ABSTRACT
The mammalian lip and primary palate form when coordinated growth and morphogenesis bring the nasal and maxillary processes into contact, and the epithelia co-mingle, remodel and clear from the fusion site to allow mesenchyme continuity. Although several genes required for fusion have been identified, an integrated molecular and cellular description of the overall process is lacking. Here, we employ single cell RNA sequencing of the developing mouse face to identify ectodermal, mesenchymal and endothelial populations associated with patterning and fusion of the facial prominences. This analysis indicates that key cell populations at the fusion site exist within the periderm, basal epithelial cells and adjacent mesenchyme. We describe the expression profiles that make each population unique, and the signals that potentially integrate their behaviour. Overall, these data provide a comprehensive high-resolution description of the various cell populations participating in fusion of the lip and primary palate, as well as formation of the nasolacrimal groove, and they furnish a powerful resource for those investigating the molecular genetics of facial development and facial clefting that can be mined for crucial mechanistic information concerning this prevalent human birth defect.
KEY WORDS: Cleft lip, Primary palate, Palate fusion, Single cell RNA sequencing, Lambdoidal junction, Nasolacrimal groove
Summary: Using single cell RNA-seq, novel cell populations are identified that are associated with fusion of the lip/primary palate, developing a model that predicts molecular and cellular behaviours during fusion.
INTRODUCTION
Development of the mammalian upper face is a multi-step process that relies on the interaction of neural crest-derived mesenchymal cells with the overlying facial ectoderm in three bilaterally paired facial prominences – the maxillary (MxP), the lateral nasal (LNP) and the medial nasal (MNP). The orderly fusion of these prominences is essential for formation of the primary palate and associated upper lip during embryogenesis. Failure of this developmental mechanism accounts for one of the most common human birth defects, clefting of the lip and/or primary palate (CL/P) – a condition that has serious consequences for speech, feeding, dentition and social interaction. This process is relatively similar in mouse and human, so that the mouse has served as the major model system for probing the genetics and cell behaviours underpinning upper lip and primary palate formation, as well as the aetiology of CL/P (Abramyan and Richman, 2015; Gritli-Linde, 2008; Juriloff and Harris, 2008; Suzuki et al., 2016).
Fusion of the upper lip and primary palate in mouse begins around embryonic day 10.5 (E10.5), when coordinated growth and morphogenesis brings the caudal edges of the LNP and MNP close together across the nasal groove at the oral side of the nasal pit. The MxP also interacts with the LNP and MNP at the base of the nasal groove via its anterior/medial aspect (Miyake et al., 1996). Prior to the onset of fusion, the surface epithelium of these prominences is a bilayer of loosely organized basal cells overlain by a highly polarized squamous layer with well-defined apical junctions (Gaare and Langman, 1977a; Millicovsky et al., 1982; Millicovsky and Johnston, 1981). However, as the prominences come together at the base of the nasal groove, the superficial epithelial cells remodel their apical contacts and begin extending processes that contact the apposing epithelia to initiate fusion (Forbes and Steffek, 1989; Gaare and Langman, 1977b, 1980; Millicovsky et al., 1982; Millicovsky and Johnston, 1981; Trasler, 1968; Trasler and Ohannessian, 1983; Reed, 1933). Over the next 24 h, epithelial fusion zips ‘up’ between the MNP and LNP to define the final shape of the nasal pit, ‘down’ between the MNP and MxP towards the oral opening to form the roof of the mouth, and ‘out’ laterally between the LNP and MxP. This three-way fusion forms the lambdoid/lambdoidal junction (LJ; Fig. 1). The seam of merging epithelia, termed the nasal fin, then clears from the fusion site so that a continuous mesenchyme bridges the LJ. The majority of the nasal fin regresses through some combination of extrusion, apoptosis, migration to the bordering epithelia and merging into the mesenchyme through an epithelial-to-mesenchymal transition (EMT) (Fitchett and Hay, 1989; Losa et al., 2018; Ferretti et al., 2011; Jiang et al., 2006). However, the dorso-posterior edge of the nasal fin remodels to establish the primitive choana, an opening that connects the oral and nasal cavities (Tamarin, 1982). Towards the eye, there is a separate junction between MxP and LNP termed the nasolacrimal groove (NLG). Here, the epithelial seam ingresses and subsequently hollows out to form the nasolacrimal duct (Lotz et al., 2006). By E12.5, these fusion and resolution processes are essentially complete: continuous layers of surface and oral epithelia form the upper lip and roof of the mouth, respectively, and enclose an uninterrupted mesenchyme that will give rise to skeletal elements, muscles and other soft tissues of the lip and primary palate.
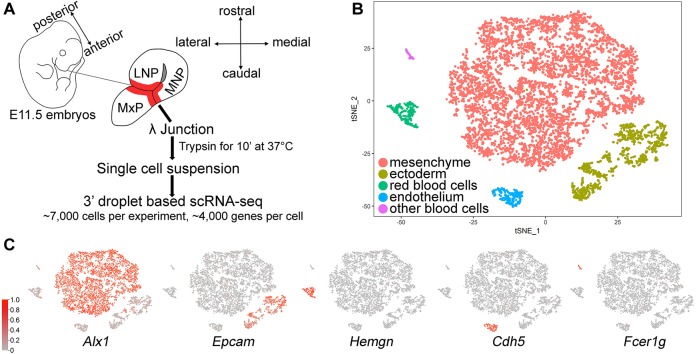
Fig. 1.
Single cell RNA-seq identification of five major cell types at the lambdoidal junction. (A) Schematic showing process of microdissection of E11.5 lambdoidal junction (red, λ junction) and scRNA-seq experiment. The double-headed arrows indicate the anatomical axes relative to whole embryo (left) or the dissected prominences (right). (B) tSNE plot of 7893 cells from the C57LJ dataset showing the five major clusters. (C) Feature plots of representative marker genes used to assign clusters shown in B, with normalized gene expression per cell (colour key on the left) superimposed on the tSNE plot. The same colour key occurs in all subsequent feature plots. LNP, lateral nasal process; MNP, medial nasal process; MxP, maxillary prominence.
Despite the prevalence of human CL/P, much of our understanding of orofacial clefting has been derived from a separate process, formation of the secondary palate. Fusion of the secondary palate occurs later in development and involves outgrowth of the palatal shelves from the MxP (Hammond et al., 2017; Li et al., 2017). Compared with the lip and primary palate, secondary palate analysis has benefited from the larger size of the fusion zones, the simpler morphology of juxtaposed epithelial seams, and the far greater number of mutant mice that present this pathology (Li et al., 2017). Nevertheless, a growing number of mouse strains have now been identified with CL/P, and human genetic analyses have identified additional affected loci. Environmental insults such as hyperthermia and disruption of retinoid balance are additional processes that can cause orofacial clefting (Peterka et al., 1994; Rhinn and Dolle, 2012; Rothman et al., 1995). A consensus is emerging that the interaction of key signalling domains involving complex and reciprocal interactions between molecules within the Fgf, Wnt, Shh and BMP pathways is essential for driving the growth, morphogenesis and fusion of the facial prominences (Diewert and Wang, 1992; Ferretti et al., 2011; Forni et al., 2013; Hu et al., 2015a; Jin et al., 2012; Losa et al., 2018; Wang et al., 1995; Xavier et al., 2016). However, the exact mechanisms by which most genes and environmental factors impact development of the lip and primary palate remain obscure. Significant gaps include the cellular diversity in and around the fusion zone, how the gene regulatory networks map onto that cellular diversity, how these programs are coordinated spatially and temporally by cell-cell signalling, and how they dictate the epithelial cell behaviours that mediate fusion and its resolution.
Although there have been previous studies examining genome-wide gene expression in the developing mouse face (Brunskill et al., 2014; Feng et al., 2009; Gong et al., 2005; Hooper et al., 2017), these have lacked the spatial resolution and sensitivity to describe behaviours of cells within and adjacent to the epithelial seams. To focus on these populations, we employed microdissection of the LJ followed by single cell RNA sequencing (scRNA-seq) to describe the genetic programs underlying fusion of the lip and primary palate. We identified groups of surface ectoderm cells representing the periderm, the dental epithelium, the NLG and a distinctive group of basal cells that appear at the fusion site and then disappear as fusion resolves. We also identified discrete populations of mesenchymal cells with distinct gene expression signatures sub-adjacent to the fusing ectoderm at the caudal tips of the LNP and MNP. The detailed gene expression and anatomical information within these scRNA-seq datasets provides a powerful resource for the study of face formation, and enables the formulation of specific new hypotheses concerning how gene regulatory networks, tissue interactions and signalling pathways mediate formation of the lip and primary palate.
RESULTS
Identification of cell and tissue populations at the LJ by scRNA-seq
Fig. S1 shows schematics of the upper mouse face at E11.5, the critical stage at which fusion and resolution is occurring between the MxP, LNP and MNP. These diagrams indicate the positions of the NLG, the LJ and other relevant landmarks with respect to the facial prominences. To investigate the cell populations associated with the regions of fusion at the LJ and NLG, we employed a combination of microdissection (shown in Fig. 1A and Fig. S1), lineage tracing and scRNA-seq. Our initial analysis used tissue microdissected from E11.5 C57BL/6J mouse embryos. Following quality assessment and filtering of this ‘C57LJ’ dataset (Fig. S2A,B), we retained 7893 cells for further analysis. The population structure is visualized in a t-distributed stochastic neighbour embedding (tSNE) plot (Fig. 1B), where each dot represents a cellular transcriptome and the distance between dots is proportional to the similarity in their transcriptomes. Clustering defined five well-separated cell populations (Fig. 1B), each described by genes whose expression is enriched within each cluster (Fig. 1, Fig. S3, Table S1). By examining gene signatures, these five populations were assigned to ectoderm, endothelium, erythrocytes and other blood cells, with the largest cluster representing mesenchyme (Fig. 1B).
To address reproducibility and begin validation of our assignments, we performed a second scRNA-seq, isolating the E11.5 LJ from mice in which an ectoderm-specific Cre transgene, Crect, activates expression of Rosa26-LacZ. Ectodermal lineages could then be identified by assessing the expression of either Cre recombinase or LacZ. This second dataset, ‘CrectLJ’, which contained 6673 cells after quality assessment, segregated into the same five clusters, with similar percentages of each cell type to the ‘C57LJ’ dataset (Fig. S2C). Moreover, comparison of expression levels for all genes between the two datasets demonstrated excellent correlation across the five cell populations (Fig. S2D). Finally, the cluster designated as ectoderm – based on expression of pan-ectoderm markers such as Epcam, Igfbp5 and Pdgfa – was confirmed to be ectoderm derived as it also expressed significant levels of transcripts corresponding to Cre recombinase and/or LacZ (Fig. S2E). Therefore, initial analyses revealed a highly reproducible population structure of cells associated with the LJ.
Three of the five clusters – the red blood cell, other blood cell and endothelial – were relatively small and compact, and had signatures associated with the developing vasculature. The red blood cell cluster had highly specific markers of erythrocytes, including genes for haem synthesis (Alas2, Alad) and globins (Hbq1b, Hba-x, Hbb-y). The ‘other blood cells’ had markers typifying the monocyte/macrophage lineage (Fcgr1, Fcgr3, Fcgr4, Cd200r1, Itgam, Csf1r and Trem2) and megakaryocytes (Vwf, Ppbp, Pf4). The endothelial cluster was characterized by markers of blood vessels (Cldn5 and Cdh5), but lacked markers of lymph vessels (Lyve1 and Prox1). To examine how this potential vasculature was distributed around the LJ, in situ hybridization was employed using the endothelial cluster markers Cldn5 and Hapln1 (Fig. S4). Both genes, but especially Cldn5, revealed an extensive branching network of cells, consistent with this cluster representing endothelial cells of the developing vasculature. Moreover, these vessels were particularly dense in the subsurface mesenchyme in the vicinity of the fusion zones, both at the tips of the prominences and at the MxP/LNP junction. Therefore, together these three smaller clusters represent cell populations responsible for the blood supply in this region. The remaining two clusters, the mesenchyme and ectoderm, were much larger with a more-complex architecture. Further analysis of these two populations required reclustering to identify specific components related to their anatomical mapping, gene expression and functional programs.
Mesenchymal cell populations in and around the LJ
Differences within the mesenchyme were partially obscured by the diversity present across all five cell populations in the initial clustering, as well as by cell cycle (Fig. S3C). To mitigate these effects and reveal sub-structure within the mesenchyme, we reanalysed the 5536 mesenchymal cells from the CrectLJ dataset after regressing out the cell cycle heterogeneity (Fig. 2). This identified nine clusters (m0-m8) that we mapped onto the midface using cluster-specific marker genes, published expression patterns and RNA in situ hybridization (Figs 2, 3, 4 Figs S5, S6, Table S2, and summarized schematically in Table S3). Although many of the markers are expressed widely in the upper face, our assignment and bioinformatics analysis of clusters is based solely upon the limited cell population within the three-dimensional tissue space defined by microdissection (heavy dashed line, Table S3).
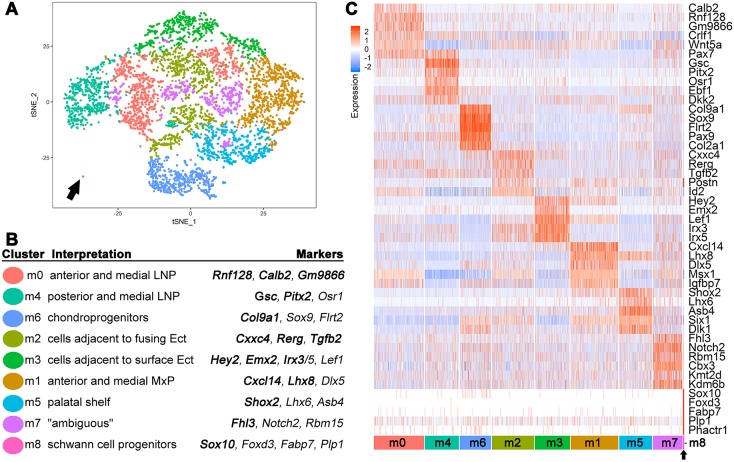
Fig. 2.
Reclustering identifies specific mesenchymal cell populations. (A) tSNE plot of reclustered mesenchymal cells from CrectLJ dataset. (B) Annotation of the re-clustered mesenchyme showing marker genes used for mapping and assignment. Genes in bold were used for in situ hybridization (Figs 3, 4, Figs S5, S6). Ect, ectoderm. (C) Heatmap representing scaled expression level (blue to red) of representative marker genes across the mesenchymal cells. Each row is a gene while each column is a cell. The bottom row demarcates the cell clusters. Black arrow indicates the smallest cluster (m8) in both A and C.
Fig. 3.
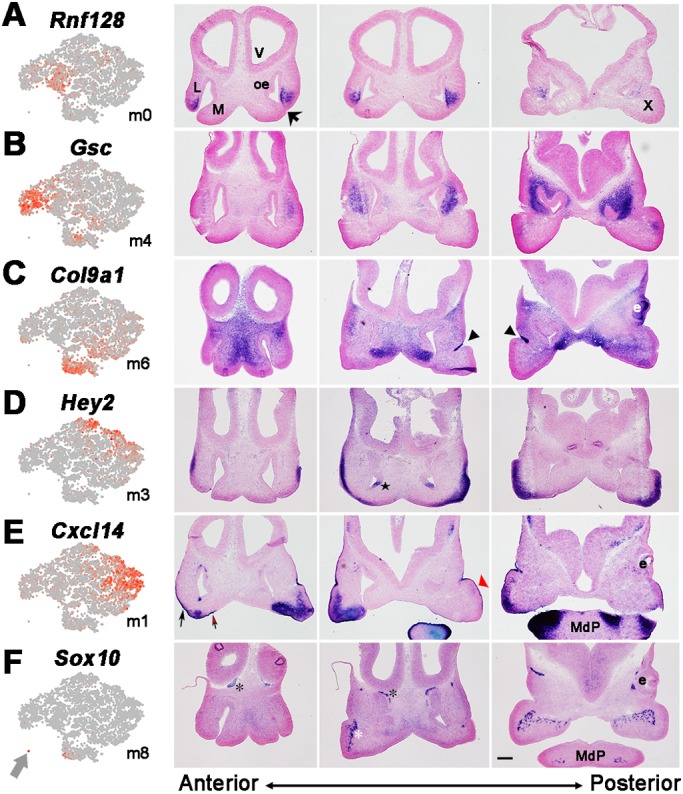
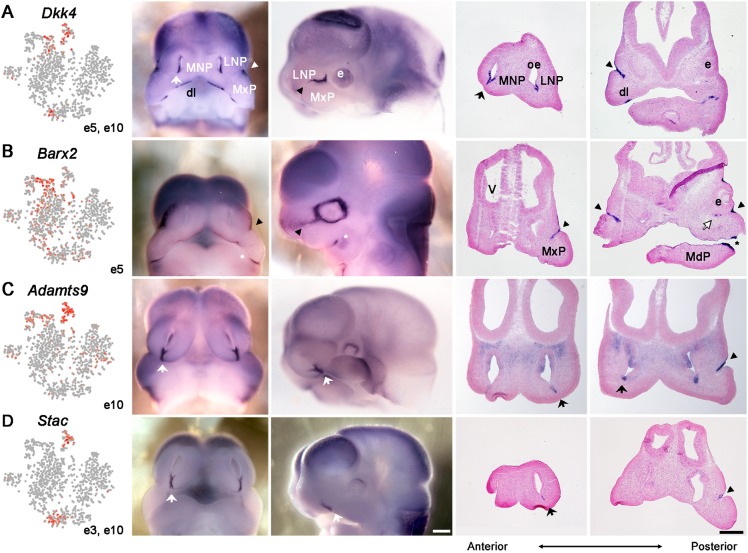
Mapping the mesenchymal clusters by in situ hybridization. Feature plots for indicated marker gene and mesenchymal cluster (left panels; grey arrow shows m8). The other three panels in each row show in situ hybridization on E11.5 frontal face sections from anterior to posterior, as indicated at the bottom. (A) Rnf128, (B) Gsc, (C) Col9a1, (D) Hey2, (E) Cxcl14 and (F) Sox10 for clusters m0, m4, m6, m3, m1 and m8, respectively. The fusing lateral and medial nasal process (large black arrow), the nasolacrimal groove (black arrowheads), the cranial nerves (white and black asterisks indicate the trigeminal and olfactory nerves, respectively) and Cxcl14 expression in the ectoderm of LNP (small black arrow), MNP (red arrow) and MxP (red arrowhead) are shown. The black star in D shows expression of Hey2 in the olfactory epithelium in the region of the developing vomeronasal organ. e, eye; L, lateral nasal process; M, medial nasal process; MdP, mandibular prominence; oe, olfactory epithelium; V, ventricle; X, maxillary prominence. Scale bar: 200 µm.
Fig. 4.
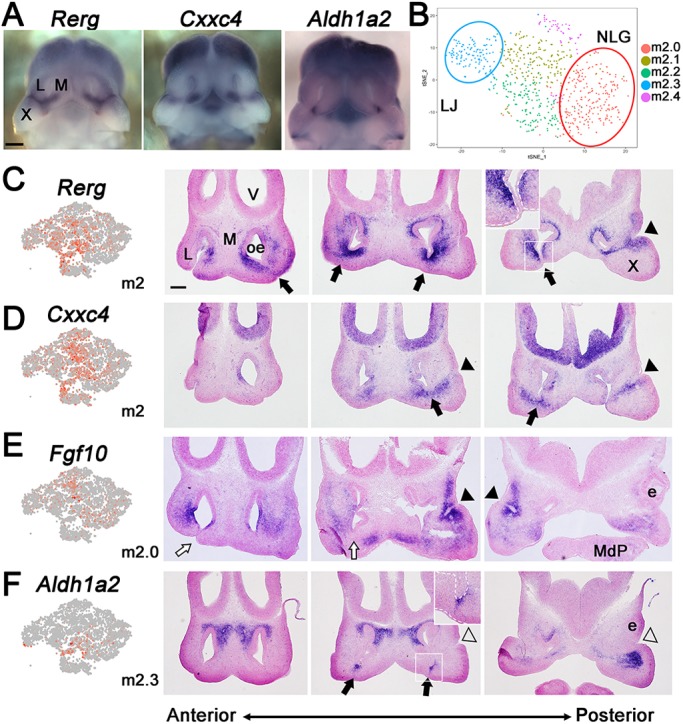
Mesenchymal cluster m2 maps adjacent to fusing epithelia. (A) Frontal view of E11.5 upper face following whole-mount in situ hybridization for the m2 markers Rerg, Cxxc4 and Aldh1a2. Scale bar: 500 µm. (B) tSNE plot of reclustered m2. (C-F) Feature plots (left panels) and in situ hybridization on E11.5 face frontal sections (three right panels, anterior to posterior) for Rerg (C), Cxxc4 (D), Fgf10 (E) and Aldh1a2 (F), representing clusters m2, m2, m2.0 and m2.3, respectively. Insets in C and F show more detailed images of the areas of fusion associated with the nasal fin (white rectangle). White dashed lines represent the boundary between ectodermal and mesenchymal layers. Black or white arrows, respectively, indicate the presence or absence of mesenchymal expression associated with the lambdoid junction and nasal fin. Black or white arrowheads, respectively, indicate the presence or absence of mesenchymal expression associated with the nasolacrimal groove. e, eye; L, lateral nasal process; M, medial nasal process; MdP, mandibular prominence; oe, olfactory epithelium; V, ventricle; X, maxillary prominence. Scale bar: 200 µm.
Most clusters mapped onto either a discrete region or tissue population within the upper face mesenchyme including: the LNP (m0, m4); MxP (m1, m5); surface ectoderm-proximal (m2, m3); chondroprogenitors (m6); and Schwann cell precursors (m8). The exception was m7, the cells of which were dispersed across the tSNE plot (Fig. 2A) and whose best marker gene, Fhl3, demonstrated widespread expression throughout the facial mesenchyme (Fig. S6). This cluster was enriched for expression of the chromatin remodelling genes Cbx3, Kdm6b, and Kmt2d, with lower expression of anatomy- or lineage-specific genes that marked the other clusters. The m7 cells were therefore ambiguous in nature but could potentially represent precursor cell populations transitioning into various differentiated cell types, or cells that were more related by cell cycle or chromatin dynamics than by anatomy.
For the more defined assignments, in situ hybridization for the m0 and m4 LNP clusters indicated that expression of the m0 markers Gm9866 and Rnf128 occurred in more anterior medial regions, whereas the m4 genes Gsc and Pitx2 mapped to the more posterior and medial parts of the LNP (Fig. 3, Figs S5 and S6) (Gaunt et al., 1993; St. Amand et al., 2000). Interestingly, we also detected an m0 subcluster, which we termed m0*, based on the expression of Calb2. The m0* cells mapped to the most anterior/distal tip of the LNP, lying adjacent to the LNP epithelia that contributes to the LJ (Figs S5 and S6). Both m0 and m4 also shared expression of the established LNP marker Pax7 (Fig. 2C and Fig. S5) (Aoto et al., 2002; Baker et al., 2016; Mansouri et al., 1996), and such overlaps can provide additional information within this scRNA-seq resource relevant to larger expression domains, regulatory interactions and potential cluster hierarchy. Clusters linked mainly to the MxP included m1, whose marker genes Cxcl14 and Lhx8 were expressed in the anterior and medial MxP mesenchyme (Fig. 3, Fig. S5) (Cesario et al., 2016), and m5, whose markers Shox2 and Lhx6 mapped to the posterior MxP-derived palatal shelves (Fig. S5) (Beverdam et al., 2001; Cesario et al., 2016; Yu et al., 2005). Cluster m6 was enriched in cells expressing Col9a1 and Sox9, and marked sites of chondroprogenitor condensation in the MNP (Fig. 3) (Mori-Akiyama et al., 2003). Cluster m8 (arrow in Fig. 2A/C) comprised 21 tightly clustered cells, likely representing Schwann cell progenitors based upon the presence of marker genes Sox10, Fabp7 and Plp1 (D'Antonio et al., 2006; Dickinson et al., 1997; Finzsch et al., 2010; Miller et al., 2010). This assignment was supported by the distribution of Sox10-expressing cells near the paths of the cranial nerves (Fig. 3). The cluster m3 markers Hey2 and Emx2 mapped to the sub-surface mesenchyme of the MxP and lateral LNP, but were less apparent in the LJ, NLG and midface (Fig. 3, Figs S5 and S6). In contrast, the m2 marker genes Rerg and Cxxc4 were expressed in close proximity to the LJ, the NLG and the choanal openings (Fig. 4A, Fig. S5).
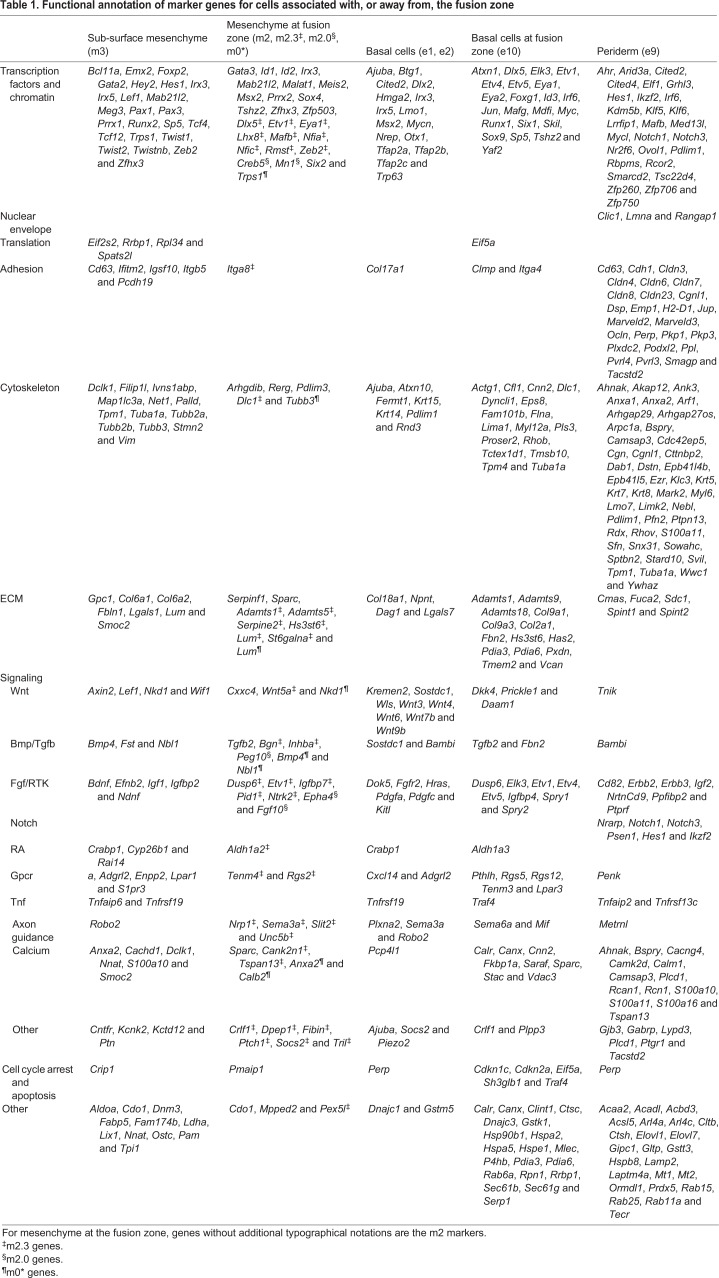
As m2 was partitioned into two separate areas on the tSNE plot (Fig. 2) and also positioned in proximity to both the LJ and NLG, it was reclustered to highlight any subpopulations and associated marker genes (Fig. 4B, Table S4). This identified subcluster m2.0 with Fgf10 as its most specific marker, and subcluster m2.3 with specific markers including Adamts5, Aldh1a2 and Inhba. In situ hybridization indicated that these subclusters represented separate regions of the fusing prominences, with the m2.0 marker Fgf10 expressed at the NLG, while Aldh1a2 mapped in the vicinity of the nasal fin and resolving choana (Fig. 4E,F). In contrast, the pan m2 markers Rerg and Cxxc4 occupied broader territories bordering the NLG, LJ and nasal fin (Fig. 4C,D). Therefore, the m2 cells, alongside m0 cells, which partly map around the LJ, are two mesenchymal populations closely associated with epithelial cells of the fusion zones. Table 1 shows the salient gene expression differences between the sub-surface mesenchyme that is either distal (m3) or proximal (m0*, m2) to the fusing seams parsed into functional groups, including transcription, adhesion, cytoskeleton and signalling. Interpreting these differences subsequently required a comparable analysis on the ectodermal cell populations.
Table 1.
Functional annotation of marker genes for cells associated with, or away from, the fusion zone
Ectodermal cell populations in and around the LJ
To identify the cells of the morphogenetically active epithelia at the LJ, the NLG and choanal openings, we selected the 943 ectodermal cells (Fig. 1), regressed out the cell cycle and tuned analytic parameters to optimize superposition of clusters with discrete groupings on the tSNE plots. This identified 12 ectodermal clusters (e0-11), each highlighted by a discrete suite of marker genes (Fig. 5, Table S5). By employing a combination of available gene expression data and in situ analysis (Figs 5, 6, Fig. S7; summarized in Table S6), these clusters could be broadly mapped to five anatomical regions: the surface ectoderm (e1, e2); the oral cavity (e3, e7); the nasal cavity (e0, e4, e11); the periderm (e9); and fusing epithelia (e5, e10). Thus, anatomy was again the organizing principal, with the exceptions of clusters e6 and e8. Notably, cells in e6 were dispersed across the tSNE plot and shared many marker genes with cluster m7, including Sox11, Cbx3, Supt16 and Kdm6b. Therefore, like the ambiguous m7 cluster, e6 may represent cells typified by a common cellular process, such as proliferation or chromatin remodelling, rather than by a particular differentiation pathway. Cluster e8 was dominated by the expression of pan-mesenchymal genes (e.g. Snai1, Alx1 and Pax3), but these cells also co-expressed markers of the ectoderm, such as Wnt3, Wnt9b, Fgf9 and Pitx1, with a few cells representing each ectodermal subtype. Thus, the e8 cluster could represent either a broad array of ectodermal cells undergoing EMT (Losa et al., 2018) or experimental artefacts where two cells of different germ layer origin were included in a single droplet for sequencing.
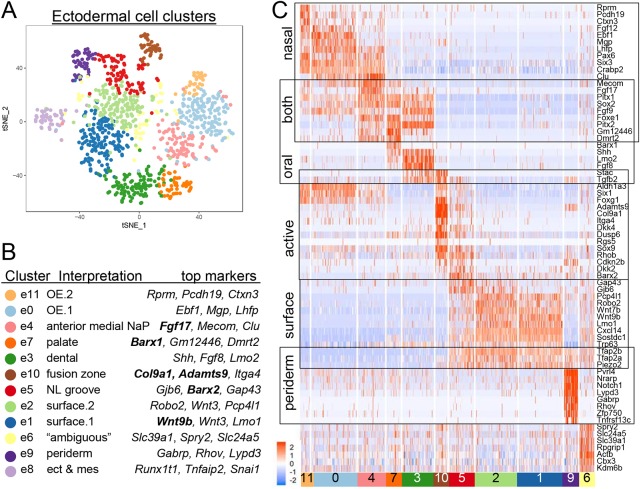
Fig. 5.
Reclustering identifies specific ectodermal cell populations. (A) tSNE plot of reclustered ectodermal cells from Fig. 1. (B) Summary of cluster-specific marker genes and interpretation/anatomical mapping. Genes in bold were used for in situ hybridization (Fig. 6, Fig. S7). (C) Heatmap representing scaled expression level (blue to red) of representative marker genes across the ectodermal population. Each row is a gene and each column is a cell. The bottom row demarcates the cell clusters (e8 cluster is omitted). The boxes indicate anatomically related groups of the clusters. ect, ectoderm; mes, mesenchyme; NL groove, nasolacrimal groove; NaP, nasal pit; OE, olfactory epithelium.
Fig. 6.
Mapping the ectodermal clusters by in situ hybridization. Feature plots for indicated marker gene and ectodermal cluster (leftmost panels). E11.5 mouse face in situ hybridization for Dkk4 (A), Barx2 (B), Adamts9 (C) and Stac (D) shown in whole-mount frontal (second panels) or lateral (third panels) view or as frontal sections (right two panels) at different anterior and posterior positions according to the large horizontal double-headed arrow. Lateral and medial nasal process fusion (solid white or black arrows), the nasolacrimal groove (arrowheads), Barx2 expression associated with MdP and MxP fusion (*) and the nasolacrimal duct (white arrow with black outline in B) are shown. The feature plots also show that Dkk4 expression was detected in e3 (marked with dl and also detailed in Fig. S7), with weaker expression of Adamts9 in e5 (overlapping the e10 domain at the NLG) and e9, the periderm. dl, dental lamina; e, eyes; LNP, lateral nasal process; MdP, mandibular prominence; MNP, medial nasal process; MxP, maxillary prominence; oe, olfactory epithelium; v, ventricles. Scale bars: 200 µm.
For the clusters with discrete mapping, e3 and e7 represented cells within the developing anterior oral cavity and shared expression of Pitx2, Shh and Sox2, which are known markers of the dental lamina (Fig. S7) (Thesleff, 2015). Examination of the specific e3 marker (Stac) indicated that this cluster represented more-anterior cells – towards to opening of the mouth – whereas the e7 marker (Barx1) positioned this cluster as more posterior (Fig. S7). e3 also contained cells with gene signatures for specialized epithelia and odontogenesis (Ctnnb1, Lef1, Sp6, Ogt, Bmp4, Bmp7, Krt14 and Krt17). In contrast, the e7 gene signature contained Krt8 and Krt18, which is indicative of a simple epithelium that will likely contribute to secondary palate rather than tooth development.
Clusters e0, e4 and e11 were derived from the nasal placode, based on their shared expression of Pax6 and Six3 (Fig. 5C) (Davis and Reed, 1996; Grindley et al., 1995). However, e4 lacked both the surface ectoderm marker Tfap2a and the olfactory epithelial marker Ebf1. Therefore, this cluster could represent cells that are positioned at the transition between surface/respiratory epithelium and olfactory epithelium. The cluster mapped anatomically to the medial side of the nasal pit based on the expression of Fgf17 (Fig. S7) and Mecom, which marks the invaginating vomeronasal organ (Perkins et al., 1991). This cluster may also have precursor cells for glandular secretion as it contained Elf5, Prss22 and Rab27b (Lapinskas et al., 2004; Donaldson et al., 2002; Konttinen et al., 1998; Chiang et al., 2011; Imai et al., 2004). The remaining placodal clusters, e0 and e11, shared the markers Ebf1, Ano1, Dmrta2, Zfhx4 and Foxg1, consistent with cells of the main olfactory epithelium (Davis and Reed, 1996; Gritli-Linde et al., 2009). Markers of differentiated olfactory sensory neurons were not present, but these clusters presumably represented precursor populations that develop from this region. Because the marker genes that distinguished clusters e0 (Ly6h, Lhfp and Gucy1a1) and e11 (Rprm, Gm266 and Ctxn3) are poorly characterized with respect to the developing olfactory epithelium, the significance of the differences between e0 and e11 remains to be determined.
The e1 and e2 clusters shared many genes (Fig. 5C), including Krt15, Perp and Trp63, which typify the basal epithelia of the developing skin. These clusters also contained several important transcription factors and signalling molecules, including Cxcl14, Sostdc1, Tfap2b, Wnt3, Wnt6, Wnt7b and Wnt9b, and examination of Cxcl14 expression confirmed widespread expression in the facial ectoderm, alongside its more localized mesenchymal expression (Fig. 3, Fig. S5). Although the profiles of these two clusters were similar, a number of genes were more prominent in e1 (Dlx1, Dlx2, Bmp4, Bambi, Rprml and Nell2) or in e2 (Lgals7, Gjb2, Gjb6, Nr2e3 and Trps1). These differences might reflect changes in the properties of the basal cells as they interact with a basement membrane during differentiation, and/or a gradient of epithelial differentiation across the prominences. The remaining three ectodermal clusters – e5, e9 and e10 – represented cell populations that are intimately associated with sites of fusion at the NLG and the LJ (e5, e10) or with the periderm (e9), and are described in more detail below.
Distinct basal cells with novel cell:cell signalling expression signatures occur at the LJ
Clusters e1, e2, e5 and e10 were all basal cell populations that formed a continuum between cells outside the fusion zones (e1/e2) to cells associated with active remodelling and fusion (e5/e10) (Fig. 5). The assignment of e5 and e10 to the fusion zones was based on in situ analysis of the shared marker Dkk4, which marked both the LJ and NLG (Fig. 6A). Markers specific for e5 (Barx2) and e10 (Adamts9, Stac and Tgfb2) were then used to refine the mapping. These in situ studies show that Barx2 was expressed in the NLG, whereas Adamts9, Stac and Tgfb2 were present in both the NLG and LJ (Fig. 6, Fig. S6 for Tgfb2) (Millan et al., 1991; Jones et al., 1997). Col9a1 was also identified as an e10 marker, and expression of this gene was apparent at the NLG, with lower expression at the LJ masked by high transcript levels in the adjacent mesenchyme (see Fig. 3). In this regard, the assignment of clusters is based on the expression levels of multiple genes across all cells analysed in the population and so variable expression is expected for any specific gene, such as Col9a1, within cells of a cluster. Although additional marker genes further distinguished e5 (Acvr2a, Dkk2, and Ddit4) from e10 (Rgs5) (Table S5), cells within these two clusters shared many notable changes from the remainder of the surface ectoderm. Specifically, expression of Cxcl14, Lmo1 and Wnt9b was reduced compared with e1/e2, whereas there was more prominent expression of several ECM components (Adamts9 and Col9a1), signalling molecules (Aldh1a3, Dkk4, Dusp6 and Tgfb2), cytoskeletal regulators (Rhob) and transcription factors (Foxg1, Junb, Six1 and Sox9). Therefore, cells in e10, and to a lesser extent in e5, are novel basal cell populations exhibiting expression profiles that suggest very different behavioural properties from the rest of the surface ectoderm, consistent with their location at critical fusion zones.
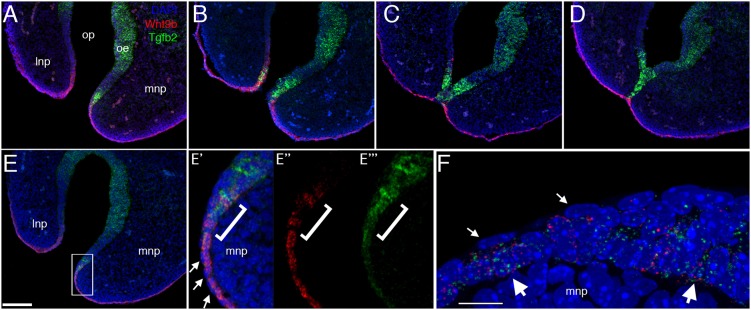
Table 1 shows the significant differences between the gene expression signatures of the basal cells that were distant (e1, e2) versus within (e10) the site of fusion. One notable difference in signalling profiles was that e10 exhibited decreased expression of genes for the Wnt ligands Wnt3, Wnt4, Wnt6, Wnt7b and Wnt9b, as well as Wls – which is required for Wnt secretion. Concurrently, e10 had increased expression of Dkk4, a secreted Wnt inhibitor, as well as an upregulation of Tgfb2. Previous studies have linked the Wnt pathway to orofacial clefting (Ferretti et al., 2011; Gong et al., 2000; Jin et al., 2012; Song et al., 2009), while Tgfb2 acts in fusion processes involving the ectoderm of the secondary palate and body wall (Dünker and Krieglstein, 2002; Sanford et al., 1997). We therefore employed RNAscope for dual labelling of the LJ epithelial seam at high resolution, with Tgfb2 as a specific marker of e10 cells and Wnt9b to mark the surface ectoderm, to determine how these two basal cell populations were distributed at the fusing LNP/MNP (Fig. 7). To investigate various stages in the fusion process, we used serial frontal sections at E11.0 to generate a pseudo-time series that included pre-fusion, fusion initiation and post-fusion resolution at the LJ. The anterior sections, near the tips of the nasal processes, are well ahead of the fusion front but are destined to fuse. The posterior sections include regions that have already fused, where the epithelial seam is regressing or has already regressed to establish continuity of the mesenchyme. In between is the fusion front, where the apposing epithelia are making contact and intermingling. We found that cells expressing either Wnt9b and/or Tgfb2 occupied the basal layer of the epithelium, adjacent to the loosely organized mesenchyme nuclei and roofed by a layer of periderm without nuclear Wnt9b/Tgfb2. Wnt9b expression covered the surface of the facial prominences, whereas Tgfb2 expression was limited to the zone bordering the olfactory epithelium. In the remainder of this paper, we will refer to the Wnt9b+ cells that occupy the basal layer of the epithelium as ‘canonical basal cells’ and the Tgfb2+ cells as ‘novel basal cells’. Tgfb2 expression appeared in the MNP about seven sections (∼100 µm) anterior to the fusion site (Fig. 7A), whereas expression was detected only in the section prior to this site (within ∼14 µm) in the LNP (Fig. 7B). At the fusion site, Tgfb2 expression marked the interacting epithelia, whereas Wnt9b-expressing cells were excluded from this region (Fig. 7C). Posterior to the fusion site, Tgfb2+ cells formed the epithelial seam (Fig. 7D), whereas Wnt9b+ cells formed the newly continuous surface epithelium. Although the Wnt9b and Tgfb2 positive cells were largely non-overlapping, there were some double-positive cells at the anterior edges of the Tgfb2+ domain (Fig. 7E), as well as a few double-positive cells within the scRNA-seq datasets. Whether these represent cells transitioning from one fate to the other remains to be investigated. The NLG also showed a similar loss of Wnt9b+ cells at the site of apposition and fusion, with a persistence of Tgfb2+ cells in this region mirroring aspects of fusion at the LJ (Fig. S8). Therefore, for both the LJ and the NLG, we conclude that cluster e10 represents novel basal cells that appear at the fusion zone. At the LJ, such cells persist as the nasal fin/epithelial seam, ultimately regressing to establish continuity of nasal and maxillary process mesenchyme.
Fig. 7.
Tgfb2 marks basal cells at the fusion zone. (A-F) RNAscope analysis of Tgfb2 (green) and Wnt9b (red) expression during fusion between the MNP and LNP, with DAPI marking nuclei (blue). (A-D) 14 μm frontal plane serial sections from tip of the nose through base of nasal pit showing (A) seven sections anterior (distal), (B) immediately anterior, (C) immediately behind (posterior) and (D) six sections posterior to the fusion front. (E) Tgfb2 expression is first detected in the MNP approximately seven or eight sections anterior (distal) to the fusion front. The boxed area in E is shown at higher magnification in E′-E″, with overlap of Tgfb2 and Wnt9b expression bracketed. (F) At higher resolution (0.5 µm optical sections), Tgfb2 and Wnt9b RNAs are seen in the same cell nuclei (large white arrows). Small white arrows in E and F show that these genes are not expressed in the flattened nuclei of the overlying periderm. Scale bars: 100 µm in A-E; 10 µm in F.
Periderm cells with altered morphology initially occur at the site of LJ fusion
Examination of cluster e9 revealed that it was equivalent to the periderm based on its expression of known marker genes, including Grhl3, Arhgap29, Sfn and Irf6 (Auden et al., 2006; de la Garza et al., 2013; Hammond et al., 2017; Kousa et al., 2017; Paul et al., 2017; Richardson et al., 2017, 2014). Cells within this cluster had a very discrete set of marker genes compared with all other ectodermal clusters (Table 1, Fig. 5C), reflecting unique adhesion and cytoskeletal properties, alongside specific suites of transcription factors. Thus, as well as being specifically marked by Gabrp, e9 also contained many genes required for the formation of desmosomes (Ppl and Pkp1), tight junctions (Cldn3, Cldn8) and focal adhesions (Nebl). Periderm appears as a protective layer on the surface of the embryo between E10 and E12, and importantly prevents premature fusion between adjacent ectodermal surfaces. We therefore examined how periderm acted during fusion at the LJ and NLG (Fig. 8 and Fig. S8). We used immunofluorescence on serial sections in a pseudo-time series at E11.0 to visualize the periderm markers Cldn3 and Grhl3, along with the basal cell marker p63. Cldn3+/Grhl3+ periderm cells covered the oral cavity and the surface of the facial processes but did not extend into the nasal cavity. Importantly, though, periderm did cover the transition zone between surface and nasal cavity epithelium (brackets in Fig. 8A,B), precisely the territory that participates in fusion. At the fusion front, there was an intermingling of Grhl3/Cldn3+ cells from the adjacent prominences, indicating that periderm potentially initiates contact. The periderm cells also lost their typical epithelial morphology (Fig. 8), a behaviour that was also observed during fusion at the NLG (Fig. S8). Posterior to the LJ fusion front, Grhl3/Cldn3+ cells were no longer observed within the nasal fin, indicating that the periderm cells were lost, while a p63+ epithelial seam persisted (Fig. 8D). We further noted that periderm nuclei at the fusion sites were rounded rather than displaying their normal flattened morphology. Volume projections confirmed that this was not a plane-of-section artefact (Movie 1). We mapped periderm nuclear morphology relative to the fusion process using serial sections. While flat periderm was associated with most surface epithelia, round periderm appeared in the nasal pit border about two to four sections ahead of the fusion front at the LJ (Fig. 8F). Therefore, in these fusion processes, the periderm cells are still present when surfaces of the facial prominences interact, but they then undergo a shift in cell morphology and are subsequently lost during establishment of a basal cell epithelial seam. Notably, reclustering of the periderm cells did not reveal consistent subpopulations with altered gene expression programs, implying that such morphological changes are driven more by post-transcriptional events.
Fig. 8.
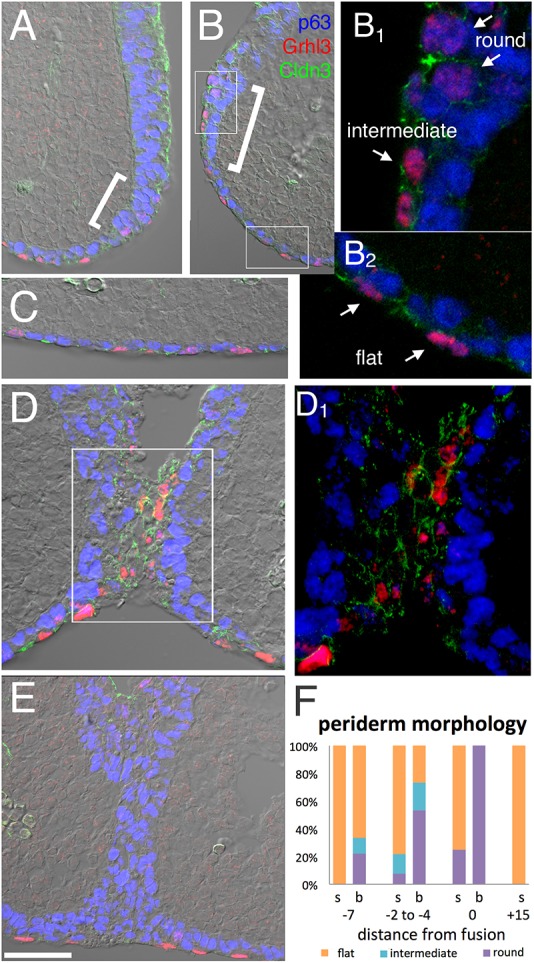
Periderm around the fusing lambdoid junction. (A-E) Immunofluorescent imaging of Grhl3 (red) in periderm nuclei, p63 (blue) in basal cell nuclei and Cldn3 (green) in cellular junctions of periderm, OE and endothelial, but not basal, cells overlaid onto a DIC image to visualize tissue organization. (A,B) The prospective fusion zone of the MNP and LNP two sections anterior (distal) to the contact point (shown in D). The brackets indicate the transition zone (the prospective fusion zone) between surface ectoderm and nasal pit ectoderm. White boxes in B indicate the surface epithelium and border epithelium, and are shown at higher magnification in B1 and B2, respectively, with arrows indicating examples of round, flat and intermediate Grhl3+ nuclei. (C) Surface epithelia near the facial midline, where p63+ basal cells and Grhl3/Cldn3+ periderm form an ordered bilayer. (D) Contact point where Cldn3/Grhl3+ cells bridge the gap, with the boxed area shown at higher magnification in D1. (E) The epithelial seam 15 sections posterior to the fusion point. Seam has p63+ cells but lacks Grhl3+ cells. (F) Quantification of periderm nuclear morphologies in surface epithelia (s) or prospective fusion zone (b), with distances from the fusion site indicated by relative section number. Scale bar: 50 µm in A,B,C,D,E.
Integration of cell signalling and cell behaviour signatures associated with the epithelial seams
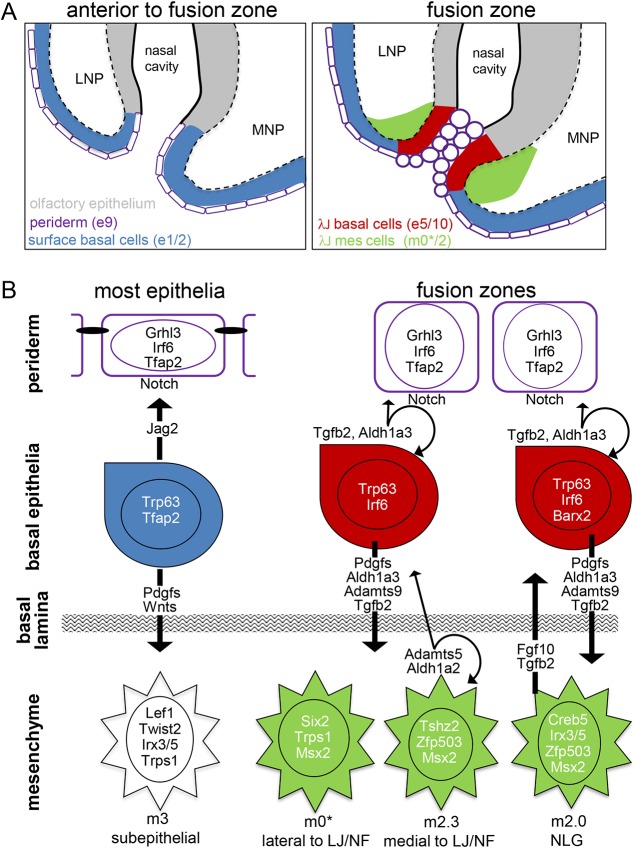
The scRNA-seq analysis identified novel cell populations in both the mesenchyme and basal cells of the ectoderm that existed around the site of fusion. These cell populations were clearly distinguished from neighbouring mesenchymal and ectodermal populations that were away from this fusion zone. Table 1 illustrates the main differences that distinguish these populations and serves to highlight potential changes in signalling interactions and cell behaviour as cells are recruited into the fusion zone. One notable event is that the basal cells switch from a Wnt-driven growth and morphogenesis program to a fusion program. This switch is accompanied by an increase of Tgfb2 expression, which is the one of the first changes detected in the basal cells of the MNP ahead of the fusion zone, an occurrence that also happens slightly later within the LNP (Fig. 7). In combination, as summarized in Fig. 9, we postulate that the expression changes in this novel basal cell population: (1) alter signalling to the overlying periderm at the junction; (2) alter signalling to the underlying mesenchyme at the junction; and (3) alter the behaviour of these basal cells in preparation for resolution of the epithelial seam by mechanisms such as EMT, apoptosis, cell extrusion and cell migration.
Fig. 9.
Model for fusion zones. (A) Diagram showing cell populations that are either anterior to (left panel) or within (right panel) the fusion zone of the lambdoid junction. The relevant tissue layers and clusters associated with the fusion front are colour coded and the white dashed lines represent the boundary between ectodermal and mesenchymal layers. Anterior to the fusion zone, the periderm has a flattened morphology. As fusion initiates, the periderm cells adopt a rounded appearance and the underlying basal ectodermal cells and adjacent mesenchymal cells adopt new gene expression programs. (B) The left schematic shows tissue away from the fusion sites. Flattened periderm connected by apical junctions (black ovals), potentially maintained by Jag2/Notch signalling. Basal cells signal to both the overlying periderm and underlying mesenchyme with a prominent Wnt signature. The right schematic shows fusing tissue at either the lambdoid junction (LJ) and nasal fin (NF) or the nasolacrimal groove (NLG). Periderm is rounded and lacks polarized junctions. The associated basal cells have largely re-tooled their signalling repertoire, replacing Wnts with Tgfb2 and with downregulating Jag2 expression, although Jag1 expression is maintained. Additionally, the basal cells express ECM remodelling enzymes such as Adamts9, which may indicate altered interaction with the basal lamina. At the transcriptional level, basal cells have lower Tfap2 and higher Irf6 levels, as well as raised Barx2 around the NLG. The mesenchyme underlying each of the fusion sites is unique, with different suites of transcription factors and signalling molecules, with Aldh1a2 at the NF and Fgf10 at the NLG. Colour coding is same as in A, except that grey hatching shows basal lamina.
DISCUSSION
This analysis provides a rich resource of gene expression information for thousands of single cells associated with the developing mammalian lip and primary palate during a critical period of fusion. By providing high-resolution anatomical mapping of specific gene expression programs onto the upper face, it complements and significantly extends previous studies on vertebrate facial development, which used techniques such as laser capture dissection, microarray analysis and bulk RNA-seq analysis (Brugmann et al., 2010; Brunskill et al., 2014; Buchtová et al., 2010; Feng et al., 2009; Gong et al., 2005; Hooper et al., 2017; Bhattacherjee et al., 2007). The data can be mined to examine multiple aspects of facial development. Specifically, at least 25 different cellular populations can be detected operating within this small but critical region, including endothelial cells and cells of the hematopoietic system that comprise an extensive vascular network associated with the fusing epithelial seams. Furthermore, the mesenchymal and ectodermal populations can both be parsed into multiple clusters, and each cluster yields specific information concerning the genetic programs operating in specific regions of the nasal and maxillary prominences associated with upper lip, NLG and primary palate development.
One critical application of this resource concerns its ability to decipher how genes associated with CL/P are expressed in relevant cell populations and how these processes are impacted by mutations or environmental insults. Information within the dataset is also relevant to interpreting other understudied aspects of facial development. Thus, a mouse knockout of Hapln1 presents with retrusion of the upper face and defects in cartilage and bone development that have been ascribed to its function in these skeletal elements (Watanabe and Yamada, 1999). However, our finding that Hapln1 is initially expressed highly in the endothelial cells of the developing vasculature indicates that it may function within this tissue to regulate craniofacial skeletal development. This conjecture is supported by the morpholino analysis in zebrafish, where hapln1b has been found to be involved in vascular development (Gomez et al., 2009). Therefore, this scRNA-seq resource provides an important database for analysis of how particular genes may act in the overall context of facial tissue populations to direct facial development and fusion.
The scRNA-seq data provide a detailed description of the cell populations associated with the fusing epithelial seams. We have identified unanticipated complexity in the epithelial and mesenchymal populations in the vicinity of the fusion sites at the LJ and NLG that we have validated by detailed anatomical mapping. We have used the gene expression profiles of the different populations to describe their behavioural and signalling potential. We highlight three key findings. First, we find that periderm is present at the fusion site, makes the contacts that potentially initiate fusion and then disappears. This periderm has an altered morphology, with cells diverging from their normal flattened appearance to a more rounded shape. As we have not detected consistent subpopulations with altered gene expression programs within the periderm cluster we have identified, we postulate that post-transcriptional events are mainly responsible for the behaviour changes accompanying fusion and subsequent removal of the periderm cells. Similar changes in periderm morphology have also been observed at the site of secondary palate fusion, where the process is controlled by Trp63 and Tgfb3 function (Hu et al., 2015b; Richardson et al., 2017). Second, we find a novel population of basal epithelial cells that appear ahead of the fusion front in the prospective fusion zone. During fusion, they intermingle to form the nasal fin, which then regresses to establish continuity of the mesenchyme. The most striking difference between canonical basal cells and the novel basal cells is a radical retooling of signalling, which could dictate the altered behaviour of periderm at the fusion zone. We also find signatures for extracellular matrix (ECM) remodelling and altered cytoskeletal dynamics in these novel basal cells that could contribute to regression of the nasal fin (Table 1). Third, we find that the mesenchyme clusters adjacent to the LJ and NLG have distinct gene expression programs that could both influence and be influenced by signalling interactions with the fusing epithelia. Despite these key observations, further studies will be needed to address how periderm and basal cells at the fusion zones may be lost through extrusion, apoptosis or migration, as many of these events are probably regulated at the level of the proteome rather than by transcriptional change, whereas the analysis of EMT will require additional studies using larger cell populations specific for the resolving epithelial seam.
The power of the scRNA-seq approach allows the identification of cell populations associated with the fusion zone, but also enables modelling of their potential regulatory behaviours and cell:cell interactions. Specifically, genes showing enriched expression within a cluster provide a description of the biological potential of those cells, including transcriptional programs, signalling competence, adhesion properties and cytoskeletal dynamics. Fig. 9 summarizes key changes in morphology, signalling interactions and transcriptional programs in the periderm, basal ectodermal cells and adjacent mesenchyme as the prominences approach, touch and fuse. Notably, many of the genes we identify within this model have prominent roles in orofacial clefting, as well as in other fusion-related processes. In this respect, there are clear changes in cell signalling potential as basal cells transition from the general surface ectoderm into the fusion zone. Outside the fusion zone, the basal cells express multiple ligands, including Jag2, which encodes an activating ligand for Notch signalling, but this gene is downregulated within the fusion zone. Previous studies have shown that loss of Jag2:Notch1 interaction within the oral cavity leads to ectopic adhesions and cleft secondary palate by shifting the periderm into a state where it is primed for fusion throughout the oral cavity (Casey et al., 2006). However, loss of Jag2 does not affect fusion of the lip and primary palate, suggesting that the oral cavity is particularly sensitive to Jag2:Notch signalling levels. Wnt signalling is an additional pathway that is altered as cells transition into the fusion zone with a major downregulation of Wnt ligand expression from the ectoderm. Concomitantly, there is also reduced expression of the canonical Wnt pathway transcriptional effector Lef1 in the mesenchyme immediately adjacent to the ectodermal fusion zones when compared with the general subsurface mesenchyme (Fig. 9, Table S2). Previous studies have observed various degrees of Wnt responsiveness in both the surface ectoderm and underlying mesenchyme of the developing mouse face using Wnt reporter constructs (Lan et al., 2006). In future, it will be important to assess whether Wnt responsiveness is altered at the fusion zone and whether there are any functional consequences caused by such an alteration. Nevertheless, considerable evidence supports the function of the Wnt signalling pathway in facial development and CL/P as inactivation of several mouse genes involved in Wnt signaling, including Lrp6, Porcn and Wnt9b, as well as those encoding Pbx transcription factors, can result in CL/P (Bankhead et al., 2015; Ferretti et al., 2011; Jin et al., 2012; Song et al., 2009). Previous studies have also shown that loss of Tgfb2 can result in cleft secondary palate, and combined loss of Tgfb2 and Tgfb3 can further result in a failure of ventral body wall closure (Dünker and Krieglstein, 2002; Sanford et al., 1997). Nevertheless, despite the increase in Tgfb2 levels as basal cells enter the fusion zone, loss of this gene has not been shown to cause CL/P in mice (Sanford et al., 1997). Therefore, although similar programs may be in place at both the site of primary and secondary palate fusion, their deployment may differ. One possibility is that Tgfb2 acts in a redundant manner at the LJ, e.g. with other members of the Tgfb superfamily, such as Bmp4 and/or Bmp7, that are also expressed in cluster e10 and are associated with orofacial clefting (Kouskoura et al., 2013; Liu et al., 2005).
Several transcription factors linked with orofacial clefting in mouse and human are also differentially expressed around the LJ and NLG, including Msx2, Trp63, Grhl3, Irf6 and Tfap2 family members (Green et al., 2015; Gritli-Linde, 2008; Kousa and Schutte, 2016; Pontoriero et al., 2008). Grhl3 and Trp63 are expressed widely throughout the periderm and basal cells, respectively, whereas other genes show more-dynamic expression profiles. Notably Irf6, which is present throughout the periderm, is also expressed specifically in the basal cells of the fusion zone. Finally, we reiterate that genes involved in orofacial clefting can be expressed in the periderm, the ectoderm or the mesenchyme, illustrating that various tissues are required for facial fusion. Similarly, such genes can be either widely expressed – possibly affecting the growth and morphogenesis required to bring the prominences in to apposition – or present in limited cell populations where they might perform more discrete functions in fusion. Further mining of this extensive scRNA-seq dataset, coupled with analysis of how these cellular programs are altered in various models of craniofacial clefting, should produce significant insight into facial development and the mechanisms of fusion of the facial prominences relevant to evolution and medicine.
MATERIALS AND METHODS
Mice
All animal experiments were performed in accordance with protocols approved by the University of Colorado Denver Animal Care and Usage Committee. Embryonic day 0.5 was considered to be noon on the day of a copulatory plug. C57BL/6J and ROSA26 LacZ reporter mice (B6.129S4-Gt(ROSA)26Sortm1Sor/J) (Soriano, 1999) were obtained from the Jackson Laboratory. Crect, an ectoderm-specific Cre recombinase line, has been described previously (Reid et al., 2011) and CD1 mice were obtained from Envigo. For the first scRNA-seq experiment, C57BL/6J mice were interbred and embryos isolated at E11.5. Those with 45-46 somites were employed for microdissection. To obtain embryos tagged with LacZ expression in the ectoderm for the second scRNA-seq experiment, male Crect mice were bred with homozygous female ROSA26 LacZ reporter mice. Embryos were isolated at E11.5 and a rapid determination of those positive for both the Crect and ROSA26 LacZ reporter alleles performed by staining for specific LacZ expression in the ectoderm. After initial isolation, the embryos were divided into sections: head, trunk and hindlimbs with tail tip. Heads were stored in ice-cold PBS prior to further dissection. Hindlimbs with tail tip were used for somite counting. The trunks were used for rapid β-gal staining as described previously (Van Otterloo et al., 2018) with modification. Briefly, trunks were fixed in 0.2% glutaraldehyde in PBS for 15 min at room temperature followed by three 5 min washes in LacZ rinse buffer [0.2 M sodium phosphate (pH 7.3), 2 mM magnesium chloride, 0.02% NP40, 0.01% sodium deoxycholate). Samples were then developed in LacZ rinse buffer containing 1 mg/ml X-Gal (Invitrogen/Life Technologies) in the dark at 37°C for ∼30 min to score the LacZ-positive embryos. Microdissection of the embryos from the entire litter (see below) occurred in parallel with β-gal staining. Four LacZ stained embryos with 45 somites were obtained from a single litter for subsequent scRNA-seq analysis.
Single cell RNA sequencing
The lambdoidal junction area (Fig. 1A, Fig. S1) was dissected in ice-cold PBS. Following microdissection, lambdoidal junction tissues from four or five embryos were pooled together and treated with 750 µl of 0.25% trypsin-EDTA (Invitrogen/Life Technologies) at 37°C for 10 min with gentle agitation. The reaction was stopped by adding 750 µl of DMEM with 10% FBS (Invitrogen/Life Technologies). The mixture was then pipetted up and down 20 times with wide orifice tips to assist in cell dispersion, and the cells were collected by centrifugation at 300 g for 3 min. Cells were washed in PBS with 0.4% BSA twice, and then filtered through a Flowmi 40 µM cell strainer (Bel-Art·H-B Instrument SP Scienceware). Subsequently, the cells were inspected by microscopy to ensure they were dissociated into a single cell suspension. Cells were then counted and immediately used for droplet derivation and library preparation with the Chromium single cell 3′ v2 chemistry (10X Genomics) according to the manufacturer's instructions. Sequencing was performed on an Illumina Hiseq 4000 with v4 chemistry. At least three lanes were used for each experiment generating ∼1000 million raw sequencing reads per dataset of which 80% were larger than Q30 with respect to quality control. All library preparation and sequencing was performed by the University of Colorado Denver Genomic and Microarray Shared Resource.
Data analysis
Cellranger toolkit version 2.0 (10X Genomics) was used for de-multiplexing, alignment to the mm10-1.2.0 transcriptome reference, and unique molecular identifier (UMI) counting with the default setting. For the second experiment involving embryos positive for both Crect and ROSA26 LacZ reporter alleles, Cre and LacZ sequences were appended to the mm10 reference genome. The output raw UMI count matrices, with cells as columns and rows as genes, were processed using the R package Seurat 2.1 (Satija et al., 2015) to filter potential doublets or damaged cells, normalize the number of UMI (nUMI)/cell, identify variable genes, regress out unwanted sources of variation, identify significant principal components of variable gene expression, reduce dimensions, cluster cells and find marker genes. Default parameters were used unless specified below. To exclude potential doublets, apoptotic or lysed cells, cells with a nUMI larger than 80,000 or a percentage of UMI representing mitochondrial-encoded genes (pct.mito) of more than 10% were filtered out. Variable genes for initial clusterings and mesenchyme reclusterings were identified using default parameters besides x.low.cutoff=0.0125, x.high.cutoff=3 and y.cutoff=0.5. For ectoderm clustering, parameters were x.low.cutoff=0.1 and y.cutoff=0.25 with all the other default settings. For the reclustering of the separate ectoderm and mesenchyme cell populations, we also regressed out cell cycle phases using the Seurat 2.1 package. For ectoderm, the G2M genelist was expanded to include additional histone, Cdk2 and Cenp genes. The number of principal components and resolution of clustering were tuned to optimize superposition of clusters with discrete groupings on the tSNE plots. For initial clustering of the LJ and CrectLJ datasets, and reclustering the mesenchymal cells, 10 principal components and resolution 0.6 were used. For reclustering of m2, 15 principal components and resolution 1.0 were used. For reclustering of the ectodermal cells, 21 principal components and resolution 2.0 were used. Marker genes for each cluster – genes with enriched expression in cells of the cluster – were calculated by the Seurat algorithm that incorporated the mean expression level inside versus outside the cluster (avg_logFC), and the percentage of cells with detectable expression inside and outside the cluster (pct.1 and pct.2) with cutoffs as avg_logFC>0.25 and adjusted P value<0.05 (Tables S1, S2, S4, and S5). We also generated specificity indexes from the ratio (pct.ratio) and difference (pct.diff) of detection inside versus outside the cluster, and used marker genes that were both highly specific and highly expressed as the top marker genes for anatomical mapping and interpretation of cluster identity.
Marker genes highlighted in Table 1 were derived using a logFC>0.25 and adjusted P value<0.05. Functional classifications for cluster marker genes in Table 1 were derived using GO terms retrieved from DAVID6.7 (Huang et al., 2009a,b) supplemented by manual annotations from the literature. As many kinases and phosphatases are not pathway specific, and few are regulated at the transcriptional level, they were excluded from Table 1.
In situ hybridization
Probes were generated by cloning a unique fragment (sequences given upon request) into a TOPO vector (Life Technologies) using cDNA synthesized from mouse embryonic mRNA as a template. cDNA was generated using the Superscript III First-Strand Synthesis System (Life Technologies), as per the manufacturer's instructions. Sequence verified plasmids were linearized and antisense probes synthesized using an appropriate DNA-dependent RNA polymerase (T7/T3/SP6) and DIG RNA labelling mix (Roche). Whole-mount in situ hybridization was performed on E11.5 embryos as described previously (Feng et al., 2009). Hybridized probe was detected using an anti-digoxigenin antibody (Roche) and signal was developed in BM Purple (Roche).
In situ hybridization on frozen sections was performed as previously described (Schmidt et al., 2018) with modifications. Briefly, fixed E11.5 embryos were embedded in OCT (Sakura Finetek), frozen, sectioned at 14 µm and fixed onto 3-aminopropyltriethoxysilane (APES) (Millipore Sigma)-treated slides. Sections were fixed, treated with proteinase K for 3 min, re-fixed, acetylated and hybridized with probe at 1 µg/ml. Hybridized probe was detected using AP-conjugated anti-digoxigenin antibody (Roche) and signal was developed in BM Purple (Roche, Basel, Switzerland). Stained slides were postfixed, and counterstained with nuclear Fast Red (Vector Laboratories), dehydrated, and mounted with Cytoseal 60 (Thomas Scientific).
RNAscope was used for multiplexed in situ hybridization. Fixed frozen serial sections were prepared as described above, and expression detected using RNAscope Probes, Multiplex Fluorescent Reagent Kit v2 (Advanced Cell Diagnostics) and TSA Plus fluorophores (Perkin Elmer) according to the manufacturers' recommendations, with following specifics: target retrieval was omitted, slides received a 15 min treatment with Protease III and TSA fluorophores were diluted 1:1500. Images were captured with a Nikon A1R scanning confocal microscope.
Immunofluorescence
Immunofluorescent labelling of fixed frozen sections was performed as previously described (Finger et al., 2017) with the following adaptations. After thawing frozen sections, antigen retrieval occurred in 10 mM sodium citrate (pH 6.0) for 8 min in a Cuisinart pressure cooker set to high, followed by endogenous peroxidase quenching for 10 min with 0.3% H2O2. Immunodetection was performed in three rounds: first, mouse anti-Grhl3 (sc-398838 at 1:5000 from Santa Cruz Biotechnology) was detected by HRP-linked anti-mouse IgG (#7076, 1:1000, Cell Signaling Technology) followed by TSA Plus Cyanine3 (1:50 for 10 min); second, rabbit anti-p63-α (#4892 at 1:10,000, Cell Signaling Technology) was detected by HRP-linked anti-rabbit IgG (#7074 at 1:1000, Cell Signaling Technology) followed by TSA Plus Cyanine5 (1:50 for 10 min); third, rabbit anti-Cldn3 (ab15102 at 1:100, Abcam) was detected with Alexa488-conjugated anti-mouse IgG Fab2 (715-546-151 at 1:400, Jackson Immunoresearch). Slides were mounted in ProLong Gold (ThermoFisher Scientific) and images were captured using a Nikon A1R confocal scanning microscope. Volumetric projections of high-resolution z-stacks and movies thereof were generated using Nikon's Elements software.
Supplementary Material
Acknowledgements
Special thanks to Irene Choi for technical assistance; to the UCD Genomics and Microarray Core Facility for performing the library preparation and single cell sequencing; to Eric Van Otterloo for comments on the manuscript; and to all our colleagues in the Cells, Stem Cells and Development Graduate Program for their insights into this work.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: H.L., T.W.; Methodology: H.L., K.L.J., T.W.; Validation: H.L., J.E.H., T.W.; Formal analysis: H.L., J.E.H., T.W.; Investigation: H.L., J.E.H., T.W.; Resources: K.L.J., T.W.; Data curation: H.L., J.E.H.; Writing - original draft: H.L., J.E.H., T.W.; Writing - review & editing: H.L., K.L.J., J.E.H., T.W.; Visualization: H.L., J.E.H., T.W.; Supervision: T.W.; Project administration: T.W.; Funding acquisition: T.W.
Funding
This work was supported by the RNA Bioscience Initiative, University of Colorado Denver (T.W.) and the National Institutes of Health (5U01DE024429 to J.E.H., K.L.J. and T.W.). Deposited in PMC for release after 12 months.
Data availability
The scRNA-seq datasets have been deposited in Facebase (www.facebase.org) under accession numbers FB00001024 (C57LJ) and FB00001039 (CrectLJ), and also in GEO under accession number GSE132462.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.174888.supplemental
References
- Abramyan J. and Richman J. M. (2015). Recent insights into the morphological diversity in the amniote primary and secondary palates. Dev. Dyn. 244, 1457-1468. 10.1002/dvdy.24338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoto K., Nishimura T., Eto K. and Motoyama J. (2002). Mouse GLI3 regulates Fgf8 expression and apoptosis in the developing neural tube, face, and limb bud. Dev. Biol. 251, 320-332. 10.1006/dbio.2002.0811 [DOI] [PubMed] [Google Scholar]
- Auden A., Caddy J., Wilanowski T., Ting S. B., Cunningham J. M. and Jane S. M. (2006). Spatial and temporal expression of the Grainyhead-like transcription factor family during murine development. Gene Expr. Patterns 6, 964-970. 10.1016/j.modgep.2006.03.011 [DOI] [PubMed] [Google Scholar]
- Baker J. L., Wood B., Karpinski B. A., LaMantia A.-S. and Maynard T. M. (2016). Testicular receptor 2, Nr2c1, is associated with stem cells in the developing olfactory epithelium and other cranial sensory and skeletal structures. Gene Expr. Patterns 20, 71-79. 10.1016/j.gep.2015.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankhead E. J., Colasanto M. P., Dyorich K. M., Jamrich M., Murtaugh L. C. and Fuhrmann S. (2015). Multiple requirements of the focal dermal hypoplasia gene porcupine during ocular morphogenesis. Am. J. Pathol. 185, 197-213. 10.1016/j.ajpath.2014.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beverdam A., Brouwer A., Reijnen M., Korving J. and Meijlink F. (2001). Severe nasal clefting and abnormal embryonic apoptosis in Alx3/Alx4 double mutant mice. Development 128, 3975-3986. 10.1046/j.1469-7580.2001.1991021715.x [DOI] [PubMed] [Google Scholar]
- Bhattacherjee V., Mukhopadhyay P., Singh S., Johnson C., Philipose J. T., Warner C. P., Greene R. M. and Pisano M. M. (2007). Neural crest and mesoderm lineage-dependent gene expression in orofacial development. Differentiation 75, 463-477. 10.1111/j.1432-0436.2006.00145.x [DOI] [PubMed] [Google Scholar]
- Brugmann S. A., Powder K. E., Young N. M., Goodnough L. H., Hahn S. M., James A. W., Helms J. A. and Lovett M. (2010). Comparative gene expression analysis of avian embryonic facial structures reveals new candidates for human craniofacial disorders. Hum. Mol. Genet. 19, 920-930. 10.1093/hmg/ddp559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunskill E. W., Potter A. S., Distasio A., Dexheimer P., Plassard A., Aronow B. J. and Potter S. S. (2014). A gene expression atlas of early craniofacial development. Dev. Biol. 391, 133-146. 10.1016/j.ydbio.2014.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchtová M., Kuo W. P., Nimmagadda S., Benson S. L., Geetha-Loganathan P., Logan C., Au-Yeung T., Chiang E., Fu K. and Richman J. M. (2010). Whole genome microarray analysis of chicken embryo facial prominences. Dev. Dyn. 239, 574-591. 10.1002/dvdy.22135 [DOI] [PubMed] [Google Scholar]
- Casey L. M., Lan Y., Cho E.-S., Maltby K. M., Gridley T. and Jiang R. (2006). Jag2-Notch1 signaling regulates oral epithelial differentiation and palate development. Dev. Dyn. 235, 1830-1844. 10.1002/dvdy.20821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesario J. M., Almaidhan A. A. and Jeong J. (2016). Expression of forkhead box transcription factor genes Foxp1 and Foxp2 during jaw development. Gene Expr. Patterns 20, 111-119. 10.1016/j.gep.2016.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang L., Ngo J., Schechter J. E., Karvar S., Tolmachova T., Seabra M. C., Hume A. N. and Hamm-Alvarez S. F. (2011). Rab27b regulates exocytosis of secretory vesicles in acinar epithelial cells from the lacrimal gland. Am. J. Physiol. Cell Physiol. 301, C507-C521. 10.1152/ajpcell.00355.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'antonio M., Michalovich D., Paterson M., Droggiti A., Woodhoo A., Mirsky R. and Jessen K. R. (2006). Gene profiling and bioinformatic analysis of Schwann cell embryonic development and myelination. Glia 53, 501-515. 10.1002/glia.20309 [DOI] [PubMed] [Google Scholar]
- Davis J. A. and Reed R. R. (1996). Role of Olf-1 and Pax-6 transcription factors in neurodevelopment. J. Neurosci. 16, 5082-5094. 10.1523/JNEUROSCI.16-16-05082.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Garza G., Schleiffarth J. R., Dunnwald M., Mankad A., Weirather J. L., Bonde G., Butcher S., Mansour T. A., Kousa Y. A., Fukazawa C. F. et al. (2013). Interferon regulatory factor 6 promotes differentiation of the periderm by activating expression of Grainyhead-like 3. J. Invest. Dermatol. 133, 68-77. 10.1038/jid.2012.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson P. J., Griffiths I. R., Barrie J. M., Kyriakides E., Pollock G. F. and Barnett S. C. (1997). Expression of the dm-20 isoform of the plp gene in olfactory nerve ensheathing cells: evidence from developmental studies. J. Neurocytol. 26, 181-189. 10.1023/A:1018584013739 [DOI] [PubMed] [Google Scholar]
- Diewert V. M. and Wang K.-Y. (1992). Recent advances in primary palate and midface morphogenesis research. Crit. Rev. Oral Biol. Med. 4, 111-130. 10.1177/10454411920040010201 [DOI] [PubMed] [Google Scholar]
- Donaldson S. H., Hirsh A., Li D. C., Holloway G., Chao J., Boucher R. C. and Gabriel S. E. (2002). Regulation of the epithelial sodium channel by serine proteases in human airways. J. Biol. Chem. 277, 8338-8345. 10.1074/jbc.M105044200 [DOI] [PubMed] [Google Scholar]
- Dünker N. and Krieglstein K. (2002). Tgfbeta2−/− Tgfbeta3−/− double knockout mice display severe midline fusion defects and early embryonic lethality. Anat. Embryol. 206, 73-83. 10.1007/s00429-002-0273-6 [DOI] [PubMed] [Google Scholar]
- Feng W., Leach S. M., Tipney H., Phang T., Geraci M., Spritz R. A., Hunter L. E. and Williams T. (2009). Spatial and temporal analysis of gene expression during growth and fusion of the mouse facial prominences. PLoS ONE 4, e8066 10.1371/journal.pone.0008066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E., Li B., Zewdu R., Wells V., Hebert J. M., Karner C., Anderson M. J., Williams T., Dixon J., Dixon M. J. et al. (2011). A conserved Pbx-Wnt-p63-Irf6 regulatory module controls face morphogenesis by promoting epithelial apoptosis. Dev. Cell 21, 627-641. 10.1016/j.devcel.2011.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger T. E., Bartel D. L., Shultz N., Goodson N. B. and Greer C. A. (2017). 5HTR3A-driven GFP labels immature olfactory sensory neurons. J. Comp. Neurol. 525, 1743-1755. 10.1002/cne.24180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzsch M., Schreiner S., Kichko T., Reeh P., Tamm E. R., Bösl M. R., Meijer D. and Wegner M. (2010). Sox10 is required for Schwann cell identity and progression beyond the immature Schwann cell stage. J. Cell Biol. 189, 701-712. 10.1083/jcb.200912142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitchett J. E. and Hay E. D. (1989). Medial edge epithelium transforms to mesenchyme after embryonic palatal shelves fuse. Dev. Biol. 131, 455-474. 10.1016/S0012-1606(89)80017-X [DOI] [PubMed] [Google Scholar]
- Forbes D. P. and Steffek A. J. (1989). Epithelial bridging of the primary palate: II. In vitro model mimics in vivo behavior. J. Craniofac. Genet. Dev. Biol. 9, 367-380. [PubMed] [Google Scholar]
- Forni P. E., Bharti K., Flannery E. M., Shimogori T. and Wray S. (2013). The indirect role of fibroblast growth factor-8 in defining neurogenic niches of the olfactory/GnRH systems. J. Neurosci. 33, 19620-19634. 10.1523/JNEUROSCI.3238-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaare J. D. and Langman J. (1977a). Fusion of nasal swellings in the mouse embryo: regression of the nasal fin. Am. J. Anat. 150, 477-499. 10.1002/aja.1001500308 [DOI] [PubMed] [Google Scholar]
- Gaare J. D. and Langman J. (1977b). Fusion of nasal swellings in the mouse embryo: surface coat and initial contact. Am. J. Anat. 150, 461-475. 10.1002/aja.1001500307 [DOI] [PubMed] [Google Scholar]
- Gaare J. D. and Langman J. (1980). Fusion of nasal swellings in the mouse embryo. DNA synthesis and histological features. Anat. Embryol. 159, 85-99. 10.1007/BF00299258 [DOI] [PubMed] [Google Scholar]
- Gaunt S. J., Blum M. and de Robertis E. M. (1993). Expression of the mouse goosecoid gene during mid-embryogenesis may mark mesenchymal cell lineages in the developing head, limbs and body wall. Development 117, 769-778. [DOI] [PubMed] [Google Scholar]
- Gomez G. A., Veldman M. B., Zhao Y., Burgess S. and Lin S. (2009). Discovery and characterization of novel vascular and hematopoietic genes downstream of etsrp in zebrafish. PLoS ONE 4, e4994 10.1371/journal.pone.0004994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S.-G., White N. J. and Sakasegawa A. Y. (2000). The Twirler mouse, a model for the study of cleft lip and palate. Arch. Oral Biol. 45, 87-94. 10.1016/S0003-9969(99)00101-6 [DOI] [PubMed] [Google Scholar]
- Gong S.-G., Gong T.-W. and Shum L. (2005). Identification of markers of the midface. J. Dent. Res. 84, 69-72. 10.1177/154405910508400112 [DOI] [PubMed] [Google Scholar]
- Green R. M., Feng W., Phang T., Fish J. L., Li H., Spritz R. A., Marcucio R. S., Hooper J., Jamniczky H., Hallgrimsson B. et al. (2015). Tfap2a-dependent changes in mouse facial morphology result in clefting that can be ameliorated by a reduction in Fgf8 gene dosage. Dis. Model. Mech. 8, 31-43. 10.1242/dmm.017616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley J. C., Davidson D. R. and Hill R. E. (1995). The role of Pax-6 in eye and nasal development. Development 121, 1433-1442. [DOI] [PubMed] [Google Scholar]
- Gritli-Linde A. (2008). The etiopathogenesis of cleft lip and cleft palate: usefulness and caveats of mouse models. Curr. Top. Dev. Biol. 84, 37-138. 10.1016/S0070-2153(08)00602-9 [DOI] [PubMed] [Google Scholar]
- Gritli-Linde A., Vaziri Sani F., Rock J. R., Hallberg K., Iribarne D., Harfe B. D. and Linde A. (2009). Expression patterns of the Tmem16 gene family during cephalic development in the mouse. Gene Expr. Patterns 9, 178-191. 10.1016/j.gep.2008.11.002 [DOI] [PubMed] [Google Scholar]
- Hammond N. L., Dixon J. and Dixon M. J. (2017). Periderm: life-cycle and function during orofacial and epidermal development. Semin. Cell Dev. Biol. (in press) 10.1016/j.semcdb.2017.08.021 [DOI] [PubMed] [Google Scholar]
- Hooper J. E., Feng W., Li H., Leach S. M., Phang T., Siska C., Jones K. L., Spritz R. A., Hunter L. E. and Williams T. (2017). Systems biology of facial development: contributions of ectoderm and mesenchyme. Dev. Biol. 426, 97-114. 10.1016/j.ydbio.2017.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D., Young N. M., Li X., Xu Y., Hallgrimsson B. and Marcucio R. S. (2015a). A dynamic Shh expression pattern, regulated by SHH and BMP signaling, coordinates fusion of primordia in the amniote face. Development 142, 567-574. 10.1242/dev.114835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L., Liu J., Li Z., Ozturk F., Gurumurthy C., Romano R.-A., Sinha S. and Nawshad A. (2015b). TGFbeta3 regulates periderm removal through DeltaNp63 in the developing palate. J. Cell. Physiol. 230, 1212-1225. 10.1002/jcp.24856 [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T. and Lempicki R. A. (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1-13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T. and Lempicki R. A. (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44-57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Imai A., Yoshie S., Nashida T., Shimomura H. and Fukuda M. (2004). The small GTPase Rab27B regulates amylase release from rat parotid acinar cells. J. Cell Sci. 117, 1945-1953. 10.1242/jcs.01048 [DOI] [PubMed] [Google Scholar]
- Jiang R., Bush J. O. and Lidral A. C. (2006). Development of the upper lip: morphogenetic and molecular mechanisms. Dev. Dyn. 235, 1152-1166. 10.1002/dvdy.20646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y.-R., Han X. H., Taketo M. M. and Yoon J. K. (2012). Wnt9b-dependent FGF signaling is crucial for outgrowth of the nasal and maxillary processes during upper jaw and lip development. Development 139, 1821-1830. 10.1242/dev.075796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones F. S., Kioussi C., Copertino D. W., Kallunki P., Holst B. D. and Edelman G. M. (1997). Barx2, a new homeobox gene of the Bar class, is expressed in neural and craniofacial structures during development. Proc. Natl. Acad. Sci. USA 94, 2632-2637. 10.1073/pnas.94.6.2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juriloff D. M. and Harris M. J. (2008). Mouse genetic models of cleft lip with or without cleft palate. Birth Defects Res. A Clin. Mol. Teratol 82, 63-77. 10.1002/bdra.20430 [DOI] [PubMed] [Google Scholar]
- Konttinen Y. T., Halinen S., Hanemaaijer R., Sorsa T., Hietanen J., Ceponis A., Xu J.-W., Manthorpe R., Whittington J., Larsson A. et al. (1998). Matrix metalloproteinase (MMP)-9 type IV collagenase/gelatinase implicated in the pathogenesis of Sjögren's syndrome. Matrix Biol. 17, 335-347. 10.1016/S0945-053X(98)90086-5 [DOI] [PubMed] [Google Scholar]
- Kousa Y. A. and Schutte B. C. (2016). Toward an orofacial gene regulatory network. Dev. Dyn. 245, 220-232. 10.1002/dvdy.24341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousa Y. A., Roushangar R., Patel N., Walter A., Marangoni P., Krumlauf R., Klein O. D. and Schutte B. C. (2017). IRF6 and SPRY4 signaling interact in periderm development. J. Dent. Res. 96, 1306-1313. 10.1177/0022034517719870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouskoura T., Kozlova A., Alexiou M., Blumer S., Zouvelou V., Katsaros C., Chiquet M., Mitsiadis T. A. and Graf D. (2013). The etiology of cleft palate formation in BMP7-deficient mice. PLoS ONE 8, e59463 10.1371/journal.pone.0059463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y., Ryan R. C., Zhang Z., Bullard S. A., Bush J. O., Maltby K. M., Lidral A. C. and Jiang R. (2006). Expression of Wnt9b and activation of canonical Wnt signaling during midfacial morphogenesis in mice. Dev. Dyn. 235, 1448-1454. 10.1002/dvdy.20723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapinskas E. J., Palmer J., Ricardo S., Hertzog P. J., Hammacher A. and Pritchard M. A. (2004). A major site of expression of the ets transcription factor Elf5 is epithelia of exocrine glands. Histochem. Cell Biol. 122, 521-526. 10.1007/s00418-004-0713-x [DOI] [PubMed] [Google Scholar]
- Li C., Lan Y. and Jiang R. (2017). Molecular and Cellular Mechanisms of Palate Development. J. Dent. Res. 96, 1184-1191. 10.1177/0022034517703580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Sun X., Braut A., Mishina Y., Behringer R. R., Mina M. and Martin J. F. (2005). Distinct functions for Bmp signaling in lip and palate fusion in mice. Development 132, 1453-1461. 10.1242/dev.01676 [DOI] [PubMed] [Google Scholar]
- Losa M., Risolino M., Li B., Hart J., Quintana L., Grishina I., Yang H., Choi I. F., Lewicki P., Khan S. et al. (2018). Face morphogenesis is promoted by Pbx-dependent EMT via regulation of Snail1 during frontonasal prominence fusion. Development 145, dev157628 10.1242/dev.157628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotz K., Proff P., Bienengraeber V., Fanghaenel J., Gedrange T. and Weingaertner J. (2006). Apoptosis as a creative agent of embryonic development of bucca, mentum and nasolacrimal duct. An in vivo study in rats. J. Craniomaxillofac. Surg. 34 Suppl. 2, 8-13. 10.1016/S1010-5182(06)60003-6 [DOI] [PubMed] [Google Scholar]
- Mansouri A., Hallonet M. and Gruss P. (1996). Pax genes and their roles in cell differentiation and development. Curr. Opin. Cell Biol. 8, 851-857. 10.1016/S0955-0674(96)80087-1 [DOI] [PubMed] [Google Scholar]
- Millan F. A., Denhez F., Kondaiah P. and Akhurst R. J. (1991). Embryonic gene expression patterns of TGF beta 1, beta 2 and beta 3 suggest different developmental functions in vivo. Development 111, 131-143. [DOI] [PubMed] [Google Scholar]
- Miller A. M., Treloar H. B. and Greer C. A. (2010). Composition of the migratory mass during development of the olfactory nerve. J. Comp. Neurol. 518, 4825-4841. 10.1002/cne.22497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millicovsky G. and Johnston M. C. (1981). Active role of embryonic facial epithelium: new evidence of cellular events in morphogenesis. J. Embryol. Exp. Morphol. 63, 53-66. [PubMed] [Google Scholar]
- Millicovsky G., Ambrose L. J. H. and Johnston M. C. (1982). Developmental alterations associated with spontaneous cleft lip and palate in CL/Fr mice. Am. J. Anat. 164, 29-44. 10.1002/aja.1001640104 [DOI] [PubMed] [Google Scholar]
- Miyake T., Cameron A. M. and Hall B. K. (1996). Detailed staging of inbred C57BL/6 mice between Theiler's [1972] stages 18 and 21 (11-13 days of gestation) based on craniofacial development. J. Craniofac. Genet. Dev. Biol. 16, 1-31. [PubMed] [Google Scholar]
- Mori-Akiyama Y., Akiyama H., Rowitch D. H. and DE Crombrugghe B. (2003). Sox9 is required for determination of the chondrogenic cell lineage in the cranial neural crest. Proc. Natl. Acad. Sci. USA 100, 9360-9365. 10.1073/pnas.1631288100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul B. J., Palmer K., Sharp J. C., Pratt C. H., Murray S. A. and Dunnwald M. (2017). ARHGAP29 mutation is associated with abnormal oral epithelial adhesions. J. Dent. Res. 96, 1298-1305. 10.1177/0022034517726079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins A. S., Mercer J. A., Jenkins N. A. and Copeland N. G. (1991). Patterns of Evi-1 expression in embryonic and adult tissues suggest that Evi-1 plays an important regulatory role in mouse development. Development 111, 479-487. [DOI] [PubMed] [Google Scholar]
- Peterka M., Tvrdek M., Likovsky Z., Peterkova R. and Fara M. (1994). Maternal hyperthermia and infection as one of possible causes of orofacial clefts. Acta Chir. Plast. 36, 114-118. [PubMed] [Google Scholar]
- Pontoriero G. F., Deschamps P., Ashery-Padan R., Wong R., Yang Y., Zavadil J., Cvekl A., Sullivan S., Williams T. and West-Mays J. A. (2008). Cell autonomous roles for AP-2alpha in lens vesicle separation and maintenance of the lens epithelial cell phenotype. Dev. Dyn. 237, 602-617. 10.1002/dvdy.21445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S. C. (1933). An embryological study of harelip in mice. Anat. Rec. 56, 101-110. 10.1002/ar.1090560202 [DOI] [Google Scholar]
- Reid B. S., Yang H., Melvin V. S., Taketo M. M. and Williams T. (2011). Ectodermal Wnt/beta-catenin signaling shapes the mouse face. Dev. Biol. 349, 261-269. 10.1016/j.ydbio.2010.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhinn M. and Dolle P. (2012). Retinoic acid signalling during development. Development 139, 843-858. 10.1242/dev.065938 [DOI] [PubMed] [Google Scholar]
- Richardson R. J., Hammond N. L., Coulombe P. A., Saloranta C., Nousiainen H. O., Salonen R., Berry A., Hanley N., Headon D., Karikoski R. et al. (2014). Periderm prevents pathological epithelial adhesions during embryogenesis. J. Clin. Invest. 124, 3891-3900. 10.1172/JCI71946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson R., Mitchell K., Hammond N. L., Mollo M. R., Kouwenhoven E. N., Wyatt N. D., Donaldson I. J., Zeef L., Burgis T., Blance R. et al. (2017). p63 exerts spatio-temporal control of palatal epithelial cell fate to prevent cleft palate. PLoS Genet. 13, e1006828 10.1371/journal.pgen.1006828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman K. J., Moore L. L., Singer M. R., Nguyen U.-S. D. T., Mannino S. and Milunsky A. (1995). Teratogenicity of high vitamin A intake. N. Engl. J. Med. 333, 1369-1373. 10.1056/NEJM199511233332101 [DOI] [PubMed] [Google Scholar]
- Sanford L. P., Ormsby I., Gittenberger-de Groot A. C., Sariola H., Friedman R., Boivin G. P., Cardell E. L. and Doetschman T. (1997). TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development 124, 2659-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R., Farrell J. A., Gennert D., Schier A. F. and Regev A. (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495-502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt L., Taiyab A., Melvin V. S., Jones K. L. and Williams T. (2018). Increased FGF8 signaling promotes chondrogenic rather than osteogenic development in the embryonic skull. Dis Model Mech 11, dmm031526 10.1242/dmm.031526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Li Y., Wang K., Wang Y.-Z., Molotkov A., Gao L., Zhao T., Yamagami T., Wang Y., Gan Q. et al. (2009). Lrp6-mediated canonical Wnt signaling is required for lip formation and fusion. Development 136, 3161-3171. 10.1242/dev.037440 [DOI] [PubMed] [Google Scholar]
- Soriano P. (1999). Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70-71. 10.1038/5007 [DOI] [PubMed] [Google Scholar]
- St. Amand T. R., Zhang Y., Semina E. V., Zhao X., Hu Y. P., Nguyen L., Murray J. C. and Chen Y. P. (2000). Antagonistic signals between BMP4 and FGF8 define the expression of Pitx1 and Pitx2 in mouse tooth-forming anlage. Dev. Biol. 217, 323-332. 10.1006/dbio.1999.9547 [DOI] [PubMed] [Google Scholar]
- Suzuki A., Sangani D. R., Ansari A. and Iwata J. (2016). Molecular mechanisms of midfacial developmental defects. Dev. Dyn. 245, 276-293. 10.1002/dvdy.24368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamarin A. (1982). The formation of the primitive choanae and the junction of the primary and secondary palates in the mouse. Am. J. Anat. 165, 319-337. 10.1002/aja.1001650308 [DOI] [PubMed] [Google Scholar]
- Thesleff I. (2015). Molecular genetics of tooth development. In Principles of Developmental Genetics, 2nd edn. Amsterdam: The Netherlands, Elsevier. [Google Scholar]
- Trasler D. G. (1968). Pathogenesis of cleft lip and its relation to embryonic face shape in A/J and C57BL mice. Teratology 1, 33-49. 10.1002/tera.1420010106 [DOI] [PubMed] [Google Scholar]
- Trasler D. G. and Ohannessian L. (1983). Ultrastructure of initial nasal process cell fusion in spontaneous and 6-aminonicotinamide-induced mouse embryo cleft lip. Teratology 28, 91-101. 10.1002/tera.1420280112 [DOI] [PubMed] [Google Scholar]
- van Otterloo E., Li H., Jones K. L. and Williams T. (2018). AP-2alpha and AP-2beta cooperatively orchestrate homeobox gene expression during branchial arch patterning. Development 145, dev157438 10.1242/dev.157438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. Y., Juriloff D. M. and Diewert V. M. (1995). Deficient and delayed primary palatal fusion and mesenchymal bridge formation in cleft lip-liable strains of mice. J. Craniofac. Genet. Dev. Biol. 15, 99-116. [PubMed] [Google Scholar]
- Watanabe H. and Yamada Y. (1999). Mice lacking link protein develop dwarfism and craniofacial abnormalities. Nat. Genet. 21, 225-229. 10.1038/6016 [DOI] [PubMed] [Google Scholar]
- Xavier G. M., Seppala M., Barrell W., Birjandi A. A., Geoghegan F. and Cobourne M. T. (2016). Hedgehog receptor function during craniofacial development. Dev. Biol. 415, 198-215. 10.1016/j.ydbio.2016.02.009 [DOI] [PubMed] [Google Scholar]
- Yu L., Gu S., Alappat S., Song Y., Yan M., Zhang X., Zhang G., Jiang Y., Zhang Z., Zhang Y. et al. (2005). Shox2-deficient mice exhibit a rare type of incomplete clefting of the secondary palate. Development 132, 4397-4406. 10.1242/dev.02013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.