ABSTRACT
Single cell biology is currently revolutionizing developmental and evolutionary biology, revealing new cell types and states in an impressive range of biological systems. With the accumulation of data, however, the field is grappling with a central unanswered question: what exactly is a cell type? This question is further complicated by the inherently dynamic nature of developmental processes. In this Hypothesis article, we propose that a ‘periodic table of cell types’ can be used as a framework for distinguishing cell types from cell states, in which the periods and groups correspond to developmental trajectories and stages along differentiation, respectively. The different states of the same cell type are further analogous to ‘isotopes’. We also highlight how the concept of a periodic table of cell types could be useful for predicting new cell types and states, and for recognizing relationships between cell types throughout development and evolution.
KEY WORDS: Cell atlas, Cell states, Cell types, Periodic table, Single cell RNA-seq
Summary: This Hypothesis proposes a periodic table that aligns cell types according to their developmental stages, connecting them to one another according to the universal axis from stem cells to differentiated cells.
Introduction
In 1665, Robert Hooke coined the term ‘cell’ to describe the micro unit of biological tissues (Hooke, 1665). Continued efforts on cells culminated in ‘cell theory’, the central tenet of which is the now well-accepted assertion of the cell as the basic unit of all living organisms (with the notable exception of viruses) (Wolpert, 1996). More importantly, cell theory proposed that all cells must arise from pre-existing cells, highlighting the heritability and fate transitions of cells (Mazzarello, 1999). Collectively, ‘cell theory’ stands today as one the greatest conceptual innovations in the life sciences.
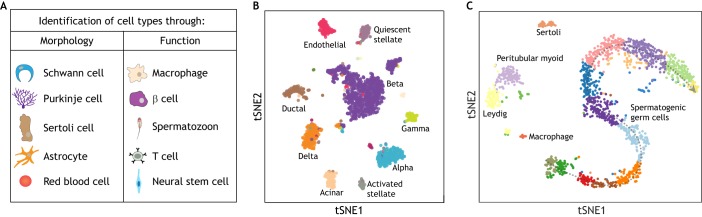
Understanding cell type has occupied a central place in research in the life sciences, with intense efforts devoted to studying how cells specialize into different cell types that exhibit distinct morphologies and carry out different functions. The sperm cell (spermatozoon) was among the very first cell types to be identified, and is easily recognizable owing to its axial filament, which facilitates motility during fertilization. Indeed, shape – or morphology – has long been used as the dominant feature for defining the identities and functions of different cell types (Wolpert, 1996). The invention and increasing optimization of microscopy technologies greatly assisted in distinguishing the anatomical properties of cells, and cell types have often been named after their morphology (e.g. astrocytes, red blood cells and muscle cells) or the name of their discoverer (e.g. Schwann, Sertoli and Purkinje cells) (Fig. 1A).
Fig. 1.
Historical and scRNA-seq approaches to identifying cell types. (A) Examples of historical identification of cell types through morphologies and cell functions. (B,C) tSNE plots of scRNA-seq data from adult human pancreatic cell populations (B) and adult human testicular cell populations (C). These data highlight the existence of discrete cell populations in the mature pancreas tissue (B) and a continuum of germ cell populations in the testicular tissue (C). The pancreas data is adapted from Baron et al. (2016) and the testis data is adapted from Xia et al. (2018 preprint).
With the development of modern biological techniques, the features used to define cell types gradually evolved to incorporate more aspects of cellular function, such as the presence of specific ‘marker’ proteins and physiological properties. The presence of cell surface proteins (Zhu and Paul, 2008), secretion molecules (Romer and Sussel, 2015), transcription factors (Tapscott et al., 1988), as well as cell functions and tissue enrichment (Fan and Rudensky, 2016), have been popular ways to identify and define cells. For example, the T cell/lymphocyte was identified by the physiological feature of its maturation in the thymus (Alberts et al., 2014). Despite this increasing understanding of cells, the definition of a cell type is generally superficial because of a lack of any standardized measurement. Such a definition may also be constrained by a lack of comprehensive understanding of the properties of a cell, leading to a general challenge for distinguishing true cell types from particular cell states of the same cell type (see also Morris, 2019 in this issue). Considering these limitations in our current understanding of cell types, there is a clear need for the development of a robust formal framework for the ‘cell type’ concept.
The rise of ‘omics’ technologies, including microarrays and high-throughput sequencing technologies (e.g. RNA-seq), have further expanded our understanding of cell types. In this context, a cell type is defined by its expressed part of the genome, i.e. transcripts, translated proteins/peptides and networks between the transcription products. High-throughput sequencing of transcriptomes across various samples has led us to appreciate that most genes have relatively broad expression patterns across multiple sample types (Melé et al., 2015). Considering this observation, the historical definition of a cell type based upon a few features or marker genes may not be sufficient to define the full gamut of cell types encoded by the genome.
In this Hypothesis article, we discuss how high-throughput sequencing technologies – with a focus on single cell transcriptomics – are revolutionizing our understanding of cell types and cell states in biological systems. We propose that it is possible to practically define cell types according to their expressed transcription factors (TFs). In addition, we propose that a ‘periodic table of cell types’ could be used as a framework for organizing the relationships between cell types and cell states encoded by a species. In this framework, the periods and groups correspond to developmental trajectories and aligned differentiation stages, respectively, and the different states of the same cell type are analogous to ‘isotopes’ of the periodic table elements. Collectively, we expect that this concept of a ‘periodic table’ of cell types will be useful for understanding cell fate transitions during development and for predicting new cell types and states in future developmental and evolutionary studies.
The single cell RNA-seq revolution
RNA-seq on tissue/organ samples such as the brain, liver, pancreas, testes or blood samples results in data that represent the average transcriptome of many different kinds of cell types, thus not faithfully characterizing any unique cell type (Kolodziejczyk et al., 2015a; Melé et al., 2015; Ozsolak and Milos, 2011). Even when pure cell populations can be isolated and processed for RNA-seq analysis, the possible cell states of that cell type may not be fully captured (Kolodziejczyk et al., 2015b). Furthermore, isolating a unique cell type requires prior knowledge about its markers, rendering it difficult to find new cell types. In contrast, high-throughput single cell RNA-seq (scRNA-seq) is able to distinguish distinct cell populations with unique transcriptomic signatures computationally when starting with a pool of cell populations containing different types and states (Gierahn et al., 2017; Klein et al., 2015; Rosenberg et al., 2018; Svensson et al., 2018). scRNA-seq has thus become a revolutionary technique, identifying diverse cells across multiple tissues, organs and species, and possibly with spatial-temporal resolution (Shapiro et al., 2013; Mayr et al., 2019 in this issue).
Over the past decade, we have witnessed many single cell transcriptomic studies revealing the cellular heterogeneity of multiple tissues and organs with unprecedented detail (Wagner et al., 2016). Using novel cell population clustering analysis and dimension-reduction visualization, a majority of cells in mature tissues have been delineated into clusters, each putatively corresponding to a specific cell type (Kolodziejczyk et al., 2015a). For example, scRNA-seq applied to pancreatic islet tissue has captured endocrine and exocrine cell types such as alpha cells, beta cells and acinar cells, and even rare populations, such as gamma cells (Baron et al., 2016; Muraro et al., 2016; Segerstolpe et al., 2016) (Fig. 1B). Cells of the same type usually have very similar transcriptomic programs, which are distinct from those of other cell types. New cell types and states can even be identified in cell populations of the whole body of a species, as has been preliminarily achieved in Caenorhabditis elegans (Cao et al., 2017) and mouse (Han et al., 2018; Tabula Muris Consortium, 2018), and the ongoing Human Cell Atlas project also aims to generate such a map of human cell types (Regev et al., 2017).
scRNA-seq raises new challenges for studies of developmental systems
Beyond identifying discrete cell populations corresponding to mature cell types and states, scRNA-seq can identify developmental trajectories in diverse systems, such as in the developing embryo, in adult stem cell populations and even during tumor progression (Behjati et al., 2018; Marioni and Arendt, 2017; Tritschler et al., 2019 in this issue). Recent applications of scRNA-seq in developmental systems include the analysis of hematopoiesis (Athanasiadis et al., 2017; Macaulay et al., 2016) and spermatogenesis (Ernst et al., 2019; Green et al., 2018; Guo et al., 2018; Hermann et al., 2018; Wang et al., 2018; Xia et al., 2018 preprint), and the analysis of embryogenesis in zebrafish, frog and mouse (Briggs et al., 2018; Cao et al., 2019; Farrell et al., 2018; Pijuan-Sala et al., 2019; Wagner et al., 2018). The increasing amount of data generated by such studies also enables us to identify minor cell types and hitherto unappreciated cell states during development, and their interactions with the neighboring environment.
However, analyzing scRNA-seq results from developmental system challenges our canonical understanding of cell types. In contrast to differentiated cell populations, which usually group into discrete populations (Fig. 1B), cells in developing systems usually appear as a continuum, as exemplified by cells during spermatogenesis (Fig. 1C). At the gene expression level, a particular cell during a stage of development will be defined by a set of genes whose expression has just initiated and is soon to diminish; as opposed to mature cell types that contain a stable set of differentially expressed (DE) genes. For example, in the mammalian testis, a full spectrum of germ cell types is seen, from spermatogonial stem cells to mature sperm cells, whereas the testicular somatic cells are mature cell types (Fig. 1C). By analyzing scRNA-seq data from both human and mouse, a continuum of cell fate transitions can be separated into specific cell populations (Chen et al., 2018; Green et al., 2018; Guo et al., 2018; Wang et al., 2018; Xia et al., 2018 preprint). Although spermatogenesis involves extremely dynamic gene expression changes, these are expected to be gradual rather than punctuated at the transcriptomic level.
The observation of a continuum of single-cell transcriptomes in developmental systems raises two important issues. First, how do we distinguish transient cell states from cell types in a developmental system? This topic is experimentally and computationally challenging because of the biological dynamics inherent to developmental systems. Second, how do we identify the developmental cell types that precede the observed mature cell types? These ‘missing’ cell states may be overlooked in scRNA-seq experiments owing to their rarity – or even absence – in the adult tissue or to the bias in current tissue dissociation methods (Clevers, 2015). A species ‘cell atlas’ is thus not complete without the inclusion of development as a key piece of biological information.
Distinguishing cell types from cell states
The analysis of scRNA-seq data also presents conceptual challenges when inferring cell types and cell states (Clevers et al., 2017; Trapnell, 2015). A typical scRNA-seq analysis involves the clustering of cells based upon the expression of differential expression (DE) genes. However, these cell clusters are not necessarily equivalent to cell types as DE genes may not only distinguish these but also capture differences in the state of the cell cycle, stress signatures, and, inevitably, technical noise (Kiselev et al., 2019). Meanwhile, the data quality may often not be sufficient to separate similar cell types, leading them to appear as a single cluster. Thus, although scRNA-seq methods have unleashed a torrent of new analysis tools, a conceptual biological approach is required for distinguishing cell types from cell states (Clevers et al., 2017).
In two recent studies, Arendt et al. proposed an evolutionary definition of a ‘cell type’ as a group of cells that share a defined core regulatory complex (CoRC) that is stable over evolutionary timescales (Arendt, 2008; Arendt et al., 2016). The CoRC is made up of a set of TFs (and their interacting factors) that together define the gene expression profiles of the cell. From this perspective, the origin of a new cell type in evolution is identifiable as an evolutionary occurrence of a unique CoRC relative to that of its sister cell types (Arendt et al., 2016). In a developmental context, the CoRC can be used to distinguish cell type identities across ontogenesis. Collectively, the CoRC concept provides a standardized definition of a cell type from an evolutionary and developmental perspective.
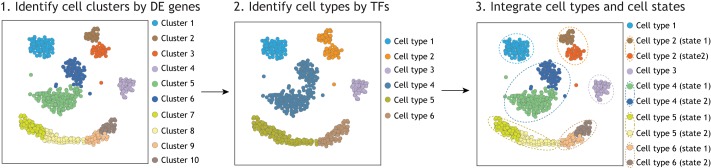
A practical approach to distinguishing cell types from cell states based upon the concept of the CoRC may be to identify the molecular signature of regulatory programs in cells based on scRNA-seq data. As there is a lack of prior knowledge for the cell-identity-defining CoRC for most cell types, one option is to use the expression profiles of TFs as a proxy for CoRCs (Lambert et al., 2018). TF expression has consistently been shown to be highly correlated with cell type identity (Graf and Enver, 2009; Lambert et al., 2018). In this framework, a cell type is expected to have a unique expression profile of TFs regardless of the state of an individual cell – whether cycling, stressed or otherwise temporally responding to its environment. Applying both the notions of cell clustering with DE genes and examining TF profiles separately, we may derive a practical approach for distinguishing cell types and cell states (Fig. 2). First, we would determine cell clusters using the DE genes, capturing both cell types and cell states (Fig. 2, left). Second, we would examine exclusively the TFs, capturing the identity of the cell types (Fig. 2, middle). Integrating these two results, the cell states within the same cell type may be distinguished (Fig. 2, right). The distinction between cell states and cell types has been observed, for example, in human pancreatic cells (Baron et al., 2016; Kiselev et al., 2019; Li et al., 2016; Segerstolpe et al., 2016) and human placental cells (Vento-Tormo et al., 2018), and in the Tabula Muris project (Han et al., 2018; Tabula Muris Consortium, 2018). Specifically, in the Tabula Muris project, the expression profile of TFs was well-utilized to distinguish cell-type identities across multiple organs, whereas other gene sets, for example cell-surface markers and RNA splicing factors, did not perform as well in distinguishing cell types (Han et al., 2018; Tabula Muris Consortium, 2018).
Fig. 2.
A framework for distinguishing cell types from cell states using scRNA-seq. Individual cell transcriptomes can be clustered using typical methods, for example based on DE genes (left), as performed by most scRNA-seq projects. At this point, the cell clusters may not be equivalent to cell types. However, a second step of cell clustering, using only informative transcription factors (TFs), can be used as an additional criterion to distinguish cell types (middle). Thus, by integrating different cell clustering tools (right), we can distinguish cell types from cell states in silico, which can be further tested by molecular and cell biology validation.
Within a population of cells corresponding to the same cell type – identified perhaps using the CoRC notion – there may be several possible states. We view a cell state as a secondary module operating in addition to the general cell type regulatory program. This may correspond, for example, to the different states observed during cell cycle. Alternatively, different cell states could reflect distinct response programs elicited following exposure to different environmental stimulations. They could involve a hypoxia signal, a general stress signature, or a cell type interaction signature in a spatial and/or temporal manner. For example, in pancreatic ductal cells, we previously observed two states – the centroacinar state and the terminal state – corresponding to the relative location of cells along the duct and the unique cells that each interacts with (Baron et al., 2016). A cell state in our view, therefore, does not reflect different progression along development or a transitional pattern, but rather corresponds to the modular addition of a regulatory program to a CoRC. Generally, a cell state exhibits dynamics across a shorter timeframe than does a cell type, and a particular stimulation leading to a cell state may be similarly invoked in other cell types.
While considering a set of TFs as a proxy for the CoRC, it should not be assumed that each TF contributes equally to defining the cell type, as previous studies have revealed hierarchical TF regulatory pathways (Lambert et al., 2018; Wilkinson et al., 2017; Yu and Gerstein, 2006). Indeed, multiple master TFs or fate-decision TFs have been identified in different cellular contexts (Lambert et al., 2018; Mullen et al., 2011). Most prominently, in embryonic stem cells (ESCs), the master regulators OCT4 (also known as POU5F1), SOX2 and NANOG together define the cell identities of ESCs, indicating that many of the TFs involved in a particular cell type can be induced because of the presence of these master TFs (Niwa, 2014, 2018). It is therefore important to deconstruct gene regulation networks to distinguish the fate-decision TFs from ‘responder’ TFs in order to further empower the classification of cell types from scRNA-seq data (Wilkinson et al., 2017). Another concern of using TFs as a tool for distinguishing cell types is the potential inconsistency with existing knowledge. Owing to a lack of standardized classification, the current definition of cell types may often involve non-TF marker gene expression. For example, the T cell lineage generates both CD4+ and CD8+ T cells, which are controlled by the Tox-Gata3-ThPOK and Runx3-MAZR TF gene sets during development, respectively (Ellmeier et al., 2013; Germain, 2002; Singer et al., 2008). However, this regulatory program of TFs may not be as apparent in mature cells, making it difficult to distinguish these cell types through TF expression profiles. This concern can be addressed by unbiased sampling of the developing cells, thereby capturing gene expression in the preceding developmental stages.
A periodic table of cell types
As we approach the ability to distinguish the full complement of cell types for a given species, an interesting – though challenging – task becomes to organize them meaningfully and informatively. Sixty-two years ago, C. H. Waddington proposed the notion of an ‘epigenetic landscape’ in his book, The Strategy of the Genes, to illustrate gene action during development and cell differentiation (Waddington, 1957). In this analogy, cells are akin to balls rolling down a landscape with different possible paths, each representing a distinct cell differentiation trajectory. Waddington's landscape effectively models local cell fate transitions and highlights the need to incorporate development in our understanding of cell types. Studying the nematode C. elegans, Sir John Sulston and colleagues deciphered the full cell lineage of embryogenesis and the somatic cells of post-embryogenesis (Sulston and Horvitz, 1977; Sulston et al., 1983). Such mapping provides an invaluable resource as it highlights the full set of differentiation trajectories in an organism. Extending this view to the gene expression level could define all possible cell types within a species. However, in the case of highly complex vertebrate species, this appears to be a bewildering task owing to the large number of cells. Recently, the Cell Ontology (CL) system has been proposed as a way of classifying cell types (Diehl et al., 2016; Osumi-Sutherland, 2017). This system, which uses an approach similar to that used in the Gene Ontology system (Ashburner et al., 2000), involves high-level abstraction and is highly attractive for synthesizing knowledge on well-studied cell types. However, we currently lack a system for informatively organizing cell types across both development and physiology (states), in terms of their connections and relationships.
The year 2019 has been designated by UNESCO as the International Year of the Periodic Table, in celebration of the 150th anniversary of the Mendeleev periodic table of elements. The periodic table was revolutionary for its vision to build connections among the elements and for leading to the prediction of new elements (Gordin, 2019). We propose that the cell types and cell fate transitions in a species – together with their corresponding cell states – can be organized in an analogous manner in a ‘periodic table of cell types’. The periodic table of cell types for a particular species would include all of the cell types in the organism, each related to one another as the elements are in the chemical periodic table: each period (row) of cell types reflects the transitions of cell fates for a particular developmental trajectory, and each group (column) of cells reflects an axis from the stem cell phase to differentiation. In an analogy to the isotopes of elements, each cell type contains multiple cell states. We believe that such a periodic table of cell types could summarize our understanding of cell fates and cell lineages, and may help to predict missing cell types during development and across species.
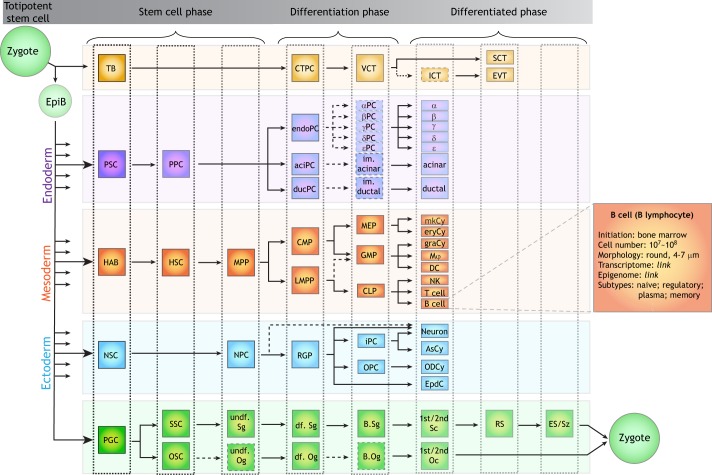
In a periodic table of cell types, each period (row) of the table represents a cell differentiation trajectory, starting from a stem cell through to fully differentiated cell types. Similar to the periods of the table of elements, in which the chemical properties transition from metal to inorganic elements, the cell types in a period are characterized by their transitioning cell fates across the developmental trajectory (Fig. 3). Each period shares three major phases from left to the right in the table: the stem cell phase, the progenitor/differentiation phase, and the differentiated phase. In contrast to the periods of elements, which exhibit incremental transitions based on electron shell filling rules, the periods of cell types may be simple or complex, i.e. uni-trajectory or complex trajectory, respectively, and this provides an order to the trajectories along their vertical axis. For example, the spermatogenic lineage can be viewed as having a simple period because of its linear (no branch) differentiation map (Fig. 3, bottom) (Kanatsu-Shinohara and Shinohara, 2013). In contrast, the hematopoietic stem cell (HSC) differentiation trajectory is an example for which a period consists of multiple branches forming various lymphoid and myeloid cell types (Athanasiadis et al., 2017; Guibentif et al., 2017; Macaulay et al., 2016; Velten et al., 2017; Zhang et al., 2018) (Fig. 3, middle). Similar complex periods can also be observed in pancreatic stem cell (PSC) differentiation (Byrnes et al., 2018; Gu et al., 2002; Jiang and Morahan, 2014; Murtaugh, 2007; Murtaugh and Kopinke, 2008; Romer and Sussel, 2015) and neural stem cell (NSC) differentiation trajectories (Artegiani et al., 2017; Homem et al., 2015; Obernier and Alvarez-Buylla, 2019; Rosenberg et al., 2018; Rowitch and Kriegstein, 2010; Zuchero and Barres, 2015) (Fig. 3, middle).
Fig. 3.
A draft periodic table of cell types. Using the mouse as an example, a periodic table of cell types can be assembled based on current knowledge. The table is formatted from left to right based on development and/or differentiation trajectories, in which each element represents a cell type in the developmental system. Each column represents a similar developmental stage of cell types across different developing trajectories. As a proof-of-principle, each embryonic layer is arranged as a single period of cell types. The dashed cell type squares and arrows indicate unclear cell identities. Each cell type can be further annotated to provide information relating to its morphology, abundance, transcriptome, epigenome and states. Note that this draft periodic table of cell types is just a summary of a limited set of cell types and is based on our current understanding of developmental programs, despite some ongoing debates. αPC, alpha progenitor cell; βPC, beta progenitor cell; γPC, gamma progenitor cell; δPC, delta progenitor cell; εPC, epsilon progenitor cell; aciPC, acinar progenitor cells; AsCy, astrocyte; B.Og, B oogonia; B.Sg, B spermatogonia; CLP, common lymphoid progenitors; CMP, common myeloid progenitors; CTPC, cytotrophoblast progenitor cell; DC, dendritic cell; d.f.Og, differentiated oogonia; d.f.Sg, differentiated spermatogonia; ducPC, ductal progenitor cells; endoPC, endocrine progenitor cells; EpdC, ependymal cell; EpiB, epiblast; eryCy, erythrocyte; ES, elongating spermatids; EVT, extravillous cytotrophoblasts. GMP, granulocyte/macrophage progenitors; graCy, granulocytes; HAB, hemangioblast; HSC, hematopoietic stem cell; ICT, interstitial cytotrophoblast; im.acinar, immature acinar cell; im.ductal, immature ductal cell; iPC, intermediate progenitor cells; LMPP, lymphoid-primed multipotent progenitors; MEP, megakaryocyte/erythrocyte progenitors; mkCy, megakaryocyte; Mφ, macrophage; MPP, multipotent progenitors; NK, natural killer cell; NPC, neural progenitor cells; NSC, neural stem cells; ODCy, oligodendrocytes; OPC, oligodendrocyte progenitor cells; OSC, oogonial stem cell; PGC, primordial germ cell; PPC, pancreatic progenitor cells; PSC, pancreatic stem cells; RGP, radial glial progenitor cells; RS, round spermatids; SCT, syncytiotrophoblast; SSC, spermtogonial stem cell; Sz, spermatozoon; TB, trophoblast; undiff.Og, undifferentiated oogonia; undiff.Sg, undifferentiated spermatogonia; VCT, villous cytotrophoblast; 1st/2nd Sc, primary/secondary spermatocytes; 1st/2nd Oc, primary/secondary oocytes.
Across periods, cell types may be aligned according to their stage of differentiation, forming different groups (columns) in the periodic table of cell types. Each group has different cell types coming from different developmental trajectories, but displaying the same degree of stemness or differentiation state in its respective period. Different cell groups may not have the same number of cell types because of the dynamic developmental trajectories. However, the cell types across different periods can be aligned using some informative ‘milestone cell types’. The milestone cell types include: (1) fetal multipotent stem cells, for example hemangioblasts (HABs) and primordial germ cells (PGCs), which form the beginning of a major period and the starting point of the ‘stem cell phase’; (2) fast-amplifying progenitor cells, which indicate the end of the ‘stem cell phase’, replicating quickly to increase cell number; (3) differentiated cells, which follow from the last round of cell division and are characteristic of the ‘differentiated phase’; (4) mature cells, representing the end point of a period. Between these milestone cell types, there may be different numbers of cell types as defined accordingly to their CoRCs. For instance, the second column contains the stem cells for generating the majority of cell types, whereas the last few groups represent the differentiated cells that make up the majority of cell types in the adult body (Fig. 3).
With such a periodic table of cell types, we can annotate each cell type with biological information, including organ system, cell morphology, cell abundance and the composition of its epigenome and transcriptome. In addition, for each cell type, the associated cell states – such as cell cycle stage or environmental stimulations – can be annotated together with their molecular and phenotypic information. As the cell type CoRC is the same for these cell states, they are classified as ‘isotopes’ of a unique cell type (Fig. 3, zoom-in legend). Moreover, instances of non-canonical cell fate transitions can be consistently incorporated into the periodic table framework by considering that the periods of the table are not necessarily the only paths across the cell types. For example, transdifferentiation has been observed in C. elegans: the Y cell in the rectum gradually becomes a motor neuron PDA (Y-to-PDA transdifferentiation) (Jarriault et al., 2008; Zuryn et al., 2014). Such non-canonical cell fate changes in vivo can also be annotated in the periodic table of C. elegans cell types.
Analogous to the composition of complex molecules from multiple chemical elements, a tissue or an organ can be seen as a collection of multiple cell types from the periodic table. Thus, the same cell type may be the integral building unit for many different tissues and/or organs. For example, fibroblasts are found in multiple organs, producing their structural framework, indicating their widespread distribution across the animal body. However, such cell types that are found in multiple tissues and/or organs may not be exactly the same, as revealed by studies into the tissue-specificity and heterogeneity of fibroblasts (Han et al., 2018), reflecting their unique tissue environment. The observation of tissue-specific variation within cells of the same type provides another example of distinct cell states, to be annotated as ‘isotopes’ of the cell type in the periodic table.
Developmental homoplasy, which refers to cell types from different lineages having similar functions (Graham, 2010), can also be evaluated in the context of a periodic table of cell types by analyzing CoRCs. For example, trunk skeletal muscle cells and cranial skeletal muscle cells are reported to have distinct genetic lineage and regulatory programs (Sambasivan et al., 2009). Despite their functional similarities for contraction, these two muscle cell types would be organized as different cell types in the periodic table. Similarly, the two motor neuron subtypes – spinal somatic motor neurons and branchiomotor/visceromotor neurons – would be assigned to different regions in the periodic table following their unique transcriptional programs defined by the Isl1-Lhx3-Hb9 and Isl1-Phox2a/2b-Tbx20 TF sets, respectively (Jessell, 2000; Mazzoni et al., 2013). However, we cannot rule out at present the possibility that cells originating from different developmental trajectories converge to identical cell types, according to their CoRCs. In this scenario, these cell populations will also converge to one cell type in the periodic table. Overall, an approach focusing on CoRCs/TFs can be applied to other systems with superficially similar cell types.
The fully defined periodic table of cell types for a given species would comprise the whole life cycle of a species, starting from a fertilized egg, the zygote, and charting its development into multiple periods of developmental trajectories (Fig. 3). Unlike the periodic table of elements, with its defined organization for all elements, the periodic table of cell types would differ across species following evolutionary divergences. For instance, the mouse periodic table would firstly segregate into two major parts: the extraembryonic periods (i.e. trophoblast lineage) and the embryonic periods (Artegiani et al., 2017; Red-Horse et al., 2004; Suryawanshi et al., 2018; Vento-Tormo et al., 2018). The embryonic periods of the table cover the organism's cell types across all developmental trajectories, as displayed in Fig. 3 by the pancreatic cell types representing the endoderm, the hematopoietic cell types representing the mesoderm, the neural cell types representing the ectoderm, and lastly the germline cell types. In a sense, the periodic table coherently aligns cell types across the vast number of developmental trajectories.
Using the periodic table of cell types to predict new cell types
One innovation of the periodic table of elements is its capability to predict the existence of unknown elements. Following the logic of the organization of the chemical elements, Dmitri Mendeleev left empty spots for the then unidentified elements, all of which were later validated experimentally. Analogously, the periodic table of cell types could be used to predict a species’ missing cell types, based on the biological logic of development and differentiation.
For example, for a given species, the periodic table of cell types may help in revealing missing cell populations in a specific developmental process. Almost all mature cells arise from a specific stem cell system, and thus an unknown stem cell or intermediate cell type in the periodic table may indicate a failure to have identified such a cell type from previous efforts, and recognizing this absence may help to answer open questions in the field. The ependymal cell, for instance, which produces the cerebrospinal fluid in the central nervous system, is thought to be directly derived from radial glial progenitors (RGPs) without progressing through an intermediate cell type (Fig. 3, NSC period) (Shah et al., 2018; Spassky et al., 2005). However, whether there is an ependymal progenitor cell type, similar to the intermediate progenitor cells that generate neurons or oligodendrocytes, is an open question in neural developmental biology. Thus, a periodic table of cell types could be used to intuitively predict that there might be an ependymal progenitor cell type (Fig. 3). Further investigation into the transcriptomic dynamics during ependymal cell maturation from RGPs would be required to test this prediction.
As some human cell types are difficult to identify for technical and/or ethical reasons (Hyun, 2010; Zhou et al., 2016), comparing the periodic tables of cell types across species may also help to predict, identify and characterize unknown or unidentified cell types in human. Different species have a mix of unique and conserved developmental trajectories, and the precise overlap between them will depend upon their evolutionary relationships. In instances in which cell types would be expected to be conserved across species, studying a cell type in one species can enable the inference of unknown and/or unidentified cell types in another. For example, although grid cells, or place cells, are found to be associated with the positioning system in the rat brain (Moser et al., 2008), they are considerably less well-studied in the human brain (Doeller et al., 2010; Jacobs et al., 2013; Staudigl et al., 2018). However, with a periodic table of cell types from rat or mouse, we may be able to predict the molecular identities of human grid cells and, therefore, better study their function. Overall, the analysis of periodic tables of cell types across species could shed light on studies of animal model systems and highlight their relevance to human biology.
Limitations of the periodic table framework
The periodic table of cell types has the advantage of revealing underlying biological connections between cell types. Although the term ‘periodic table’ has been mentioned in a biological context on different occasions in the past (Dobrott et al., 2019; Regev et al., 2017; https://www.alleninstitute.org/what-we-do/brain-science/news-press/press-releases/allen-institute-brain-science-database-release-nearly-doubles-mouse-brain-cell-data), it was referred to as an assortment of identified cell types. In contrast, the periodic table of cell types that we describe here logically organizes a species’ cell types – including the developing ones – in an aligned manner such that each cell type is anchored by its developmental trajectory (period) as well as its differentiation stage (group). Similar to the periodic table of elements, which is invariant according to the rules of proton number and electron shell filling, the periodic table of cell types would be unique for a given species according to its developmental program. However, because of the evolutionary complexity of biological systems, different species would encode varied periodic tables of cell types following their evolutionary backgrounds. Even conserved sister species may contain different cell types (Arendt et al., 2016), leading to slightly different periodic tables between species. Thus, whereas the organizational principle of the periodic table of elements is the proton number and electron shell filling rules, the underlying logic of the periodic table of cell types is the developmental program encoded by a given species.
Because of its simplicity, however, the table should be taken as just a summary of our existing knowledge of cell types. As our understanding of cell types increases, the cell type repertoire within the table may also evolve, similar to the evolution of the periodic table of elements. Currently, single cell transcriptomic data is our best mode of delineating cell types, although it may produce biased results. Technical advances in studying single cell regulatory programs, for example chromatin accessibility and epigenetic modifications, promise to further help in determining cell identities (Corces et al., 2016; Shema et al., 2019; reviewed by Ludwig and Bintu, 2019 in this issue). In addition, it has to be acknowledged that the examination of minor cell populations during development may yield debatable results, based on our current knowledge of cell types and technical resolution. Thus, a periodic table of cell types also needs to be maintained with caution and with revised knowledge of cell identities and cell fate transitions during development. For example, despite being subjected to substantial studies, the cellular lineages involved in the development of the mammalian hematopoietic system are still heavily debated, requiring lineage tracing studies with single cell-resolution in the future (Jacobsen and Nerlov, 2019; Zhang et al., 2018; McKenna and Gagnon, 2019 in this issue). As such, evolving knowledge, uncertainties and controversies should be considered when building a periodic table of cell types.
The periodic table of cell types does come with important limitations. The table will not be able to reveal detailed cellular lineage information as compared with single cell-resolution lineage trees (as represented by the Sulston lineage tree of C. elegans). Deciphering cellular lineage information for a developing system is a fundamental goal in developmental biology, especially for understanding early embryonic development. For these specific and early developmental phases, lineage tracing continues to reveal important developmental knowledge. However, it appears to be impossible for complex organisms and may indeed not be necessary to trace the cellular lineage of all cells. A periodic table of cell types aims to greatly simplify the cellular lineage tree of development in a logical manner, while distilling the essential developmental information and connections among different cells. Thus, the periodic table of cell types for a species would cover the most significant developmental fate transitions of cells, whereas the lineage trees of specific developmental stages or tissues and/or organs would work in parallel to re-assemble lineage information at single cell resolution.
Building periodic tables of cell types
To demonstrate the possibility of constructing a periodic table of cell types for a given species, we propose to begin with the well-studied model organism, C. elegans. Coupling the recent advent of scRNA-seq and cellular lineage tracing, we can now align molecular identities (i.e. single cell transcriptomes; Cao et al., 2017) to the Sulston cell lineage tree in order to reveal the transcriptomic signatures of all cell types in C. elegans. Recent efforts in this direction have begun, and we expect to have a much better understanding of the molecular identities of all cells across C. elegans development. However, it should be noted that, despite its simplicity, C. elegans harbors a complex cell lineage tree, with 1341 cells occurring overall during embryogenesis of the hermaphrodite to produce the 671 cells of the hatched larvae (Sulston and Horvitz, 1977; Sulston et al., 1983). Nonetheless, building a periodic table of cell types means that cells of the same type will be collapsed, thus consolidating the lineage tree into an informative and manageable table. Such efforts require single cell transcriptomic analysis across developmental stages, capturing all cell types present during development. Classifying the cells of different developmental trajectories into aligned groups will require a quantitative assessment of differentiation stages, which remains a significant computational challenge, in particular for defining TF networks.
Although the C. elegans periodic table would be a useful proof-of-principle, a more significant goal would be to build a periodic table of human cell types. The Human Cell Atlas project presents a wonderful initiative for furthering our understanding of human cell types (Regev et al., 2017). The current efforts of Human Cell Atlas project focus on the cell types present in the adult human. However, theoretically, such an effort will not be able to reveal the full developmental history of most adult cell types. Moreover, although current efforts of the project can capture some of the adult stem cell types, most of them are likely remnants of fetal stem cells, and the similarity between fetal and adult stem cell populations remains to be tested. This highlights the necessity for a human ‘developing cell’ atlas, spanning the fertilized egg to the adult body, in order to build a comprehensive periodic table of human cell types.
Considering the strong ethical challenges of studying developing cell types in humans, starting from a vertebrate animal model, such as the mouse, is a more favorable initiative. Indeed, a Mouse Cell Atlas was recently published, with ∼4 million cells sampled, covering major tissue and organs of the whole mouse body (Han et al., 2018). A similar effort was also reported by the Tabula Muris project (Tabula Muris Consortium, 2018). With these resources, we can already start to build the ‘differentiated phase’ and some of the ‘stem cell phase’ and ‘differentiation phase’ of the cell type table. Moreover, two recent studies mapped developing cells in mouse embryos during gastrulation and early organogenesis, thereby providing a Mouse Organogenesis Cell Atlas (Cao et al., 2019; Pijuan-Sala et al., 2019). Such resources greatly increase our understanding of the cell fate transitions occurring during embryonic development in mammals. Further efforts into the integrative analysis of the Mouse Cell Atlas and Mouse Organogenesis Cell Atlas are definitely necessary, as most of the analyses are still in their infancy and lack comprehensive experimental validation. Ultimately, we expect it will not be long before we have a clear delineation of mouse developmental cell types, enabling the construction of a mouse cell type periodic table and thus shedding light on the theoretical table of human cell types.
Conclusions and perspectives
Here, we have highlighted how cell types can be distinguished by analyzing their regulatory programs, as revealed by single cell transcriptomics data. Notably, we propose a periodic table of cell types as an approach to move beyond a mere parts list of the cell types of a species. Currently, there is no single resource to organize the full gamut of cell types in a complex species. The periodic table provides a scalable and easily accessible framework for assembling a comprehensive collection of cell types and cell states, explicitly by considering development. Moreover, the analogy of cell states as isotopes provides a natural way to classify the many states now observable using single cell technologies. Another advantage of the periodic table of cell types is that it considers the highly important but usually underrepresented stem cell and progenitor cell populations harbored by a species. Most importantly, the periodic table aligns cell types according to their developmental stage, thereby connecting them to one another according to the universal axis of stem cells to differentiated cells. Overall, a periodic table of cell types will be useful for recognizing the relationships and dependencies amongst cell types and may lead to interesting new biology.
By characterizing cell types and informatively organizing them into a periodic table, we will also be in a better position to understand the origin and evolution of cell types. Such a table may also aid biomedical investigations, for example by systematically uncovering cell types that could be targeted for functional studies with perturbations. We can also couple cell-type information with other biological information, such as chromatin accessibility and epigenome states (Kelsey et al., 2017; Shema et al., 2019), to understand the regulatory programs of development and differentiation. Collectively, we expect a periodic table of cell types to provide a novel perspective for both basic science and clinical applications.
Acknowledgements
We thank Maayan Baron for providing figure materials used in Fig. 1B. We also thank Dalia Barkley and the anonymous reviewers for constructive comments and suggestions to the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
This work was supported by a grant from National Institutes of Health/National Institute of Allergy and Infectious Diseases to I.Y. (R01-AI143290). Deposited in PMC for release after 12 months.
References
- Alberts B., Johnson A., Lewis J., Morgan D., Raff M., Roberts K. and Walter P. (2014). The innate and adaptive immune systems. In Molecular Biology of the Cell, 6th edn, pp. 1297-1342. Garland Science. [Google Scholar]
- Arendt D. (2008). The evolution of cell types in animals: emerging principles from molecular studies. Nat. Rev. Genet. 9, 868-882. 10.1038/nrg2416 [DOI] [PubMed] [Google Scholar]
- Arendt D., Musser J. M., Baker C. V. H., Bergman A., Cepko C., Erwin D. H., Pavlicev M., Schlosser G., Widder S., Laubichler M. D. et al. (2016). The origin and evolution of cell types. Nat. Rev. Genet. 17, 744-757. 10.1038/nrg.2016.127 [DOI] [PubMed] [Google Scholar]
- Artegiani B., Lyubimova A., Muraro M., van Es J. H., van Oudenaarden A. and Clevers H. (2017). A single-cell RNA sequencing study reveals cellular and molecular dynamics of the hippocampal neurogenic niche. Cell Rep. 21, 3271-3284. 10.1016/j.celrep.2017.11.050 [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T. et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25-29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiadis E. I., Botthof J. G., Andres H., Ferreira L., Lio P. and Cvejic A. (2017). Single-cell RNA-sequencing uncovers transcriptional states and fate decisions in haematopoiesis. Nat. Commun. 8, 2045 10.1038/s41467-017-02305-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron M., Veres A., Wolock S. L., Faust A. L., Gaujoux R., Vetere A., Ryu J. H., Wagner B. K., Shen-Orr S. S., Klein A. M. et al. (2016). A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 3, 346-360.e4. 10.1016/j.cels.2016.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behjati S., Lindsay S., Teichmann S. A. and Haniffa M. (2018). Mapping human development at single-cell resolution. Development 145, dev152561 10.1242/dev.152561 [DOI] [PubMed] [Google Scholar]
- Briggs J. A., Weinreb C., Wagner D. E., Megason S., Peshkin L., Kirschner M. W. and Klein A. M. (2018). The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 360, eaar5780 10.1126/science.aar5780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes L. E., Wong D. M., Subramaniam M., Meyer N. P., Gilchrist C. L., Knox S. M., Tward A. D., Ye C. J. and Sneddon J. B. (2018). Lineage dynamics of murine pancreatic development at single-cell resolution. Nat. Commun. 9, 3922 10.1038/s41467-018-06176-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Packer J. S., Ramani V., Cusanovich D. A., Huynh C., Daza R., Qiu X., Lee C., Furlan S. N., Steemers F. J. et al. (2017). Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661-667. 10.1126/science.aam8940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D. M., Hill A. J., Zhang F., Mundlos S., Christiansen L., Steemers F. J. et al. (2019). The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496-502. 10.1038/s41586-019-0969-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zheng Y., Gao Y., Lin Z., Yang S., Wang T., Wang Q., Xie N., Hua R., Liu M. et al. (2018). Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell Res. 28, 879-896. 10.1038/s41422-018-0074-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. (2015). What is an adult stem cell? Science 350, 1319-1320. 10.1126/science.aad7016 [DOI] [PubMed] [Google Scholar]
- Clevers H., Rafelski S., Elowitz M., Klein A., Shendure J., Trapnell C., Lein E., Lundberg E., Uhlen M., Martinez-Arias A. et al. (2017). What is your conceptual definition of “cell type” in the context of a mature organism? Cell Syst. 4, 255-259. 10.1016/j.cels.2017.03.006 [DOI] [PubMed] [Google Scholar]
- Corces M. R., Buenrostro J. D., Wu B., Greenside P. G., Chan S. M., Koenig J. L., Snyder M. P., Pritchard J. K., Kundaje A., Greenleaf W. J. et al. (2016). Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 48, 1193-1203. 10.1038/ng.3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl A. D., Meehan T. F., Bradford Y. M., Brush M. H., Dahdul W. M., Dougall D. S., He Y., Osumi-Sutherland D., Ruttenberg A., Sarntivijai S. et al. (2016). The Cell Ontology 2016: enhanced content, modularization, and ontology interoperability. J. Biomed. Semantics 7, 44 10.1186/s13326-016-0088-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrott C. I., Sathyamurthy A. and Levine A. J. (2019). Decoding cell type diversity within the spinal cord. Curr. Opin. Physiol. 8, 1-6. 10.1016/j.cophys.2018.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doeller C. F., Barry C. and Burgess N. (2010). Evidence for grid cells in a human memory network. Nature 463, 657-661. 10.1038/nature08704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellmeier W., Haust L. and Tschismarov R. (2013). Transcriptional control of CD4 and CD8 coreceptor expression during T cell development. Cell Mol. Life Sci. 70, 4537-4553. 10.1007/s00018-013-1393-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst C., Eling N., Martinez-Jimenez C. P., Marioni J. C. and Odom D. T. (2019). Staged developmental mapping and X chromosome transcriptional dynamics during mouse spermatogenesis. Nat. Commun. 10, 1251 10.1038/s41467-019-09182-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X. and Rudensky A. Y. (2016). Hallmarks of tissue-resident lymphocytes. Cell 164, 1198-1211. 10.1016/j.cell.2016.02.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell J. A., Wang Y., Riesenfeld S. J., Shekhar K., Regev A. and Schier A. F. (2018). Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360, eaar3131 10.1126/science.aar3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain R. N. (2002). T-cell development and the CD4-CD8 lineage decision. Nat. Rev. Immunol. 2, 309-322. 10.1038/nri798 [DOI] [PubMed] [Google Scholar]
- Gierahn T. M., Wadsworth M. H., Hughes T. K., Bryson B. D., Butler A., Satija R., Fortune S., Love J. C. and Shalek A. K. (2017). Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods 14, 395-398. 10.1038/nmeth.4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordin M. D. (2019). Ordering the elements. Science 363, 471-473. 10.1126/science.aav7350 [DOI] [PubMed] [Google Scholar]
- Graf T. and Enver T. (2009). Forcing cells to change lineages. Nature 462, 587-594. 10.1038/nature08533 [DOI] [PubMed] [Google Scholar]
- Graham A. (2010). Developmental homoplasy: convergence in cellular differentiation. J. Anat. 216, 651-655. 10.1111/j.1469-7580.2010.01232.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C. D., Ma Q., Manske G. L., Shami A. N., Zheng X., Marini S., Moritz L., Sultan C., Gurczynski S. J., Moore B. B. et al. (2018). A comprehensive roadmap of murine spermatogenesis defined by single-cell RNA-seq. Dev. Cell 46, 651-667.e10. 10.1016/j.devcel.2018.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu G., Dubauskaite J. and Melton D. A. (2002). Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development 129, 2447-2457. [DOI] [PubMed] [Google Scholar]
- Guibentif C., Rönn R. E., Böiers C., Lang S., Saxena S., Soneji S., Enver T., Karlsson G. and Woods N.-B. (2017). Single-cell analysis identifies distinct stages of human endothelial-to-hematopoietic transition. Cell Rep. 19, 10-19. 10.1016/j.celrep.2017.03.023 [DOI] [PubMed] [Google Scholar]
- Guo J., Grow E. J., Mlcochova H., Maher G. J., Lindskog C., Nie X., Guo Y., Takei Y., Yun J., Cai L. et al. (2018). The adult human testis transcriptional cell atlas. Cell Res. 28, 1141-1157. 10.1038/s41422-018-0099-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Wang R., Zhou Y., Fei L., Sun H., Lai S., Saadatpour A., Zhou Z., Chen H., Ye F. et al. (2018). Mapping the mouse cell atlas by microwell-seq. Cell 173, 1307 10.1016/j.cell.2018.05.012 [DOI] [PubMed] [Google Scholar]
- Hermann B. P., Cheng K., Singh A., Roa-De La Cruz L., Mutoji K. N., Chen I.-C., Gildersleeve H., Lehle J. D., Mayo M., Westernströer B. et al. (2018). The mammalian spermatogenesis single-cell transcriptome, from spermatogonial stem cells to spermatids. Cell Rep. 25, 1650-1667.e8. 10.1016/j.celrep.2018.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homem C. C. F., Repic M. and Knoblich J. A. (2015). Proliferation control in neural stem and progenitor cells. Nat. Rev. Neurosci. 16, 647-659. 10.1038/nrn4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooke R. (1665). Micrographia. Royal Society.
- Hyun I. (2010). The bioethics of stem cell research and therapy. J. Clin. Invest. 120, 71-75. 10.1172/JCI40435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J., Weidemann C. T., Miller J. F., Solway A., Burke J. F., Wei X.-X., Suthana N., Sperling M. R., Sharan A. D., Fried I. et al. (2013). Direct recordings of grid-like neuronal activity in human spatial navigation. Nat. Neurosci. 16, 1188-1190. 10.1038/nn.3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen S. E. W. and Nerlov C. (2019). Haematopoiesis in the era of advanced single-cell technologies. Nat. Cell Biol. 21, 2-8. 10.1038/s41556-018-0227-8 [DOI] [PubMed] [Google Scholar]
- Jarriault S., Schwab Y. and Greenwald I. (2008). A Caenorhabditis elegans model for epithelial-neuronal transdifferentiation. Proc. Natl. Acad. Sci. USA 105, 3790-3795. 10.1073/pnas.0712159105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessell T. M. (2000). Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 1, 20-29. 10.1038/35049541 [DOI] [PubMed] [Google Scholar]
- Jiang F.-X. and Morahan G. (2014). Pancreatic stem cells remain unresolved. Stem Cells Dev. 23, 2803-2812. 10.1089/scd.2014.0214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M. and Shinohara T. (2013). Spermatogonial stem cell self-renewal and development. Annu. Rev. Cell Dev. Biol. 29, 163-187. 10.1146/annurev-cellbio-101512-122353 [DOI] [PubMed] [Google Scholar]
- Kelsey G., Stegle O. and Reik W. (2017). Single-cell epigenomics: Recording the past and predicting the future. Science 358, 69-75. 10.1126/science.aan6826 [DOI] [PubMed] [Google Scholar]
- Kiselev V. Y., Andrews T. S. and Hemberg M. (2019). Challenges in unsupervised clustering of single-cell RNA-seq data. Nat. Rev. Genet. 20, 273-282. 10.1038/s41576-018-0088-9 [DOI] [PubMed] [Google Scholar]
- Klein A. M., Mazutis L., Akartuna I., Tallapragada N., Veres A., Li V., Peshkin L., Weitz D. A. and Kirschner M. W. (2015). Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187-1201. 10.1016/j.cell.2015.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziejczyk A. A., Kim J. K., Svensson V., Marioni J. C. and Teichmann S. A. (2015a). The technology and biology of single-cell RNA sequencing. Mol. Cell 58, 610-620. 10.1016/j.molcel.2015.04.005 [DOI] [PubMed] [Google Scholar]
- Kolodziejczyk A. A., Kim J. K., Tsang J. C. H., Ilicic T., Henriksson J., Natarajan K. N., Tuck A. C., Gao X., Bühler M., Liu P. et al. (2015b). Single cell RNA-sequencing of pluripotent states unlocks modular transcriptional variation. Cell Stem Cell 17, 471-485. 10.1016/j.stem.2015.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S. A., Jolma A., Campitelli L. F., Das P. K., Yin Y., Albu M., Chen X., Taipale J., Hughes T. R. and Weirauch M. T. (2018). The human transcription factors. Cell 172, 650-665. 10.1016/j.cell.2018.01.029 [DOI] [PubMed] [Google Scholar]
- Li J., Klughammer J., Farlik M., Penz T., Spittler A., Barbieux C., Berishvili E., Bock C. and Kubicek S. (2016). Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO Rep. 17, 178-187. 10.15252/embr.201540946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig C. H. and Bintu L. (2019). Mapping chromatin modifications at the single cell level. Development 146, dev170217 10.1242/dev.170217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay I. C., Svensson V., Labalette C., Ferreira L., Hamey F., Voet T., Teichmann S. A. and Cvejic A. (2016). Single-cell RNA-sequencing reveals a continuous spectrum of differentiation in hematopoietic cells. Cell Rep. 14, 966-977. 10.1016/j.celrep.2015.12.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni J. C. and Arendt D. (2017). How single-cell genomics is changing evolutionary and developmental biology. Annu. Rev. Cell Dev. Biol. 33, 537-553. 10.1146/annurev-cellbio-100616-060818 [DOI] [PubMed] [Google Scholar]
- Mayr U., Serra D. and Liberali P. (2019). Exploring single cells in space and time during tissue development, homeostasis and regeneration. Development 146, dev176727 10.1242/dev.176727 [DOI] [PubMed] [Google Scholar]
- Mazzarello P. (1999). A unifying concept: the history of cell theory. Nat. Cell Biol. 1, E13-E15. 10.1038/8964 [DOI] [PubMed] [Google Scholar]
- Mazzoni E. O., Mahony S., Closser M., Morrison C. A., Nedelec S., Williams D. J., An D., Gifford D. K. and Wichterle H. (2013). Synergistic binding of transcription factors to cell-specific enhancers programs motor neuron identity. Nat. Neurosci. 16, 1219-1227. 10.1038/nn.3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A. and Gagnon J. A. (2019). Recording development with single cell dynamic lineage tracing. Development 146, dev169730 10.1242/dev.169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melé M., Ferreira P. G., Reverter F., DeLuca D. S., Monlong J., Sammeth M., Young T. R., Goldmann J. M., Pervouchine D. D., Sullivan T. J. et al. (2015). The human transcriptome across tissues and individuals. Science 348, 660-665. 10.1126/science.aaa0355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris S. A. (2019). The evolving concept of cell identity in the single cell era. Development 146, dev169748 10.1242/dev.169748 [DOI] [PubMed] [Google Scholar]
- Moser E. I., Kropff E. and Moser M.-B. (2008). Place cells, grid cells, and the brain's spatial representation system. Annu. Rev. Neurosci. 31, 69-89. 10.1146/annurev.neuro.31.061307.090723 [DOI] [PubMed] [Google Scholar]
- Mullen A. C., Orlando D. A., Newman J. J., Lovén J., Kumar R. M., Bilodeau S., Reddy J., Guenther M. G., DeKoter R. P. and Young R. A. (2011). Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell 147, 565-576. 10.1016/j.cell.2011.08.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraro M. J., Dharmadhikari G., Grün D., Groen N., Dielen T., Jansen E., van Gurp L., Engelse M. A., Carlotti F., de Koning E. J. P. et al. (2016). A single-cell transcriptome atlas of the human pancreas. Cell Syst. 3, 385-394.e3. 10.1016/j.cels.2016.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh L. C. (2007). Pancreas and beta-cell development: from the actual to the possible. Development 134, 427-438. 10.1242/dev.02770 [DOI] [PubMed] [Google Scholar]
- Murtaugh L. C. and Kopinke D. (2008). Pancreatic stem cells. In: StemBook (ed. The Stem Cell Research Community). Harvard Stem Cell Institute; 10.3824/stembook.1.3.1 [DOI] [PubMed] [Google Scholar]
- Niwa H. (2014). The pluripotency transcription factor network at work in reprogramming. Curr. Opin. Genet. Dev. 28, 25-31. 10.1016/j.gde.2014.08.004 [DOI] [PubMed] [Google Scholar]
- Niwa H. (2018). The principles that govern transcription factor network functions in stem cells. Development 145, dev157420 10.1242/dev.157420 [DOI] [PubMed] [Google Scholar]
- Obernier K. and Alvarez-Buylla A. (2019). Neural stem cells: origin, heterogeneity and regulation in the adult mammalian brain. Development 146, dev156059 10.1242/dev.156059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi-Sutherland D. (2017). Cell ontology in an age of data-driven cell classification. BMC Bioinformatics 18, 558 10.1186/s12859-017-1980-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F. and Milos P. M. (2011). RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 12, 87-98. 10.1038/nrg2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijuan-Sala B., Griffiths J. A., Guibentif C., Hiscock T. W., Jawaid W., Calero-Nieto F. J., Mulas C., Ibarra-Soria X., Tyser R. C. V., Ho D. L. L. et al. (2019). A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490-495. 10.1038/s41586-019-0933-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Red-Horse K., Zhou Y., Genbacev O., Prakobphol A., Foulk R., McMaster M. and Fisher S. J. (2004). Trophoblast differentiation during embryo implantation and formation of the maternal-fetal interface. J. Clin. Invest. 114, 744-754. 10.1172/JCI200422991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev A., Teichmann S. A., Lander E. S., Amit I. and Benoist C. (2017). Science forum: the human cell atlas. eLife 6, e27041 10.7554/eLife.27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer A. I. and Sussel L. (2015). Pancreatic islet cell development and regeneration. Curr. Opin Endocrinol. Diabetes Obes. 22, 255-264. 10.1097/MED.0000000000000174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg A. B., Roco C. M., Muscat R. A., Kuchina A., Sample P., Yao Z., Graybuck L. T., Peeler D. J., Mukherjee S., Chen W. et al. (2018). Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360, 176-182. 10.1126/science.aam8999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowitch D. H. and Kriegstein A. R. (2010). Developmental genetics of vertebrate glial-cell specification. Nature 468, 214-222. 10.1038/nature09611 [DOI] [PubMed] [Google Scholar]
- Sambasivan R., Gayraud-Morel B., Dumas G., Cimper C., Paisant S., Kelly R. G. and Tajbakhsh S. (2009). Distinct regulatory cascades govern extraocular and pharyngeal arch muscle progenitor cell fates. Dev. Cell 16, 810-821. 10.1016/j.devcel.2009.05.008 [DOI] [PubMed] [Google Scholar]
- Segerstolpe Å., Palasantza A., Eliasson P., Andersson E.-M., Andréasson A.-C., Sun X., Picelli S., Sabirsh A., Clausen M., Bjursell M. K. et al. (2016). Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 24, 593-607. 10.1016/j.cmet.2016.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P. T., Stratton J. A., Stykel M. G., Abbasi S., Sharma S., Mayr K. A., Koblinger K., Whelan P. J. and Biernaskie J. (2018). Single-cell transcriptomics and fate mapping of ependymal cells reveals an absence of neural stem cell function. Cell 173, 1045-1057.e9. 10.1016/j.cell.2018.03.063 [DOI] [PubMed] [Google Scholar]
- Shapiro E., Biezuner T. and Linnarsson S. (2013). Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev. Genet. 14, 618-630. 10.1038/nrg3542 [DOI] [PubMed] [Google Scholar]
- Shema E., Bernstein B. E. and Buenrostro J. D. (2019). Single-cell and single-molecule epigenomics to uncover genome regulation at unprecedented resolution. Nat. Genet. 51, 19-25. 10.1038/s41588-018-0290-x [DOI] [PubMed] [Google Scholar]
- Singer A., Adoro S. and Park J.-H. (2008). Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat. Rev. Immunol. 8, 788-801. 10.1038/nri2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky N., Merkle F. T., Flames N., Tramontin A. D., García-Verdugo J. M. and Alvarez-Buylla A. (2005). Adult ependymal cells are postmitotic and are derived from radial glial cells during embryogenesis. J. Neurosci. 25, 10-18. 10.1523/JNEUROSCI.1108-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudigl T., Leszczynski M., Jacobs J., Sheth S. A., Schroeder C. E., Jensen O. and Doeller C. F. (2018). Hexadirectional modulation of high-frequency electrophysiological activity in the human anterior medial temporal lobe maps visual space. Curr. Biol. 28, 3325-3329.e4. 10.1016/j.cub.2018.09.035 [DOI] [PubMed] [Google Scholar]
- Sulston J. E. and Horvitz H. R. (1977). Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110-156. 10.1016/0012-1606(77)90158-0 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G. and Thomson J. N. (1983). The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64-119. 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- Suryawanshi H., Morozov P., Straus A., Sahasrabudhe N., Max K. E. A., Garzia A., Kustagi M., Tuschl T. and Williams Z. (2018). A single-cell survey of the human first-trimester placenta and decidua. Sci. Adv. 4, eaau4788 10.1126/sciadv.aau4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson V., Vento-Tormo R. and Teichmann S. A. (2018). Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 13, 599-604. 10.1038/nprot.2017.149 [DOI] [PubMed] [Google Scholar]
- Tabula Muris Consortium (2018). Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367-372. 10.1038/s41586-018-0590-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott S. J., Davis R. L., Thayer M. J., Cheng P. F., Weintraub H. and Lassar A. B. (1988). MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science 242, 405-411. 10.1126/science.3175662 [DOI] [PubMed] [Google Scholar]
- Trapnell C. (2015). Defining cell types and states with single-cell genomics. Genome Res. 25, 1491-1498. 10.1101/gr.190595.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler S., Büttner M., Fischer D. S., Lange M., Bergen V., Lickert H. and Theis F. J. (2019). Concepts and limitations for learning developmental trajectories from single cell genomics. Development 146, dev170506 10.1242/dev.170506 [DOI] [PubMed] [Google Scholar]
- Velten L., Haas S. F., Raffel S., Blaszkiewicz S., Islam S., Hennig B. P., Hirche C., Lutz C., Buss E. C., Nowak D. et al. (2017). Human haematopoietic stem cell lineage commitment is a continuous process. Nat. Cell Biol. 19, 271-281. 10.1038/ncb3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vento-Tormo R., Efremova M., Botting R. A., Turco M. Y., Vento-Tormo M., Meyer K. B., Park J.-E., Stephenson E., Polański K., Goncalves A. et al. (2018). Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563, 347-353. 10.1038/s41586-018-0698-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington C. H. (1957). The Strategy of the Genes: a Discussion of Some Aspects of Theoretical Biology. Allen & Unwin. [Google Scholar]
- Wagner A., Regev A. and Yosef N. (2016). Revealing the vectors of cellular identity with single-cell genomics. Nat. Biotechnol. 34, 1145-1160. 10.1038/nbt.3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D. E., Weinreb C., Collins Z. M., Briggs J. A., Megason S. G. and Klein A. M. (2018). Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981-987. 10.1126/science.aar4362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Liu X., Chang G., Chen Y., An G., Yan L., Gao S., Xu Y., Cui Y., Dong J. et al. (2018). Single-Cell RNA sequencing analysis reveals sequential cell fate transition during human spermatogenesis. Cell Stem Cell 23, 599-614.e4. 10.1016/j.stem.2018.08.007 [DOI] [PubMed] [Google Scholar]
- Wilkinson A. C., Nakauchi H. and Göttgens B. (2017). Mammalian transcription factor networks: recent advances in interrogating biological complexity. Cell Syst. 5, 319-331. 10.1016/j.cels.2017.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpert L. (1996). The evolution of “the cell theory”. Curr. Biol. 6, 225-228. 10.1016/S0960-9822(02)00463-3 [DOI] [PubMed] [Google Scholar]
- Xia B., Yan Y., Baron M., Wagner F., Barkley D., Chiodin M., Kim S. Y., Keefe D. L., Alukal J. P., Boeke J. D. et al. (2018). Widespread transcriptional scanning in the testes modulates gene evolution rates. BioRxiv. 10.1101/282129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. and Gerstein M. (2006). Genomic analysis of the hierarchical structure of regulatory networks. Proc. Natl. Acad. Sci. USA 103, 14724-14731. 10.1073/pnas.0508637103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Gao S., Xia J. and Liu F. (2018). Hematopoietic hierarchy - an updated roadmap. Trends Cell Biol. 28, 976-986. 10.1016/j.tcb.2018.06.001 [DOI] [PubMed] [Google Scholar]
- Zhou F., Li X., Wang W., Zhu P., Zhou J., He W., Ding M., Xiong F., Zheng X., Li Z. et al. (2016). Tracing haematopoietic stem cell formation at single-cell resolution. Nature 533, 487-492. 10.1038/nature17997 [DOI] [PubMed] [Google Scholar]
- Zhu J. and Paul W. E. (2008). CD4 T cells: fates, functions, and faults. Blood 112, 1557-1569. 10.1182/blood-2008-05-078154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchero J. B. and Barres B. A. (2015). Glia in mammalian development and disease. Development 142, 3805-3809. 10.1242/dev.129304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuryn S., Ahier A., Portoso M., White E. R., Morin M.-C., Margueron R. and Jarriault S. (2014). Transdifferentiation. Sequential histone-modifying activities determine the robustness of transdifferentiation. Science 345, 826-829. 10.1126/science.1255885 [DOI] [PubMed] [Google Scholar]