ABSTRACT
RNA epigenetics has received a great deal of attention in recent years, and the reversible N6-methyladenosine (m6A) modification on messenger RNAs (mRNAs) has emerged as a widespread phenomenon. The vital roles of m6A in diverse biological processes are dependent on many RNA-binding proteins (RBPs) with ‘reader’ or ‘nonreader’ functions. Moreover, m6A effector proteins affect cellular processes, such as stem cell differentiation, tumor development and the immune response by controlling signal transduction. This review provides an overview of the interactions of m6A with various RBPs, including the ‘reader’ proteins (excluding the YT521-B homology (YTH) domain proteins and the heterogeneous nuclear ribonucleoproteins (hnRNPs)), and the functional ‘nonreader’ proteins, and this review focuses on their specific RNA-binding domains and their associations with other m6A effectors. Furthermore, we summarize key m6A-marked targets in distinct signaling pathways, leading to a better understanding of the cellular m6A machinery.
KEYWORDS: M6a reader, RNA-binding protein, protein domain, signaling pathway
Introduction
Over a hundred forms of posttranscriptional modifications have been identified in regulatory RNA modifications [1,2]. Among them, N6-methyladenosine (m6A) is the most prevalent and abundant internal RNA modification in both coding and noncoding (nc) RNAs. The influences of m6A on RNA metabolism and processing relate to almost all fundamental aspects, including RNA stability (half-life), maturation, nuclear export, and translational efficiency [3]. Meanwhile, m6A is implicated in many cellular processes, including meiosis, differentiation of various stem cells, DNA damage response (DDR), and germ cell and neuronal development [4–10]. The recent findings about the impact of m6A on the human circadian clock and on hippocampus-dependent learning and memory have expanded the known biological functions of m6A, providing inspirations for follow-up studies concerning the circadian field and brain activity [11–14].
The dynamic interplay among three effectors termed ‘writers’, ‘erasers’ and ‘readers’ determines the effects of m6A on RNA metabolism and cellular processes under normal or stress conditions [15]. RNA m6A is installed by methyltransferases called ‘writers’ that are composed of the core complex Methyltransferase Like 3 (Mettl3), Mettl14 and Wilms Tumor 1 Associated Protein (WTAP), and other subunits of the writer complex, including Virilizer (Kiaa1429), RNA binding Motif Protein 15 (RBM15) and a zinc-finger protein Zc3h13 (zinc finger CCCH domain-containing protein 13) [15–17]. Recent studies have characterized Mettl16 (a Mettl3 homolog) as a writer of precursor (pre) mRNAs and some ncRNAs, including U6 small nuclear (sn) RNAs and long noncoding (lnc) RNAs [18,19]. Intriguingly, the E3 ubiquitin ligase HAKAI, identified as one of the core constituents of the plant writer complex, is likely to have a similar role in mammalian RNA methylation [20]. In addition, Fat mass and obesity-associated gene (FTO) and α-ketoglutarate-dependent dioxygenase alkB homolog 5 (Alkbh5) were successively reported to display m6A demethylation activities as m6A ‘erasers’ [21]. The effects of m6A-modified transcripts are achieved by directly recruiting specific ‘reader’ proteins to m6A sites. As the most common m6A readers, the YTH domain-containing proteins, are evolutionarily conserved and are independent of the cell type [22]. In the nucleus, Ythdc1 regulates splicing by binding to pre-RNAs, while in the cytoplasm, both Ythdf2-directed mRNA decay and Ythdf1/3 and Ythdc2-controlled translation enhancing machineries are well established [23,24]. In addition, accumulating evidence has confirmed that hnRNPs containing the RNA recognition motif (RRM) domains also have reading capabilities. The binding of hnRNPG and hnRNPC to m6A sites is dependent on m6A-mediated structural alteration of mRNAs [25,26]. However, whether hnRNPA2/B1 can act as an m6A reader is controversial. Alarcon et al. demonstrated that hnRNPA2/B1 can mediate RNA splicing and micro (mi) RNA processing by directly binding to m6A [27]. Nevertheless, m6A was reported to induce RNA unfolding and might therefore promote hnRNPA2/B1 access to certain RNA sites instead of direct m6A recognition [28].
The proper modulation of mRNA behavior relies on interactions with substantial RNA-binding proteins (RBPs) and the involvement of various signaling pathways. In this review, we provide a summary of the newly identified RBPs that act as either m6A readers or the functional factors associated with m6A-induced RNA metabolism or biological processes, illustrating their effects and binding features (Table 1). Moreover, considering that variant m6A-marked targets are included in critical signaling pathways, we summarize the research progress with respect to the roles of m6A methylation in the cellular signal transduction network.
Table 1.
RNA-binding proteins act as either novel m6A readers or functional factors in m6A-induced RNA biology.
| RBPs | m6A reader | Binding site | Functions | Targets | Interacting molecules | References |
|---|---|---|---|---|---|---|
| eIF3 | + | 5ʹUTR | induces eIF4E-independent mRNA translation | [33] | ||
| IGF2BPs | + | 3ʹUTR | stabilizes RNA; impairs miRNA-induced RNA decay | MYC, Actin, Lin28, SRF | miRNAs | [39,43–45] |
| Prrc2a | + | CDS | stabilizes RNA | olig2 | [46] | |
| FMR1 | + | 3ʹUTR | inhibits translation; stabilizes RNA | Ythdf2 | [47–49] | |
| eIF3 | - | 5ʹUTR | aids Ythdf1 or Mettl3 to promote translation | Ythdf1, Mettl3 | [37,38] | |
| ABCF1 | - | 5ʹUTR | aids eIF4F-independent mRNA translation | Hsp70 | Mettl3 | [34] |
| HuR/ELAVL1 | - | 3ʹUTR | stabilizes RNA | IGFBPs, FOXM1, SOX2 | miRNAs | [50–53,55–57] |
| RNAPII | - | regulate translation efficiency | PCIF1, MTC | [36,59] | ||
| XRN1 | - | aids Ythdc2 to stabilize RNA | Ythdc2 | [60] | ||
| DDX3 | - | aids Alkbh5 to erase m6A | Alkbh5, AGO2 | [61] | ||
| DDX46 | - | regulates nuclear export | MAVS, TRAF3, TRAF6 | Alkbh5 | [62] | |
| RdRp-3D | - | aids viral replication | Mettl3 | [63,64] | ||
| PARP | - | 5ʹUTR | aids DDR | Mettl3, Pol κ | ||
| MazF | - | cleaves specific RNA sequence without m6A | [65] | |||
| CEBPZ | - | CCAAT-box | activates transcription | SP1 | Mettl3 | [73] |
| SRSF2 | - | ESE | regulates pre-mRNA alternative splicing | RUNXITI | FTO | [68,69] |
| SRSF3 | - | 3ʹUTR | regulates pre-mRNA alternative splicing; export adaptor | Ythdc1 | [7,70] | |
| NXF1 | - | export receptor | Ythdc1 | [70] | ||
| TREX | - | regulates nuclear export | Ythdc1 | [74] | ||
| G3BP1 | - | repelled by m6A and promotes mRNA stability | Ythdf1 | [37,49] |
m6a reader proteins
In eukaryotic cells, mounting evidence has indicated the presence of distinct m6A ‘reader’ proteins aside from the well-characterized YTH proteins and hnRNPs. These readers recognize and bind directly to m6A sites by different protein domains and play various roles in RNA metabolism.
eIF3
Generally, eukaryotic cells initiate translation by relying on a cap-dependent scanning mechanism. For most cellular mRNAs, protein synthesis begins with recognition of the 5ʹ 7-methylguanylate (m7G) cap by eukaryotic initiation factor 4E (eIF-4E). eIF-4E recruits a scaffold protein, eIF-4G, and a helicase, eIF-4A, to form the heterotrimeric eIF-4F complex. eIF-4F then recruits the 43S preinitiation complex, containing eIF3, a 40S ribosomal subunit, and a ternary complex (TC, including eIF2, GTP, and methionine-loaded initiator transfer RNA) [29,30]. Nevertheless, in the absence of eIF-4E, cells rely on alternative mechanisms for a substantial amount of mRNA translation with the help of eIF3. Lee et al. revealed that eIF3d (a subunit of the eIF3 complex) can initiate specialized translation by its previously unknown cap-binding activity [31]. An eIF4GI homolog, termed DAP5, facilitated cap-dependent translation via binding directly to eIF3d [32]. Moreover, recent studies have demonstrated that m6A modifications in the 5ʹ untranslated region (5ʹUTR) can be directly bound by eIF3 to initiate translation independent of the m7G cap and by eIF-4E, which is incompatible with the readily employed cap-independent mechanisms characterized by internal ribosome entry sites (IRESs) (Figure 1(a)) [33].
Figure 1.
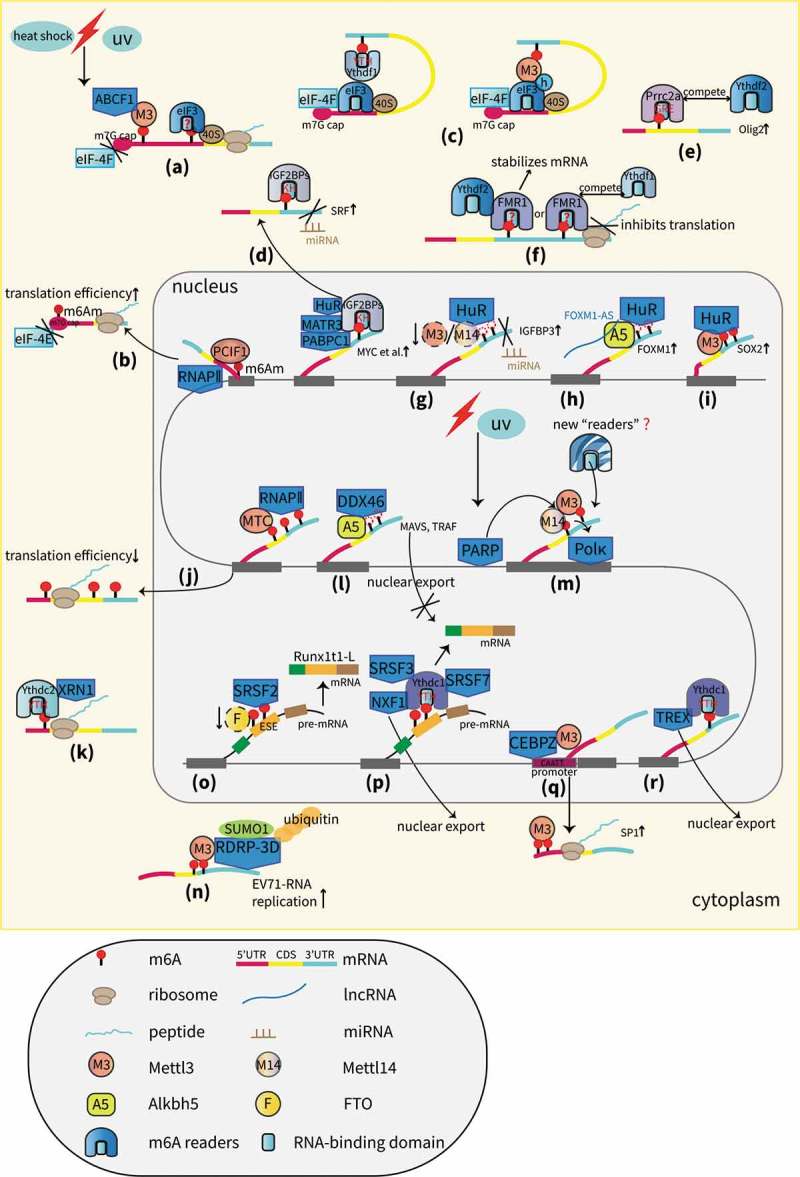
A proposed model of RBPs in the m6A field. (a) Cap-independent translation is mediated by m6A and requires the eIF3 (reader protein), ABCF1, and Mettl3 proteins that bind to internal m6A residues but not to the m7G cap [33,34]. (b) PCIF1 interacts with RNAPII and regulates the N6-methylation of m6Am, which promotes the translation of capped mRNAs [36]. (c) Ythdf1 or Mettl3 promotes translation with the help of eIF3 and mRNA circularization [37,38]. (d) IGF2BPs promote the translation of m6A-mRNAs either by stabilizing targets, such as MYC with the aid of mRNA stabilizers, including HuR, MATR3 and PABPC1 or by antagonizing miRNA-directed mRNA repression [39,45]. (e) Prrc2a stabilizes olig2 mRNA by binding to m6A sites in the CDS and competes with Ythdf2 to regulate RNA stability [46]. (f) FMR1 stabilizes m6A mRNAs by interacting with Ythdf2 or inhibits translation by competing with Ythdf1 [48,49]. (g) HuR stabilizes IGFBP3 mRNA by preventing miRNA targeting [53]. (h) HuR promotes the stability of the demethylated mRNA of FOXM1, which is mediated by Alkbh5, and the cooperation between Alkbh5 and FOXM1-AS.[105] (i) Mettl3 stabilizes the mRNA of SOX2 by recruiting HuR [57]. (j) Slow or paused RNAPII dynamics facilitate MTC binding and m6A deposition, leading to reduced translation efficiency [59]. (k) Ythdc2 stabilizes m6A-mRNAs by interacting with XRN1 [60]. (l) Alkbh5 inhibits the nuclear export of several antiviral transcripts by recruiting DDX46 [62]. (m) Mettl3, recruited by PARP, promotes m6A deposition and subsequent Pol κ binding accompanied by Mettl14, leading to cell survival from DDR [9]. (n) Mettl3 interacts with RdRp-3D and regulates the sumoylation and ubiquitination of RdRp-3D that can promote viral replication [63]. (o) The loss of FTO promotes m6A deposition and the SRSF2 binding ability, leading to the increased inclusion of target exons [69]. (p) Ythdc1 recruits SRSF3 and SRSF7 to promote exon inclusion; Ythdc1 recruits SRSF3 and NXF1 to promote nuclear export [7,70]. (q) CEBPZ recruits Mettl3 to gene promoter regions to augment their translation [73]. (r) TREX interacts with Ythdc1 to promote nuclear export [74].
Evidence has confirmed that m6A modifications located in the 5ʹUTR enable cap-independent translation initiation, especially under stress conditions [34,35]. The results from Coots et al. suggested that Mettl3 directly binds to internal m6A but not to the m7G cap. Moreover, the ATP-binding cassette F1 protein (ABCF1) can not only recruit the ternary complex (TC) that acts as a vital component during translation initiation but also participate in the self-regulation of Mettl3 mRNA translation (Figure 1(a)) [34]. Aside from the internal m6A modifications, N6, 2ʹ-O-dimethyladenosine (m6Am) that is present at the start nucleotide of mRNAs was newly identified to upregulate cap-dependent translation. N6-methylation of m6Am is installed by a new m6A writer called PCIF1, a cap-specific adenosine methyltransferase (CAPAM), and regulates cap-binding proteins independent of eIF4E (Figure 1(b)) [36]. Whether eIF3 is involved in the promotion of m6Am-induced translation needs to be further clarified.
In addition, studies have shown that eIF3 can also act as a nonreader RBP in translation control. Wang et al. revealed that Ythdf1, an m6A reader, promotes translation initiation through a Ythdf1-eIF3 axis, which is based on mRNA looping controlled by eIF-4G and the RNA-independent interaction of Ythdf1 with eIF3 [37]. Moreover, Mettl3 can enhance translation through its interaction with eIF3h (a subunit of eIF3) only when it binds to specific mRNA sites near the stop codon. Mechanistically, this specific interaction mediates polyribosome conformation and mRNA circularization, which is critical for the oncogenic role of Mettl3 to facilitate oncogene translation and tumorigenesis (Figure 1(c)) [38]. These findings expand our understanding of the complex role of eIF3 and the functions of individual subunits.
IGF2BPs
The insulin-like growth factor 2 binding proteins (IGF2BPs, including IGF2BP1/2/3) are generally recognized as IGF2 modifiers in a wide variety of cell types. Recently, Huang et al. showed that IGF2BPs recognize m6A-binding sites through their K homology (KH) domains. Acting as m6A readers, IGF2BPs promote the stability of thousands of potential target mRNAs with the help of cofactors, including human antigen R (HuR), matrin 3 (MATR3) and poly(A) binding protein cytoplasmic 1 (PABPC1), and play oncogenic roles in cancers by improving the expression of oncogenes, such as MYC (Figure 1(d)) [39]. Intriguingly, the results from two distinct studies of acute myeloid leukemia (AML) showed that the high level of m6A in the target mRNAs of genes, including MYB, MYC, Bcl-2 and PTEN, promotes transcript stability and protein expression regulated by Mettl3 and Mettl14, respectively [40,41]. It remains possible that IGF2BPs could be a potential reader since MYC is the typical target of IGF2BPs in an m6A-dependent manner and PTEN mRNA is recognized as a target of IGF2BP1 [39,42]. Further studies are needed to verify this hypothesis.
In addition, IGF2BP1 has been corroborated to enhance the expression of oncogenic factors by antagonizing miRNA-directed repression in cancers [43,44]. In accordance with this conclusion, a novel report identified a conserved mRNA as an IGF2BP1 target in cancers transcribed by Serum response factor (SRF). IGF2BP1 can directly bind to SRF-mRNA in the 3ʹUTR in an m6A-dependent manner and can promote its stability and gene expression by impairing the miRNA-induced decay of SRF transcripts. Therefore, IGF2BP1 indirectly stimulates the expression of a set of SRF-mediated genes, including PDLIM7 and FOXK1, leading to tumor progression [45]. This IGF2BP1-induced regulation of SRF mRNA is irrelevant to HuR (Figure 1(d)).
Other reader proteins
Proline rich coiled-coil 2A (Prrc2a) is recognized as a new m6A reader in neural cells controlling oligodendrocyte specification and myelination. One key target of Prrc2a is oligodendrocyte lineage transcription factor 2 (olig2), which has a well-conserved m6A motif (GGACU) in the coding sequence (CDS) region. Prrc2a specifically binds to and stabilizes the methylated mRNA of olig2 at the CDS region by its GRE domain, the P2 fragment of Prrc2a that contains the enriched glycine, arginine and glutamic acid residues. Furthermore, Prrc2a competed with Ythdf2 to dynamically control RNA stability (Figure 1(e)). The function of Prrc2a can be reversed by FTO [46].
In addition, researchers have identified leucine-rich pentatricopeptide repeat-containing protein (LRPPRC) and fragile X mental retardation protein 1 (FMR1) as two potential readers by developing a chemical proteomics approach [47]. A later study indicated that FMR1 binds to m6A sites in a manner that is dependent on the RNA sequence and the secondary structure. FMR1 competes with Ythdf1 for mRNA binding and represses the translation of target mRNAs by binding to ribosomes, whereas the results from Zhang et al. found an adverse effect of FMRP (encoded by the FMR1 gene) [48,49]. FMRP maintains the stability of m6A-marked mRNAs, most likely through an association with Ythdf2 in an RNA-independent manner, thus contributing to the molecular pathogenesis of Fragile X syndrome (FXS) [48]. Given that there are no YTH domains in FMR1, the precise mechanism by which FMR1 recognizes m6A transcripts remains to be explored (Figure 1(f)).
m6a-associated nonreader RBPs
Reader proteins bind directly to m6A; however, increasing studies have found that some RBPs interact with m6A-containing transcripts without recognizing the m6A base directly. In many cases, these nonreader RBPs could be recruited by different m6A effectors and could play significant roles in RNA biology. These findings provide us with a better understanding of the m6A regulatory network.
HuR
As a well-established RNA stabilizer protein, HuR has been identified in thousands of transcripts. HuR contains three RRM domains and is mainly located in the 3ʹUTR of mRNA, nearing the miRNA binding sites [50]. Considering that the enrichment of RNA methylation is generally at the 3ʹUTR and that HuR can be pulled down by m6A-containing RNAs, researchers investigated whether the presence of m6A affects HuR binding to RNA and the role of HuR in regulating m6A-mediated targets.
Earlier research findings from Dominissini et al. showed that ELAV-like RNA binding protein 1 (ELAVL1, also known as HuR) is significantly associated with m6A-containing transcripts [51]. Subsequent studies have shown that HuR participates in m6A-mediated stem cell development, probably by regulating crucial naïve pluripotency-promoting transcripts, such as Nanog, sex determining region Y-box 2 (SOX2) and IGFBPs [52–54]. Wang et al. indicated that in Mettl3/Mettl14-knockdown (KD) mouse embryonic stem cells (mESCs), the loss of m6A on RNA transcripts, particularly those encoding developmental regulators, enhances the recruitment of HuR. HuR binding reportedly suppresses the inhibitory effect of miRNAs by competing for 3ʹUTR binding sites [55]. As exemplified by IGFBP3, a direct target of some miRNAs, HuR promotes mRNA stability by preventing miRNA targeting (Figure 1(g)) [53]. Furthermore, Zhang et al. demonstrated that in glioma stem-like cells (GSCs), HuR has significant effects on the regulation of FOXM1 (Forkhead box protein M1), a vital transcription factor like SOX2. By binding to the Alkbh5-mediated unmethylated 3ʹUTR of FOXM1 pre-mRNA, HuR promotes its protein expression. Meanwhile, a long noncoding RNA (lncRNA) antisense to FOXM1, termed FOXM1-AS, also plays an indispensable role in Alkbh5-induced GSC self-renewal since it facilitates the interaction between Alkbh5 and FOXM1 nascent transcripts (Figure 1(h)) [56]. Nevertheless, HuR has also been shown to favor binding with the m6A-enriched sites on RNAs. Reports have revealed that Mettl3 can methylate SOX2 mRNA at the 3ʹUTR in GSCs, enhancing its stability by recruiting HuR. The increase of SOX2 is vital for the maintenance of GBM stem-like properties (Figure 1(i)) [57].
Thus, HuR plays a vital role in m6A-containing mRNA stabilization and target translation. Intriguingly, Wang et al. suggested that the spacing between HuR and the m6A sites can be essential for their binding [53]. However, Chen et al. indicated that most of the HuR-binding sites are far from the m6A sites [58]. In addition, the RNA sequence recognized by HuR is different from the m6A-containing sequence [23,58]. Therefore, HuR may interact with m6A indirectly by being recruited by other proteins, such as m6A effectors, or may recognize m6A modifications by the unidentified HuR motif.
Functional enzymes
Various functional RBPs, such as polymerases, helicases and ribonucleases, have been implicated in m6A-directed RNA metabolism; some enzymes are recruited and/or interact with m6A effectors to contribute to diverse cellular processes. Slobodin et al. identified the interaction of RNA polymerase II (RNAPII) with the methyltransferase complex (MTC) during transcription and confirmed that m6A links transcription with translation. The impediment of RNAPII elongation dynamics triggers a suboptimal rate of transcription, leading to the enhancement of the interaction between RNAPII and MTC and a higher m6A deposition on mRNAs. Consequently, the translation efficiency (TE) is reduced (Figure 1(j)) [59]. Moreover, the above-mentioned PCIF1 is recruited to the early elongation complex of RNAPII by interacting with the serine-5-phosphorylated carboxyl-terminal domain (CTD) of RNAPII; this leads to the increased TE of capped mRNAs (Figure 1(b)) [36]. In addition, a report revealed that the m6A reader Ydhdc2 regulates an RNA-independent interaction with the 5ʹ-3ʹ exoribonuclease XRN1 by its ankyrin repeats within the helicase core, suggesting a potential role of Ythdc2 to mediate mRNA stability (Figure 1(k)) [60]. Previous studies have identified DDX3, a family member of DEAD-box RNA helicases, as a novel partner of Alkbh5 and Argonaute2 (AGO2) to modulate the m6A demethylation of mRNAs and miRNAs [61]. Subsequently, Zheng et al. uncovered that DDX46 is recruited by Alkbh5 in the nucleus to erase the m6A tags on several antiviral transcripts, including MAVS, TRAF3, and TRAF6, which substantially inhibits their nuclear export and expression levels (Figure 1(l)). Thus, DDX46 repressed type 1 interferon production after viral infection, acting as a negative regulator of the innate antiviral response [62]. Another interesting finding from Xiang et al. demonstrated that ultraviolet (UV) irradiation can induce the rapid and transient m6A methylation in the 5ʹUTR of many poly(A)+ RNAs, including transcripts localized at DNA damage sites, which are regulated by Mettl3, Mettl14 and FTO. In addition, poly (ADP-ribose) polymerase (PARP), the crucial early regulator of DDR, functions upstream of m6A by recruiting Mettl3 to the damage sites. The accumulation of m6A facilitates UV-responsive DDR and cell survival through the recruitment of the repair and translation synthesis DNA polymerase (Pol κ). However, whether a new ‘reader’ protein exists to induce m6A-induced Pol κ recruitment remains enigmatic. Together, m6A RNA, Mettl3 and Pol κ may constitute a novel pathway against UV-induced DDR separately from canonical repair factors (Figure 1(m)) [9]. Recently, a study showed that the m6A modifications on enterovirus 71 (EV71) RNA are regulated by the cytoplasmic expression of host Mettl3 and FTO and can modulate viral replication. In addition, RNA-dependent RNA polymerase 3D (RdRp-3D), the vital replication promoter of EV71, is stabilized by Mettl3 protein that induces sumoylation and ubiquitination of RdRp-3D . However, this interaction is irrelevant to viral m6A modifications (Figure 1(n)) [63,64].
On the other hand, m6A modifications affect some frequently used enzymes in biotechnology. Imanishi et al. provided a convenient method to assess the activities of m6A effectors (demethylases and methyltransferases). They identified an ACA sequence-specific endoribonuclease called MazF to be the m6A-sensitive RNA cleavage enzyme that distinguishes adenosine (A) from m6A; in addition, MazF also cleaves RNAs with a 5ʹ-ACA-3ʹ sequence rather than a 5ʹ-(m6A) CA-3ʹ sequence [65]. Moreover, Potapov et al. developed a novel method for detecting the accuracy of RNA synthesis and reverse transcription. The modified RNA bases, including 5-hydroxymethyluridine and m6A, increase the combined error rate of T7 RNA polymerase and reverse transcriptases and decrease the fidelity of enzymes [1].
Splicing factors
Alternative splicing (AS) within the nuclear speckle is a vital cellular process during the processing of pre-mRNAs into mature mRNAs [66]. The localization of m6A effectors can regulate not only the exonic or intronic m6A modifications but also the binding of different trans-regulatory splicing factors to cis-regulatory RNA elements [67]. For instance, Zhao et al. found m6A enrichment in alternatively spliced exons that were adjacent to intronic 5ʹ/3ʹ splice sites. The m6A sites are highly overlapped with cis-acting elements termed exonic splicing enhancers (ESEs), which can be recognized by serine/arginine-rich splicing factor 2 (SRSF2), a specific regulator of AS. In preadipocytes, the loss of FTO promotes m6A deposition on the mRNA of the adipogenic regulatory factor termed Runt-related TF 1(RUNX1T1) and increases the binding capacity of SRSF2, leading to the increased inclusion of target exons and poor preadipocyte differentiation (Figure 1(o)) [68,69]. In addition, Ythdc1 recruits the pre-mRNA processing factors SRSF3 and SRSF7 to facilitate exon inclusion and to regulate alternative polyadenylation in oocytes, apparently in an m6A-dependent manner (Figure 1(p)) [7]. Ythdc1 is also responsible for the nuclear export of methylated mRNAs in HeLa cells by facilitating RNA binding to both the export adaptor SRSF3 and the receptor NXF1 (Figure 1(p)) [70]. Other reports have also revealed that the loss of Alkbh5 strongly impacts the nuclear speckle localization of some splicing factors and that WTAP interacts genetically with early-acting splicing regulators in Drosophila, including Snf, U2AF50 and U5-40K; this indicates that these regulators have potential roles in splicing [71,72].
Other nonreader RBPs
Barbieri et al. unveiled that the CCAAT/enhancer-binding protein CEBPZ, known as a strong transcriptional activator, recruits Mettl3 to the promoters of a specific set of active genes, such as SP1, leading to their augmented translation by relieving ribosome stalling at specific codons (Figure 1(q)). The overexpression of SP1 then activates the oncogene c-Myc and facilitates AML cell growth [73]. Another report indicated that the m6A writer complex recruits the TRanscription-Export (TREX) protein complex, which plays a major role in controlling the nuclear export of m6A-containing mRNAs. TREX also stimulates the interaction with Ythdc1 (Figure 1(r)) [74].
Intriguingly, modified mRNAs with m6A residues also regulate mRNA processing by repelling several RBPs. The GTPase-activating protein-binding protein 1 (G3BP1), identified as a mRNA stabilizer, has a preference for unmodified mRNAs. The binding of G3BP1 to mRNAs inhibits methylation by competing with the m6A machinery in the nucleus while aiding the formation of RNPs in the cytoplasm [49]. However, an interaction exists between G3BP1 (known as a stress granule marker) and Ythdf1 in the Ythdf1-mediated translational control system, which is contingent on the presence of bound mRNA [37]. Additionally, Ythdc1 is reported to block the binding of SRSF10 (driving exon exclusion) to mRNAs in oocytes [75].
The effects of m6a on regulating signal transduction
Dynamic mRNA methylation in the form of m6A affects stem cell differentiation, tumor development and the immune response by controlling distinct signaling pathways.
Akt signaling pathway
The Akt (also known as the protein kinase B, PKB) pathway is frequently activated in different cancers via mutations in oncogenes and is highly related to cell proliferation and survival [76,77]. A recent study from Liu et al. revealed that the reduction in m6A methylation caused by either Mettl14 mutation or Mettl3 downregulation facilitates endometrial cancer cell proliferation by controlling the expression of key enzymes in the Akt signaling pathway. Mechanistically, the decreased m6A levels inhibits the Ythdf1-promoted translation of PHLPP2, a negative regulator of Akt, and the Ythdf2-promoted degradation of the mammalian target of rapamycin complex 2 (mTORC2), a positive Akt regulator [78]. Moreover, the overexpression of wild-type Mettl3 in AML cells promotes the translation efficiency of c-MYC, Bcl2 and PTEN in an m6A-dependent manner, leading to proliferation stimulation coupled with apoptosis and the differentiation inhibition of myeloid malignancies. Conversely, m6A depletion activates the PI3K/Akt signaling pathway (Figure 2) [41].
Figure 2.
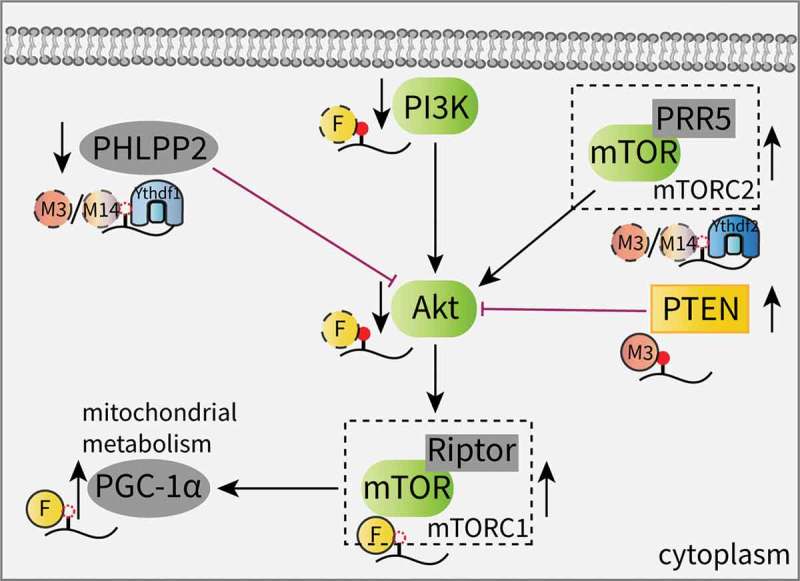
Effects of m6A on the AKT signaling pathway.
In addition, evidence has recognized the functional roles of m6A effectors in stem cells and their link to the Akt pathway. In adult neural stem cells (aNSCs), the loss of FTO decreases neuronal proliferation and differentiation. Increased m6A levels on transcripts involved in the brain derived neurotrophic factor (BDNF) pathway, such as phosphatidylinositol 3-kinase (PI3K), Akt and mTOR, were found to promote mRNA degradation and decreased gene expression. This impairment of the BDNF/Akt signaling cascade results in neurogenesis deficits and impaired learning and memory [56]. Another study showed that FTO contributes to skeletal muscle differentiation by affecting mitochondrial biogenesis. FTO activates mTOR, which is dependent on demethylase activity, and subsequently upregulates the expression of peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PGC-1α), a primary transcriptional coactivator for mitochondrial biogenesis, without altering the mRNA stability; this suggests that the FTO-mediated mTOR-PGC-1α-mitochondria axis is involved in this regulation (Figure 2) [79].
JAK/STAT signaling pathway
Janus kinase (JAK) and signal transducer and activator of transcription (STAT) proteins have effects on tumorigenesis, cancer progression, immune responses, and stem cell differentiation. A majority of ligands, including cytokines and growth factors (GFs), achieve their regulatory role by activating the JAK/STAT pathway [80].
As potent antiviral cytokines, interferons (IFNs) are essential to the innate immune response and can be divided into three major subfamilies: type I (IFN-α and IFN-β), type II (IFN-γ) and type III (IFN-λ) [81,82]. IFNs stimulate the JAK/STAT pathway to activate the expression of hundreds of IFN-stimulated genes (ISGs) against viral infections [83]. Winkler et al. revealed that m6A can serve as a negative regulator of the type I IFN response by controlling the IFNA and IFNB mRNAs. Following infection, m6A deletion in response to Mettl3 or Ythdf2 depletion stabilizes the IFNB (encoding IFN-β) mRNA and sustains IFN-β production, leading to a strong antiviral response [84]. Similar results from Rubio et al. validated that after an infection with a dsDNA virus or human cytomegalovirus (HCMV), Mettl14 and Alkbh5 control IFN-β production and the subsequent JAK/STAT signaling pathway in an m6A-dependent manner [85]. However, the previously mentioned interaction between DDX46 and Alkbh5 initiates the demethylation of antiviral genes, which inhibits IFN-β production (Figure 3). This finding suggests that m6A may act as a positive regulator of the type I IFN response [62].
Figure 3.
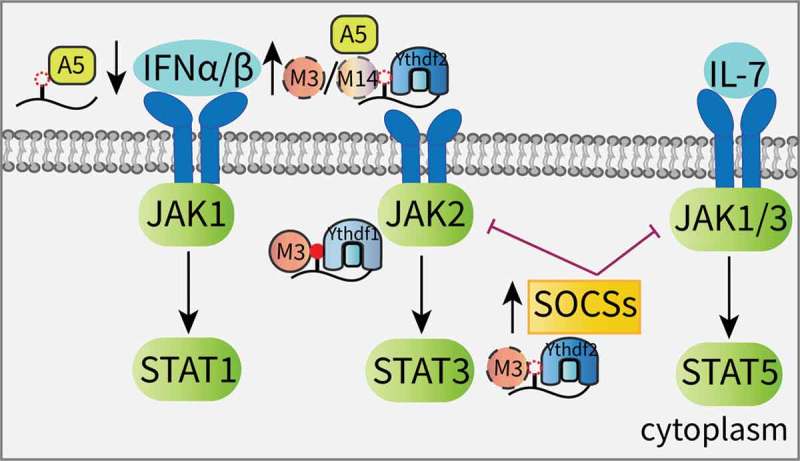
Effects of m6A on the JAK/STAT signaling pathway.
The suppressor of cytokine signaling (SOCS) family of proteins are key negative regulators of the JAK signaling cascade, and their transcripts are always identified as m6A targets that are modulated by the Mettl3-Ythdf2 axis [86–88]. Due to this mechanism, knocking down Mettl3 in liver cancer promotes the expression of SOCS2, and the subsequent inactivation of STAT5 impedes tumorigenesis [87]. Furthermore, Mettl3 deletion attenuates the degradation of the SOCS family genes and blocks Interleukin-7 (IL-7)/STAT5 signaling, depriving T-cells of their proliferation and differentiation abilities [88]. In stem cell research, JAK2 and SOCS3 in porcine-induced PSCs (piPSCs) have been identified as targets of the Mettl3-Ythdf1 axis and the Mettl3-Ythdf2 axis, respectively. Increased m6A methylation triggers the self-renewal of piPSCs [86]. In addition, the enrichment of several negative regulators of the JAK/STAT cascade are found in Ythdf2-deficient neural stem/progenitor cells (NSPCs), suggesting that Ythdf2 contributes to mouse neuroprotection and neurite outgrowth, at least partly, by JAK pathway activation (Figure 3) [89].
p53 signaling pathway
As a canonical tumor suppressor, p53 is capable of preventing eukaryotic cells from DNA damage or deficient oxygenation [90]. The application of m6A sequencing (m6A-seq) established that the silencing of Mettl3 can result in the altered expression and alternative splicing of a group of genes involved in the p53 signaling pathway, leading to p53 pathway modulation and apoptosis [51]. Another study from Kwok et al. also revealed a strong association between the genetic alternations of the m6A effectors, including the m6A writers, erasers, and readers, and p53 mutations in AML patients. The copy number variations (CNVs) of these m6A modifiers and the mutations of p53 can both lead to poor survival [91]. These conclusions inspire further research into the role of m6A regulators in different tumor types and the correlation with the p53 pathway and its downstream targets.
Other signaling pathways
Many signaling molecules associated with stem cell differentiation and tumor development have been recognized as m6A targets and are controlled by m6A modifiers. The transforming growth factor-β (TGF-β) pathway is well known to maintain the pluripotency of human pluripotent stem cells (PSCs) since the key members Activin and Nodal activate the intracellular effector SMAD2/3 (drosophila mothers against decapentaplegic protein 2/3) to bind to and activate the transcriptional regulators that promote pluripotency. Intriguingly, Bertero et al. revealed a novel mechanism by which TGF-β signaling can induce neuroectoderm differentiation by regulating m6A. Phosphorylated SMAD2/3 interacts with the Mettl3-Mettl14-WTAP (M-M-W) complex and promotes m6A deposition on transcripts, including Nanog, which also acts as a SMAD2/3 target. The subsequent decay of these mRNAs allows rapid exit from pluripotency [92]. Moreover, hematopoietic stem and progenitor cells (HSPCs) are essential for the maintenance of hematopoietic function in the human body, and m6A is also involved in the development of both hematopoietic and leukemic stem cells [40,93]. During vertebrate embryogenesis, Notch1a is a key regulator of endothelial-to-hematopoietic transition (EHT), which generates the earliest HSPC. The m6A-forming enzyme Mettl3 represses Notch 1a expression by Ythdf2-mediated mRNA decay, thereby facilitating EHT and HSPC specification by the Notch signaling pathway [94,95]. In addition, Wu et al. demonstrated that reduced levels of m6A modifications in bone marrow mesenchymal stem cells (MSCs) impede parathyroid hormone receptor-1(Pth1r) translation and repress their anabolic responses to parathyroid hormone (PTH) during bone accrual. The inhibited PTH/Pth1r signaling axis attenuates cyclic adenosine monophosphate (cAMP) accumulation and blocks the activation of the protein kinase A (PKA) pathway, leading to increased marrow adiposity [96].
In addition to the stem cell field, Cheng et al. identified the oncogenic role of Mettl3 in bladder cancer (BCa). The overexpression of Mettl3 increases the levels of m6A on the mRNA targets, including AF4/FMR2 family member 4 (AFF4), IKBKB, RELA (known as two key regulators of the NF-κB pathway) and MYC. In addition, AFF4 can bind to the MYC promoter directly, enhancing its expression. This study identifies a novel AFF4/NF-κB/MYC signaling network associated with m6A [97].
Conclusions and perspectives
In eukaryotes, one of the most critical sites for controlling gene expression at the posttranscriptional level is the 3ʹUTR, which is recognized by abundant RBPs and miRNAs [98]. Our review summarizes some vital RBPs that recognize the m6A-containing transcripts at the 3ʹUTR, such as IGF2BPs and HuR, and their functions. Various functional RBPs, such as polymerases, helicases and ribonucleases, have been implicated in m6A-directed RNA metabolism; some enzymes are recruited and/or interact with m6A effectors to contribute to diverse cellular processes. While some regulators have potential roles in splicing. Moreover, multiple RNA-binding domains of RBPs are responsible for interactions between m6A-containing RNA and proteins [99]. In addition to the YTH domain of YTH proteins and the RRM domain of hnRNPs, several new protein domains, including KH from IGF2BPs and GRE from Prrc2a, were identified [39,46]. The KH domain is an evolutionarily conserved single-stranded (ss) RNA-binding domain found in several proteins, including FMR1. Further work is required to investigate the structures of other KH domain proteins and their relationship to m6A [100].
In addition, the regulation of diverse signaling pathways affects different cellular processes. Since m6A has broad roles in RNA biology, most of its targets tend to be essential molecules in cellular signal transduction. Identifying novel m6A-associated pathway molecules contributes to a deeper understanding of the m6A regulatory system. Collectively, this review represents a rich resource that explains the m6A-related RBPs and pathway molecules, shedding light on the complex interplay among m6A, m6A interactors and RNA metabolism; together, this implies the potential role of m6A machinery in cellular signal transduction. Since only a subset of the RNA-protein interactions are truly related to their specific functions [101], seeking novel m6A readers and functional RBPs still has long way to go.
Funding Statement
This work was supported by the National Natural Science Foundation, China [grant number 81672362];National Natural Science Foundation of China [81672362].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Potapov V, Fu X, Dai N, et al. Base modifications affecting RNA polymerase and reverse transcriptase fidelity. Nucleic Acids Res. 2018;46:5753–5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Jaffrey SR, Kharas MG.. Emerging links between m(6)A and misregulated mRNA methylation in cancer. Genome Med. 2017;9:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wang S, Chai P, Jia R, et al. Novel insights on m(6)A RNA methylation in tumorigenesis: a double-edged sword. Mol Cancer. 2018;17:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wang Y, Li Y, Yue M, et al. N(6)-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat Neurosci. 2018;21:195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wang CX, Cui GS, Liu X, et al. METTL3-mediated m6A modification is required for cerebellar development. PLoS Biol. 2018;16:e2004880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tang C, Klukovich R, Peng H, et al. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3’-UTR mRNAs in male germ cells. Proc Natl Acad Sci USA. 2018;115:E325–e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kasowitz SD, Ma J, Anderson SJ, et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 2018;14:e1007412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Huang X, Broxmeyer HE. m(6)A reader suppression bolsters HSC expansion. Cell Res. 2018;28:875–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Xiang Y, Laurent B, Hsu CH, et al. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017;543:573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wojtas MN, Pandey RR, Mendel M, et al. Regulation of m(6)A transcripts by the 3’–>5’ RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol Cell. 2017;68(374–87):e12. [DOI] [PubMed] [Google Scholar]
- [11].Kruttner S, Caroni P. m(6)A-epitranscriptome modulates memory strength. Cell Res. 2019;29:4–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhong X, Yu J, Frazier K, et al. Circadian clock regulation of hepatic lipid metabolism by modulation of m(6)A mRNA methylation. Cell Rep. 2018;25(1816–28):e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shi H, Zhang X, Weng YL, et al. m(6)A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature. 2018;563:249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Fustin JM, Kojima R, Itoh K, et al. Two Ck1δ transcripts regulated by m6A methylation code for two antagonistic kinases in the control of the circadian clock. Proc Natl Acad Sci USA. 2018;115:5980–5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yang Y, Hsu PJ, Chen YS, et al. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28:616–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wen J, Lv R, Ma H, et al. Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol Cell. 2018;69(1028–1038):e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Knuckles P, Lence T, Haussmann IU, et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the <ins style="background: springGreen">m 6 A machinery component Wtap/Fl(2)d. Genes Dev. 2018;32:415–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Warda AS, Kretschmer J, Hackert P, et al. Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18:2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Pendleton KE, Chen B, Liu K, et al. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169(824–835):e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ruzicka K, Zhang M, Campilho A, et al. Identification of factors required for <ins style="background: springGreen">m 6 A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 2017;215:157–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wu Y, Zhou C, Yuan Q. Role of DNA and RNA N6-adenine methylation in regulating stem cell fate. Curr Stem Cell Res Ther. 2018;13:31–38. [DOI] [PubMed] [Google Scholar]
- [22].Wang S, Sun C, Li J, et al. Roles of RNA methylation by means of N(6)-methyladenosine (m(6)A) in human cancers. Cancer Lett. 2017;408:112–120. [DOI] [PubMed] [Google Scholar]
- [23].Liao S, Sun H, Xu C. YTH Domain: A Family of N(6)-methyladenosine (m(6)A) Readers. Genomics Proteomics Bioinf. 2018;16:99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Patil DP, Pickering BF, Jaffrey SR. Reading m(6)A in the Transcriptome: m(6)A-binding proteins. Trends Cell Biol. 2018;28:113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Liu N, Zhou KI, Parisien M, et al. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017;45:6051–6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Liu N, Dai Q, Zheng G, et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Alarcon CR, Goodarzi H, Lee H, et al. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell.. 2015;162:1299–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wu B, Su S, Patil DP, et al. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat Commun. 2018;9:420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Marintchev A, Edmonds KA, Marintcheva B, et al. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136:447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lee AS, Kranzusch PJ, Doudna JA, et al. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature. 2016;536:96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].de la Parra C, Ernlund A, Alard A, et al. A widespread alternate form of cap-dependent mRNA translation initiation. Nat Commun. 2018;9:3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Meyer KD, Patil DP, Zhou J, et al. 5’ UTR m(6)A promotes cap-independent translation. Cell. 2015;163:999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Coots RA, Liu XM, Mao Y, et al. m(6)A facilitates eIF4F-Independent mRNA translation. Mol Cell. 2017;68(504–514):e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhou J, Wan J, Gao X, et al. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Akichika S, Hirano S, Shichino Y, et al. Cap-specific terminal N <ins style="background: springGreen">6-methylation of RNA by an RNA polymerase II–associated methyltransferase. Science. 2019;363. [DOI] [PubMed] [Google Scholar]
- [37].Wang X, Zhao BS, Roundtree IA, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Choe J, Lin S, Zhang W, et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561:556–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Huang H, Weng H, Sun W, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Weng H, Huang H, Wu H, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell. 2018;22:191–205,e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Vu LP, Pickering BF, Cheng Y, et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23:1369–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Huang X, Zhang H, Guo X, et al. Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) in cancer. J Hematol Oncol. 2018;11:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Busch B, Bley N, Muller S, et al. The oncogenic triangle of HMGA2, LIN28B and IGF2BP1 antagonizes tumor-suppressive actions of the let-7 family. Nucleic Acids Res. 2016;44:3845–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Muller S, Bley N, Glass M, et al. IGF2BP1 enhances an aggressive tumor cell phenotype by impairing miRNA-directed downregulation of oncogenic factors. Nucleic Acids Res. 2018;46:6285–6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Müller S, Glaß M, Singh AK, et al. IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic Acids Res. 2019;47:375–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wu R, Li A, Sun B, et al. A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 2019;29:23–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Arguello AE, DeLiberto AN, Kleiner RE. RNA chemical proteomics reveals the N(6)-methyladenosine (m(6)A)-regulated protein-RNA interactome. J Am Chem Soc. 2017;139:17249–17252. [DOI] [PubMed] [Google Scholar]
- [48].Zhang F, Kang Y, Wang M, et al. Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum Mol Genet. 2018;27:3936–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Edupuganti RR, Geiger S, Lindeboom RGH, et al. N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat Struct Mol Biol. 2017;24:870–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lebedeva S, Jens M, Theil K, et al. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell. 2011;43:340–352. [DOI] [PubMed] [Google Scholar]
- [51].Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. [DOI] [PubMed] [Google Scholar]
- [52].Geula S, Moshitch-Moshkovitz S, Dominissini D, et al. ins style="background: springGreen">m 6 A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347::1002–1006. [DOI] [PubMed] [Google Scholar]
- [53].Wang Y, Li Y, Toth JI, et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Batista PJ, Molinie B, Wang J, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Meisner NC, Filipowicz W. Properties of the regulatory RNA-binding protein HuR and its role in controlling miRNA repression. Adv Exp Med Biol. 2011;700:106–123. [DOI] [PubMed] [Google Scholar]
- [56].Li L, Zang L, Zhang F, et al. Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum Mol Genet. 2017;26:2398–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Visvanathan A, Patil V, Arora A, et al. Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018;37:522–533. [DOI] [PubMed] [Google Scholar]
- [58].Chen K, Lu Z, Wang X, et al. High-resolution <ins style="background: springGreen">N 6 -Methyladenosine (<ins style="background: springGreen">m 6 A) map using photo-crosslinking-assisted <ins style="background: springGreen">m 6 A Sequencing. Angew Chem Int Ed Engl. 2015;54:1587–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Slobodin B, Han R, Calderone V, et al. Transcription impacts the efficiency of mRNA translation via co-transcriptional N6-adenosine methylation. Cell. 2017;169:326–337,e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kretschmer J, Rao H, Hackert P, et al. The <ins style="background: springGreen">m 6 A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5′–3′ exoribonuclease XRN1. Rna. 2018;24:1339–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Shah A, Rashid F, Awan HM, et al. The DEAD-Box RNA Helicase DDX3 Interacts with <ins style="background: springGreen">m 6 A RNA Demethylase ALKBH5. Stem Cells Int. 2017;2017:8596135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Zheng Q, Hou J, Zhou Y, et al. The RNA helicase DDX46 inhibits innate immunity by entrapping m(6)A-demethylated antiviral transcripts in the nucleus. Nat Immunol. 2017;18:1094–1103. [DOI] [PubMed] [Google Scholar]
- [63].Hao H, Hao S, Chen H, et al. N6-methyladenosine modification and METTL3 modulate enterovirus 71 replication. Nucleic Acids Res. 2019;47:362–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Liu Y, Zheng Z, Shu B, et al. SUMO modification stabilizes enterovirus 71 polymerase 3D to facilitate viral replication. J Virol. 2016;90:10472–10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Imanishi M, Tsuji S, Suda A, et al. Detection of N(6)-methyladenosine based on the methyl-sensitivity of MazF RNA endonuclease. Chem Commun. 2017;53:12930–12933. [DOI] [PubMed] [Google Scholar]
- [66].Baralle FE, Giudice J. Alternative splicing as a regulator of development and tissue identity. Nat Rev Mol Cell Biol. 2017;18:437–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Braunschweig U, Gueroussov S, Plocik AM, et al. Dynamic integration of splicing within gene regulatory pathways. Cell. 2013;152:1252–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Ben-Haim MS, Moshitch-Moshkovitz S, Rechavi G. FTO: linking m6A demethylation to adipogenesis. Cell Res. 2015;25:3–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Zhao X, Yang Y, Sun BF, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Roundtree IA, Luo GZ, Zhang Z, et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife. 2017;6:pii:e31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Penn JK, Graham P, Deshpande G, et al. Functioning of the Drosophila Wilms’-tumor-1-associated protein homolog, Fl(2)d, in Sex-lethal-dependent alternative splicing. Genetics. 2008;178:737–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Barbieri I, Tzelepis K, Pandolfini L, et al. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature. 2017;552:126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Lesbirel S, Viphakone N, Parker M, et al. The m(6)A-methylase complex recruits TREX and regulates mRNA export. Sci Rep. 2018;8:13827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Xiao W, Adhikari S, Dahal U, et al. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol Cell. 2016;61:507–519. [DOI] [PubMed] [Google Scholar]
- [76].Manning BD, Toker A. AKT/PKB Signaling: Navigating The Network. Cell. 2017;169:381–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. [DOI] [PubMed] [Google Scholar]
- [78].Liu J, Eckert MA, Harada BT, et al. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol. 2018;20:1074–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Wang X, Huang N, Yang M, et al. FTO is required for myogenesis by positively regulating mTOR-PGC-1alpha pathway-mediated mitochondria biogenesis. Cell Death Dis. 2017;8:e2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Quintas-Cardama A, Verstovsek S. Molecular pathways: jak/STAT pathway: mutations, inhibitors, and resistance. Clin Cancer Res. 2013;19:1933–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Zanoni I, Granucci F, Broggi A. Interferon (IFN)-lambda takes the helm: immunomodulatory roles of type III IFNs. Front Immunol. 2017;8:1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Raftery N, Stevenson NJ. Advances in anti-viral immune defence: revealing the importance of the IFN JAK/STAT pathway. Cell Mol Life Sci. 2017;74:2525–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Nan Y, Wu C, Zhang YJ. Interplay between Janus Kinase/signal transducer and activator of transcription signaling activated by type I interferons and viral antagonism. Front Immunol. 2017;8:1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Winkler R, Gillis E, Lasman L, et al. m(6)A modification controls the innate immune response to infection by targeting type I interferons. Nat Immunol. 2018;20:173–182. [DOI] [PubMed] [Google Scholar]
- [85].Rubio RM, Depledge DP, Bianco C, et al. RNA m(6) A modification enzymes shape innate responses to DNA by regulating interferon beta. Genes Dev. 2018;32:1472–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Wu R, Liu Y, Zhao Y, et al. m(6)A methylation controls pluripotency of porcine induced pluripotent stem cells by targeting SOCS3/JAK2/STAT3 pathway in a YTHDF1/YTHDF2-orchestrated manner. Cell Death Dis. 2019;10:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Chen M, Wei L, Law CT, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–2270. [DOI] [PubMed] [Google Scholar]
- [88].Li HB, Tong J, Zhu S, et al. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548:338–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Li M, Zhao X, Wang W, et al. Ythdf2-mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol. 2018;19:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Chen J, Liu S, Hu X. Long non-coding RNAs: crucial regulators of gastrointestinal cancer cell proliferation. Cell Death Discov. 2018;4:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Kwok CT, Marshall AD, Rasko JE, et al. Genetic alterations of m(6)A regulators predict poorer survival in acute myeloid leukemia. J Hematol Oncol. 2017;10:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Bertero A, Brown S, Madrigal P, et al. The SMAD2/3 interactome reveals that TGFbeta controls m(6)A mRNA methylation in pluripotency. Nature. 2018;555:256–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Li Z, Qian P, Shao W, et al. Suppression of m(6)A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res. 2018;28:904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Lv J, Zhang Y, Gao S, et al. Endothelial-specific m(6)A modulates mouse hematopoietic stem and progenitor cell development via Notch signaling. Cell Res. 2018;28:249–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Zhang C, Chen Y, Sun B, et al. m(6)A modulates haematopoietic stem and progenitor cell specification. Nature. 2017;549:273–276. [DOI] [PubMed] [Google Scholar]
- [96].Wu Y, Xie L, Wang M, et al. Mettl3-mediated m(6)A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat Commun. 2018;9:4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Cheng M, Sheng L, Gao Q, et al. The m(6)A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-kappaB/MYC signaling network. Oncogene. 2019. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- [98].Wahle E, Ruegsegger U. 3’-End processing of pre-mRNA in eukaryotes. FEMS Microbiol Rev. 1999;23:277–295. [DOI] [PubMed] [Google Scholar]
- [99].Wang X, Huang J, Zou T, et al. Human m(6)A writers: two subunits, 2 roles. RNA Biol. 2017;14:300–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Valverde R, Edwards L, Regan L. Structure and function of KH domains. Febs J. 2008;275:2712–2726. [DOI] [PubMed] [Google Scholar]
- [101].Konig J, Zarnack K, Rot G, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]


