Abstract
PrankWeb is an online resource providing an interface to P2Rank, a state-of-the-art method for ligand binding site prediction. P2Rank is a template-free machine learning method based on the prediction of local chemical neighborhood ligandability centered on points placed on a solvent-accessible protein surface. Points with a high ligandability score are then clustered to form the resulting ligand binding sites. In addition, PrankWeb provides a web interface enabling users to easily carry out the prediction and visually inspect the predicted binding sites via an integrated sequence-structure view. Moreover, PrankWeb can determine sequence conservation for the input molecule and use this in both the prediction and result visualization steps. Alongside its online visualization options, PrankWeb also offers the possibility of exporting the results as a PyMOL script for offline visualization. The web frontend communicates with the server side via a REST API. In high-throughput scenarios, therefore, users can utilize the server API directly, bypassing the need for a web-based frontend or installation of the P2Rank application. PrankWeb is available at http://prankweb.cz/, while the web application source code and the P2Rank method can be accessed at https://github.com/jendelel/PrankWebApp and https://github.com/rdk/p2rank, respectively.
INTRODUCTION
The field of structural biology has recently experienced enormous progress in all aspects of structural determination and, as a result, 3D structures of proteins are becoming increasingly available. Indeed, structural genomics consortia are now able to solve protein structures with no known function (1), the information acquired from 3D coordinates for such proteins being used to annotate the proteins. An important clue for predicting protein function is the identification of ligands or small molecules that can bind to the protein. Ligands and other small molecules can either be determined directly within the protein’s 3D structure or a 3D structure of the protein can be used to predict ligand binding sites, and thus help to annotate the protein.
A range of protein ligand binding site prediction approaches have been developed over recent years, including a number that are provided as a web service (Table 1). Fpocket (2), SiteHound (3), ConCavity (4), POCASA (5), MetaPocket 2.0 (6), FTSite (7) and bSiteFinder (8) all support online visualization using Jmol (9), a Java-based molecular structure viewer. Due to known security risks, however, Java applets are no longer supported in modern web browsers and these websites can now be considered outdated. A simple solution to the Jmol issue is to use JSmol (10), a JavaScript replacement for Jmol. This is the avenue taken by 3DLigandSite (11), COFACTOR (12,13), COACH (14) ISMBLAB-LIG (15) and LIBRA (16). Though JSmol supports complex visualization options, it suffers from performance issues due to inefficiencies introduced when migrating Jmol code from Java to JavaScript. Fpocket uses OpenAstex (17), another Java based visualizer; however, this project suffers from the same problems as Jmol and now appears to have been discontinued as we were unable to find an active resource. Relatively few of the web servers support visualization via modern WebGL-based viewers, such as LiteMol (18), NGL (19,20) and PV (21). As an example, NGL supports visualizations in DoGSite (22) and DeepSite (23); however, while it is possible to view 3D structures in NGL, the DeepSite and DoGSite websites lack the option to customize protein, ligand and binding site visualizations. Similarly, GalaxySite (24) only offers minimal 3D cartoon visualization of the protein and its ligands via the PV viewer. In response to this situation, we recently developed P2Rank (25), a state-of-the-art method for protein ligand binding site prediction. Here, we describe PrankWeb, an online web server providing an interactive interface for the P2Rank method. PrankWeb serves as an intuitive tool for ligand binding site prediction and its immediate visual analysis by displaying the prediction as a combination of the protein’s 3D structure, its sequence and a list of binding pockets. It allows users to display protein ligand binding sites and conservation as structural and sequence views and to customize the visualization style. As PrankWeb’s visualization is based on LiteMol and Protael (26), it runs on all modern browsers with no additional plugins.
Table 1.
Availability of web-based tools for structure-based ligand binding site prediction introduced since 2009
| Name | Year | Type | Stand-alone | Online Visualization | Offline visualization | Source code |
|---|---|---|---|---|---|---|
| SiteHound (3) | 2009 | Energetic | Yes | Jmol | PyMOLb, Chimerab | Yes |
| ConCavity (4) | 2009 | Conservation | Yes | Jmol | PyMOL | Yes |
| Fpocket (2) | 2010 | Geometric | Yes | Jmol, OpenAstex | PyMOL, VMD | Yes |
| 3DLigandSite (11) | 2010 | Template | — | JSmol | PyMOL | — |
| POCASA (5) | 2010 | Geometric | — | Jmol | — | — |
| DoGSite (22) | 2010 | Geometric | — | NGL | — | — |
| MetaPocket 2.0 (6) | 2011 | Consensus | — | Jmol | PyMOL | — |
| FTSite (7) | 2012 | Energetic | — | Jmol, static | PyMOL | — |
| COFACTOR(12,13) | 2012, 2017 | Template | Yes | JSmol | — | — |
| COACH (14) | 2013 | Template | Yes | JSmol | — | — |
| eFindSite (27)a | 2014 | Template | Yes | — | PyMOL, VMD, Chimera | Yes |
| GalaxySite (24) | 2014 | Template/docking | — | PV, static | — | — |
| bSiteFinder (8) | 2016 | Template | — | Jmol | — | — |
| ISMBLab-LIG (15) | 2016 | Machine learning | — | JSmol & sequence | — | — |
| LIBRA-WA (16) | 2017 | Template | Yes | JSmol | — | — |
| DeepSite (23) | 2017 | Machine learning | — | NGL | — | — |
| PrankWeb (P2Rank) | this work | Machine learning | Yes | LiteMol & Proteal | PyMOL | Yes |
aIn the process of setting up a new interface.
bOnly data files provided.
MATERIALS AND METHODS
P2Rank
P2Rank (25), the backend of PrankWeb, is a template-free, machine learning-based method for ligand binding site prediction employing random forests (28) to predict ligandability of points on the solvent accessible surface of a protein. These points represent potential locations of binding ligand contact atoms and are described by a feature vector calculated from the local geometric neighbourhood. The feature vector consists of physico-chemical and geometric properties calculated from the surrounding atoms and residues (e.g. hydrophobicity, aromaticity or surface protrusion). PrankWeb also introduces a new model that includes information derived from residue sequence evolutionary conservation scores (see Supplementary Information for computation of conservation scores). Points with high predicted ligandability are clustered and ranked according to a ranking function based on the cumulative score of the cluster.
P2Rank is able to use different pre-trained models with varying feature vectors. PrankWeb exposes two such models, the default P2Rank model (without conservation) and a new model that uses conservation information (P2Rank+Conservation). Both models were trained on a relatively small but diverse dataset of protein ligand complexes (25,29).
As a template-free method, P2Rank does not share the limitations of template-based methods that are unable to predict truly novel sites with no analogues in their template libraries of known protein–ligand complexes. As such, P2Rank should be particularly beneficial for predicting novel allosteric sites for which template-based methods are generally less effective (25). Another advantage of P2Rank is its ability to work directly with multi-chain structures and predict binding sites formed near the chain interfaces.
We compared the predictive performance of P2Rank with several competing algorithms using two datasets: COACH420 (14), which contains 420 single-chain complexes, and HOLO4K (25), which contains 4009 multi-chain structures (see Table 2). The default model used by PrankWeb (P2Rank+Conservation) clearly outperformed the other methods, as did the original P2Rank model (without conservation) in most cases. Many of the methods listed in Table 1 are hard to compare using larger datasets as, unlike PrankWeb, they do not expose REST APIs; consequently, batch processing is hindered by slow running times, with results only being deliverable by email or captcha. For a description of the evaluation methodology and more detailed results, see the Supplementary Material. Possible reasons why P2Rank requires less training data and performs better than methods based on more modern machine learning approaches (e.g. DeepSite) are discussed in (25).
Table 2.
Benchmark on COACH420 and HOLO4K datasets
| COACH420 | HOLO4K | |||
|---|---|---|---|---|
| Top-n | Top-(n+2) | Top-n | Top-(n+2) | |
| Fpocket 1.0 | 56.4 | 68.9 | 52.4 | 63.1 |
| Fpocket 3.1 | 42.9 | 56.9 | 54.9 | 64.3 |
| SiteHounda | 53.0 | 69.3 | 50.1 | 62.1 |
| MetaPocket 2.0a | 63.4 | 74.6 | 57.9 | 68.6 |
| DeepSitea | 56.4 | 63.4 | 45.6 | 48.2 |
| P2Rank | 72.0 | 78.3 | 68.6 | 74.0 |
| P2Rank+Cons.b | 73.2 | 77.9 | 72.1 | 76.7 |
Comparing identification success rate [%] measured by the DCA criterion (distance from pocket center to closest ligand atom) with 4 Å threshold considering only pockets ranked at the top of the list (n is the number of ligands in the considered structure).
aFailed to produce predictions for some of the input proteins. Here we display calculated success rates based only on those protein subsets for which the corresponding method was finished successfully.
bP2Rank with conservation (the default prediction model of PrankWeb).
Prediction speeds varied greatly between tools, ranging from under one second (Fpocket, P2Rank) to >10 h (COACH) for prediction on one average sized protein (2500 atoms). We have previously shown that P2Rank (without conservation) is the second fastest of the tools presently available (25). While PrankWeb provides little overhead to prediction speed, use of the model with conservation may take a few minutes if conservation scores need to be calculated from scratch (see Conservation pipeline section in the Supplementary Material).
Web server
PrankWeb allows users to predict and visualize the protein ligand binding sites and contrast these with both highly conserved areas and actual ligand binding sites.
To carry out the prediction, users can either upload a PDB file or provide a PDB ID, in which case PrankWeb will download and store the corresponding PDB file from the PDB database (30). In addition to selecting what protein to analyze, users can also specify whether evolutionary conservation should be included in the prediction process, which in turn determines which of the two pre-trained models will be used.
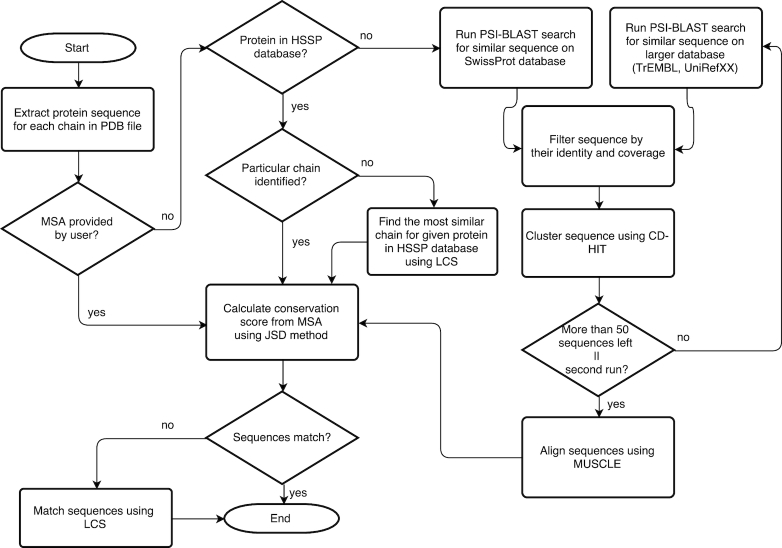
Conservation scores are calculated using the Jensen-Divergence method (31) from a multiple sequence alignment (MSA) file, which can come from three sources: (i) users can specify their own alignment file, (ii) if a protein’s PDB code is provided, PrankWeb uses MSA from the HSSP (32) database or (iii) where no MSA is provided and no MSA is found in HSSP, the MSA is computed using PrankWeb’s own conservation pipeline, which utilizes UniProt (33), PSI-Blast (34), MUSCLE (35) and CD-HIT (36). This process is depicted in Figure 2 and described in detail in the Supplementary Material.
Figure 2.
Flow diagram illustrating conservation loading workflow and conservation pipeline.
After specification of the input, the submitted data is sent via a REST API to the server, which then starts the prediction pipeline. The user is provided with a URL address from which progress of the prediction process can be tracked and results inspected once the process finishes.
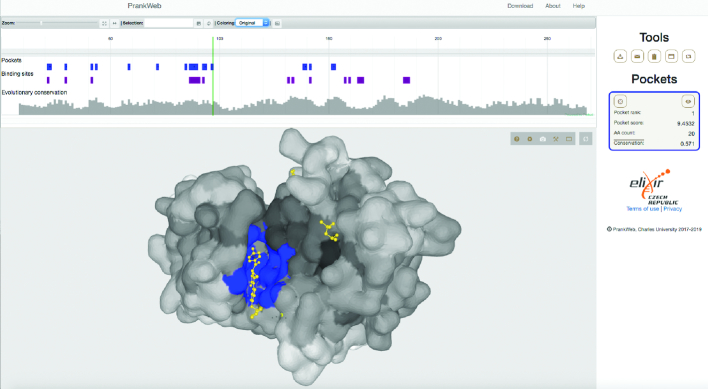
On the results page, PrankWeb utilizes LiteMol for visualization of 3D structural information and Protael for sequence visualization. Figure 1 displays the predicted binding sites of dasatinib (a drug used for treatment of chronic myelogenous leukemia) bound to the kinase domain of human LCK (PDB ID 3AD5). The sequence and structure plugins are synchronized so that the user can easily locate a sequence position in the structure and vice versa. The sequence view comprises predicted pockets, computed conservation and binding sites (if present in the PDB file). The side panel displays information about the identified pockets and a toolbar allowing the user to (i) download all inputs and calculated results, (ii) share the results page link or (iii) switch between visualization modes. PrankWeb comes with three predefined 3D model renderings (protein surface, cartoon and atoms) and the predicted binding sites and conservation scores are color coded. Conservation is displayed in grayscale (darker denoting more conserved residues) and binding sites are color-highlighted. When the conservation score is not available, the protein surface is white. If conservation analysis is chosen, the user can contrast the positions of putative active sites with conservation scores of the respective positions. In cases where the preset modes do not suffice, one can completely customize the 3D visualization using LiteMol’s advanced user interface or the PyMOL visualization script for offline inspection.
Figure 1.
An example of PrankWeb output. The figure shows a predicted ligand binding site (blue colour) on the surface of human Lck kinase (3AD5) The actual ligand binding pose of dasatinib is shown in yellow. The second small molecule in the figure is dimethyl sulfoxide. The figure also shows a sequence view of the protein with binding sites and conservation scores indicated (top panel). The right panel shows a summary of the binding sites and provides tools to modify the view or to download the results.
PrankWeb consists of a Java backend, REST API and a Typescript frontend, the backend being based on the WildFly (37) web server and the P2Rank application, while the frontend uses the Protael, LiteMol and Bootstrap.js libraries to provide an interactive user interface on top of the REST API. All source code is available under Apache License 2.0 at GitHub (https://github.com/jendelel/PrankWebApp). The GitHub website also includes documentation for developers on how to use our REST API and how to deploy their own version of the server.
DISCUSSION
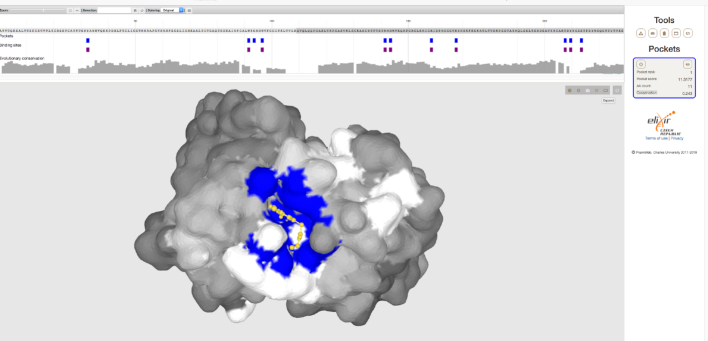
PrankWeb has been shown to provide correct predictions, even in cases where other methods have failed. Nghan et al. (7) mentions three cases (i.e. the glucose/galactose receptor (1GCG, 1GCA), purine nucleoside phosphorylase (1ULA,1ULB) and mouse FV antibody fragment (1A6U,1A6W)) where their FTSite method was unable to identify a ligand binding site with their best ranked prediction. PrankWeb, on the other hand, correctly identified the binding site as best ranked in both apo and holo structure in all three cases. Figure 3 shows the predicted ligand binding site of the holo structure (1GCA) on the interface of two imunoglobulin subunits, together with the experimentally solved structure of 4-HYDROXY-5-IODO-3-NITROPHENYLACETYL-EPSILON-AMINOCAPROIC ACID ANION (NIP). The 3D structure of NIP appears in the PDB just once, however, which makes it difficult to train its binding.
Figure 3.
Prediction of a ‘difficult' pocket. The authors of the FTSite method describe three structures for which their method failed. This figure shows a PrankWeb prediction for one of these, the structure of mouse immunoglobulin (1a6w). The prediction is indicated by the blue colour and the actual ligand is in yellow.
It should be noted that the current version of PrankWeb is aimed at discovering the binding sites of small biological ligands. None of the models employed by PrankWeb has been trained on other ligand types, such as metallic ion ligands or peptides. Such tasks would be better served by models trained on specialized datasets. We plan to build on our current work by including such models into PrankWeb in the future (38).
CONCLUSION
Here, we present PrankWeb, a new web interface for P2Rank, a state-of-the-art ligand binding prediction method. PrankWeb allows users to quickly carry out predictions and visually inspect the results. PrankWeb also contains a pipeline for computation of conservation scores, which are included in the ligand binding site prediction and the results of structure-sequence visualization. PrankWeb not only provides a user-friendly interface it also serves as a REST API, enabling developers to use PrankWeb as a service. Both PrankWeb and P2Rank are open sourced on GitHub and freely available.
Supplementary Material
ACKNOWLEDGEMENTS
We greatly appreciate access given to computing and storage facilities owned by parties and projects contributing to the MetaCentrum National Grid Infrastructure, as provided under the programme ‘Projects of Large Research, Development, and Innovation Infrastructures' (CESNET LM2015042).
Notes
Present address: Lukas Jendele, Department of Computer Science, ETH Zurich, Switzerland.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the ELIXIR CZ Research Infrastructure Project [MEYS Grant LM2015047] and by the Grant Agency of Charles University [1556217].
Conflict of interest statement. None declared.
REFERENCES
- 1. Skolnick J., Fetrow J.S., Kolinski A.. Structural genomics and its importance for gene function analysis. Nat. Biotechnol. 2000; 18:283. [DOI] [PubMed] [Google Scholar]
- 2. Schmidtke P., Le Guilloux V., Maupetit J., Tufféry P.. Fpocket: online tools for protein ensemble pocket detection and tracking. Nucleic Acids Res. 2010; 38:W582–W589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hernandez M., Ghersi D., Sanchez R.. SITEHOUND-web: a server for ligand binding site identification in protein structures. Nucleic Acids Res. 2009; 37:W413–W416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Capra J.A., Laskowski R.A., Thornton J.M., Singh M., Funkhouser T.A.. Predicting protein ligand binding sites by combining evolutionary sequence conservation and 3D structure. PLoS Comput. Biol. 2009; 5:e1000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yu J., Zhou Y., Tanaka I., Yao M.. Roll: a new algorithm for the detection of protein pockets and cavities with a rolling probe sphere. Bioinformatics. 2010; 26:46–52. [DOI] [PubMed] [Google Scholar]
- 6. Zhang Z., Li Y., Lin B., Schroeder M., Huang B.. Identification of cavities on protein surface using multiple computational approaches for drug binding site prediction. Bioinformatics. 2011; 27:2083. [DOI] [PubMed] [Google Scholar]
- 7. Ngan C.-H., Hall D.R., Zerbe B.S., Grove L.E., Kozakov D., Vajda S.. FTSite: high accuracy detection of ligand binding sites on unbound protein structures. Bioinformatics. 2012; 28:286–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gao J., Zhang Q., Liu M., Zhu L., Wu D., Cao Z., Zhu R.. bSiteFinder, an improved protein-binding sites prediction server based on structural alignment: more accurate and less time-consuming. J. Cheminf. 2016; 8:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tully S.P., Stitt T.M., Caldwell R.D., Hardock B.J., Hanson R.M., Maslak P.. Interactive web-based pointillist visualization of hydrogenic orbitals using jmol. J. Chem. Educ. 2013; 90:129–131. [Google Scholar]
- 10. Hanson R.M., Prilusky J., Renjian Z., Nakane T., Sussman J.L.. JSmol and the next-generation web-based representation of 3 D molecular structure as applied to proteopedia. Isr. J. Chem. 2013; 53:207–216. [Google Scholar]
- 11. Wass M.N., Kelley L.A., Sternberg M.J.. 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res. 2010; 38:W469–W473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Roy A., Yang J., Zhang Y.. COFACTOR: an accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Res. 2012; 40:W471–W477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang C., Freddolino P.L., Zhang Y.. COFACTOR: improved protein function prediction by combining structure, sequence and protein–protein interaction information. Nucleic Acids Res. 2017; 45:W291–W299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang J., Roy A., Zhang Y.. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics. 2013; 29:2588–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jian J.-W., Elumalai P., Pitti T., Wu C.Y., Tsai K.-C., Chang J.-Y., Peng H.-P., Yang A.-S.. Predicting ligand binding sites on protein surfaces by 3-dimensional probability density distributions of interacting atoms. PLoS One. 2016; 11:e0160315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Toti D., Viet Hung L., Tortosa V., Brandi V., Polticelli F.. LIBRA-WA: a web application for ligand binding site detection and protein function recognition. Bioinformatics. 2017; 34:878–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hartshorn M.J. AstexViewer TM†: a visualisation aid for structure-based drug design. J. Comput. Aid. Mol. Des. 2002; 16:871–881. [DOI] [PubMed] [Google Scholar]
- 18. Sehnal D., Deshpande M., Vareková R.S., Mir S., Berka K., Midlik A., Pravda L., Velankar S., Koča J.. LiteMol suite: interactive web-based visualization of large-scale macromolecular structure data. Nat. Methods. 2017; 14:1121–1122. [DOI] [PubMed] [Google Scholar]
- 19. Rose A.S., Hildebrand P.W.. NGL viewer: a web application for molecular visualization. Nucleic Acids Res. 2015; 43:W576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rose A.S., Bradley A.R., Valasatava Y., Duarte J.M., Prlić A., Rose P.W.. Web-based molecular graphics for large complexes. Proc. 21st Int. Conf. Web3D Technology. 2016; NY: ACM; 185–186. [Google Scholar]
- 21. Biasini M. pv: v1.8.1. 2015; 10.5281/zenodo.20980. [DOI] [Google Scholar]
- 22. Volkamer A., Griewel A., Grombacher T., Rarey M.. Analyzing the topology of active sites: On the prediction of pockets and subpockets. J. Chem. Inf. Model. 2010; 50:2041–2052. [DOI] [PubMed] [Google Scholar]
- 23. Jiménez J., Doerr S., Martínez-Rosell G., Rose A.S., Fabritiis G.D.. DeepSite: protein-binding site predictor using 3D-convolutional neural networks. Bioinformatics. 2017; 33:3036–3042. [DOI] [PubMed] [Google Scholar]
- 24. Heo L., Shin W.-H., Lee M.S., Seok C.. GalaxySite: ligand-binding-site prediction by using molecular docking. Nucleic Acids Res. 2014; 42:W210–W214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Krivák R., Hoksza D.. P2Rank: machine learning based tool for rapid and accurate prediction of ligand binding sites from protein structure. J. Cheminf. 2018; 10:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sedova M., Jaroszewski L., Godzik A.. Protael: protein data visualization library for the web. Bioinformatics. 2016; 32:602–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Feinstein W.P., Brylinski M.. eFindSite: Enhanced Fingerprint-Based virtual screening against predicted ligand binding sites in protein models. Mol. Inf. 2014; 33:135–150. [DOI] [PubMed] [Google Scholar]
- 28. Ho T.K. Random decision forests. Proc. 3rd Int. Conf. Document Analysis and Recognition. 1995; 1:IEEE; 278–282. [Google Scholar]
- 29. Chen K., Mizianty M., Gao J., Kurgan L.. A critical comparative assessment of predictions of protein-binding sites for biologically relevant organic compounds. Structure. 2011; 19:613–621. [DOI] [PubMed] [Google Scholar]
- 30. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E.. The protein data bank. Nucleic Acids Res. 2000; 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Capra J.A., Singh M.. Predicting functionally important residues from sequence conservation. Bioinformatics. 2007; 23:1875–1882. [DOI] [PubMed] [Google Scholar]
- 32. Joosten R.P., te Beek T.A., Krieger E., Hekkelman M.L., Hooft R.W., Schneider R., Sander C., Vriend G.. A series of PDB related databases for everyday needs. Nucleic Acids Res. 2011; 39:D411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. The UniProt Consortium UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2018; 47:D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J.. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32:1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li W., Godzik A.. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006; 22:1658. [DOI] [PubMed] [Google Scholar]
- 37. WildFly Homepage · WildFly http://wildfly.org/.
- 38. Krivák R., Jendele L., Hoksza D.. Peptide-Binding site prediction from protein structure via points on the solvent accessible surface. Proc. 2019 ACM Int. Conf. Bioinformatics. 2018; ACM; 645–650. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.