ABSTRACT
Epigenetic resetting in germ cells during development de-represses transposable elements (TEs). piRNAs protect fetal germ cells by targeted mRNA destruction and deposition of repressive epigenetic marks. Here, we provide the first evidence for an active piRNA pathway and TE repression in germ cells of human fetal testis. We identify pre-pachytene piRNAs with features of secondary amplification that map most abundantly to the long interspersed element type 1 (L1) family of TEs. L1-ORF1p expression is heterogeneous in fetal germ cells, peaks at mid-gestation and declines concomitantly with increases in piRNAs, nuclear localization of HIWI2 and an increase in H3K9me3. Surprisingly, the same cells with accumulation of L1-ORF1p display highest levels of HIWI2 and H3K9me3. Conversely, the earliest germ cells with high levels of L1-ORF1p express low levels of the chaperone HSP90α. We propose that a subset of germ cells resists L1 expression, whereas L1-expressing germ cells activate the repression pathway that leads to epigenetic silencing of L1 via H3K9me3.
KEY WORDS: LINE-1, piRNA, PIWI, Transposon, Human fetal germ cells, Epigenetic reprogramming
Summary: Expression of L1 transposons in germ cells of the human fetal testis is developmentally regulated and heterogeneous, and corresponds with upregulation of PIWI/piRNA repression machinery and accumulation of H3K9me3.
INTRODUCTION
Tasked with the vital role of species propagation, germ cells must be able to faithfully transmit heritable units of information between generations. A potential risk to genome integrity during this transmission arises in the developing embryo as the primordial germ cells (PGCs) undergo epigenetic reprogramming, characterized by both DNA demethylation and global resetting of histone marks (Gkountela et al., 2015; Guo et al., 2015; Tang et al., 2015, 2016). This rearrangement of the epigenetic landscape primes germ cells for meiotic entry in fetal ovaries, and mitotic arrest and differentiation in the fetal testis (Messerschmidt et al., 2014), yet comes at the risk of reactivating transposable elements (TEs), which are normally silenced by repressive epigenetic marks. The greatest threat comes from retrotransposons, which comprise ∼40% of the human genome (Kazazian and Moran, 2017). Although most TEs are mutated and inactive, some remain capable of retrotransposition in the human genome, mostly stemming from the evolutionarily young long interspersed element type 1 (L1), SVA and Alu families (Kazazian and Moran, 2017). L1-mediated integration of active TEs at random genomic sites is potentially dangerous, and has been associated with over 100 disease mutations in humans (Hancks and Kazazian, 2016). Their activity in humans is high, with new L1 and Alu integrations observed in ∼1/100 and 1/20 human births, respectively (Hancks and Kazazian, 2012), although not as high as other mammalian species such as mice, where new L1 insertions occur in 1/8 births (Richardson et al., 2017).
In germ cells, the activity of TEs is mollified by an evolutionarily conserved genome defense system made up of PIWI proteins and PIWI-interacting RNAs (piRNAs) collectively called the PIWI-piRNA pathway. piRNAs that are 26-32 nt complex with PIWI-family proteins to directly target retrotransposons for mRNA degradation in the cytoplasm, while also leading to nuclear silencing at endogenous genomic loci through the deposition of DNA methylation and histone modifications (Iwasaki et al., 2015; Ernst et al., 2017). The PIWI-piRNA pathway is required for male fertility, and genetic mutations in this pathway invariably lead to meiotic catastrophe and failure to make mature sperm, while displaying increased expression of TEs along with decreased DNA methylation and loss of the repressive Histone 3 lysine 9 trimethyl (H3K9me3) mark at TE genomic loci (Aravin et al., 2007, 2009; Carmell et al., 2007; Pezic et al., 2014). Elevated retrotransposition has also been observed in piRNA-deficient mice using a transgenic model for L1 mobilization (Newkirk et al., 2017). Germ cells deploy the PIWI-piRNA pathway at two stages during development: in pre-pachytene fetal germ cells and postnatally in pachytene stage germ cells (Czech and Hannon, 2016; Iwasaki et al., 2015). In mice, expression of the effector proteins PIWIL2 (MILI) and PIWIL4 (MIWI2) in fetal germ cells regulates transposons at multiple levels. TE transcripts are processed into primary piRNAs, which are subjected to MILI-mediated amplification, generating antisense secondary piRNAs. The latter are loaded onto MIWI2, resulting in their nuclear localization and the subsequent epigenetic silencing of TEs (Aravin et al., 2008; De Fazio et al., 2011; Kuramochi-Miyagawa et al., 2008; Manakov et al., 2015; Pezic et al., 2014). An array of co-factors is involved in the PIWI-piRNA pathway, including the inducible chaperone Hsp90 (HSP90AA1/HSP90α in mammals), which assists in piRNA biogenesis, loading of piRNAs onto PIWI-family proteins and transposon repression (Gangaraju et al., 2011; Ichiyanagi et al., 2014; Izumi et al., 2013; Specchia et al., 2010; Xiol et al., 2012).
Most studies of TE repression in developing mammalian germ cells have been conducted in mice due to the limited availability and ethical considerations of studying human fetal gonads. However, it was recently shown that epigenetic reprogramming via global DNA demethylation and histone modification is conserved in human fetal cells (Gkountela et al., 2013; Guo et al., 2015; Tang et al., 2015, 2016), as is the accompanying activation of retrotransposon transcript expression (Gkountela et al., 2015; Guo et al., 2015; Tang et al., 2015) and upregulation of PIWI-piRNA pathway genes (Tang et al., 2015). The expression of PIWI proteins and piRNAs has furnished evidence of an active piRNA pathway in adult human testis and in fetal human ovaries (Ha et al., 2014; Roovers et al., 2015; Williams et al., 2015). Notably, the human piRNAs identified are derived from piRNA clusters and resemble the composition of mouse pachytene piRNAs rather than fetal pre-pachytene piRNAs, which are largely transposon-derived (Aravin et al., 2007, 2008; Kuramochi-Miyagawa et al., 2008). In the human fetal testis, piRNA expression has been difficult to detect due to low abundance (Gainetdinov et al., 2017; Williams et al., 2015), and reported localization of PIWIL2 (HILI) and PIWIL4 (HIWI2) in the cytoplasm does not support nuclear silencing by this pathway (Gomes Fernandes et al., 2018).
In this study, we generate the most comprehensive picture to date of the dynamic expression of transposons, their derivative piRNAs and PIWI family proteins in humans. This constitutes the first evidence for activity of the PIWI-piRNA pathway in the human fetal testis. We identify pre-pachytene TE-derived piRNAs that contain the signature of secondary biogenesis. L1 expression increases concomitantly with L1-derived piRNAs and PIWI-piRNA machinery in fetal germ cells during mid-gestation; conversely, we find that L1 levels decline during development, as levels of the repressive histone mark H3K9me3 increase. However, we observe surprising heterogeneity at the single cell level that suggests only a subset of human fetal germ cells express L1 and deploy the PIWI-piRNA pathway to repress TEs via H3K9me3-mediated silencing, whereas other germ cells remain resistant.
RESULTS
Dynamic expression and subcellular localization of PIWI homologs in male fetal germ cells
The piRNA pathway is regulated by the PIWI family of conserved RNA-binding proteins, which is expressed almost exclusively in germ cells. In mouse fetal testis, biogenesis as well as secondary amplification of pre-pachytene piRNAs depends upon MILI, which turns on at approximately embryonic day (E) 12.5, shortly after the onset of sex differentiation (Aravin et al., 2008). In human fetal testis, we detected protein expression of HILI from gestational week (GW) 11 to 22, which corresponds to the period of sex-specific differentiation. HILI was found in the more differentiated population termed advanced germ cells (AGCs; Gkountela et al., 2013) marked by VASA (Fig. 1A) as well as in the co-existing more primitive population of PGCs defined by the pluripotency marker OCT4 (Fig. S1A). Initially low levels of HILI at GW11 progressively increased through development, and protein was distributed throughout the cytoplasm as well as in puncta beginning at GW13, consistent with reported localization of MILI to pi-bodies, or the intermitochondrial cement, in mice (Aravin et al., 2009). HIWI (PIWIL1) protein expression was not observed in fetal testes from GW10 to GW22 (Fig. S1B), as expected based on its known adult-specific expression pattern in human (Gomes Fernandes et al., 2018) and mouse (Fig. S1C) (Deng and Lin, 2002). HIWI2 was first detected in a small subset of VASA+ AGCs at GW14, and in an increased proportion of AGCs from GW18 onwards (Fig. 1B). The subcellular localization of HIWI2 underwent a dramatic shift from predominantly cytoplasmic before GW18 to nuclear in most AGCs by GW21-22 (Fig. 1C). In mice, MIWI2 is predominantly nuclear by E16.5-E17.5 (Aravin et al., 2008, 2009; Kuramochi-Miyagawa et al., 2010; Shoji et al., 2009), but remains cytoplasmic in fetal germ cells of piRNA pathway mutants, including Mili (Piwil2) Mael and Mov10l1 (Aravin et al., 2009; Shoji et al., 2009; Zheng et al., 2010). Indeed, we observed a cytoplasmic to nuclear progression of MIWI2 from E16.5 to E17.5 in mouse fetal testes (Fig. S1D). Nuclear localization of PIWIL4 homologs in other species requires piRNAs and co-factors such as HSP90α, and leads to transcriptional silencing of transposons at endogenous genomic loci (Fu and Wang, 2014; Ichiyanagi et al., 2014; Iwasaki et al., 2015). Accordingly, the dynamic localization of HIWI2 from the cytoplasm to the nucleus between GW18 and 21 suggests that piRNAs are produced and transported to the nucleus for similar epigenetic functions in male fetal germ cells of humans.
Fig. 1.
Expression of PIWI proteins in human fetal testis across development. (A) Expression of HILI and VASA at gestational week (GW) 11, 13, 16, 18 and 21, counterstained with DAPI (white, in merge images). Scale bars: 20 μm. Arrowheads indicate germ cells with HILI foci. A total of 12 embryos were analyzed across all time points. (B) Expression of HIWI2 and VASA at GW14, 16, 18, 21 and 22, counterstained with DAPI. Scale bars: 20 μm. A total of 11 embryos were analyzed across all time points. (C) Relative percentages of VASA+ germ cells with HIWI2 localization in cytoplasm or nucleus, or both, at indicated time points, with scoring examples on the right. Two-tailed Fisher's exact test was performed on ‘nuclear’ and grouped ‘non-nuclear’ categories across two time points; ****P<0.0001, n.s., not significant. Scale bars: 5 μm.
Evidence of transposon-derived primary and secondary piRNAs in fetal testis
We next looked for evidence of piRNAs across developmental timepoints spanning GW13-22. Small RNA sequencing was performed on bulk fetal testis tissue from eight samples at a depth of 35-40 million reads and processed according to our custom pipeline (Fig. S2). Trimmed 18-40 nt reads were first aligned to a curated list of human TE consensus sequences obtained from the GIRI Repbase database (Bao et al., 2015), and the remainder were aligned to the human genome (hg38). An overall alignment rate of >97% was obtained across all samples (Table S1). At all timepoints, the majority of reads (∼75%) corresponded to miRNAs, with smaller fractions derived from tRNAs, snoRNAs and protein-coding transcripts (Fig. 2A, left panel; Fig. S3A; Table S1). Strikingly, fewer than 1% of reads for all samples mapped to Repbase, and these resolved into two size classes: 27 nt and 22 nt (Fig. S3B). Reads comprising the 22 nt peak showed extremely low sequence diversity, with the vast majority mapping to the L2C retrotransposon (data not shown). The 27 nt peak was highest at GW20, and likely corresponds to putative TE-derived piRNAs, which range from 26-32 nt in other species. As our small RNA populations were derived from bulk fetal testis tissue without cell type or biochemical enrichment, we focused on the 26-32 nt small reads for the remainder of the analysis (Fig. S2). This increased the fraction of Repbase-mapping reads to ∼6% in the 26-32 nt RNAs but not in 18-22 nt small RNAs, which were mostly miRNAs (Fig. 2A, middle and right panels). The number of Repbase-mapping reads was dynamic over development, with a maximum at GW20-21 (Fig. 2B, Table S1), corresponding to the window of HIWI2 nuclear translocation. Hallmarks of pre-pachytene piRNAs were identified in these 26-32 nt Repbase-mapping reads, including a bias toward 1st position uridine (Fig. 2C, Fig. S3C) – a key signature of primary piRNA biogenesis (Iwasaki et al., 2015). Additionally, an adenine bias at the 10th position of mapped 26-32 nt reads at all timepoints with the exception of the earliest, GW13A (Fig. 2C, Fig. S3D), suggests active secondary piRNA biogenesis, also known as ‘ping-pong’ amplification (Czech and Hannon, 2016). Corroborating the specificity of Repbase-mapping reads, nucleotide distribution analysis on miRNA-annotated reads demonstrated a reduced, yet still apparent, 5′ U bias (Fig. S3E), similar to miRNAs in fetal ovary and adult testis (Williams et al., 2015), and no adenine bias at position 10 (Fig. S3F). Another signature of ping-pong biogenesis, ten nucleotide overlaps between reverse complementary piRNAs, was detected in all samples to varying extents, with the exception of GW13A, and most strongly at GW20 (Fig. 2D). Together, these analyses identify small RNAs in human fetal testes with defining characteristics of pre-pachytene piRNAs, although at much lower abundance than found in mice or human fetal ovaries (Roovers et al., 2015; Vasiliauskaitė et al., 2017; Williams et al., 2015).
Fig. 2.
Identification of transposon derived piRNAs in the human fetal testis. (A) Relative percentages of different RNA subtypes present in gestational week (GW) 20 sample after size filtering for 18-40 nt, 26-32 nt and 18-22 nt small RNAs. Values in bold italic highlight Repbase-mapped reads, which include putative piRNAs. (B) Normalized number of reads (26-32 nt) mapped to human transposon consensus sequences in Repbase database across all samples. (C) Relative nucleotide distribution at a given read position of pooled Repbase-mapping reads (26-32 nt) at GW20. (D) Ping-pong overlap signatures calculated on pooled Repbase-mapping reads (26-32 nt). (E) Top transposable elements mapped to by Repbase-mapping reads (26-32 nt) at GW20. (F) Distribution of read sizes mapping to the L1HS transposon across all samples. Positive and negative values represent reads that map to the positive and negative strand, respectively.
Among the Repbase-mapping reads, the L1 family was the most abundantly represented class of repeat elements at GW20 (Fig. 2E). This result is consistent with the derivation of piRNAs from actively transcribed TEs, given that L1 elements are known to be active in humans (Hancks and Kazazian, 2012). The evolutionarily youngest L1 family (Smit et al., 1995), L1HS, was the most highly represented element in all samples (with the exception of GW13A), followed by ‘L1’ elements, which here represent the broad primate-specific L1PA clade (Kojima, 2018) (Table S2). Reads were mapped to both the forward and reverse strand of L1HS, particularly at later stages (Fig. 2F), consistent with a progressive ping-pong amplification process. Analysis of other highly represented classes of TEs revealed different strand-specific biases, yet consistently showed GW20 as the sample with the largest number of relative mapping reads (Fig. S3G-I). We conclude that pre-pachytene piRNAs are produced and amplified in the human fetal testes, with a large fraction derived from TEs, which is similar to the developing mouse testis (Aravin et al., 2007, 2008).
L1 transposons are expressed in AGCs coordinately with the transposon repression network at the single cell level
The prevalence of L1-derived piRNAs and the possibility of piRNA-mediated nuclear silencing prompted a closer examination of the relationship between transposons and the transposon-repression network in an unbiased, population-wide manner. We took advantage of a recently published single cell RNA-seq dataset from human fetal testis tissue (Li et al., 2017) to assess the heterogeneity of transposon-derived and gene-coding transcripts. By mapping to repeat elements (based on RepeatMasker annotations) as well as the genome, we obtained profiles for expression of TEs as well as genes for each cell (Fig. S4). We focused on GW19 in light of the high level of piRNA expression and dynamic HIWI2 localization. t-distributed stochastic neighbor embedding (t-SNE) analysis based on gene expression profiles identified three distinct clusters corresponding to PGCs, AGCs and somatic support cells based on known lineage- and stage-specific markers (Fig. 3A,B, Fig. S5A-C). As anticipated, PGC and AGC clusters were distinguished by high expression of OCT4 (POU5F1) and VASA (DDX4), respectively (Fig. 3B). The machinery of primary piRNA biogenesis [MAEL and HILI (PIWIL2)] was expressed in both PGCs and AGCs, though higher in the latter, whereas HIWI2 mRNA was largely limited to AGCs (Fig. 3B,C). This developmental delay in transcription of HIWI2 as compared with HILI recapitulates the sequential expression of the proteins observed in the fetal testis tissues (Fig. 1A,B). With a few exceptions, TE expression was dramatically elevated in AGCs compared with PGCs, and absent in the soma (Fig. 3B,C). The most highly expressed TEs in AGCs belong to the L1 clade, specifically L1HS, L1PA2 and L1PA3. This corresponds well with the abundance of piRNAs derived from the L1 clade (Fig. 2E). Other highly expressed TEs include members of the Alu family, especially the evolutionarily young AluYa5 and AluYb8 subfamilies, which are known to be active in humans (Batzer and Deininger, 2002). Paired correlation analyses between L1HS, and either HILI or HIWI2 both yielded positive correlations (Fig. 3D,E). Together, this analysis shows that at the single cell level, the expression of genes of the transposon repression network is upregulated during germ cell development in concert with the expression of transposons (Fig. 3C).
Fig. 3.
Single cell sequencing reveals dramatic upregulation of L1 family retrotransposons in advanced germ cells. (A) tSNE clustering on transcriptome reference was used to generate three distinct cell populations in the GW19 dataset. (B) Violin plots showing expression of germ cell markers (top), transposon repression genes (middle) and L1 family retrotransposons (bottom) in AGCs, PGCs and soma. (C) Heatmap displaying the most differentially expressed transposons sorted by myAUC score (top), and expression of germ cell markers (middle) and transposon repression genes (bottom). (D-F) Pairwise correlation analysis between L1HS and HILI (D) and HIWI2 (E) in GCs; and HSP90α (F) in AGCs (i), PGCs (ii) and combined GCs (ii). Pearson's correlation coefficient scores and P-values are listed above graph.
Dynamic relationships between L1 and the repression pathway at transcript and protein level
To confirm the correlation between L1 transcripts and the transposon repression network, we performed immunostaining. Using a previously validated antibody against the L1-ORF1p RNA-binding protein (Rodić et al., 2014), we detected expression primarily in VASA+ AGCs starting at GW16, whereas earlier it was found very sparsely and at low levels (Fig. S6). L1-ORF1p was restricted to the cytoplasm and concentrated in granules, some of which colocalized with HILI foci (Fig. 4A). However, L1-ORF1p granules were distinct from those containing HIWI2 (Fig. 4B). As suggested by the heatmap in Fig. 3C, a small subset of PGCs express high levels of L1 transcript but not the transposon repression network. We similarly observed rare instances of robust L1-ORF1p expression in OCT4+ PGCs (Fig. 4C, top panel) and in cells lacking VASA and harboring low levels of OCT4 (Fig. 4C, bottom panel). It is not clear whether these cells are transitioning between PGCs and AGCs or whether they will successfully turn on the repression machinery, but this observation raises the issue of whether transposon protein expression precedes that of HIWI2 and piRNA biogenesis.
Fig. 4.
L1 expression in advanced germ cells. (A) L1 and HILI immunofluorescence in a GW16 human testis. Arrowheads indicate puncta of L1/HILI colocalization. (B) Expression of L1 and HIWI2 at GW17. Scale bars: 20 μm. Arrows indicate regions of mutually exclusive staining between L1 and HIWI2 puncta. (C) L1 expression at GW21 (top panel) and GW18 (bottom panel), co-stained with OCT4 or a pan-GC marker (OCT4 and VASA), respectively. (Top) Arrowheads indicate an L1+, OCT4+ germ cell; arrows highlight a L1+, OCT4− cell. (Bottom) Arrows indicate L1+ and pan-GC negative cell. (D) L1+/HIWI2low germ cells (arrows), a HIWI2high/L1low germ cell (arrowheads) and double-positive AGCs (stars) in human fetal testis at GW19 and GW22. (E) xy scatter plots of HIWI2 versus L1 fluorescence intensities (left panels) and HIWI2 versus VASA (right panels), with Pearson's correlation coefficients and corresponding P values. For GW19 (top), n=526 VASA+ objects, counted from three stitched xy sections. For GW22 (bottom), n=472 VASA+ objects, from two stitched xy sections. (F) L1high/HSP90αlow cells (arrows), HSP90αhigh/L1low cells (arrowheads) and double-positive cells (stars) in human fetal testis at GW16-22. (G) xy scatter plots of HSP90α versus L1 fluorescence intensities (left) and HSP90α versus VASA fluorescence intensities (right). Pearson's correlation coefficients and corresponding P values are indicated. For GW16 (top), n=768 VASA+ objects, counted from three stitched xy sections. For GW19 (middle), n=1809 VASA+ objects, counted from four stitched xy sections. For GW22 (bottom), n=2098 VASA+ objects, counted from three stitched xy sections. Scale bars: 20 μm in A,B,D,F; 10 μm in C. Dashed red boxes in E and G indicate the ‘arms’ of single-positive cell populations.
Examination of the relative expression of L1-ORF1p, HIWI2 and VASA in AGCs revealed a more dynamic relationship during fetal development than previously identified in the single cell RNA-seq analysis. The majority of L1-ORF1p expressing AGCs at GW22 co-expressed HIWI2 with significant correlation that approached the r value between HIWI2 and VASA (Fig. 4D,E). Earlier, however, at GW19, we observed VASA+ cells with exclusive expression of either L1-ORF1p or HIWI2 and within the AGC population, the correlation between the two proteins was lacking at this timepoint (Fig. 4D,E). Compared with the high correlation between L1 and HIWI2 transcripts in single cell RNA-seq at GW19 (Fig. 3E, Fig. S5D,E), these results indicate a delay in the coordinated expression of HIWI2 and L1-ORF1p proteins during fetal development. It is likely that the VASA single-positive cells at GW22 (Fig. 4E, right) represent newly differentiating PGCs that have not yet turned on HIWI2.
HSP90α is a chaperone protein, with an inducible isoform enriched in germ cells, which has been implicated in transposon repression and piRNA biogenesis in flies and mice (Gangaraju et al., 2011; Ichiyanagi et al., 2014; Izumi et al., 2013; Specchia et al., 2010; Xiol et al., 2012). Levels of HSP90α transcript were high but variable in AGCs and PGCs (Fig. 3B,C), with a negative correlation observed between HSP90α and L1HS in AGCs but not PGCs (Fig. 3F). Consistent with this, immunofluorescence revealed that a significant number of AGCs at GW16 and GW19 expressed exclusively L1-ORF1p or HSP90α proteins but not both (Fig. 4F, top and middle panels); accordingly, the XY scatter plots for L1-ORF1p and HSP90α intensities displayed ‘arms’ at the extreme end of each axis of AGCs with mutually exclusive expression (dashed red boxes in Fig. 4G). This relationship changed by GW22, when most AGCs expressing L1-ORF1p also expressed HSP90α (Fig. 4F, bottom panel), and a highly significant correlation was found (Fig. 4G). Together, these observations suggest that three AGC subpopulations co-exist at GW16 – L1-high, HSP90α-high and those low for both – and by GW22 a subpopulation of HSP90α single positive AGCs persist, but the majority of L1+ cells also express moderate to high levels of HSP90α.
Declining L1 expression and increasing H3K9me3 during fetal development
Thus far, we have characterized expression of key proteins of the PIWI-piRNA pathway (HILI and HIWI2) and confirmed expression of piRNAs and retrotransposons, yet it remains unclear whether the PIWI-piRNA pathway represses retrotransposons in human fetal testis. Although we observed increasing cytoplasmic to nuclear relocalization of HIWI2 by GW21 (Fig. 1C), examination of subsequent events is limited by the availability of samples after GW22. We returned to the published single cell RNA-seq dataset of human fetal germ cells (Li et al., 2017) to examine the expression of TEs at GW25, again mapping reads to repeats as well as the genome. Distinct populations of somatic cells, PGCs and AGCs persisted at GW25, and TEs were largely absent from somatic cells and PGCs, but highly expressed in AGCs (Fig. S7). Comparing the scaled expression of L1 family transposons in AGCs between GW19 and GW25 revealed a modest but statistically significant decrease (Fig. 5, Table 1). This decline in L1 transcript at GW25 follows the upregulation of the transposon repression network, increased piRNA expression and nuclear translocation of HIWI2, suggesting that the PIWI-piRNA pathway may play an active role to silence TEs in developing AGCs.
Fig. 5.
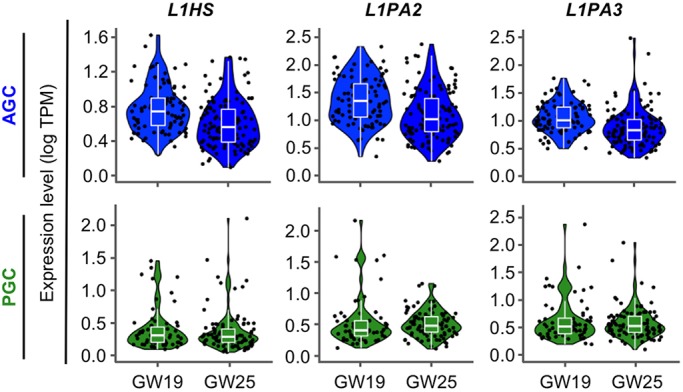
Decrease in L1 transcript levels as development progresses. Expression of L1 transcripts from the single cell RNA-sequencing dataset at GW19 and GW25. Violin plots showing expression of L1 family retrotransposons in AGCs (top) and PGCs (bottom), with median and quartile levels displayed in the box plots.
Table 1.
L1 transcript expression from the single cell dataset in Fig. 5

To further examine the relationship between the PIWI-piRNA pathway and L1 by immunostaining, we turned to a xenograft model (Mitchell et al., 2010; Tharmalingam et al., 2018), in which human fetal testis implanted under the kidney capsule of immunocompromised mice will vascularize and continue to grow. Fragments of two testes, GW16 and GW17, were grafted into separate mice and allowed to grow from 1 to 4 months (Fig. 6A, Fig. S8A). We confirmed the continued development of fetal germ cells in grafts by examining HIWI2 localization. For both testes, there was a notable shift in HIWI2 localization. Although non-transplanted GW17 samples showed predominantly cytoplasmic localization of HIWI2 (∼88% of cells), this frequency decreased in xenotransplanted samples after 4 or 8 weeks (41% and 60%; Fig. 6B,C). Similarly, the non-transplanted GW16 sample was enriched for cytoplasmic rather than nuclear HIWI2 in AGCs (∼70% versus 7%, respectively), but nuclear localization greatly increased after 5 and 16 weeks engraftment up to 41% and 55% of AGCs (Fig. S8B,C). This change in HIWI2 distribution suggests that grafted tissues continue to develop to an equivalent of GW22 given the similar frequency of HIWI2 distribution, or that HIWI2 does not egress from the cytoplasm in all germ cells.
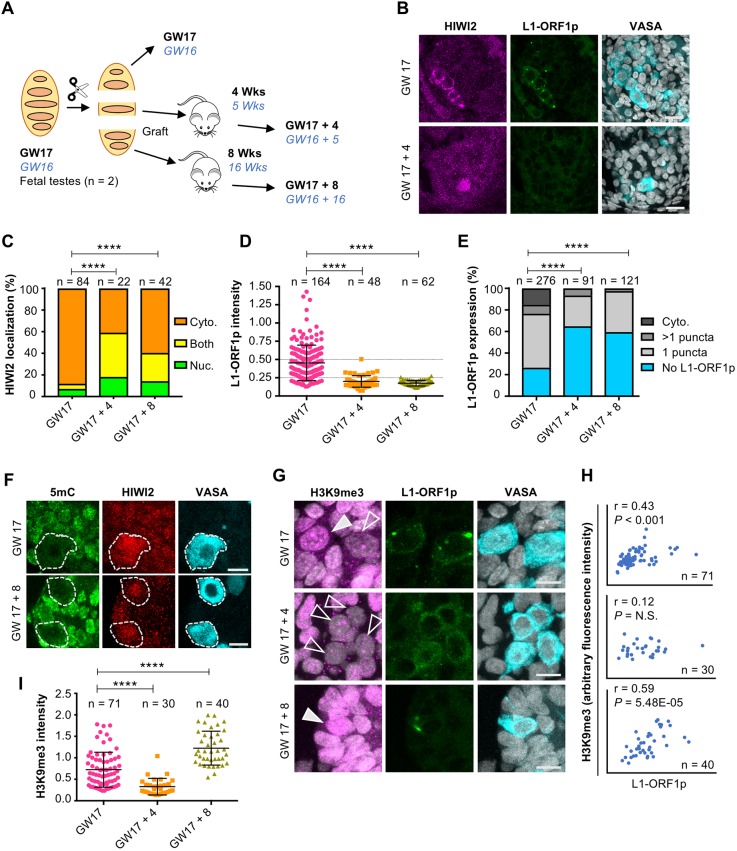
Fig. 6.
Evidence for PIWI-piRNA pathway activity in fetal testis. (A) Schematic of xenotransplant model with timepoints examined for n=2 biological repeats. The GW17 sample is shown, whereas the GW16 sample (blue italic text) is depicted in Fig. S8. (B) Immunostaining for HIWI2, L1-ORF1p and VASA (shown in merge with DAPI) at GW17 (no xenograft; top) and GW17+4 weeks xenograft (bottom). Scale bars: 20 μm. (C) Quantification of HIWI2 subcellular localization in VASA+ cells of xenografted testes. Two-tailed Fisher's exact test was performed on ‘nuclear’ and grouped ‘non-nuclear’ categories across two time points. (D) Quantification of L1-ORF1p intensity normalized to VASA; Mann-Whitney U-tests were used to test significance. (E) Manual scoring of L1-ORF1p expression in VASA+ cells grouped into one of four categories: cytoplasmic staining; punctate cytoplasmic staining (either one puncta or multiple puncta); or no L1-ORF1p expression. Two-tailed Fisher's exact test was performed on the categories ‘no L1-ORF1p’ versus all grouped ‘L1-ORF1p-expressing’ cells across two time points to determine significance. (F) Immunostaining for 5mC, HIWI2 and VASA in fetal testis at GW17 and xenografts. VASA+ cells are outlined. Scale bars: 10 μm. (G) Immunostaining for H3K9me3, L1-ORF1p and VASA (shown in merge with DAPI) at GW17 and xenografts. Filled and empty arrowheads indicate high and low H3K9me3, respectively. Scale bars: 10 μm. (H) xy scatter plots of H3K9me3 and L1-ORF1p fluorescence intensities with Pearson's correlation coefficients, P values and number of cells counted. (I) Quantification of H3K9me3 intensity normalized to VASA, measured in Volocity. Mann-Whitney U-tests were used to test significance. For all graphs, ****P<0.0001; n.s., not significant; n represents number of individual cells counted per time point.
We next examined the persistence of L1 expression in germ cells of xenografted testes. Similar to the decline observed in vivo from GW16 to GW22 (Fig. S6), L1-ORF1p attenuated significantly over the first 4 weeks following transplant, with levels decreased by ∼50% in individual AGCs (Fig. 6D) and ∼2/3 of VASA+ cells devoid of L1-ORF1p (Fig. 6E). A similar decline was observed in the GW16 xenograft sample after 16 but not 5 weeks (Fig. S8B,D,E). As the vast majority of AGCs at this timepoint have entered mitotic arrest (Li et al., 2017; Mitchell et al., 2010) (data not shown), a decrease in the frequency of L1-ORF1p+ cells must arise through degradation of protein, in addition to locus-specific silencing or transcript degradation. To determine whether the decrease in L1-ORF1p levels correlates with an increase in DNA methylation over time, we performed immunostaining for 5-methylcytosine (5mC). While robust methylation was detected in surrounding somatic cells, we were unable to detect 5mC in VASA+ AGCs at GW17-22 or in xenografted tissues (Fig. 6F, Figs S8F, S9). This is consistent with previous reports of low global DNA methylation levels at GW26 (Guo et al., 2017), suggesting that DNA remethylation occurs at later timepoints.
The piRNA-guided deposition of H3K9me3 marks has emerged as an additional mechanism for transcriptional silencing of retrotransposons (Pezic et al., 2014). Thus, we examined this repressive histone modification by immunofluorescence. Many VASA+ cells harbored few or no H3K9me3 puncta at GW16/17 or in the xenografts aged 4 or 5 weeks (Fig. 6G, Fig. S8G). Following prolonged engraftment, we observed a significant increase in the levels in AGCs by GW17+8 and GW16+16 (Fig. 6G,I; Fig. S8G,I). The parallel trends of decreasing L1-ORF1p and rising H3K9me3 repressive mark would seem to be functionally related. However, unexpectedly, on a per cell basis, we observed that high levels of H3K9me3 were associated with high levels of L1-ORF1p (Fig. 6H, Fig. S8H). Thus, we conclude that the developmental trends of L1-ORF1p and H3K9me3 in human AGCs parallel those described in mouse at the population level, but that our analysis of single cells opens up the possibilities that either nuclear silencing operates independently of L1 protein degradation, or that some level of L1 protein will persist in human AGCs. Our results also raise the possibility that the TE nuclear silencing mechanism via H3K9me3 is exclusively activated in the subset of AGCs that initially express high levels of L1 protein.
Our observations raise the possibility that expression of L1 precedes that of the repressive pathway. Unresolved issues include the means by which the PIWI-piRNA network becomes activated as well as how TEs remain adequately repressed in PGCs given their demethylated state. The arginine methyltransferase PRMT5 has been implicated in TE repression in mouse PGCs by depositing repressive H2A/H4R3me2 marks on retrotransposons (Kim et al., 2014) and subsequent modification/activation of PIWI-family proteins (Vagin et al., 2009). We detected PRMT5 expression in fetal testes across development. Intriguingly, at GW13, when PGCs outnumber AGCs, PRMT5 could be detected in the cytoplasm and the nucleus, whereas it was exclusively cytoplasmic by GW17 (Fig. S10). In mouse male germ cells, such a nuclear to cytoplasmic translocation is observed by E11.5, signifying the transition from the role of PRMT5 in histone modification to methylation of PIWI proteins (Kim et al., 2014; Vagin et al., 2009). Together, these results indicate that conserved mammalian mechanisms regulate TEs in the human testes. However, our analysis at the single cell level suggests that TE expression, and the resulting activation of the PIWI-piRNA pathway, occur in a subset of germ cells, whereas others are resistant.
DISCUSSION
In preparation for gametogenesis, PGCs undergo genome-wide epigenetic reprogramming; the removal of silencing marks that normally suppress TEs leaves the germline vulnerable to DNA breaks or transposition. Our characterization of L1 expression in human fetal germ cell development (Figs 3–6, Fig. S6) underscores the need for the PIWI-piRNA pathway to protect germ cells from TE activity and maintain germline integrity. In this study, we describe the rise and fall of L1 expression in germ cells of the human fetal testis and provide evidence in support of an active TE repression network, including key PIWI proteins, pre-pachytene piRNAs and crucial co-factors, such as HSP90α.
The fetal PIWI-piRNA pathway in mice is defined by PIWI family protein expression, TE-derived piRNAs and piRNA-dependent deposition of repressive epigenetic marks (Ernst et al., 2017). We show that these features are conserved in the human fetal testis by characterizing: (1) the composition, temporal expression pattern and sub-cellular localization of PIWI-family proteins (Fig. 1); and (2) the presence of transposon-derived piRNAs that feature a secondary amplification signature (Fig. 2). In mouse germ cells, MIWI2 is expressed from E15 to P3 (Aravin et al., 2008), during which time it is required for deposition of the epigenetic silencing marks 5mC (Aravin et al., 2008; Kuramochi-Miyagawa et al., 2008) and H3K9me3 (Pezic et al., 2014) in order to fully deplete L1-ORF1p by P2 (Aravin et al., 2009). Similarly, we observed a decline in the expression of L1 transcripts (Fig. 5, Table 2) and L1-ORF1p that coincided with rising levels of H3K9me3 in AGCs from GW17 onwards (Fig. 6, Figs S6, S8). Given the limitations of working with human clinical samples, we could not test whether this was directly attributable to the PIWI-piRNA pathway or to the specific sites of H3K9me3 acquisition in the genome. However, our cumulative results suggest that the fetal PIWI-piRNA pathway restricts TE expression in human fetal germ cells.
Table 2.
Samples used for small RNA analysis

Our study provides the most comprehensive characterization of the PIWI-piRNA pathway in the human fetal testis to date. In addition to confirming and expanding upon the reported expression of HILI and HIWI2 in the human fetal testis (Gomes Fernandes et al., 2018), we observed dynamic localization of HIWI2 during human development, with transition from the cytoplasm to the nucleus occurring between GW18 and GW21 (Fig. 1). Recent studies in human fetal ovary and adult testis showed that piRNAs were derived from genomic piRNA clusters (Ha et al., 2014; Roovers et al., 2015; Williams et al., 2015). Previous efforts to identify piRNAs from fetal testis were confounded by low abundance, dearth of samples and conflicting analyses (Gainetdinov et al., 2017; Williams et al., 2015). Here, we characterized piRNAs from eight fetal testes samples across a wide developmental range (GW13-22) and identified TE-derived piRNAs with a peak expression at GW20, which corresponds with the timing of nuclear HIWI2 translocation and further supports an active PIWI-piRNA pathway that represses TEs. This notion is corroborated by the finding that high L1 transcript correlates with lower expression of PIWI-piRNA pathway genes, including HIWI2, in boys at risk of infertility (Hadziselimovic et al., 2015). Additionally, mechanistic studies in human iPS cells showed that HILI can directly modulate L1 activity (Marchetto et al., 2013).
Human fetal germ cell development is marked by heterogeneity at the level of transcript, protein and cell state. For example, whereas PGCs in mice acquire markers of male differentiation and undergo mitotic arrest from E13.5 to E14.5 (Western et al., 2008), in humans the transition from PGC to AGC is spread across weeks and results in both populations within the same gonad (Gkountela et al., 2013). We observed heterogeneity between cells in the expression of transcripts for TEs and the TE repression network (Figs 3,5, Fig. S7). Similarly, we observed heterogeneity in the levels of L1-ORF1p, HIWI2 and HSP90α proteins (Fig. 4). Notably, heterogenous L1-ORF1p expression was previously observed in fetal oocytes in mice (Malki et al., 2014). HSP90 is a highly expressed molecular chaperone, constituting ∼1% of soluble cytoplasmic protein content (Buchner, 1999). A germ cell-specific inducible isoform, HSP90α, has been implicated in regulating the abundance of L1-ORF1p in mouse, most likely at the translational or post-translational stage (Ichiyanagi et al., 2014). A physical interaction between HSP90α and the second open reading frame of L1 (ORF2p) was recently revealed by proteomics (Taylor et al., 2018). In the fetal testes at GW16-19, we observed a pattern of reciprocal expression between HSP90α and L1-ORF1p, with AGCs positive for one or the other, but not both proteins (Fig. 4F); however, by GW22 and later, the expression of L1-ORF1p was diminished overall (Fig. 6D, Fig. S8D) and cells retaining L1-ORF1p now simultaneously expressed HSP90α (Fig. 4F). A similar trajectory was found with HIWI2 and L1-ORF1p, in which AGCs at GW19 were primarily single positive, but express both proteins at similar levels by GW22 (Fig. 4E). Based on these observations, we propose a model whereby a subset of early AGCs marked by high levels of HSP90α at ∼GW16 is resistant to the expression of transposons (Fig. 7). After this point, we propose that AGC development diverges and HSP90αlow AGCs become susceptible to expression of L1-ORF1p, which leads to the upregulation of the PIWI-piRNA pathway and transposon silencing. Based on the coincidence of sustained L1-ORF1p and high levels of HIWI2 and H3K9m3 in later AGCs, we suggest that silencing occurs through piRNA-guided deposition of H3K9me3 in the nucleus (Fig. 6G, Fig. S8G). Although we did not detect 5mC in AGCs in xenografted testes after 2-4 months (Fig. 6F, Fig. S8F), DNA remethylation at TE loci could be undetectable by immunofluorescence or else could occur later in development. By contrast, we propose that HSP90αhigh AGCs are protected from initial transposon expression and it remains to be determined if they ever express transposons or upregulate the transposon repression/PIWI-piRNA pathway. Our data suggest that suppression of TEs earlier in development in PGCs, prior to the expression of PIWI genes, could occur via other mechanisms such as PRMT5-mediated nuclear silencing (Fig. S10), as previously shown in the mouse (Kim et al., 2014).
Fig. 7.
Coordinated transposon repression in GCs of the human fetal testis. (A) (Top) Summary of the expression pattern of PIWI-piRNA pathway components and L1 protein in developing germ cells in the human fetal testis at indicated timepoints and in aged xenograft samples. (Bottom) Relationships between L1 expression (protein, except where indicated) and expression of PIWI-piRNA pathway factors and H3K9me3 at indicated developmental stages. By GW22, many L1-expressing AGCs also express components of the repression network. (B) Model for transposon expression and repression in developing AGCs. See Discussion for details.
The PIWI-piRNA pathway is often considered the immune system of the germline. In this paradigm, TEs are viewed as a threat to developing germ cells that must be immediately silenced by the PIWI-piRNA pathway to prevent their mobilization and subsequent genome disruption. However, our single cell analyses reveal that the expression of TEs (Figs 3,4, Fig. S7) precedes that of the transposon repression network, and thus cells experience a window of TE expression prior to repression. This observation is consistent with previous studies that have shown consistent L1 transcript expression from GW4-19 (Guo et al., 2015), where the earliest timepoint, GW4, substantially predates the onset of the repression pathway. Could this early expression of TEs play a functional role during germ cell development? Recent studies argue in favor of this idea, by providing evidence that the L1 transcript is essential for early embryonic development in mice and may function in the remodeling of the epigenetic landscape (Jachowicz et al., 2017; Percharde et al., 2018). In addition, both L1-ORF1p and L1-ORF2p were previously observed in a single GW18 testis (Ergün et al., 2004). As we observe the onset of HIWI2 and HSP90α ∼1 month later than the first L1-ORF1p (Fig. 7), it is tempting to speculate that initial expression of TEs is required to ‘prime’ the activation of the repression network. This issue, and whether L1 plays any other functional roles during germ cell development will be the subject of future investigations.
MATERIALS AND METHODS
Human sample collection and processing
Human fetal specimens were collected free of patient identifiers after elective termination of pregnancy (Committee on Human Research, University of California, San Francisco, IRB# 12-08813 and 16-11909). All fetal samples were transported on ice for gonad microdissection. Age of specimen (referred to as GW – gestational week in text) was determined by heel-toe measurement. Gonads were left in RPMI 1640 media supplemented with L-glutamine at 4°C from a few hours to overnight, and for up to two nights for some samples. Specimens were either flash frozen for molecular biology analysis or fixed for immunofluorescent staining. For staining, tissues were fixed overnight in 2% PFA in PBS solution at 4°C with gentle agitation. Tissues were then washed, incubated overnight in 30% sucrose at 4°C, embedded and frozen in OCT solution (Tissue-Tek). Embedded tissues were cryosectioned at 8 μm.
Animals and xenograft model
Wild-type embryos were generated by mating CD1 females with homozygous Oct4-ΔPE-GFP males (MGI:3057158, multiple-copy transgene insertion; Anderson et al., 1999). Embryos were dissected from timed matings and staged using anatomical landmarks from E16.5 to E18.5.
Immunodeficient CD1 nud/nud homozygous (Charles River) mice were used as host for the xenograft. Prior to surgery, human fetal testes were cut into four fragments and held in ice-cold RPMI 1640 medium until xenografting, with one fragment set aside for fixation. Hosts were anesthetized, a 2 cm back midline incision was made to exteriorize each kidney and a 2 mm opening was made in the renal capsule. One fetal testis fragment was xenografted into the left kidney capsule of each host. The peritoneum was closed with c-9 silk surgical sutures (Henry Schein), and the incision was stapled closed with stainless steel wound clips (MikRon). Following surgery, animals were monitored daily for food and water intake, and any obvious signs of stress. All mouse work was carried out under the University of California, San Francisco, Institutional Animal Care and Use Committee guidelines in an approved facility of the Association for Assessment and Accreditation of Laboratory Animal Care International.
Immunofluorescence
We performed immunofluorescent staining on sectioned slides using our general staining protocol. Slides were washed three times with 1× PBS for 5 min at room temperature to dissolve OCT. Samples were permeabilized with PBT (1× PBS+0.5% Triton X -100) for 10 min at room temperature, followed by three short rinses with 1× PBS, and then blocked for 1 h at room temperature with fresh blocking buffer (10% heat inactivated donkey serum in 1× PBS+0.1% Triton X-100). Subsequent primary antibody incubations were performed in humidified chambers at 4°C overnight, with primary antibodies (see Table S3) diluted appropriately in blocking buffer. The next day, slides were washed three times in 1× PBS for 5 min at room temperature, and then treated with secondary antibodies (1:200) and DAPI nuclear counterstain (1:1000) for 1 h at room temperature. Slides were washed three times in 1× PBS, and then mounted with Vectashield Antifade Mounting Medium (Vector Laboratories, H-1000). Some antibodies required antigen retrieval with either sodium citrate buffer (pH 6.0) or Tris-EDTA buffer (pH 8.8) (see Table S3). Antigen retrieval conditions consisted of boiling slides in appropriate solution for 10 min, followed by washes in 1× PBS and processing according to the general staining protocol. 5meC antibody staining required Tris-EDTA (pH 8.8) antigen retrieval followed by PBT permeabilization and incubation in 3.5 N HCl for 1 h prior to the general staining protocol. Xenograft samples incubated with a primary antibody from a mouse host required antigen retrieval and blocking with Mouse on Mouse blocking reagent (Vector Laboratories, MKB-2213). Following our staining protocols, images were acquired from a Keyence fluorescence microscope and/or Leica Sp5 upright confocal microscope w/AOBS.
Immunofluorescence quantification
Automated fluorescence intensity quantification
Images were obtained from Keyence microscope (model BZ-X710) using a 20× objective and stitched with Keyence imaging software. Background was subtracted on FIJI software (Schindelin et al., 2012) using a rolling ball radius of 25 pixels and disabled smoothing. VASA+ objects corresponding to single cells were identified using Volocity version 6.3. Our VASA+ object detection protocol included the following modules: ‘Find objects’ in the VASA channel, ‘Close’ objects for two iterations, followed by ‘Fill holes in objects’ and separate touching objects at a size of 600 μm2. Next, VASA objects were excluded below and above size thresholds (<599 μm2 and >2405 μm2). Finally, VASA+ objects were filtered for mean VASA channel intensity >45 and shape factor >0.413. Identified objects were manually inspected and non-cellular objects were manually deleted. A variation of this protocol was used to find VASA+ objects from images acquired from the Leica Sp5 confocal microscope using a 63× oil objective.
Manual scoring for HIWI2 localization
Z-stack images through the entire section width (8 μm) were obtained on a Leica Sp5 confocal microscope using a 63× oil objective. Z-stacks were opened on FIJI imaging software and VASA+ cells were manually scored for HIWI2 subcellular localization. HIWI2 localization was compared with VASA (cytoplasmic marker) and DAPI (nuclear marker) and assigned to either of three categories – nuclear (where the majority of HIWI2 appeared overlapped with DAPI staining), cytoplasmic (where HIWI2 staining was excluded from DAPI and overlapped with VASA) or both (where HIWI2 staining appeared equally distributed between the compartments). See Fig. 1C for representative examples of cells scored for each localization phenotype.
Small RNA-sequencing of human fetal testis
All samples were processed and sequenced by Quick Biology. Total RNA extractions were performed on bulk fetal testis tissue using Trizol, and quality control was performed on total RNA using the Agilent Total RNA Nano Bioanalyzer kit. Resulting RNA integrity number (RIN) scores, are provided in Table 2. Libraries were prepared using the QIAseq miRNA Library Kit, and library quality was assessed using the Agilent Bioanalyzer High Sensitivity DNA assay. Paired-end sequencing was performed on an Illumina HiSeq 4000.
Small RNA-sequencing data analysis
All samples were sequenced to a depth of approximately 35-40 million reads in order to meet the minimum read requirement for small RNA-seq data of 30 million as outlined by the ENCODE consortium (www.encodeproject.org/rna-seq/small-rnas/#restrictions). Raw reads in fastq files were trimmed to remove the adapter sequence (AACTGTAGGCACCATCAAT) using cutadapt version 1.8.1 (Martin, 2011), and reads were subsequently size filtered to 18-40 nt using cutadapt. FastQC (version 0.11.5) was utilized to broadly assess read quality. Trimmed reads were aligned to Repbase (version 22.11), a collection of consensus sequences of known human transposons, curated by GIRI: the Genetic Information Research Institute (Bao et al., 2015). Alignments were performed with bowtie version 1.2.2 (Langmead et al., 2009) using the following options: -v 1, -M 1, –best, –strata, signifying a maximum of one mismatch between the read and reference sequence, with multimappers being randomly assigned to only one location.
All reads that did not align to Repbase were subsequently aligned (with bowtie version 1.2.2 and same options) to the human genome (GRCh38) in order to determine the contribution of other RNA types to the total small RNA pool. Aligned reads were intersected with genomic features and counted using the htseq-count function of HTSeq version 0.9.1 (Anders et al., 2015), with the following parameters: -f sam -t exon -s yes -i gene_type -m intersection-nonempty –nonunique all -a 0. The reference file containing genomic feature coordinates was a custom GTF file generated by merging the GENCODE 28 comprehensive gene annotation list with GENCODE 28 predicted tRNA genes. Feature counts for miRNAs, tRNAs, snoRNAs and protein-coding transcripts were combined with the number of Repbase-mapping reads to generate a breakdown of RNA subtypes. For a comprehensive explanation of RNA subtypes and relative percentages, see Table S1.
Repbase-mapped reads were further analyzed for known piRNA signatures, including a 5′ uridine bias and 10th position adenine bias, by calculating the nucleotide frequency distribution per base position along the read for all pooled Repbase-mapped reads of a given sample. Read overlap signatures, which are characteristic of the piRNA ping-pong biogenesis cycle, were calculated on the MISSISSIPPI Galaxy server (mississippi.snv.jussieu.fr) using the ‘Small RNA Signatures’ tool (Galaxy Version 3.1.0), developed by Antoniewski (2014). Transposable element annotations and transposable element-specific read size distributions were calculated directly from alignment files post Repbase mapping.
Single cell RNA-sequencing reanalysis
Bioinformatic pipeline for single-cell RNA-seq data
Adapter trimmed reads were downloaded from Gene Expression Omnibus (GEO). FastQC (Conesa et al., 2016) was used initially for a broad quality control check. Using UMI-Tools (Smith et al., 2017), we determined real cells from background noise using the cell ID/UMI information in read 2. Cell IDs/UMI information from read 2 was extracted and appended to the read name. Using BBDuk (sourceforge.net/projects/bbmap/), reads were quality trimmed to Q10 (Mbandi et al., 2014; Mills, 2014). For our transcriptome alignment, we used the default STAR/2.9.3a parameters (Dobin et al., 2013) to align to reference genome GRCh38.91 human transcriptome (Ensembl) with mitochondrial annotations added. To maintain the presence of multimappers in our human repeat reference alignment, the following STAR parameters were used: –outFilterMultimapNmax 100, –winAnchorMultimapNmax 100, –outSAMmultNmax 100 and –outFilterMismatchNmax 3 (Ge, 2017). After specific gene or repeat element annotations were respectively assigned to the aligned files using Subread (Liao et al., 2014), UMI-Tools was used to count the unique reads per annotated gene/repeat element per cell. Finally, count matrices of the human repeat reference alignment and transcriptome were concatenated, providing standard single cell RNA-sequencing data on the transcriptome in conjunction with novel count data of repeat elements at a single cell level.
Clustering and differential gene expression analysis
Briefly, the concatenated count matrix was read into R/3.4.4 for analysis with the Seurat/2.1 suite of tools (Satija et al., 2015). Beginning with 196 cells from GW19 and 389 cells from GW25, we filtered on a 0.06% mitochondrial gene expression threshold and an nGene value of 3000. We were left with 191 cells from GW19 and 363 cells from GW25. 24,432 genes and 1225 repeat elements were expressed across all 191 cells from GW19. 24,507 genes and 1320 repeat elements were expressed across all 363 cells from GW19. Using Seurat, 782 and 637 variable genes were identified from the three defined clusters of somatic cells, primordial germ cells and advanced germ cells in GW19 and GW25, respectively. Transcriptome markers for the primordial and advanced germ cell populations were determined using the FindMarkers Seurat function and the ‘roc’ test with a minimum cellular detection threshold of 0.25 for each population. Additionally, transposon expression data alone were appended as an assay to the Seurat object, enabling us to ascertain which repeat elements were markers for each germ cell population.
Statistics
Experimental data were compared using Student's t-test and Fisher's exact test as noted in the figure legends. The significance of Pearson's correlation coefficient was determined with regression analysis.
Supplementary Material
Acknowledgements
We are grateful to Brandon Sigamony, Daniel Nguyen, Suping Peng and other members of the Laird lab for their feedback and support, as well as to Drs Gerald Cunha and Miguel Ramalho-Santos for comments on the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: B.R., D.J.L.; Methodology: B.R., S.A.C., R.G.J., L.J.M., P.Y., W.A.; Software: S.A.C., R.G.J., P.Y.; Validation: P.Y., D.J.L.; Formal analysis: B.R., S.A.C., R.G.J.; Investigation: B.R., J.S., M.C., W.A., D.J.L.; Resources: J.S., M.C., L.B., D.J.L.; Data curation: S.A.C., R.G.J.; Writing - original draft: B.R., S.A.C., D.J.L.; Writing - review & editing: B.R., S.A.C., R.G.J., L.J.M., P.Y., W.A., D.J.L.; Supervision: L.B., D.J.L.; Project administration: D.J.L.; Funding acquisition: L.B., D.J.L.
Funding
This work was funded by the National Institutes of Health (R21ES023297, R01GM122902 and R01ES028212 to D.J.L.; R21HD080143 and R21OD017965 to W.A.; T32HD7263-31 to B.R.; 1F31HD096840 to R.G.J.; K12DK083021 to L.B.; and 5T32HD007470 to S.A.C.), by the California Institute of Regenerative Medicine (EDUC2-08391 to L.J.M.), by the Markl Faculty Fund for Cancer Research (W.A.) and by the Escher Fund for Autism (D.J.L.). Deposited in PMC for release after 12 months.
Data availability
The human fetal samples obtained for this study were de-identified and explicit consent to share raw data that could contain genetic variants permitting identification is not permissible under our CHR protocol. Table 1 contains a breakdown of read counts for gene features in each individual sample.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.171157.supplemental
References
- Anders S., Pyl P. T. and Huber W. (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166-169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R., Fässler R., Georges-Labouesse E., Hynes R. O., Bader B. L., Kreidberg J. A., Schaible K., Heasman J. and Wylie C. (1999). Mouse primordial germ cells lacking beta1 integrins enter the germline but fail to migrate normally to the gonads. Development 126, 1655-1664. [DOI] [PubMed] [Google Scholar]
- Antoniewski C. (2014). Computing siRNA and piRNA overlap signatures. Methods Mol. Biol. 1173, 135-146. 10.1007/978-1-4939-0931-5_12 [DOI] [PubMed] [Google Scholar]
- Aravin A. A., Sachidanandam R., Girard A., Fejes-Tóth K. and Hannon G. J. (2007). Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316, 744-747. 10.1126/science.1142612 [DOI] [PubMed] [Google Scholar]
- Aravin A. A., Sachidanandam R., Bourc'his D., Schaefer C., Pezic D., Toth K. F., Bestor T. and Hannon G. J. (2008). A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31, 785-799. 10.1016/j.molcel.2008.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A. A., van der Heijden G. W., Castañeda J., Vagin V. V., Hannon G. J. and Bortvin A. (2009). Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet. 5, e1000764 10.1371/journal.pgen.1000764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W., Kojima K. K. and Kohany O. (2015). Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11 10.1186/s13100-015-0041-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer M. A. and Deininger P. L. (2002). Alu repeats and human genomic diversity. Nat. Rev. Genet. 3, 370-379. 10.1038/nrg798 [DOI] [PubMed] [Google Scholar]
- Buchner J. (1999). Hsp90 & Co. - a holding for folding. Trends Biochem. Sci. 24, 136-141. 10.1016/S0968-0004(99)01373-0 [DOI] [PubMed] [Google Scholar]
- Carmell M. A., Girard A., van de Kant H. J. G., Bourc'his D., Bestor T. H., de Rooij D. G. and Hannon G. J. (2007). MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 12, 503-514. 10.1016/j.devcel.2007.03.001 [DOI] [PubMed] [Google Scholar]
- Conesa A., Madrigal P., Tarazona S., Gomez-Cabrero D., Cervera A., McPherson A., Szcześniak M. W., Gaffney D. J., Elo L. L., Zhang X. et al. (2016). A survey of best practices for RNA-seq data analysis. Genome Biol. 17, 13 10.1186/s13059-016-0881-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B. and Hannon G. J. (2016). One loop to rule them all: the ping-pong cycle and piRNA-guided silencing. Trends Biochem. Sci. 41, 324-337. 10.1016/j.tibs.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fazio S., Bartonicek N., Di Giacomo M., Abreu-Goodger C., Sankar A., Funaya C., Antony C., Moreira P. N., Enright A. J. and O'Carroll D. (2011). The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480, 259-263. 10.1038/nature10547 [DOI] [PubMed] [Google Scholar]
- Deng W. and Lin H. (2002). miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell 2, 819-830. 10.1016/S1534-5807(02)00165-X [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M. and Gingeras T. R. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15-21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergün S., Buschmann C., Heukeshoven J., Dammann K., Schnieders F., Lauke H., Chalajour F., Kilic N., Strätling W. H. and Schumann G. G. (2004). Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J. Biol. Chem. 279, 27753-27763. 10.1074/jbc.M312985200 [DOI] [PubMed] [Google Scholar]
- Ernst C., Odom D. T. and Kutter C. (2017). The emergence of piRNAs against transposon invasion to preserve mammalian genome integrity. Nat. Commun. 8, 1411 10.1038/s41467-017-01049-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q. and Wang P. J. (2014). Mammalian piRNAs: biogenesis, function, and mysteries. Spermatogenesis 4, e27889 10.4161/spmg.27889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gainetdinov I., Skvortsova Y., Kondratieva S., Funikov S. and Azhikina T. (2017). Two modes of targeting transposable elements by piRNA pathway in human testis. RNA 23, 1614-1625. 10.1261/rna.060939.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangaraju V. K., Yin H., Weiner M. M., Wang J., Huang X. A. and Lin H. (2011). Drosophila Piwi functions in Hsp90-mediated suppression of phenotypic variation. Nat. Genet. 43, 153-158. 10.1038/ng.743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S. X. (2017). Exploratory bioinformatics investigation reveals importance of “junk” DNA in early embryo development. BMC Genomics 18, 200 10.1186/s12864-017-3566-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkountela S., Li Z., Vincent J. J., Zhang K. X., Chen A., Pellegrini M. and Clark A. T. (2013). The ontogeny of cKIT+ human primordial germ cells proves to be a resource for human germ line reprogramming, imprint erasure and in vitro differentiation. Nat. Cell Biol. 15, 113-122. 10.1038/ncb2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkountela S., Zhang K. X., Shafiq T. A., Liao W.-W., Hargan-Calvopiña J., Chen P.-Y. and Clark A. T. (2015). DNA demethylation dynamics in the human prenatal germline. Cell 161, 1425-1436. 10.1016/j.cell.2015.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes Fernandes M., He N., Wang F., Van Iperen L., Eguizabal C., Matorras R., Roelen B. A. J. and Chuva de Sousa Lopes S. M. (2018). Human-specific subcellular compartmentalization of P-element induced wimpy testis-like (PIWIL) granules during germ cell development and spermatogenesis. Hum. Reprod. 33, 258-269. 10.1093/humrep/dex365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Yan L., Guo H., Li L., Hu B., Zhao Y., Yong J., Hu Y., Wang X., Wei Y. et al. (2015). The transcriptome and DNA methylome landscapes of human primordial germ cells. Cell 161, 1437-1452. 10.1016/j.cell.2015.05.015 [DOI] [PubMed] [Google Scholar]
- Guo H., Hu B., Yan L., Yong J., Wu Y., Gao Y., Guo F., Hou Y., Fan X., Dong J. et al. (2017). DNA methylation and chromatin accessibility profiling of mouse and human fetal germ cells. Cell Res. 27, 165-183. 10.1038/cr.2016.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha H., Song J., Wang S., Kapusta A., Feschotte C., Chen K. C. and Xing J. (2014). A comprehensive analysis of piRNAs from adult human testis and their relationship with genes and mobile elements. BMC Genomics 15, 545 10.1186/1471-2164-15-545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadziselimovic F., Hadziselimovic N. O., Demougin P., Krey G. and Oakeley E. (2015). Piwi-pathway alteration induces LINE-1 transposon derepression and infertility development in cryptorchidism. Sex. Dev. 9, 98-104. 10.1159/000375351 [DOI] [PubMed] [Google Scholar]
- Hancks D. C. and Kazazian H. H. (2012). Active human retrotransposons: variation and disease. Curr. Opin. Genet. Dev. 22, 191-203. 10.1016/j.gde.2012.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks D. C. and Kazazian H. H. (2016). Roles for retrotransposon insertions in human disease. Mob. DNA 7, 9 10.1186/s13100-016-0065-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichiyanagi T., Ichiyanagi K., Ogawa A., Kuramochi-Miyagawa S., Nakano T., Chuma S., Sasaki H. and Udono H. (2014). HSP90α plays an important role in piRNA biogenesis and retrotransposon repression in mouse. Nucleic Acids Res. 42, 11903-11911. 10.1093/nar/gku881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki Y. W., Siomi M. C. and Siomi H. (2015). PIWI-interacting RNA: its biogenesis and functions. Annu. Rev. Biochem. 84, 405-433. 10.1146/annurev-biochem-060614-034258 [DOI] [PubMed] [Google Scholar]
- Izumi N., Kawaoka S., Yasuhara S., Suzuki Y., Sugano S., Katsuma S. and Tomari Y. (2013). Hsp90 facilitates accurate loading of precursor piRNAs into PIWI proteins. RNA 19, 896-901. 10.1261/rna.037200.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jachowicz J. W., Bing X., Pontabry J., Bošković A., Rando O. J. and Torres-Padilla M.-E. (2017). LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat. Genet. 49, 1502-1510. 10.1038/ng.3945 [DOI] [PubMed] [Google Scholar]
- Kazazian H. H. and Moran J. V. (2017). Mobile DNA in health and disease. N. Engl. J. Med. 377, 361-370. 10.1056/NEJMra1510092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Günesdogan U., Zylicz J. J., Hackett J. A., Cougot D., Bao S., Lee C., Dietmann S., Allen G. E., Sengupta R. et al. (2014). PRMT5 protects genomic integrity during global DNA demethylation in primordial germ cells and preimplantation embryos. Mol. Cell 56, 564-579. 10.1016/j.molcel.2014.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K. K. (2018). Human transposable elements in Repbase: genomic footprints from fish to humans. Mob. DNA 9, 2 10.1186/s13100-017-0107-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S., Watanabe T., Gotoh K., Totoki Y., Toyoda A., Ikawa M., Asada N., Kojima K., Yamaguchi Y., Ijiri T. W. et al. (2008). DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 22, 908-917. 10.1101/gad.1640708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S., Watanabe T., Gotoh K., Takamatsu K., Chuma S., Kojima-Kita K., Shiromoto Y., Asada N., Toyoda A., Fujiyama A. et al. (2010). MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 24, 887-892. 10.1101/gad.1902110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M. and Salzberg S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Dong J., Yan L., Yong J., Liu X., Hu Y., Fan X., Wu X., Guo H., Wang X. et al. (2017). Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell 20, 858-873.e4. 10.1016/j.stem.2017.03.007 [DOI] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K. and Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923-930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Malki S., van der Heijden G. W., O'Donnell K. A., Martin S. L. and Bortvin A. (2014). A role for retrotransposon LINE-1 in fetal oocyte attrition in mice. Dev. Cell 29, 521-533. 10.1016/j.devcel.2014.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manakov S. A., Pezic D., Marinov G. K., Pastor W. A., Sachidanandam R. and Aravin A. A. (2015). MIWI2 and MILI have differential effects on piRNA biogenesis and DNA methylation. Cell Rep. 12, 1234-1243. 10.1016/j.celrep.2015.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto M. C. N., Narvaiza I., Denli A. M., Benner C., Lazzarini T. A., Nathanson J. L., Paquola A. C. M., Desai K. N., Herai R. H., Weitzman M. D. et al. (2013). Differential L1 regulation in pluripotent stem cells of humans and apes. Nature 503, 525-529. 10.1038/nature12686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Mbandi S. K., Hesse U., Rees D. J. G. and Christoffels A. (2014). A glance at quality score: implication for de novo transcriptome reconstruction of Illumina reads. Front. Genet. 5, 17 10.3389/fgene.2014.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt D. M., Knowles B. B. and Solter D. (2014). DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 28, 812-828. 10.1101/gad.234294.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills L. (2014). Common file formats. Curr. Protoc. Bioinformatics 45, A.1B.1-A.1B.18. [DOI] [PubMed] [Google Scholar]
- Mitchell R. T., Saunders P. T. K., Childs A. J., Cassidy-Kojima C., Anderson R. A., Wallace W. H. B., Kelnar C. J. H. and Sharpe R. M. (2010). Xenografting of human fetal testis tissue: a new approach to study fetal testis development and germ cell differentiation. Hum. Reprod. 25, 2405-2414. 10.1093/humrep/deq183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newkirk S. J., Lee S., Grandi F. C., Gaysinskaya V., Rosser J. M., Vanden Berg N., Hogarth C. A., Marchetto M. C. N., Muotri A. R., Griswold M. D. et al. (2017). Intact piRNA pathway prevents L1 mobilization in male meiosis. Proc. Natl. Acad. Sci. USA 114, E5635-E5644. 10.1073/pnas.1701069114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percharde M., Lin C.-J., Yin Y., Guan J., Peixoto G. A., Bulut-Karslioglu A., Biechele S., Huang B., Shen X. and Ramalho-Santos M. (2018). A LINE1-nucleolin partnership regulates early development and ESC identity. Cell 174, 391-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezic D., Manakov S. A., Sachidanandam R. and Aravin A. A. (2014). piRNA pathway targets active LINE1 elements to establish the repressive H3K9me3 mark in germ cells. Genes Dev. 28, 1410-1428. 10.1101/gad.240895.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S. R., Gerdes P., Gerhardt D. J., Sanchez-Luque F. J., Bodea G.-O., Muñoz-Lopez M., Jesuadian J. S., Kempen M.-J. H. C., Carreira P. E., Jeddeloh J. A. et al. (2017). Heritable L1 retrotransposition in the mouse primordial germline and early embryo. Genome Res. 27, 1395-1405. 10.1101/gr.219022.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodić N., Sharma R., Sharma R., Zampella J., Dai L., Taylor M. S., Hruban R. H., Iacobuzio-Donahue C. A., Maitra A., Torbenson M. S. et al. (2014). Long interspersed element-1 protein expression is a hallmark of many human cancers. Am. J. Pathol. 184, 1280-1286. 10.1016/j.ajpath.2014.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roovers E. F., Rosenkranz D., Mahdipour M., Han C.-T., He N., Chuva de Sousa Lopes S. M., van der Westerlaken L. A. J., Zischler H., Butter F., Roelen B. A. J. et al. (2015). Piwi proteins and piRNAs in mammalian oocytes and early embryos. Cell Rep. 10, 2069-2082. 10.1016/j.celrep.2015.02.062 [DOI] [PubMed] [Google Scholar]
- Satija R., Farrell J. A., Gennert D., Schier A. F. and Regev A. (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495-502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji M., Tanaka T., Hosokawa M., Reuter M., Stark A., Kato Y., Kondoh G., Okawa K., Chujo T., Suzuki T. et al. (2009). The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev. Cell 17, 775-787. 10.1016/j.devcel.2009.10.012 [DOI] [PubMed] [Google Scholar]
- Smit A. F. A., Tóth G., Riggs A. D. and Jurka J. (1995). Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J. Mol. Biol. 246, 401-417. 10.1006/jmbi.1994.0095 [DOI] [PubMed] [Google Scholar]
- Smith T., Heger A. and Sudbery I. (2017). UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 27, 491-499. 10.1101/gr.209601.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specchia V., Piacentini L., Tritto P., Fanti L., D'Alessandro R., Palumbo G., Pimpinelli S. and Bozzetti M. P. (2010). Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature 463, 662-665. 10.1038/nature08739 [DOI] [PubMed] [Google Scholar]
- Tang W. W. C., Dietmann S., Irie N., Leitch H. G., Floros V. I., Bradshaw C. R., Hackett J. A., Chinnery P. F. and Surani M. A. (2015). A unique gene regulatory network resets the human germline epigenome for development. Cell 161, 1453-1467. 10.1016/j.cell.2015.04.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W. W. C., Kobayashi T., Irie N., Dietmann S. and Surani M. A. (2016). Specification and epigenetic programming of the human germ line. Nat. Rev. Genet. 17, 585-600. 10.1038/nrg.2016.88 [DOI] [PubMed] [Google Scholar]
- Taylor M. S., Altukhov I., Molloy K. R., Mita P., Jiang H., Adney E. M., Wudzinska A., Badri S., Ischenko D., Eng G. et al. (2018). Dissection of affinity captured LINE-1 macromolecular complexes. eLife 7, e30094 10.7554/eLife.30094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharmalingam M. D., Jorgensen A. and Mitchell R. T. (2018). Experimental models of testicular development and function using human tissue and cells. Mol. Cell. Endocrinol. 468, 95-110. 10.1016/j.mce.2017.12.011 [DOI] [PubMed] [Google Scholar]
- Vagin V. V., Wohlschlegel J., Qu J., Jonsson Z., Huang X., Chuma S., Girard A., Sachidanandam R., Hannon G. J. and Aravin A. A. (2009). Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 23, 1749-1762. 10.1101/gad.1814809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiliauskaitė L., Vitsios D., Berrens R. V., Carrieri C., Reik W., Enright A. J. and O'Carroll D. (2017). A MILI-independent piRNA biogenesis pathway empowers partial germline reprogramming. Nat. Struct. Mol. Biol. 24, 604-606. 10.1038/nsmb.3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western P. S., Miles D. C., van den Bergen J. A., Burton M. and Sinclair A. H. (2008). Dynamic regulation of mitotic arrest in fetal male germ cells. Stem Cells 26, 339-347. 10.1634/stemcells.2007-0622 [DOI] [PubMed] [Google Scholar]
- Williams Z., Morozov P., Mihailovic A., Lin C., Puvvula P. K., Juranek S., Rosenwaks Z. and Tuschl T. (2015). Discovery and characterization of piRNAs in the human fetal ovary. Cell Rep. 13, 854-863. 10.1016/j.celrep.2015.09.030 [DOI] [PubMed] [Google Scholar]
- Xiol J., Cora E., Koglgruber R., Chuma S., Subramanian S., Hosokawa M., Reuter M., Yang Z., Berninger P., Palencia A. et al. (2012). A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol. Cell 47, 970-979. 10.1016/j.molcel.2012.07.019 [DOI] [PubMed] [Google Scholar]
- Zheng K., Xiol J., Reuter M., Eckardt S., Leu N. A., McLaughlin K. J., Stark A., Sachidanandam R., Pillai R. S. and Wang P. J. (2010). Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc. Natl. Acad. Sci. USA 107, 11841-11846. 10.1073/pnas.1003953107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.