Abstract
As the amount of genomic variation data increases, tools that are able to score the functional impact of single nucleotide variants become more and more necessary. While there are several prediction servers available for interpreting the effects of variants in the human genome, only few have been developed for other species, and none were specifically designed for species of veterinary interest such as the dog. Here, we present Fido-SNP the first predictor able to discriminate between Pathogenic and Benign single-nucleotide variants in the dog genome. Fido-SNP is a binary classifier based on the Gradient Boosting algorithm. It is able to classify and score the impact of variants in both coding and non-coding regions based on sequence features within seconds. When validated on a previously unseen set of annotated variants from the OMIA database, Fido-SNP reaches 88% overall accuracy, 0.77 Matthews correlation coefficient and 0.91 Area Under the ROC Curve.
INTRODUCTION
One of the major challenges in medical genetics is to identify the functional effects of coding and non-coding single nucleotide variants (SNVs) to develop personalized medicine (1). For the human genome, several methods were implemented to interpret and score the impact of genomic variations (2,3). Most methods need protein sequences and score single amino acid changes (4). Few other methods also score the impact of non-coding variants: CADD (5), FATHMM (6) and PhD-SNPg (7).
Despite the numerous tools available for predicting the impact of genomic variations in the human genome, only few methods are available to score the effect of genomic variants in other species (8,9) and none are specifically designed for the dog genome. However, biology, disease presentation, and clinical response of many diseases in dog often mimic the human counterpart (10,11). Dogs in many cases spontaneously develop diseases, such as tumors, at a rate comparable to humans, as opposite to transgenic laboratory animal models in which cancers have to be implanted (10–13). This offers several advantages over other animal systems for mapping genes relevant to human disease (10–13). Here, we present Fido-SNP the first machine learning classifier to predict the effect of SNVs in the dog genome. Presentation of the methods and data sets as well as the assessment of Fido-SNP performances are in agreement with the guidelines reported in Vihinen 2012 (14).
METHOD OUTLINE
Fido-SNP is a method for predicting the impact of single nucleotide variants (SNVs) in the dog genome. Fido-SNP is not retrained on canine genome, but extends PhD-SNPg, which was previously developed (7) to accomplish the same task in the human genome. PhD-SNPg is a binary classifier based on machine learning, which classifies human genomic variants as either Pathogenic or Benign. It classifies both coding and non-coding variants and it is based on the Gradient Boosting method implemented through the scikit-learn package (15). Fido-SNP inherits the trained prediction model from PhD-SNPg, which was trained on a set of ∼35,800 annotated human variants derived from the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/), ∼68% of which are Pathogenic.
Like PhD-SNPg, Fido-SNP only uses features derived from DNA sequences and conservation scores. Fido-SNP transforms each input SNV into a 35-element vector, of which the first 25 elements encode the sequence window of five nucleotides (the generic base N is also considered) around the SNV site (5 × 5 = 25 positions); the remaining 10 elements encode the PhyloP conservation scores for each genomic position within the 5-nucleotide-long window (five elements for the PhyloP4 and five elements for the PhyloP11 conservation scores) (16). With this input vector, Fido-SNP predicts the probability that the SNV is Pathogenic.
No retraining was done to implement Fido-SNP method, since it exploits PhD-SNPg predictor, previously trained on human variation data. Fido-SNP extends PhD-SNPg to the dog genome by implementing the two following adaptations:
the computation of the conservation scores for each position of the dog genome
the optimization of the classification threshold.
The first step was achieved by computing the conservation scores through the PhyloP program for each position of the dog genome. The threshold optimization task was achieved by maximizing the discrimination between a set of potentially pathogenic variants (1,479 human variants in highly conserved loci) and a set of potentially neutral variants (∼3 million variants from dbSNP) in the dog genome.
Data sets
A crucial point for the development of a machine learning method for predicting the impact of genetic variants on a specific organism is the selection of a gold standard data set of annotated variants. Unfortunately, large and curated data sets containing such information are only available for few species, not including the dog. To overcome this lack of information, we selected a set of common single nucleotide variants (SNVs) in the human and the dog genomes that are potentially Pathogenic in both species (hd-pathogenic) assuming that Pathogenic SNVs in highly conserved loci are more likely to be functionally deleterious across different species. The data set initially contained ∼24,000 human Pathogenic variants annotated in the ClinVar database (17) and used to train PhD-SNPg. From this set, we selected the subset of highly conserved loci across a 5-nucleotide window sequence, which corresponds to the window sequence taken as input by PhD-SNPg. Conservation is established on the human 100-way alignment from UCSC. In detail, the variants in the hd-pathogenic set are selected using the following criteria:
the 5-nucleotide window sequence around the variant locus in human and dog are the same
the conservation of the reference allele in the variant locus is >95%
the average conservation of the nucleotides in the 5-nucleotide window sequence around the variant locus is greater than 95%
the alternative allele is not observed in the variant locus for any species in the human 100-way multiple sequence alignment
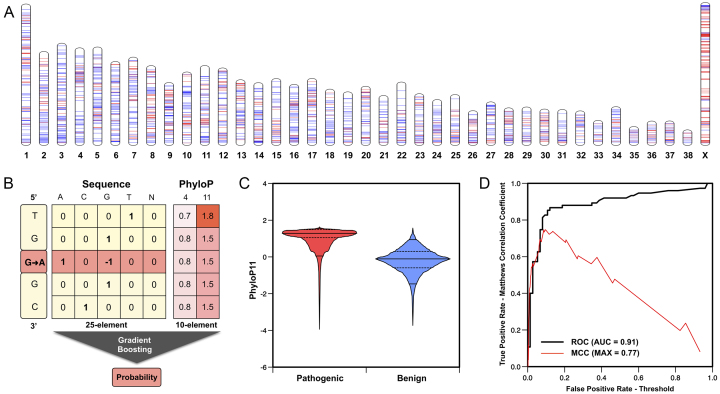
After this filtering procedure, we obtained a data set consisting of 2,359 variants. From this set we selected the variants for which the dog PhyloP11 conservation score is available, thus eliminating the genomic positions for which the conservation score is not computable as there are not enough aligned sequences. This second filter produced a set of 1,479 possible pathogenic variants in the dog genome, 20% of which are in chromosome X (Figure 1A) .
Figure 1.
(A) Distribution of the variants in the hd-pathogenic data set and an equal number of potentially benign SNVs from dbSNP (build 146) along the dog chromosomes. (B) Schematic view of the Fido-SNP algorithm and its input features. (C) Distribution of the PhyloP11 score for the potentially pathogenic mutated loci in the hd-pathogenic data set and a random set of variants from dbSNP. (D) Receiver Operator Characteristic (ROC) curve (black) obtained on the validation data set (dog-omia) and Matthews correlation coefficient (MCC) at different classification thresholds (red).
A second data set (dog-omia) for the validation of Fido-SNP was extracted from the Online Mendelian Inheritance in Animals (OMIA) database (18). OMIA is a catalogue of variations associated with genetic disorders in 244 animal species, including the dog. From the 319 disease-associated dog variants available in OMIA, we selected the 75 single nucleotide variants (dog-omia) which trait is Pathogenic (It is considered a to be defect) and for which the PhyloP11 conservation score is available. These 75 SNVs are found in 67 genes.
For the optimization and validation of Fido-SNP algorithm we collected a set of possible Benign variants from the dbSNP database (19). From the initial set of ∼5.6 million dog variants in the dbSNP build 146, we extracted ∼4.6 million SNVs. Of these, the PhyloP11 conservation score is available for ∼3 million SNVs (dbsnp-benign).
The performance of Fido-SNP has been tested on a balanced data set composed of the 75 Pathogenic dog SNVs (dog-omia) and of an equal number of possible Benign variants, randomly selected from dbsnp-benign.
722Dogs is a data set derived from 722 dog whole genomes available in the dog genome project (https://research.nhgri.nih.gov/dog_genome). This data set provides us with 6,038,693 SNVs after mapping on our multiple alignments and minor allele frequency filtering (MAF>5%). 722Dogs is used as a source of possible neutral SNVs.
Finally, Lym168 is a manually curated data set of 168 SNVs associated to canine lymphomas from 85 dogs (nine different breeds) (12), where a significant enrichment in pathogenic variations is expected.
A summary of the data sets before and after filtering procedures is reported in Supplementary Table S1.
Method features and prediction output
Fido-SNP is an untrained tool based on the previously developed PhD-SNPg, which is a program based on the Gradient Boosting algorithm (7). The two methods differ in two major aspects: (i) the reference genome (human versus dog), (ii) the different prediction threshold used to define pathogenicity. The input for Fido-SNP is the same as for PhD-SNPg, and includes features from the 5-nucleotide sequence centered on the variant locus. Briefly, the input consists of a 35-element vector, of which 25 elements encode for the sequence and variation and 10 elements encode for the PhyloP4 and PhyloP11 conservation scores. These selection parameters were established during the PhD-SNPg optimization, where different nucleotide window sizes were tested (7).
For the implementation of Fido-SNP we computed the PhyloP scores using an in-house computational facility to assemble the pairwise alignments between the dog and other species available online at the UCSC repository (following the instructions provided through personal communications by the UCSC team).
The output returned by Fido-SNP is a probabilistic score (s) between 0 and 1. This value is calculated by rescaling the output of the PhD-SNPg core method by multiplying all the scores below 0.1 by 5 (hence expanding the range of predicted benign SNVs) and multiplying all the scores greater than 0.1 by 5/9 (hence reducing the range of predicted pathogenic SNVs). This is necessary since the model is transferred from the human to the dog genome and hence is biased by the different number of aligned sequences, which compress the prediction output. The rescaling factor restores the output signal. A schematic view of Fido-SNP algorithm is shown in Figure 1B.
Conservation scores
Conservation scores for each position of the dog genome were computed through the PhyloP program of the PHAST package (16). This package computes conservation scores on multiple sequence alignments of genome sequences. To calculate the PhyloP scores in Fido-SNP, we considered the genomes of 10 different species. We built two different conservation scores, PhyloP4 and PhyloP11, which were obtained by aligning the dog reference genome to 3 (human, rat and mouse) and 10 (Human, Chimpanzee, Mouse, Rat, Cow, Panda, Marmoset, Cat, Horse and Opossum) genomes from other species, respectively. These scores replace the PhyloP7 and PhyloP100 conservation indexes used in PhD-SNPg.
The multiple sequence alignments of the genomic sequences from these species were built using the TBA/Multiz program (20) on the basis of the pairwise sequence alignments between the dog and the other species. We adopted the alignment pipeline suggested by Kent and colleagues (21), that is summarized at the following URL https://goo.gl/vyrKTd. In the case of the canfam2 assembly, pairwise alignments were downloaded from the UCSC repository, whereas for the canfam3 assembly they were calculated in-house though the LAST alignment program (http://last.cbrc.jp). All the genome sequences for the computation of the alignments were downloaded from UCSC. To calculate the multiple sequence alignments we assembled the pairwise alignments using a phylogenetic tree. A representation of this tree is shown in Supplementary Figure S1. The tree was derived from the phylogenetic tree used to calculate the UCSC hg38 100-way alignment (https://goo.gl/4euFfM). Finally, the dog 4-way and 11-way alignments were used to calculate the PhyloP4 and PhyloP11 conservation scores. A representation of the distributions of the PhyloP11 scores for potentially pathogenic loci in the hd-pathogenic data set and the randomly selected benign variant loci from the dbsnp-benign data set is shown in Figure 1C. The alignment procedures were the most demanding computations of this work: For each chromosome, the alignments required about 12 hours of single core computations using 100GB of RAM and 100GB of disk space for temporary files. The total amount of computation required about one month of computer time. The calculated PhyloP scores in bigWig format are available online at http://fidosnp.bca.unipd.it/ucsc and http://snps.biofold.org/Fido-SNP/ucsc.
Method optimization and performance
The performance of the Fido-SNP algorithm is estimated (without retraining) on a balanced data set consisting of (i) SNVs associated or potentially associated with pathogenic phenotypes in dogs and (ii) an equal number of potentially benign variants randomly selected from dbSNP (dbsnp-benign data set). For a better generalization of the predictive models, we randomly sampled dog variants from the dbsnp-benign data set 10 times. Thus, the Fido-SNP performances are averaged on 10 balanced data sets, which differ only in their subset of potentially benign variants. For each data set, we calculated the performance measures defined in the supplementary materials.
The lack of large data sets of annotated dog variants does not allow for the training of a dog-specific algorithm. Assuming that the basic mechanism of functional loss upon nucleotide variation is common to all species, we optimized the human model implanted in PhD-SNPg to predict the impact of variants in the dog genome. It is worth noting that no retraining of the original PhD-SNPg model was performed, only output rescaling was applied to fine-tune Fido-SNP. Rescaling was necessary since the difference between the human and dog genome alignments used for calculating the conservation scores requires the calibration of the prediction threshold. For this purpose, we used the hd-pathogenic data set to find the optimal threshold for the binary classification task by maximizing the Matthews correlation coefficient (Equation 2 in Supplementary Materials). The optimal classification threshold was then used to score the performance of Fido-SNP on previously unseen data extracted from the OMIA database (dog-omia). To verify the robustness of our results we also performed a 3-fold cross-validation procedure on a data set composed of the dog-omia SNVs and an equal number of variants from dbsnp-benign. With this procedure we determined the optimal classification threshold on 2/3 of the previous data set and applied this threshold to the remaining third of the SNVs.
The analysis of the results shows that in the optimization process Fido-SNP achieved 87% overall accuracy and 0.77 Matthews correlation coefficient for a classification threshold of 0.09 (Table 1). Approximating the classification threshold to 0.10, we observed that Fido-SNP achieved the same level of performance on the dog-omia and hd-pathogenic data sets in terms of overall accuracy (Q2), Matthews correlation coefficient (MCC), and area under the Receiver Operating Characteristic curve (AUC) (22). The AUCs on both data sets reached a value of 0.91. Consistent results are obtained with a 3-fold cross validation procedure on the dog-omia data set. An example of the ROC curve and Matthews correlation coefficient values obtained on a balanced data set composed of dog-omia SNVs and an equal number of randomly selected benign variants from dbsnp-benign is shown in Figure 1D.
Table 1.
Average performance of Fido-SNP on the hd-pathogenic and dog-omia data sets. All data contains pathogenic variants and an equal number of potentially benign variants randomly selected from dbSNP. Optimized performance of Fido-SNP obtained maximizing the MCC on the hd-pathogenic set. Performance on the validation set (dog-omia) considering a classification threshold of 0.1
| Data set | Threshold | Q2 | TNR | NPV | TPR | PPV | MCC | AUC |
|---|---|---|---|---|---|---|---|---|
| hd-pathogenic | 0.09±0.02 | 0.87±0.04 | 0.91±0.04 | 0.86±0.01 | 0.85±0.01 | 0.91±0.04 | 0.77±0.04 | 0.91±0.01 |
| dog-omia | 0.10±0.01 | 0.88±0.02 | 0.92±0.03 | 0.85±0.01 | 0.84±0.01 | 0.92±0.03 | 0.77±0.04 | 0.91±0.01 |
| dog-omia* | 0.11±0.03 | 0.87±0.04 | 0.92±0.05 | 0.84±0.05 | 0.82±0.07 | 0.92±0.05 | 0.75±0.08 | 0.91±0.04 |
*Performance of Fido-SNP on the dog-omia data set using a 3-fold cross-validation procedure. The performance measures are defined in Supplementary Materials. The values are computed using the canfam3 assembly.
We performed two further tests on the 722Dogs and Lym168 data sets. On the 6,038,693 SNVs contained in 722Dogs, Fido-SNP predicts as pathogenic only 7.6% of them. This is in line with the idea that most of these SVNs should be benign. Fido-SNP predictions on Lym168 are reported in Table 2, where for comparison we add the predictions made with SIFT (23) on both Lym168 and dog-omnia data sets. Fido-SNP predictions cover higher genome fractions including non-coding regions at a good level of performance.
Table 2.
Comparison between Fido-SNP and SIFT predictions on dog-omia and Lym168 data sets
| Data set | Method | Pathogenic | Predicted SNVs |
|---|---|---|---|
| Lym168 | Fido-SNP | 119 (78.8%) | 168/168 (100.0%) |
| SIFT | 70 (51.1%) | 137/168 (70.8%) | |
| dog-omia | Fido-SNP | 64 (85.3%) | 75/75 (100.0%) |
| SIFT | 43 (84.3%) | 51/75 (68.0%) |
SERVER DETAILS
Predicting the impact of single nucleotide variants
The Fido-SNP web-server predicts the impact of a single nucleotide variant based on input provided in comma-separated value (CSV) text or variant calling format (VCF). For each SNV the CSV input is composed of four elements: the chromosome, the position, and the reference and alternative alleles. For example, the variation of a Guanine to Adenine in chromosome 28, position 13,677,911 is represented by 28,13677911,G,A. Multiple SNVs can be provided by copy/pasting a list of variants as separate rows in the input box. For formatting reasons, the VCF input format should be provided by uploading a file containing a header starting with a hashtag (#) and followed by the identifiers of at least 5 columns (CHROM, POS, ID, REF, ALT) separated by a tab character. After the header line, each SNV is indicated in a separated row. If the variant's ID in the third column is missing or not available a dot sign (.) can be used.
When the list of SNVs is provided, either in CSV or VCF format, the server analyzes each variant and checks if the reference allele corresponds to the allele reported in the selected version of the dog genome (canfam2 or canfam3). This task is performed using the twoBitToFa program (24), which quickly extracts a portion of the dog genome from a sequence file in binary format. A window sequence of five nucleotides centred around the variant locus is used to generate the 25-element vector encoding the sequence information. If the nucleotide in the input matches the reference allele, the server extracts the corresponding conservation scores (PhyloP4 and PhyloP11) for the 5-nucleotide windows. The pre-calculated conservation scores are collected using the bigWigToBedGraph program (24). The PhyloP4 and PhyloP11 scores are used to generate a 10-element vector which contains the conservation features. After this step, the 35-element vector encoding the sequence and conservation features is given as input to the Gradient Boosting algorithm which returns the prediction output described above. In the final step of the prediction task, the Fido-SNP server annotates the input variants using Annovar (25). Annovar finds the possible effect on the amino acid sequence of the longest matching transcript corresponding to the variant region.
Input interface
The web interface of Fido-SNP consists of a ‘textarea’ box where the SNVs are provided in either CSV or format. CSV and VCF files, in either standard text or gzipped format, can be uploaded using the ‘Browse’ button below the ‘textarea’ box. When the list of SNVs is provided, the appropriate input format can be selected using the ‘Input Type’ buttons (CSV or VCF). A second group of buttons (Assembly) is used to indicate the dog reference genome (canfam2 or canfam3) to which the SNVs are referred. An example of inputs in CSV format can be found by clicking the ‘chr,pos,ref,alt’ hyperlink located at the top of the web interface. Although an example of VCF-like input is linked in the ‘Help’ web page, the usage of the ‘textarea’ box for the VCF input format is discouraged.
On the bottom of the Fido-SNP web page, the e-mail box (optional) is available to receive Fido-SNP output by e-mail.
Prediction output
Fido-SNP web server takes input in two different formats (CSV, VCF) containing the single nucleotide variants at the DNA level. It returns an output containing the probability that a given SNV is Pathogenic.
This probability is rescaled ∼0.1 (Equation 1 in Supplementary Materials) that represents the optimized threshold for discriminating between Pathogenic and Benign SNVs in the dog genome. In addition to the pathogenicity prediction, Fido-SNP also returns the estimated False Discovery Rate (FDR) and the PhyloP11 conservation score for the dog genome. When the SNV is located in coding regions, it also provides the RefSeq (26) code of the transcript and the HGNC gene product (27) through the HUGO website (https://genenames.org). This annotation process is performed using Annovar tool (25).
CONCLUSIONS
Fido-SNP is the first web server that specifically predicts the impact of single-nucleotide variations, both coding and non-coding, in the dog genome. Fido-SNP achieves a very good performance in the classification of pathogenic variants, limited only by the presence of unaligned regions in the dog genome that prevent the calculation of the PhyloP11 conservation score. We estimate that, on average, Fido-SNP returns predictions on ∼68% of the SNVs in dbsnp-benign data set. From a computational point of view, Fido-SNP is light and fast and constitutes a very useful resource for veterinary medicine applications. It can also be seen as a proof-of-concept that the knowledge acquired through the high level of annotation of the human genome can be transferred and exploited to boost prediction performances in other species.
DATA AVAILABILITY
The Fido-SNP web server is freely available at: http://fidosnp.bca.unipd.it/ and http://snps.biofold.org/fido-snp/ Fido-SNP scripts are available at https://github.com/biofold/Fido-SNP. All the data sets used in this work are available online at http://fidosnp.bca.unipd.it/method.html and http://snps.biofold.org/fido-snp/method.html
Supplementary Material
ACKNOWLEDGEMENTS
We thank Matteo Tuzzato for the technical support and Morgan Smits for improving the English language of the manuscript. EC acknowledge the Institute for Mathematical Modeling of Biological Systems at the University of Düsseldorf (Germany) for providing computational support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
SID-2017 from Padova University, delivered (to P.F.); FFABR grant from the Ministry of Education, Universities and Research (MIUR) (to E.C.); Italian Ministry for Education, University and Research under the programme ‘Dipartimenti di Eccellenza 2018–2022’ [D15D18000410001 to P.F.]; L.M. has been supported by EBA-PRISM, an Israel-Italy collaborative project between the Israel Ministry of Science and Technology and the Italian Ministry of Foreign Affairs and International Cooperation. Funding for open access charge: SID-2017.
Conflict of interest statement. None declared.
REFERENCES
- 1. Fernald G.H., Capriotti E., Daneshjou R., Karczewski K.J., Altman R.B.. Bioinformatics challenges for personalized medicine. Bioinformatics. 2011; 27:1741–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Niroula A., Vihinen M.. Variation interpretation predictors: principles, types, performance, and choice. Hum. Mutat. 2016; 37:579–597. [DOI] [PubMed] [Google Scholar]
- 3. Capriotti E., Nehrt N.L., Kann M.G., Bromberg Y.. Bioinformatics for personal genome interpretation. Brief. Bioinform. 2012; 13:495–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Capriotti E., Ozturk K., Carter H.. Integrating molecular networks with genetic variant interpretation for precision medicine. Wiley Interdiscip. Rev. Syst. Biol. Med. 2018; 11:e1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J.. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014; 46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shihab H.A., Rogers M.F., Gough J., Mort M., Cooper D.N., Day I.N., Gaunt T.R., Campbell C.. An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics. 2015; 31:1536–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Capriotti E., Fariselli P.. PhD-SNPg: a webserver and lightweight tool for scoring single nucleotide variants. Nucleic Acids Res. 2017; 45:W247–W252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gross C., de Ridder D., Reinders M.. Predicting variant deleteriousness in non-human species: applying the CADD approach in mouse. BMC Bioinformatics. 2018; 19:373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reeb J., Hecht M., Mahlich Y., Bromberg Y., Rost B.. Predicted molecular effects of sequence variants link to system level of disease. PLoS Comput. Biol. 2016; 12:e1005047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aresu L., Ferraresso S., Marconato L., Cascione L., Napoli S., Gaudio E., Kwee I., Tarantelli C., Testa A., Maniaci C. et al.. New molecular and therapeutic insights into canine diffuse large B cell lymphoma elucidates the role of the dog as a model for human disease. Haematologica. 2018; haematol.2018.207027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hernandez B., Adissu H.A., Wei B.R., Michael H.T., Merlino G., Simpson R.M.. Naturally occurring canine melanoma as a predictive comparative oncology model for human mucosal and other triple wild-type melanomas. Int. J. Mol. Sci. 2018; 19:E394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bushell K.R., Kim Y., Chan F.C., Ben-Neriah S., Jenks A., Alcaide M., Fornika D., Grande B.M., Arthur S., Gascoyne R.D. et al.. Genetic inactivation of TRAF3 in canine and human B-cell lymphoma. Blood. 2015; 125:999–1005. [DOI] [PubMed] [Google Scholar]
- 13. Ostrander E.A., Kruglyak L.. Unleashing the canine genome. Genome Res. 2000; 10:1271–1274. [DOI] [PubMed] [Google Scholar]
- 14. Vihinen M. Guidelines for reporting and using prediction tools for genetic variation analysis. Hum. Mutat. 2013; 34:275–282. [DOI] [PubMed] [Google Scholar]
- 15. Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V. et al.. Scikit-learn: machine learning in python. J. Mach. Learn. Res. 2011; 12:2825–2830. [Google Scholar]
- 16. Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A.. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010; 20:110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J. et al.. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016; 44:D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nicholas F.W. Online Mendelian Inheritance in Animals (OMIA): a comparative knowledgebase of genetic disorders and other familial traits in non-laboratory animals. Nucleic Acids Res. 2003; 31:275–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K.. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001; 29:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D. et al.. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004; 14:708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kent W.J., Baertsch R., Hinrichs A., Miller W., Haussler D.. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:11484–11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baldi P., Brunak S., Chauvin Y., Andersen C.A., Nielsen H.. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics. 2000; 16:412–424. [DOI] [PubMed] [Google Scholar]
- 23. Vaser R., Adusumalli S., Leng S.N., Sikic M., Ng P.C.. SIFT missense predictions for genomes. Nat. Protoc. 2016; 11:1–9. [DOI] [PubMed] [Google Scholar]
- 24. Kent W.J., Zweig A.S., Barber G., Hinrichs A.S., Karolchik D.. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010; 26:2204–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang K., Li M., Hakonarson H.. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010; 38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pruitt K.D., Tatusova T., Maglott D.R.. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007; 35:D61–D65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gray K.A., Yates B., Seal R.L., Wright M.W., Bruford E.A.. Genenames.org: the HGNC resources in 2015. Nucleic Acids Res. 2015; 43:D1079–D1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Fido-SNP web server is freely available at: http://fidosnp.bca.unipd.it/ and http://snps.biofold.org/fido-snp/ Fido-SNP scripts are available at https://github.com/biofold/Fido-SNP. All the data sets used in this work are available online at http://fidosnp.bca.unipd.it/method.html and http://snps.biofold.org/fido-snp/method.html