. A., Gennert D., Schier A. F. and Regev A. (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495-502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scionti I., Hayashi S., Mouradian S., Girard E., Esteves de Lima J., Morel V., Simonet T., Wurmser M., Maire P., Ancelin K. et al. (2017). LSD1 controls timely MyoD expression via MyoD core enhancer transcription. Cell Rep. 18, 1996-2006. 10.1016/j.celrep.2017.01.078 [DOI] [PubMed] [Google Scholar]
- Seale P., Sabourin L. A., Girgis-Gabardo A., Mansouri A., Gruss P. and Rudnicki M. A. (2000). Pax7 is required for the specification of myogenic satellite cells. Cell 102, 777-786. 10.1016/S0092-8674(00)00066-0 [DOI] [PubMed] [Google Scholar]
- Sherwood R. I., Christensen J. L., Conboy I. M., Conboy M. J., Rando T. A., Weissman I. L. and Wagers A. J. (2004). Isolation of adult mouse myogenic progenitors: functional heterogeneity of cells within and engrafting skeletal muscle. Cell 119, 543-554. 10.1016/j.cell.2004.10.021 [DOI] [PubMed] [Google Scholar]
- Shi S. Y., Lu S. Y., Sivasubramaniyam T., Revelo X. S., Cai E. P., Luk C. T., Schroer S. A., Patel P., Kim R. H., Bombardier E. et al. (2015). DJ-1 links muscle ROS production with metabolic reprogramming and systemic energy homeostasis in mice. Nat. Commun. 6, 7415 10.1038/ncomms8415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sincennes M.-C., Brun C. E. and Rudnicki M. A. (2016). Concise review: epigenetic regulation of myogenesis in health and disease. Stem Cells Transl. Med. 5, 282-290. 10.5966/sctm.2015-0266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solanas G., Peixoto F. O., Perdiguero E., Jardi M., Ruiz-Bonilla V., Datta D., Symeonidi A., Castellanos A., Welz P. S., Caballero J. M. et al. (2017). Aged stem cells reprogram their daily rhythmic functions to adapt to stress. Cell 170, 678-692.e620. 10.1016/j.cell.2017.07.035 [DOI] [PubMed] [Google Scholar]
- Sousa-Victor P., Gutarra S., García-Prat L., Rodriguez-Ubreva J., Ortet L., Ruiz-Bonilla V., Jardi M., Ballestar E., Gonzalez S., Serrano A. L. et al. (2014). Geriatric muscle stem cells switch reversible quiescence into senescence. Nature 506, 316-321. 10.1038/nature13013 [DOI] [PubMed] [Google Scholar]
- Spalding K. L., Bhardwaj R. D., Buchholz B. A., Druid H. and Frisen J. (2005). Retrospective birth dating of cells in humans. Cell 122, 133-143. 10.1016/j.cell.2005.04.028 [DOI] [PubMed] [Google Scholar]
- Stuart J. M., Segal E., Koller D. and Kim S. K. (2003). A gene-coexpression network for global discovery of conserved genetic modules. Science 302, 249-255. 10.1126/science.1087447 [DOI] [PubMed] [Google Scholar]
- Tabula Muris Consortium. et al. (2018). Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367-372. 10.1038/s41586-018-0590-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A. H. and Rando T. A. (2014). Induction of autophagy supports the bioenergetic demands of quiescent muscle stem cell activation. EMBO J. 33, 2782-2797. 10.15252/embj.201488278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney M. T., Aydogdu T., Sala D., Malecova B., Gatto S., Puri P. L., Latella L. and Sacco A. (2014). STAT3 signaling controls satellite cell expansion and skeletal muscle repair. Nat. Med. 20, 1182-1186. 10.1038/nm.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosic M., Allen A., Willmann D., Lepper C., Kim J., Duteil D. and Schule R. (2018). Lsd1 regulates skeletal muscle regeneration and directs the fate of satellite cells. Nat. Commun. 9, 366 10.1038/s41467-017-02740-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N. J., Livak K. J., Mikkelsen T. S. and Rinn J. L. (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381-386. 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink S. C., Sage F., Vertesy A., Spanjaard B., Peterson-Maduro J., Baron C. S., Robin C. and van Oudenaarden A. (2017). Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods 14, 935-936. 10.1038/nmeth.4437 [DOI] [PubMed] [Google Scholar]
- van Velthoven C. T. J., de Morree A., Egner I. M., Brett J. O. and Rando T. A. (2017). Transcriptional profiling of quiescent muscle stem cells in vivo. Cell Rep. 21, 1994-2004. 10.1016/j.celrep.2017.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma M., Asakura Y., Murakonda B. S. R., Pengo T., Latroche C., Chazaud B., McLoon L. K. and Asakura A. (2018). Muscle satellite cell cross-talk with a vascular niche maintains quiescence via VEGF and notch signaling. Cell Stem Cell 23, 530-543.e539. 10.1016/j.stem.2018.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. B., Biérinx A.-S., Gnocchi V. F. and Zammit P. S. (2010). Dynamics of muscle fibre growth during postnatal mouse development. BMC Dev. Biol. 10, 21 10.1186/1471-213X-10-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yennek S., Burute M., Théry M. and Tajbakhsh S. (2014). Cell adhesion geometry regulates non-random DNA segregation and asymmetric cell fates in mouse skeletal muscle stem cells. Cell Rep. 7, 961-970. 10.1016/j.celrep.2014.04.016 [DOI] [PubMed] [Google Scholar]
- Zammit P. S., Heslop L., Hudon V., Rosenblatt J. D., Tajbakhsh S., Buckingham M. E., Beauchamp J. R. and Partridge T. A. (2002). Kinetics of myoblast proliferation show that resident satellite cells are competent to fully regenerate skeletal muscle fibers. Exp. Cell Res. 281, 39-49. 10.1006/excr.2002.5653 [DOI] [PubMed] [Google Scholar]
- Zhang X., Li T., Liu F., Chen Y., Yao J., Li Z., Huang Y. and Wang J. (2019). Comparative analysis of droplet-based ultra-high-throughput single-cell RNA-seq systems. Mol. Cell 73, 130-142.e5. 10.1016/j.molcel.2018.10.020 [DOI] [PubMed] [Google Scholar]
- Zheng J., Lu Z., Kocabas F., Bottcher R. T., Costell M., Kang X., Liu X., Deberardinis R. J., Wang Q., Chen G.-Q. et al. (2014). Profilin 1 is essential for retention and metabolism of mouse hematopoietic stem cells in bone marrow. Blood 123, 992-1001. 10.1182/blood-2013-04-498469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zismanov V., Chichkov V., Colangelo V., Jamet S., Wang S., Syme A., Koromilas A. E. and Crist C. (2016). Phosphorylation of eIF2alpha is a translational control mechanism regulating muscle stem cell quiescence and self-renewal. Cell Stem Cell 18, 79-90. 10.1016/j.stem.2015.09.020 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.