Abstract
ProSNEx (Protein Structure Network Explorer) is a web service for construction and analysis of Protein Structure Networks (PSNs) alongside amino acid flexibility, sequence conservation and annotation features. ProSNEx constructs a PSN by adding nodes to represent residues and edges between these nodes using user-specified interaction distance cutoffs for either carbon-alpha, carbon-beta or atom-pair contact networks. Different types of weighted networks can also be constructed by using either (i) the residue-residue interaction energies in the format returned by gRINN, resulting in a Protein Energy Network (PEN); (ii) the dynamical cross correlations from a coarse-grained Normal Mode Analysis (NMA) of the protein structure; (iii) interaction strength. Upon construction of the network, common network metrics (such as node centralities) as well as shortest paths between nodes and k-cliques are calculated. Moreover, additional features of each residue in the form of conservation scores and mutation/natural variant information are included in the analysis. By this way, tool offers an enhanced and direct comparison of network-based residue metrics with other types of biological information. ProSNEx is free and open to all users without login requirement at http://prosnex-tool.com.
INTRODUCTION
Proteins mediate a great number of functions in a living cell. The folded state (i.e. the shape) plays a major role in determining thermostability (1–3), dynamics (4,5) and thereby the function of the protein. The shape, in turn, is highly dependent on the sequence of amino acid residues, their physicochemical properties and the various types of chemical interactions (bonded or non-bonded) they are involved in within the folded state (5–12). Sequence conservation and protein stability as well as dynamics have been found to be closely connected in several protein families (6,7,13–17).
In order to better understand the role played by each residue in shaping protein function, it is proper to consider the protein structure as a network of amino acids that interact with each other in the three-dimensional space via various chemical interactions. In the past few years, network formalism has been a popular approach for studying individual proteins as well as protein complexes with the aim of understanding the underlying structural organization within the protein structure and elucidating the importance and functional roles of individual residues (18–23). In this approach, a Protein Structure Network (PSN) is constructed by taking residues within the structure as nodes and adding edges between them using distance cutoffs or other more advanced criteria. Weights can also be given to edges or residues to construct weighted networks in order to emphasize the interaction strength, e.g. by using force-field based interaction energies, atom-atom contacts or pairwise residue dynamic coupling. Dynamic coupling between residues can be obtained by constructing Dynamical Cross-Correlation Maps (DCCMs) from Molecular Dynamics (MD) simulation trajectories (24–29). If interaction energies are used for edge weight assignment, the network becomes a ‘Protein Energy Network’ (PEN) (30–33). Once the network is constructed, an analysis of residue-based local or global network metrics including centrality measures can be useful to detect non-evident functions of residues such as relative importances for protein stability (19,30,34–36), allosteric communication between parts of protein (37–42) and other family-specific functions (e.g. catalytics sites in enzymes) (19,43–45).
The network approach is very useful for a fast initial characterization of protein dynamics as well. To this end, Elastic Network Models (ENMs) have found widespread usage among researchers (46–52). In an ENM, usually a selected set of atoms from each residue (e.g. carbon-alpha atoms) are connected to each other with hypothetical springs, yielding a harmonic interaction network. A Normal Mode Analysis (NMA) is then performed to generate harmonic vibrational modes of motion and flexibility profiles. Despite the assumption of harmonic motion around the native protein structure, flexibility predictions by the ENM approach has been found to correlate well with the global cooperative modes of motions generated from experimentally elucidated conformation ensembles or atomistic MD simulations (52). Following the first application by Tirion on all-atom systems (53), it has been also found that reducing the resolution by coarse-graining the protein structure and using only alpha carbons to represent the topology of the protein structure is sufficiently enough for accurate predictions. There are two basic types of ENMs: the Gaussian Network Model (GNM) (47) and the Anisotropic Network Model (ANM) (48,50,54). In GNM, fluctuations are assumed to be isotropic (i.e. distributed equally along different directions in the coordinate system) whereas in ANM, directionality of motions, and thus the three-dimensionality, is included.
Taking only a single protein structure and cutoff distances as input, simple PSN and ENM approaches are fast and powerful for getting valuable initial insights regarding the structural and dynamical behavior of a protein. To this end, several freely accessible web-services have been offered to researchers for the construction, visualization and analysis of PSNs and ENMs. For PSNs, NAPS (55) server offers an analysis based on a variety of weighted and unweighted network types. RING 2.0 (56) offers a similar service in addition to identifying different types of chemical interactions between residues and the results can be visualized by using Cytoscape (57). Similarly, RINalyzer (58) and structureViz integrate Cytoscape (57) and UCSF Chimera (59) to create and visualize RINS. CSU (60), PSN-Ensemble (61), NetworkAnalyzer (62) and PyMOL (63) plug-ins xPyder (64) and PyInteraph (65) can also be used for similar purposes. The Protein Contact Atlas (66) offers a rich visualization interface for examining different types of contacts within protein structures, including residue-centric network metrics alongside mapping of custom metrics such as sequence conservation or thermodynamics stability changes that can be supplied by the user. VERMONT 2.0 (67) is another web-server for performing network analysis on protein structures while integrating additional features such as sequence conservation, residue physicochemical parameters and solvent accessibility. For ENM-based protein dynamics analysis, elNemo (68), the ANM 2.1 (69,70), iGNM 2.0 (71) and DynOmics ENM (72) servers are available. Bio3D-web also offers user-friendly interface for performing NMA (Bio3D-NMAweb) on single protein structures (73). DynaMut server offers NMA-based prediction of stability changes upon amino-acid mutations (74). WebPSN web-server combines ENM and NMA approaches for finding allosteric pathways in protein structures (75).
In a typical research workflow involving an investigation of relationships between sequence, structure, dynamics and function of a given protein of interest; all relevant features, such as conservation scores for the amino-acid sequence, residue-centric network metrics, flexibility profiles and other functional annotations must be obtained to facilitate the analysis. Researches with sufficient technical expertise may choose to utilize customized workflows by utilizing multiple software packages for this purpose. For non-experts, however, the options are limited. Although some of the aforementioned web-servers offer an integration of more than one type of feature to some extent, there is currently no web-service available offering extensive comparative analysis features involving PSN analysis, sequence conservation and protein flexibility profiles as well as other functional annotations.
Aiming to fill this gap, we have developed the ProSNEx (Protein Structure Network Explorer) web-server. ProSNEx offers enhanced PSN-based protein structure analysis by integrating sequence conservation profiles, protein annotations and flexibility profiles from different types of ENM into a single user interface.
MATERIALS AND METHODS
Protein structure networks and elastic network models
ProSNEx offers the construction of several types of unweighted and weighted networks. Unweighted networks can be constructed by selecting one of the four methods for edge assignment between nodes (carbon-alpha, carbon-beta, atom-pair interaction and interaction strength). In the carbon-alpha and carbon-beta networks, only carbon-alpha or carbon-beta atom positions from the input PDB structure are used to specify the edges in the network, respectively. Edges between nodes are added only if two atoms are closer than the user-specified threshold (cutoff) distance. In the atom-pair interaction network, edges between nodes are added if at least one atom from a residue is closer than the user-specified distance threshold to at least one atom from another residue. In the interaction strength network, edges between nodes are added only if the interaction strength is higher than the user-specified threshold. The interaction strength calculations are based on (34).
Once the network edge assignment type is selected, ProSNEx can also assign weights to edges, yielding a weighted network. Here, four options are offered to the user. Using either one of the first two options; the user can construct a weighted PSN by extracting weights from DCCMs calculation from of GNM/ANM simulations or the NMA method of Wako et al. (76–78). In the latter case, normalized and time-averaged cross-correlations, as reported at the Promode-elastic database of PDB Japan, are used (77,79). Hence, this option is not available for custom PDB files. The GNM and ANM calculations are performed by ProSNEx server.
In the third option, the user is given the option to choose average force-field based interaction energies from a MD simulation trajectory as edge weights to construct a PEN. Here, average interaction energies in the file format returned by gRINN (31) is accepted. In the last option, the interaction strength values are taken from (80).
Following network construction, global and local network metrics are calculated. Specifically, average degree, path length, network density, clustering coefficient as well as node (residue)-centric metrics including node degrees, betweenness-centrality and closeness-centrality are calculated and reported. Additionally, shortest path between two selected nodes and k-cliques are calculated as well.
Sequence conservation profiles, protein annotations and interatomic interactions
If available, ProSNEx retrieves sequence conservation from the CONSURF database (CONSURF-DB), which includes conservation scores calculated by the CONSURF method (81–83). If the user supplies a custom PDB file instead of a PDB access code or if no CONSURF score is available at CONSURF-DB for the given PDB access code, an option to import conservation scores in the format returned by the CONSURF server is available following network construction. ProSNEx also retrieves annotations (if available) from the Uniprot database, including sequence variants, mutagenesis experiment results, etc. (84,85). Finally, interatomic interactions within the input structure are retrieved by using Arpeggio and annotated on the network structure (86).
RESULTS
Use case: investigating the sequence–structure–dynamics relationships in TEM-1 β-lactamase
We demonstrate the use of ProSNEx server for investigating the sequence–structure–dynamics relationships in a TEM-1 β-lactamase enzyme from Escherichia coli.
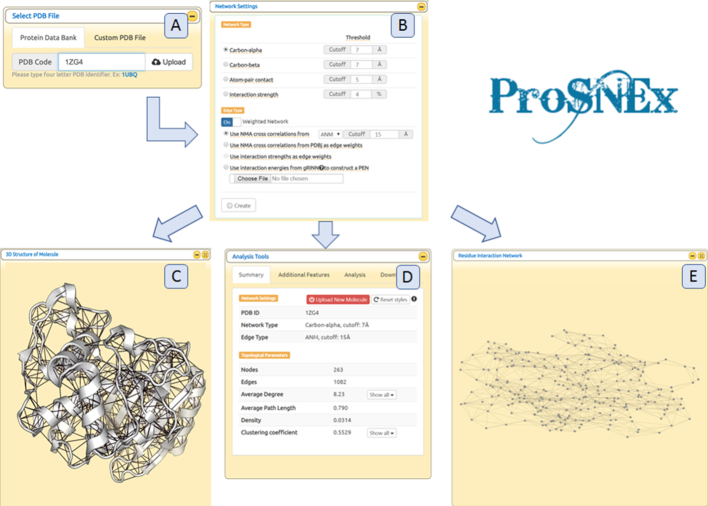
ProsNEx main page includes a simple interface for entering a PDB code, selection of protein chains (if applicable) and network settings, respectively. (Figure 1A-B). Starting from a TEM-1β-lactamase structure (PDB code: 1ZG4), a weighted PSN (carbon-alpha network, threshold distance: 7 Å) was constructed using cross-correlations from an ANM simulation (threshold distance: 15 Å) as edge weights.
Figure 1.
(A) Select PDB window. (B) Network settings specification window (C–E) 3D structure of the molecule, Analysis Tools and Residue Interaction Network windows.
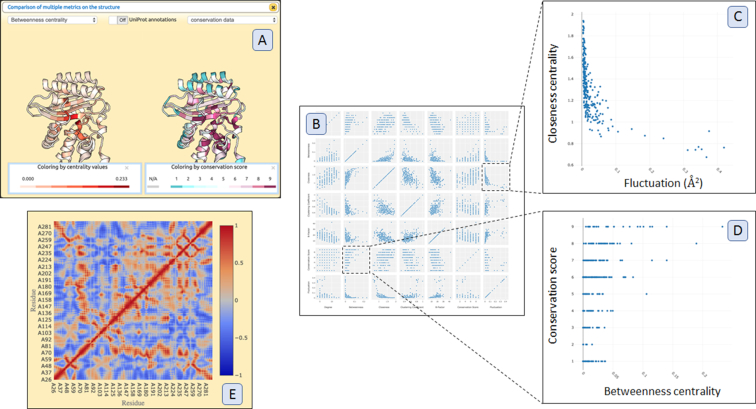
Figure 1C–E gives an overview of outputs from ProSNEx. Upon finishing calculations, the tool presents three major windows for displaying the 3D structure of the input molecule (Figure 1C), a 2D network representation (Figure 1E) and a window titled ‘Analysis Tools’ (Figure 1D). The Analysis Tools window is the access point for investigating further features included in ProSNEx analysis. In Figure 2, a collection of plots, all accessible from the Analysis Tools window, is given. Figure 2C and Figure 2C shows two scatter plots of particular interest: in the first one, the closeness centrality is seen to be correlated to fluctuation profiles from ANM, confirming its usefulness as a rigidity descriptor (87). In Figure 2D, high betweenness-centrality values are seen to coincide well with highly conserved residues, highlighting a relationship between sequence evolution and dynamic cross-talk within the ß-lactamase structure.
Figure 2.
(A) Comparison of multiple node (residue) metrics on protein structure. (B) An ‘all-in-one’ Scatter Plot shows scatter plots between degree, betweenness-centrality, closeness-centrality, fluctuations (if NMA is used for edge-weight assignment), sequence conservation scores, B-factors and clustering coefficients. (C, D) Selected scatter plots between closeness-centrality and fluctuation and betweenness-centrality and sequence conservation scores. (E) Cross correlation plot when NMA is used for edge-weight assignment.
IMPLEMENTATION
The tool is implemented in JavaScript and jQuery. The web-interface is built using Bootstrap CSS style. PSN is constructed and analysed using by JSNetworkX (88) library. Cytoscape.js (57) framework is used for network visualization. PV is used for protein structure visualization (89). For GNM and ANM calculations, ProDy is used (90).
CONCLUSION
We have developed ProSNEx server which provides a novel and enhanced analysis of protein structures using network formalism. The tool is designed to be very user friendly and easily adaptable for all researchers in the field of protein structural biology. The major novelty of the tool lies in the presence of features such as: (i) comparison of multiple single residue metrics from network analysis as well as other additional information such as sequence conservation scores and Uniprot annotations and (ii) usage of dynamic cross-correlations between pairs of amino acids from NMA in the weighted PSNs.
DATA AVAILABILITY
ProSNEx is free and open to all users without login requirement. The server does not store any data submitted by the user. It is compatible with major web browser including Chrome, Firefox and Safari.
ACKNOWLEDGEMENTS
O.S. conceived the idea of server; R.M.A. designed the user interface, constructed network analysis computations and retrieval of external data; O.S. constructed the backend GNM and ANM computations; P.O. wrote the help documents; O.S. and P.O. provided critical input on analysis methods. O.S. and P.O. wrote the manuscript. P.O. supervised the project and oversaw the manuscript preparation. We acknowledge the helpful discussion with Fatma Gizem Avcı on the structural details of TEM-1 β-lactamase. TUBITAK 218M861 is acknowledged.
FUNDING
Funding for open access charge: Self funded.
Conflict of interest statement. None declared.
REFERENCES
- 1. Robertson A.D., Murphy K.P.. Protein structure and the energetics of protein stability. Chem. Rev. 1997; 97:1251–1268. [DOI] [PubMed] [Google Scholar]
- 2. Baldwin R.L. Energetics of protein folding. J. Mol. Biol. 2007; 371:283–301. [DOI] [PubMed] [Google Scholar]
- 3. Argos P., Rossmann M.G., Grau U.M., Zuber H., Frank G., Tratschin J.D.. Thermal stability and protein structure. Biochemistry. 1979; 18:5698–5703. [DOI] [PubMed] [Google Scholar]
- 4. Hinsen K. Analysis of domain motions by approximate normal mode calculations. Proteins Struct. Funct. Genet. 1998; 33:417–429. [DOI] [PubMed] [Google Scholar]
- 5. Tiwari S.P., Reuter N.. Conservation of intrinsic dynamics in proteins — what have computational models taught us. Curr. Opin. Struct. Biol. 2018; 50:75–81. [DOI] [PubMed] [Google Scholar]
- 6. Maguid S., Fernandez-Alberti S., Echave J.. Evolutionary conservation of protein vibrational dynamics. Gene. 2008; 422:7–13. [DOI] [PubMed] [Google Scholar]
- 7. Liu Y., Bahar I.. Sequence evolution correlates with structural dynamics. Mol. Biol. Evol. 2012; 29:2253–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shakhnovich E. Protein folding thermodynamics and dynamics: where physics, chemistry, and biology meet. Chem. Rev. 2006; 106:1559–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gilson A.I., Marshall-Christensen A., Choi J.-M., Shakhnovich E.I.. The role of evolutionary selection in the dynamics of protein structure evolution. Biophys. J. 2017; 112:1350–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bastolla U., Demetrius L.. Stability constraints and protein evolution: the role of chain length, composition and disulfide bonds. Protein Eng. Des. Sel. 2005; 18:405–415. [DOI] [PubMed] [Google Scholar]
- 11. DePristo M.A., Weinreich D.M., Hartl D.L.. Missense meanderings in sequence space: a biophysical view of protein evolution. Nat. Rev. Genet. 2005; 6:678–687. [DOI] [PubMed] [Google Scholar]
- 12. Sikosek T., Chan H.S.. Biophysics of protein evolution and evolutionary protein biophysics. J. R. Soc. Interface. 2014; 11:20140419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wilke C.O. Bringing molecules back into molecular evolution. PLoS Comput. Biol. 2012; 8:e1002572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Echave J. Why are the low-energy protein normal modes evolutionarily conserved. Pure Appl. Chem. 2012; 84:1931–1937. [Google Scholar]
- 15. Tripathi S., Wang Q., Zhang P., Hoffman L., Waxham M.N., Cheung M.S.. Conformational frustration in calmodulin-target recognition. J. Mol. Recognit. 2015; 28:74–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Parra R.G., Espada R., Verstraete N., Ferreiro D.U.. Structural and energetic characterization of the ankyrin repeat protein family. PLoS Comput. Biol. 2015; 11:e1004659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dib L., Salamin N., Gfeller D.. Polymorphic sites preferentially avoid co-evolving residues in MHC class I proteins. PLoS Comput. Biol. 2018; 14:e1006188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brysbaert G., Mauri T., de Ruyck J., Lensink M.F.. Identification of key residues in proteins through centrality analysis and flexibility prediction with RINspector. Curr. Protoc. Bioinforma. 2019; 65:e66. [DOI] [PubMed] [Google Scholar]
- 19. Amitai G., Shemesh A., Sitbon E., Shklar M., Netanely D., Venger I., Pietrokovski S.. Network analysis of protein structures identifies functional residues. J. Mol. Biol. 2004; 344:1135–1146. [DOI] [PubMed] [Google Scholar]
- 20. Bhattacharyya M., Ghosh S., Vishveshwara S.. Protein structure and function: looking through the network of side-chain interactions. Curr. Protein Pept. Sci. 2016; 17:4–25. [DOI] [PubMed] [Google Scholar]
- 21. Grewal R.K., Roy S.. Modeling proteins as residue interaction networks. Protein Pept. Lett. 2015; 22:923–933. [DOI] [PubMed] [Google Scholar]
- 22. Salamanca Viloria J., Allega M.F., Lambrughi M., Papaleo E.. An optimal distance cutoff for contact-based Protein Structure Networks using side-chain centers of mass. Sci. Rep. 2017; 7:2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Papaleo E. Integrating atomistic molecular dynamics simulations, experiments, and network analysis to study protein dynamics: strength in unity. Front. Mol. Biosci. 2015; 2:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sethi A., Eargle J., Black A.A., Luthey-Schulten Z.. Dynamical networks in tRNA:protein complexes. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:6620–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sethi A., Tian J., Derdeyn C.A., Korber B., Gnanakaran S.. A mechanistic understanding of allosteric immune escape pathways in the HIV-1 envelope glycoprotein. PLoS Comput. Biol. 2013; 9:e1003046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Feher V.A., Durrant J.D., Van Wart A.T., Amaro R.E.. Computational approaches to mapping allosteric pathways. Curr. Opin. Struct. Biol. 2014; 25:98–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Van Wart A.T., Durrant J., Votapka L., Amaro R.E.. Weighted implementation of suboptimal paths (WISP): An optimized algorithm and tool for dynamical network analysis. J. Chem. Theory Comput. 2014; 10:511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vanwart A.T., Eargle J., Luthey-Schulten Z., Amaro R.E.. Exploring residue component contributions to dynamical network models of allostery. J. Chem. Theory Comput. 2012; 8:2949–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bradley M.J., Chivers P.T., Baker N.A.. Molecular dynamics simulation of the Escherichia coli NikR protein: equilibrium conformational fluctuations reveal interdomain allosteric communication pathways. J. Mol. Biol. 2008; 378:1155–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vijayabaskar M.S., Vishveshwara S.. Comparative analysis of thermophilic and mesophilic proteins using Protein Energy Networks. BMC Bioinformatics. 2010; 11(Suppl. 1):S49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Serçinoğlu O., Ozbek P.. gRINN: a tool for calculation of residue interaction energies and protein energy network analysis of molecular dynamics simulations. Nucleic Acids. Res. 2018; 46:W554–W562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sladek V., Tokiwa H., Shimano H., Shigeta Y.. Protein residue networks from energetic and geometric data: are they identical. J. Chem. Theory Comput. 2018; 14:6623–6631. [DOI] [PubMed] [Google Scholar]
- 33. Vijayabaskar M.S., Vishveshwara S.. Interaction energy based protein structure networks. Biophys. J. 2010; 99:3704–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brinda K.V., Vishveshwara S.. A network representation of protein structures: implications for protein stability. Biophys. J. 2005; 89:4159–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Atilgan C., Okan O.B., Atilgan A.R.. Network-based models as tools hinting at nonevident protein functionality. Annu. Rev. Biophys. 2012; 41:205–225. [DOI] [PubMed] [Google Scholar]
- 36. Taylor N.R. Small world network strategies for studying protein structures and binding. Comput. Struct. Biotechnol. J. 2013; 5:e201302006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tse A., Verkhivker G.M.. Molecular dynamics simulations and structural network analysis of c-Abl and c-Src Kinase core proteins: capturing allosteric mechanisms and communication pathways from residue centrality. J. Chem. Inf. Model. 2015; 55:1645–1662. [DOI] [PubMed] [Google Scholar]
- 38. Tse A., Verkhivker G.M.. Molecular determinants underlying binding specificities of the ABL kinase inhibitors: combining alanine scanning of binding hot spots with network analysis of residue interactions and coevolution. PLoS One. 2015; 10:e0130203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tse A., Verkhivker G.M.. Exploring molecular mechanisms of paradoxical activation in the BRAF kinase dimers: atomistic simulations of conformational dynamics and modeling of allosteric communication networks and signaling pathways. PLoS One. 2016; 11:e0166583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stetz G., Tse A., Verkhivker G.M.. Dissecting structure-encoded determinants of allosteric Cross-Talk between Post-Translational Modification Sites in the Hsp90 Chaperones. Sci. Rep. 2018; 8:6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ermakova E., Kurbanov R.. Effect of ligand binding on the dynamics of trypsin. Comparison of different approaches. J. Mol. Graph. Model. 2014; 49:99–109.24642055 [Google Scholar]
- 42. Ghosh A., Sakaguchi R., Liu C., Vishveshwara S., Hou Y.-M.. Allosteric communication in cysteinyl tRNA synthetase: a network of direct and indirect readout. J. Biol. Chem. 2011; 286:37721–37731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fossépré M., Leherte L., Laaksonen A., Vercauteren D.P.. Understanding the Structure and Dynamics of Peptides and Proteins Through the Lens of Network Science. 2018; John Wiley & Sons, Ltd; 105–161. [Google Scholar]
- 44. del Sol A., Fujihashi H., Amoros D., Nussinov R.. Residues crucial for maintaining short paths in network communication mediate signaling in proteins. Mol. Syst. Biol. 2006; 2:2006.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yan W., Zhou J., Sun M., Chen J., Hu G., Shen B.. The construction of an amino acid network for understanding protein structure and function. Amino Acids. 2014; 46:1419–1439. [DOI] [PubMed] [Google Scholar]
- 46. Sanejouand Y.-H. Elastic Network Models: Theoretical and Empirical Foundations. 2013; Totowa: Humana Press; 601–616. [DOI] [PubMed] [Google Scholar]
- 47. Bahar I., Atilgan A.R., Erman B.. Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Fold. Des. 1997; 2:173–181. [DOI] [PubMed] [Google Scholar]
- 48. Atilgan A.R., Durell S.R., Jernigan R.L., Demirel M.C., Keskin O., Bahar I.. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys. J. 2001; 80:505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bahar I., Atilgan A.R., Demirel M.C., Erman B.. Vibrational dynamics of folded proteins: significance of slow and fast motions in relation to function and stability. Phys. Rev. Lett. 1998; 80:2733. [Google Scholar]
- 50. Haliloglu T., Bahar I., Erman B.. Gaussian dynamics of folded proteins. Phys. Rev. Lett. 1997; 79:3090–3093. [Google Scholar]
- 51. Chennubhotla C., Rader A.J., Yang L.-W., Bahar I.. Elastic network models for understanding biomolecular machinery: from enzymes to supramolecular assemblies. Phys. Biol. 2005; 2:S173–S180. [DOI] [PubMed] [Google Scholar]
- 52. Orellana L., Rueda M., Ferrer-Costa C., Lopez-Blanco J.R., Chacón P., Orozco M.. Approaching elastic network models to molecular dynamics flexibility. J. Chem. Theory Comput. 2010; 6:2910–2923. [DOI] [PubMed] [Google Scholar]
- 53. Tirion M.M. Large amplitude elastic motions in proteins from a single-parameter, atomic analysis. Phys. Rev. Lett. 1996; 77:1905–1908. [DOI] [PubMed] [Google Scholar]
- 54. Doruker P., Atilgan A.R., Bahar I.. Dynamics of proteins predicted by molecular dynamics simulations and analytical approaches: Application to-Amylase inhibitor. Proteins. 2000; 40:512–524. [PubMed] [Google Scholar]
- 55. Chakrabarty B., Parekh N.. NAPS: Network analysis of protein structures. Nucleic Acids Res. 2016; 44:W375–W382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Piovesan D., Minervini G., Tosatto S.C.E.. The RING 2.0 web server for high quality residue interaction networks. Nucleic Acids Res. 2016; 44:W367–W374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T.. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Doncheva N.T., Klein K., Domingues F.S., Albrecht M.. Analyzing and visualizing residue networks of protein structures. Trends Biochem. Sci. 2011; 36:179–182. [DOI] [PubMed] [Google Scholar]
- 59. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 60. Sobolev V., Sorokine A., Prilusky J., Abola E., Edelman M.. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999; 15:327–332. [DOI] [PubMed] [Google Scholar]
- 61. Bhattacharyya M., Bhat C.R., Vishveshwara S.. An automated approach to network features of protein structure ensembles: An automated approach to network features of protein structure ensembles. Protein Sci. 2013; 22:1399–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Doncheva N.T., Assenov Y., Domingues F.S., Albrecht M.. Topological analysis and interactive visualization of biological networks and protein structures. Nat. Protoc. 2012; 7:670–685. [DOI] [PubMed] [Google Scholar]
- 63. Schrodinger LLC The PyMOL Molecular Graphics System. 2015; Version 1.8. [Google Scholar]
- 64. Pasi M., Tiberti M., Arrigoni A., Papaleo E.. xPyder: A PyMOL plugin to analyze coupled residues and their networks in protein structures. J. Chem. Inf. Model. 2012; 52:1865–1874. [DOI] [PubMed] [Google Scholar]
- 65. Tiberti M., Invernizzi G., Lambrughi M., Inbar Y., Schreiber G., Papaleo E.. PyInteraph: a framework for the analysis of interaction networks in structural ensembles of proteins. J. Chem. Inf. Model. 2014; 54:1537–1551. [DOI] [PubMed] [Google Scholar]
- 66. Kayikci M., Venkatakrishnan A.J., Scott-Brown J., Ravarani C.N.J., Flock T., Babu M.M.. Visualization and analysis of non-covalent contacts using the Protein Contacts Atlas. Nat. Struct. Mol. Biol. 2018; 25:185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fassio A.V., Martins P.M., Guimarães S. da S., Junior S.S.A., Ribeiro V.S., de Melo-Minardi R.C., Silveira S.de A.. Vermont: a multi-perspective visual interactive platform for mutational analysis. BMC Bioinformatics. 2017; 18:403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Suhre K., Sanejouand Y.-H.. ElNemo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids. Res. 2004; 32:W610–W614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Eyal E., Lum G., Bahar I.. The Anisotropic Network Model web server at 2015 (ANM 2.0). Bioinformatics. 2015; 31:1487–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Eyal E., Yang L.-W., Bahar I.. Anisotropic network model: systematic evaluation and a new web interface. Bioinformatics. 2006; 22:2619–2627. [DOI] [PubMed] [Google Scholar]
- 71. Li H., Chang Y.Y., Yang L.W., Bahar I.. iGNM 2.0: the Gaussian network model database for biomolecular structural dynamics. Nucleic Acids. Res. 2016; 44:D415–D422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Li H., Chang Y.Y., Lee J.Y., Bahar I., Yang L.W.. DynOmics: Dynamics of structural proteome and beyond. Nucleic Acids. Res. 2017; 45:W374–W380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Skjærven L., Jariwala S., Yao X.-Q., Grant B.J.. Online interactive analysis of protein structure ensembles with Bio3D-web. Bioinformatics. 2016; 32:btw482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rodrigues C.H., Pires D.E., Ascher D.B.. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids. Res. 2018; 46:W350–W355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Seeber M., Felline A., Raimondi F., Mariani S., Fanelli F.. WebPSN: a web server for high-throughput investigation of structural communication in biomacromolecules. Bioinformatics. 2015; 31:779–781. [DOI] [PubMed] [Google Scholar]
- 76. Wako H., Endo S.. Normal mode analysis as a method to derive protein dynamics information from the Protein Data Bank. Biophys. Rev. 2017; 9:877–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wako H., Kato M., Endo S.. ProMode: a database of normal mode analyses on protein molecules with a full-atom model. Bioinformatics. 2004; 20:2035–2043. [DOI] [PubMed] [Google Scholar]
- 78. Wako H., Endo S.. Normal mode analysis based on an elastic network model for biomolecules in the Protein Data Bank, which uses dihedral angles as independent variables. Comput. Biol. Chem. 2013; 44:22–30. [DOI] [PubMed] [Google Scholar]
- 79. Kinjo A.R., Bekker G.-J., Suzuki H., Tsuchiya Y., Kawabata T., Ikegawa Y., Nakamura H.. Protein Data Bank Japan (PDBj): updated user interfaces, resource description framework, analysis tools for large structures. Nucleic Acids Res. 2017; 45:D282–D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kannan N., Vishveshwara S.. Identification of side-chain clusters in protein structures by a graph spectral method. J. Mol. Biol. 1999; 292:441–464. [DOI] [PubMed] [Google Scholar]
- 81. Goldenberg O., Erez E., Nimrod G., Ben-Tal N.. The ConSurf-DB: pre-calculated evolutionary conservation profiles of protein structures. Nucleic Acids. Res. 2009; 37:D323–D327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Armon A., Graur D., Ben-Tal N.. ConSurf: an algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information11Edited by F. Cohen. J. Mol. Biol. 2001; 307:447–463. [DOI] [PubMed] [Google Scholar]
- 83. Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., Ben-Tal N.. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016; 44:W344–W350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. UniProt Consortium, T.U. The universal protein resource (UniProt). Nucleic Acids Res. 2008; 36:D190–D195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Jubb H.C., Higueruelo A.P., Ochoa-Montaño B., Pitt W.R., Ascher D.B., Blundell T.L.. Arpeggio: a web server for calculating and visualising interatomic interactions in protein structures. J. Mol. Biol. 2017; 429:365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hu G., Di Paola L., Liang Z., Giuliani A.. Comparative study of elastic network model and protein contact network for protein complexes: the hemoglobin case. Biomed Res. Int. 2017; 2017:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kling F. 2014; Jsnetworkx: A javascript port of the networkx graph library.
- 89. Biasini M. PV - WebGL-based protein viewer. 2014; doi:10.5281/ZENODO.12620. [Google Scholar]
- 90. Bakan A., Meireles L.M., Bahar I.. ProDy: protein dynamics inferred from theory and experiments. Bioinformatics. 2011; 27:1575–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
ProSNEx is free and open to all users without login requirement. The server does not store any data submitted by the user. It is compatible with major web browser including Chrome, Firefox and Safari.