Abstract
Drug combination therapy has the potential to enhance efficacy, reduce dose-dependent toxicity and prevent the emergence of drug resistance. However, discovery of synergistic and effective drug combinations has been a laborious and often serendipitous process. In recent years, identification of combination therapies has been accelerated due to the advances in high-throughput drug screening, but informatics approaches for systems-level data management and analysis are needed. To contribute toward this goal, we created an open-access data portal called DrugComb (https://drugcomb.fimm.fi) where the results of drug combination screening studies are accumulated, standardized and harmonized. Through the data portal, we provided a web server to analyze and visualize users’ own drug combination screening data. The users can also effectively participate a crowdsourcing data curation effect by depositing their data at DrugComb. To initiate the data repository, we collected 437 932 drug combinations tested on a variety of cancer cell lines. We showed that linear regression approaches, when considering chemical fingerprints as predictors, have the potential to achieve high accuracy of predicting the sensitivity of drug combinations. All the data and informatics tools are freely available in DrugComb to enable a more efficient utilization of data resources for future drug combination discovery.
INTRODUCTION
The current cancer treatment is still largely based on a ‘one size fits all’ approach, resulting in limited efficacy due to the heterogeneity between the patients. Molecular diagnostics, histopathology and imaging techniques help stratify and monitor patients, but they provide limited support to guide treatment selection, especially for patients with recurrent cancers. NGS (Next Generation Sequencing) technologies and other omics profiling have revealed the intrinsic heterogeneity in cancer, partly explaining why patients respond differently to the same therapy (1). Even when there is an initial treatment response, cancer cells can easily develop drug resistance by the emerging activation of compensating or bypassing pathways (2). To reach effective and sustained clinical responses, many cancer patients who become resistant to standard treatments urgently need new multi-targeted drug combinations, which can effectively inhibit the cancer cells and block the emergence of drug resistance, while selectively incurring minimal effects on healthy cells (3). Although many new drugs are being developed, there is little information to guide the selection of effective combinations, as well as the identification of patients that would benefit from such combinatorial therapies. Recently, high-throughput drug combination screening techniques have been successfully applied for the functional testing of cancer cell lines or patient-derived samples, with several important hits being made (4). However, the exponentially increasing number of possible drug combinations makes a pure experimental approach quickly unfeasible, even with automated drug screening instruments (5). Therefore, data integration approaches to predict and annotate the drug combination effects at the systems level becomes a necessary route (6). Recent efforts included the use of network-based modeling to predict drug combinations (7). However, the size of drug combination data utilized for training such complex models has been often limited. To guide the patient stratification, biomarker discovery and treatment selection, a number of data collection, standardization and harmonization challenges need to be solved before the promise of personalized drug combinations is ultimately met (8,9).
To help achieve these goals, we present DrugComb (https://drugcomb.fimm.fi/), a web-based data portal that aims to harmonize and standardize drug combination screen data for cancer cell lines. In particular, we focused on the common experimental designs where drug pairs were crossed at different doses, forming a dose–response matrix. We provided computational tools via a web server that allow users to visualize, analyze and annotate such drug combination dose–response data. These tools can be used for the determination of drug combination sensitivity and synergy, such that the most promising drug combinations can be efficiently prioritized for the downstream experimentation. Furthermore, to facilitate a crowdsourcing effort, we provided data submission tools to encourage users to share and redistribute their data in a standardized manner. Through the web server, we established a data curation pipeline to collect datasets from several major drug combination studies, covering 437 923 drug combination experiments with 7 423 800 data points across 93 human cancer cell lines. We provided the sensitivity and synergy scores for these drug combinations, and showed that these scores can be predicted by linear regression models using the structural information of the compounds. The mechanisms of action of drug combinations can be further illustrated from drug–target interaction profiles provided by major pharmacology databases including STITCH (10), PubChem (11) and ChEMBL (12). The harmonized DrugComb data can be readily linked with genomic, transcriptomic and proteomic profiles of the cancer cells, which are available in major cancer cell line databases such as CCLE (13), GDSC (14), COSMIC (15), CTRP (16) and MCLP (17).
DrugComb is designed to be a major source of information that can be findable, assessable, interoperable and reusable (FAIR) for drug combination research, as there is currently lack of open-access services and repositories containing harmonized results of drug combinations studies. Furthermore, the analysis of drug combinations, especially in terms of their efficacy and synergy, as well as their mechanisms of action, were largely missing. With the help of data curation and analysis tools provided by DrugComb, we expect that the users may benefit from such efforts and be willing to form a community with a critical mass, so that more datasets can be collectively curated and centrally deposited. Ultimately, such a drug combination community shall lead to a consensus on the essential information that is needed to conform to the FAIR principle of research data (18). Furthermore, we expect that DrugComb will make an ideal testbed for more advanced machine learning algorithms to predict and prioritize the most effective drug combinations, which may ultimately lead to a cost-effective treatment decision support tool for the rational design of personalized drug combinations. DrugComb prioritizes the collection and dissemination of high-throughput screen data related to drug combinations to enable a better understanding, validation, and prediction of synergistic drug combinations for individual cancer cell lines. This one-stop workflow proposed by DrugComb makes it a unique tool in cancer drug discovery research.
In this manuscript, we described major components of DrugComb, including a web server with a variety of data analysis tools, as well as a database repository that shall facilitate the curation and standardization of the major drug combination studies. Such a data integration pipeline can be further developed into a protocol that may be adopted by a wider drug combination screen community. Furthermore, we reported the initial results of the drug combination prediction as a case study, and highlighted the potential of machine learning techniques to improve the efficiency of drug combination discovery. To facilitate the use of web server and the interpretation of the data analysis results, a step-by-step user guide was provided in the web site. Future aspects of DrugComb development were also discussed in Conclusions.
DATA PORTAL COMPONENTS
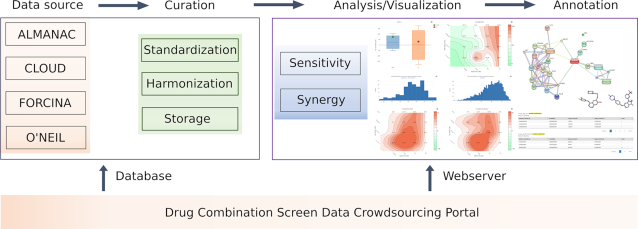
The DrugComb data portal includes two major components, the web server and the database (Figure 1). The web server, mainly available at the Analysis page (https://drugcomb.fimm.fi/analysis/), consists of a pipeline that generates the numeric and graphical results of drug combination sensitivity and synergy analyses for users’ own experimental data. Furthermore, a registered user may also submit the proprietary data via the Contribution page (https://drugcomb.fimm.fi/contribute/), which will be evaluated by the administrator for its appropriateness to be deposited in the database. The experimental protocols that have been implemented to produce the data are compulsory for a valid data deposit, as such information is critical to evaluate and adjust the potential batch effect (19). The database, retrievable at the Home page, harbors the curated drug combination screen datasets as well as their associated data analysis results. To facilitate the annotation of these drug combinations, we utilized third party APIs to access (i) chemical–protein association networks in the STITCH database, (ii) molecular structural information in the PubChem database and (iii) ligand-based target predictions in the ChEMBL database. All the data visualization functionalities are built using Javascript. Computational backend employs MariaDB for the database, while R, Python and PHP routines are used for the drug combination sensitivity and synergy analyses.
Figure 1.
Overview of DrugComb portal and the workflow. Drug combination screen data can be uploaded by users or from the literature. Data curation includes standardization of compound and cell line names, harmonization of drug effects as percentage inhibitions compared to the DMSO negative control, and a simplified file format to facilitate data storage in the database. The web server aims to analyze the curated data to determine and visualize the sensitivity and synergy of drug combinations. External tools are provided for a network-centric representation of mechanisms of action of drug combinations, skeletal view of drug molecules, as well as predicted drug–target interactions.
Computational tools
We designed, developed and integrated a set of tools that facilitate the data processing and analysis tasks in drug combination screening research. A user needs to upload an input file that should contain information about the compounds and the cell lines, including names, concentrations and drug effects in the unit of percentage of inhibition (% inhibition) of cancer cells. Furthermore, a unique identifier, termed block id, is needed to differentiate the same drug combination—cell line pair that has been repeated in multiple experiments. The output of the web server consists of sensitivity and synergy scores that are summarized in a table which can be further linked to more detailed graphical results. For example, the drug combination sensitivity score (CSS) is determined as the average area under curve (AUC) for the combinations’ dose–response with one compound fixed at the IC50 concentration (In press, doi:10.1371/journal.pcbi.1006752). CSS summarizes the dose–responses of a drug combination using a metric of % inhibition, which could then be readily compared to its monotherapy drug responses. The difference between CSS and the sum of AUCs of the monotherapy dose–response curves, termed as S score, is used to evaluate the synergy of a drug combination at their IC50 concentrations. To assess the degree of drug-drug interactions over the full dose–response matrix, we provided reference models to determine the expected effect of non-interaction. Currently four commonly-used reference models were utilized, including Bliss independence (BLISS), Highest single agent (HSA), Loewe additivity (LOEWE), and Zero interaction potency (ZIP) (20–22). Depending on whether the drug combination response is greater, identical or less than what is predicted by a reference model, we may classify the drug combination at a specific dose level as synergistic, additive or antagonistic respectively (23). As these four reference models are based on a distinctive set of empirical or biological assumptions, which might lead to different quantification of the degree of interaction, we therefore provided the results of all of them for users’ discretion (24). However, we recommend that only if a drug combination that achieves a higher synergy score in all the models (i.e. S, BLISS, HSA, LOEWE, ZIP) as well as a higher sensitivity score (CSS) should be prioritized for deeper validations.
Web server implementation
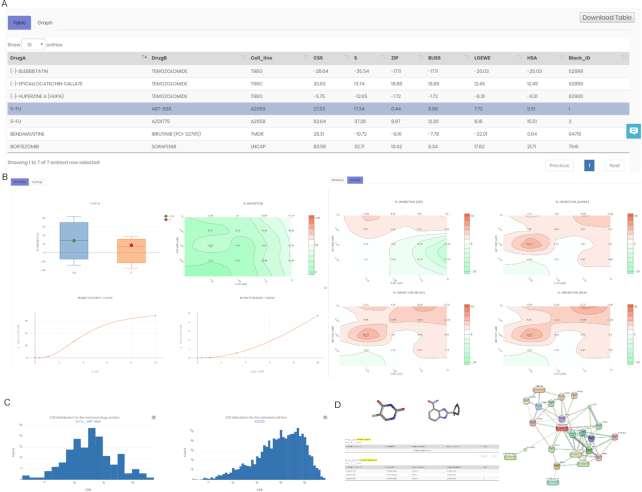
To start the DrugComb data analysis pipeline, a comma-separated values (csv) file compliant with a specific format needs to be uploaded. The input file must contain information about cell line names, drug names, concentrations and drug combination responses measured in the unit of % inhibition. A template file is provided in the Analysis page to facilitate the preparation of input data. The web server will produce the data analysis results in two panels: Table and Graph (Figure 2A and B). The Table panel is the default display which provides summary information about the sensitivity and synergy scores for the drug combination-cell line pairs. The graphical results are displayed under the Graph panel, which can be activated after selecting a drug combination in the Table panel. This Graph panel contains two tabs including Sensitivity and Synergy. The Sensitivity tab provides the results on drug combination sensitivity, including the CSS-S box plots, dose–response matrix in the unit of % inhibition, as well as monotherapy dose–response curves. The Synergy tab contains drug combination synergy landscapes over the dose matrix, determined by the four reference models explained earlier. The computational engine of the web server is extended from the R package synergyfinder (25), while the details on the analytical methods can be found in online documentation.
Figure 2.
Examples of the DrugComb analysis results. (A) The Table view summarizes the web server results for a selected set of drug combinations, including the 5-FU (fluorouracil) and ABT-888 (veliparib) combination in the A2058 cell line (melanoma). (B) The Graph view shows sensitivity (left panel) and synergy (right panel) of the selected drug combination-cell line pair. Sensitivity panel includes CSS-S boxplots as well as the combination dose–response matrix and monotherapy dose–response curves. Synergy panel shows drug synergy landscapes determined using the ZIP, BLISS, LOEWE and HSA reference models. (C) Histograms of drug combination sensitivity scores (CSS) of 5-FU and ABT-888 combination across all the cell lines (left) and across all drug combinations for the A2058 line (right). (D) Annotation for 5-FU and ABT-888 about their chemical structures, drug–target profiles and protein–protein interaction networks obtained from PubChem, ChEMBL and STITCH databases.
Database content
DrugComb aims at a free access to standardized drug combination screening results. Utilizing the computational tools that are available in the web server, we managed to collect and curate high-throughput drug combination screen data involving 2276 drugs tested in 437 932 combinations for 93 cancer cell lines from 10 different tissues. The sources of the data include: i) The NCI ALMANAC dataset (26), ii) The ONEIL dataset (27), iii) The FORCINA dataset (28) and iv) The CLOUD dataset (29) (Table 1). To make the datasets comparable, we standardized the % viability values, determined as the ratio between the counts for cells treated with drugs and cells treated with DMSO as negative control, measured at the end time point. The drug effects were then represented as % inhibition, defined as 100 – % viability. The data curation aims to determine a full dose–response matrix where the monotherapy and combination doses were matched. More specifically, in the ALMANAC dataset screenings have been performed in two stages. In the first stage drugs were screened in single doses on the full NCI60 cell panel to efficiently capture compounds with anti-proliferative activity. Compounds with above-threshold effects were subsequently screened in the drug combination stage, for which two different screening protocols were utilized, resulted in full dose–response matrices of 6 × 4 and 4 × 4 sizes. For the ONEIL dataset the cell viability was measured as the ratio of the exponential growth rate for cells treated with a drug versus DMSO. The experiment was designed so that the monotherapy and the drug combinations were tested separately. However, the concentrations that were tested in the monotherapy screen were not identical to those in the combination screen. We thus utilized the four-parameter log-logistic model, available in the R drc package (30), to estimate the monotherapy responses at the concentrations tested in the combination screen. For the Forcina dataset, the % viability values were determined using the cell counts at the time of 96 h, even though the data for other intermediate time points were also available. For the CLOUD dataset, we fitted a 4-parameter log-logistic model similar for the ONEIL dataset to estimate the % inhibition values for those drug combinations for which the single drug effects were not reported.
Table 1.
The data statistics of the studies curated in DrugComb
| Study | Number of drugs | Number of drug combinations | Number of cell lines | Number of tissues | Size of the full dose–response matrix |
|---|---|---|---|---|---|
| ALMANAC | 103 | 303 737 | 60 | 10 | 4 × 4 or 6 × 4 |
| ONEIL | 38 | 92 208 | 39 | 6 | 5 × 5 |
| FORCINA | 1818 | 1818 | 1 | 1 | 2 × 2 |
| CLOUD | 283 | 40 160 | 1 | 1 | 2 × 2 |
The number of drug combinations was counted as one experiment where a drug combination has been tested for a particular cell line. For the ONEIL study, there are 583 unique drug combinations, where all of them have been tested in each of 39 cell lines, and therefore 583 × 39 = 22 737 drug combinations. All the drug combinations have been repeated multiple times including 22 422 drug combinations repeated four times while 315 drug combinations repeated eight times. Therefore, the total number of drug combination experiments sum up to 22 422 × 4 + 315 × 8 = 92 208 drug combinations. All the other studies have not provided the drug combinations that have been replicated on the exactly same concentrations.
For the curated drug combinations, DrugComb reported the analysis results provided by the computational tools as described earlier, and also the distributions of CSS scores for a given drug combination and a given cell line (Figure 2C). In addition, multiple views on their annotations from third-party databases were also made directly available under the Annotation panel (Figure 2D). For example, STITCH can provide a network-centric view on the drug–target interactions for a drug combination, while ChEMBL and PubChem can provide the most up-to-date information on their potential mechanisms of actions and signaling pathways. Information shown in the Annotation panel should allow for further exploration of the mechanisms of action for a selected drug combination, which can be further validated using experimental techniques, such as CRISPR-Cas9 or RNAi genetic screens (31,32).
We provided flexible query options to navigate the repository of harmonized drug combination data and their analysis results, which may encourage users to contribute their own screening results, thus promoting a community-driven ecosystem for data sharing and redistribution. A data contribution module (https://drugcomb.fimm.fi/contribute/) is therefore provided to allow users to upload their curated datasets for which the reporting of sufficient information on the experimental procedures is mandatory.
DrugComb is built using PHP 7.2.11 for server-side data processing, Javascript ECMAScript 2015 for the frontend and Plotly library 1.40.0 for the generation of the interactive visualizations. Data is stored in MariaDB 10.1.37 with RMariaDB 1.0.6.9000 as the driver for interfacing with R. Software development tools including Python 3.6.7, numpy 1.14.1, pandas 0.23.4, scikit-learn 0.20.2, RDkit 2018.03.4, R version 3.5.1, synergyfinder 1.8.0 and tidyverse 1.2.1 are used in the analytical pipelines. Linux distribution CentOS-7 with the kernel 3.10.0 64-bit running on four processor cores and 64 Gb of RAM is used for hosting the web service on the in-house computational cluster. API-based access to PubChem is performed according to https://pubchemdocs.ncbi.nlm.nih.gov/pug-rest, to STITCH using https://www.stitchdata.com/docs/stitch-connect/api, and ChEMBL using https://www.ebi.ac.uk/chembl/api/data/docs.
CASE STUDIES
Here we present three case studies that have been performed on the curated data in DrugComb. The first case study involved a descriptive analysis of the dataset, where drugs and cell lines were clustered according to their mechanisms of action and tissue of origin. The second case study aimed to analyze the reproducibility of drug combination screen data. This was done via the comparison of the CSS values of replicates found across and within the study sources. The third case study employed linear regression to predict the CSS values using chemical descriptors of the drug molecules, demonstrating the potential of machine learning methods.
Annotations of drugs and cell lines
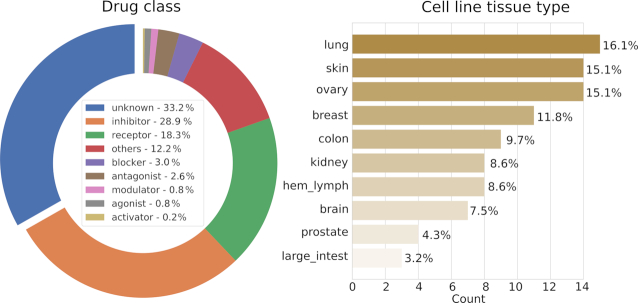
To retrieve the mechanisms of actions of the 2276 drugs in DrugComb, their chemical identifiers were queried from major databases including STITCH, PubCHEM, ChEMBL, DrugBank (33) and KEGG (34). These identifiers were then used for retrieving the pharmacological action information that is available in these databases. We followed the compound classification used in ChEMBL to manually determine the mechanism type, yielding the following categories with their proportions: inhibitor (28.09%), receptor (18.34%), blocker (2.98%), antagonist (2.54%), modulator (0.83%), agonist (0.79%) and activator (0.22%) (Figure 3A). In addition, 12.21% of drugs have been labeled as ‘other’ as their mechanisms of action are not common enough to be placed in new categories. Notably, the remaining 33.22% of drugs do not have well-documented mechanisms of action and hence have been labeled as ‘unknown’. To understand the mechanisms of action of these drug combinations, it becomes imperative to obtain more information on their unannotated constituent compounds. For example, MK-4541 was tested in 5,772 combinations across six cancer tissues, while its pharmacology information remains unknown in those major databases. We did a literature survey and found that MK-4541 has been reported to selectively modulate androgen receptor (AR), acting as an AR agonist (35). Therefore, we expected that more compounds may be annotated similarly by searching the literature which has yet been curated. A more systematic annotation may be achieved via the DrugTargetCommons platform (https://drugtargetcommons.fimm.fi/), where the crowdsourcing efforts are utilized for extracting quantitative bioactivity values of drug–target interactions from the literature (36). For the 93 cancer cell lines, their annotations have been obtained from the Cellosaurus database (37) to determine their tissues of origin. All together 10 distinct tissues were present with lung cancer (16.13%), ovary cancer (15.05%) and skin cancer (15.05%) being the most common ones (Figure 3B). It can be seen that all the major cancer tissue types except for liver and stomach cancers are well represented in DrugComb, and thus demonstrating the general relevance of the existing data.
Figure 3.
Classification of drugs and cell lines and their proportions in DrugComb. Drugs were classified according to the mechanism types, with 33.3% of which (n = 756) do not have well-documented mechanisms of action from major databases. Cell lines were classified according to the tissue of origin. hem_lymph: hematopoietic and lymphoid tissue; large_intest: large intestine.
Reproducibility of drug combination screens
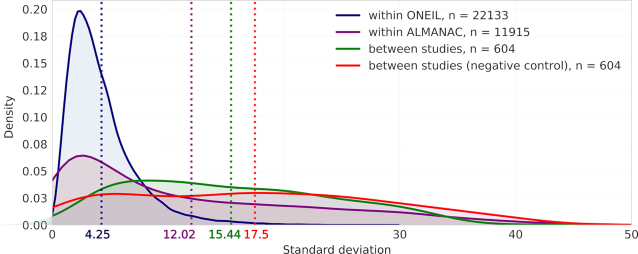
Experimental reproducibility, in particular levels of inter-laboratory concordance in the drug response phenotypes has been reported to be an issue in cancer drug screening (38). Since DrugComb aims to provide standardized results of drug combination screens, assessment of inter- and intra-study data reproducibility is of high importance. The reproducibility was evaluated using standard deviation (sd) of CSS values, which is determined for each unique drug pair and cell line combination. We chose to evaluate the CSS reproducibility as CSS indicates the average % inhibition of a drug combination and therefore makes the replicates comparable even though they were done in different concentrations. For example, Temozolomide and Adm hydrochloride combination has been tested twice in the MALME-3M cell line within the ALMANAC study (denoted as block_id 402838 and 426170 in DrugComb), but their concentrations were different (for 402838, temozolomide has been tested using 1, 10 and 100 μM while in 426170 temozolomide has been tested using 0.2, 2 and 20 μM; Adm hydrochloride has been tested using 0.001, 0.01, 0.1, 1 and 10 μM in 402 838 while using 0.005, 0.05 and 0.5 μM in 426170). These two experiments were still considered as replicates when evaluating the variation of CSS scores. Altogether 34 936 drug-pair-cell-line combinations were replicated, while the majority of them were found either from only within the ONEIL study (n = 22 133) or from only within the ALMANAC study (n = 11 915). In contrast, the number of replicated drug combinations across the ONEIL and the ALMANAC studies is relatively few (n = 604). On the other hand, the drug combinations that were tested in the FORCINA and the CLOUD studies were not replicated, as FORCINA and CLOUD involve single cell lines of T98G and KBM-7 separately, that were not tested elsewhere. The average sd for within-study replicates is 4.25 and 12.02 for ONEIL and ALMANAC respectively, both of which are smaller than that (average sd 15.44) for their between-study replicates (P < 10−30, Wilcoxon rank-sum test, Figure 4). The higher reproducibility of ONEIL compared to ALMANAC is expected, as the ONEIL study consisted of a standardized experiment design that involves only technical replicates while the ALMANAC study collected data from multiple labs that differed in their experimental designs, and therefore may be confounded by multiple factors or batch effects (Table 1). On the other hand, for each of the n = 604 drug-pair-cell-line combinations that were replicated between ONEIL and ALMANAC, we fixed the drug-pair and picked up randomly one cell line from ONEIL and one cell line from ALMANAC, and considered the sd of the CSS values as the negative control for the between-study reproducibility. The average sd for such ‘negative control’ replicates is 17.5 which is significantly higher (P < 10−4, Wilcoxon signed-rank paired test), suggesting a satisfactory reproducibility of the between-study replicates (Figure 4).
Figure 4.
Replicability of drug combinations between and within studies represented as the distribution of the standard deviations of the Drug combination sensitivity scores (CSS). Mean standard deviations for each of the kernel density plots are shown under their corresponding dotted lines.
Prediction accuracy of drug combination sensitivity
In this case study we aimed to evaluate the prediction accuracy of machine learning algorithms on the drug combination sensitivity (CSS) data. We considered the fingerprint information of the drug combinations as the predictors and utilized the root mean squared error (RMSE) to evaluate the prediction accuracy. To generate the fingerprint vectors for a drug combination, canonical SMILES for the constituent drugs were obtained from PubChem and then were converted to 2048 fingerprint bits using Rdkit python module (version 2018.03.4), where each bit corresponds to the presence or absence of a particular structural feature. The drug combination fingerprints were generated using the bitwise averaging of the single drug fingerprints (39). More specifically, the presence of a structural feature in both drugs yields 2 in the combination fingerprint, while presence only in one yields 1 and lack in both yields 0. These 3-bit arrays were then used as features in the machine learning algorithms. For each cell line, we fit a linear regression model on the 80% of drug combinations using a nested cross-validation and then test its prediction accuracy on the remaining 20% data. As a control, we utilized an additive model to predict CSS, which is the sum of average %inhibition from the two single drugs. The use of such an additive model was to reflect the baseline prediction assuming that the average %inhibition of a drug combination is simply the sum of their individual drug effects.
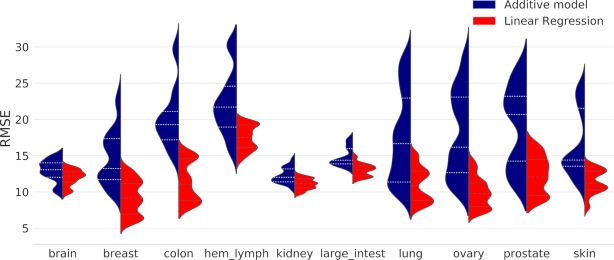
As shown in Figure 5, we found that the prediction accuracy is higher for the linear regression model than the additive model across all the tissue types, suggesting that the drug combination fingerprints carry predictive features for explaining the sensitivity. However, all the tissues exhibited multi-modality in the distribution of RMSE, suggesting that the prediction accuracies varied across different cell lines and drug combinations. As a future step more advanced non-linear machine learning methods such as deep learning may be tested (40). Furthermore, molecular information of the cell lines may worth exploring for the discovery of predictive biomarkers for drug combinations.
Figure 5.
Performance of predicting CSS using linear regression as compared to the additive model. The RMSE for each cell line was grouped as according to its tissue type. Dashed lines within each density plot indicate interquartile range.
COMPARISON TO EXISTING DATA PORTALS
To the best of our knowledge, the existing data portals that cover partially drug combination screen data analysis and collection included DeepSynergy (http://shiny.bioinf.jku.at/DeepSynergy/), DrugCombdb (http://drugcombdb.denglab.org) (unpublished, https://www.biorxiv.org/content/10.1101/477547v2) and SynergyFinder (https://synergyfinder.fimm.fi/) (41). DeepSynergy provides a deep learning machine learning model that was trained on the ONEIL data and has been shown to predict new drug combinations with superior accuracy compared to conventional machine learning approaches. However, DeepSynergy did not provide the web service for the sensitivity and synergy analyses of the drug combination screen data. Furthermore, the deep learning model was trained only with the ONEIL dataset, and thus may become suboptimal when predicting a drug combination in an untested cell line. DrugCombdb is a database that harbors the concurrent screening data for 105k drug combinations. While the dataset has been collected via deep curation, it has not been analyzed with the drug combination sensitivity and synergy tools either. Therefore, both DeepSynergy and DrugCombdb provided limited web-server functionality to analyze drug combination screen data. In contrast, DrugComb provided the web-server that builds on our recent informatics approaches to assess both the sensitivity and synergy level of drug combinations, and therefore may potentially help the interpretations of the DrugCombdb data as well as contributing to the training data that is needed for DeepSyerngy and other advanced machine learning models. SynergyFinder is our recent web application for the drug combination screen data analysis. However, the focus of SynergyFinder is to analyze the degree of interactions in a drug combination screen, while the functionality of analyzing the sensitivity of drug combinations is missing. Furthermore, SynergyFinder does not provide the data curation and annotation functionality. In contrast, DrugComb provides the functionality of both a web-server and a database that have become integral components for establishing a major portal for drug combination data standardization and harmonization. On the other hand, there exist web servers to predict the side effects of drug-drug interactions including DDI-CPI (42). Therefore, linking DrugComb with DDI-CPI may provide a more comprehensive view about the efficacy and side effects of a given drug combination. Taken together, DrugComb is well positioned to provide complementary resources that can be connected with these existing tools for a more systematic and more community-driven effort for future drug combination prediction and network modelling development (43).
CONCLUSIONS
How to make cancer treatment more personalized and more effective remains one of the grand challenges in the healthcare system. Drug combinations may provide enhanced efficacy to combat the cancer drug resistance and therefore may provide more sustainable treatment options for the patients. To accelerate the discovery of personalized multi-targeted drug combinations, knowledge-bases to curate, annotate and interpret the drug combination screen data are needed. The DrugComb portal provides free-access web server to analyze high-throughput drug combination screen data and thus makes it possible to develop a community-driven data repository that allows for the testing of machine learning algorithms. Future efforts include the collection of molecular profiles for cancer cell lines from the LINCS program (www.lincsproject.org), such that more predictive features may be extracted from the cellular genetic or epigenetic context. This may lead to the identification of biomarkers which can be used to stratify the patients for a rational selection of drug combinations. On the other hand, the curated drug combination screen data may also help define more accurate cancer cell dependency models that are being developed at Cell Model Passports (44) and DepMap (https://depmap.org). Furthermore, efficient statistical methods need to be developed for evaluating the significance of drug combination experimental data, which shall demonstrate that the drug combination predictions can be reliably translated into treatment suggestions. With the data analysis and data contribution tools that are made freely available in DrugComb, we encourage more cancer researchers to participate the crowdsourcing efforts of drug combination data generation and harmonization. In the long run, we envisage DrugComb to be a major portal to provide widely applicable informatics tools to predict, test and understand drug combinations, not only for cancer cell lines but also for patient-derived samples, so that it may lead to novel, more effective and safe treatments compared to the current cytotoxic and single-targeted therapies.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the authors of the ALAMANC, ONEIL, FORCINA and CLOUD studies for making their drug combination data fully accessible.
Authors contributions: J.T., B.Z. and J.A. designed the study. J.T. and B.Z. wrote the manuscript. J.A. engineered the web server. B.Z., S.Z., W.W., W.W., Y.W., J.S., A.M., A.P. performed data analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Research Council (ERC) starting grant DrugComb (Informatics approaches for the rationale selection of personalized cancer drug combinations) [No. 716063]; European Commission H2020 EOSC-life (Providing an open collaborative space for digital biology in Europe [No. 824087]; Academy of Finland Research Fellow grant [No. 317680]; China Scholarship Council grant [No. 201706740080]; Finland's EDUFI Fellowship [No. TM-18-10928]. Funding for open access charge: European Research Council (ERC) starting grant agreement DrugComb [No. 716063].
Conflict of interest statement. None declared.
REFERENCES
- 1. Lawrence M.S., Stojanov P., Mermel C.H., Robinson J.T., Garraway L.A., Golub T.R., Meyerson M., Gabriel S.B., Lander E.S., Getz G.. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014; 505:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gottesman M.M., Lavi O., Hall M.D., Gillet J.P.. Toward a better understanding of the complexity of cancer drug resistance. Annu. Rev. Pharmacol. Toxicol. 2016; 56:85–102. [DOI] [PubMed] [Google Scholar]
- 3. Hanahan D. Rethinking the war on cancer. Lancet. 2014; 383:558–563. [DOI] [PubMed] [Google Scholar]
- 4. Crystal A.S., Shaw A.T., Sequist L.V., Friboulet L., Niederst M.J., Lockerman E.L., Frias R.L., Gainor J.F., Amzallag A., Greninger P. et al.. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014; 346:1480–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tang J., Aittokallio T.. Network pharmacology strategies toward multi-target anticancer therapies: from computational models to experimental design principles. Curr. Pharm. Des. 2014; 20:23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lord C.J., Tutt A.N., Ashworth A.. Synthetic lethality and cancer therapy: lessons learned from the development of PARP inhibitors. Annu. Rev. Med. 2015; 66:455–470. [DOI] [PubMed] [Google Scholar]
- 7. Cheng F., Kovács I.A., Barabási A.-L.. Network-based prediction of drug combinations. Nat. Commun. 2019; 10:1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tang J. Informatics approaches for predicting, understanding, and testing cancer drug combinations. Methods Mol. Biol. 2017; 1636:485–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Scarlett U.K., Chang D.C., Murtagh T.J., Flaherty K.T.. High-throughput testing of novel-novel combination therapies for cancer: an idea whose time has come. Cancer Discov. 2016; 6:956–962. [DOI] [PubMed] [Google Scholar]
- 10. Szklarczyk D., Santos A., von Mering C., Jensen L.J., Bork P., Kuhn M.. STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016; 44:D380–D384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B. et al.. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019; 47:D1102–D1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gaulton A., Hersey A., Nowotka M., Bento A.P., Chambers J., Mendez D., Mutowo P., Atkinson F., Bellis L.J., Cibrian-Uhalte E. et al.. The ChEMBL database in 2017. Nucleic Acids Res. 2017; 45:D945–D954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D. et al.. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012; 483:603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang W., Soares J., Greninger P., Edelman E.J., Lightfoot H., Forbes S., Bindal N., Beare D., Smith J.A., Thompson I.R. et al.. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013; 41:D955–D961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., Boutselakis H., Cole C.G., Creatore C., Dawson E. et al.. COSMIC: The Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2019; 47:D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Seashore-Ludlow B., Rees M.G., Cheah J.H., Cokol M., Price E.V., Coletti M.E., Jones V., Bodycombe N.E., Soule C.K., Gould J. et al.. Harnessing connectivity in a Large-Scale Small-Molecule sensitivity dataset. Cancer Discov. 2015; 5:1210–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li J., Zhao W., Akbani R., Liu W., Ju Z., Ling S., Vellano C.P., Roebuck P., Yu Q., Eterovic A.K. et al.. Characterization of human cancer cell lines by Reverse-phase protein arrays. Cancer Cell. 2017; 31:225–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wilkinson M.D., Dumontier M., Aalbersberg I.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.W., da Silva Santos L.B., Bourne P.E. et al.. The FAIR guiding principles for scientific data management and stewardship. Sci. Data. 2016; 3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leek J.T., Scharpf R.B., Bravo H.C., Simcha D., Langmead B., Johnson W.E., Geman D., Baggerly K., Irizarry R.A.. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 2010; 11:733–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berenbaum M.C. What is synergy. Pharmacol. Rev. 1989; 41:93–141. [PubMed] [Google Scholar]
- 21. Loewe S. The problem of synergism and antagonism of combined drugs. Arzneimittel-Forschung. 1953; 3:285–290. [PubMed] [Google Scholar]
- 22. Yadav B., Wennerberg K., Aittokallio T., Tang J.. Searching for drug synergy in complex Dose-Response landscapes using an interaction potency model. Comput. Struct. Biotechnol. J. 2015; 13:504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tallarida R.J. Quantitative methods for assessing drug synergism. Genes Cancer. 2011; 2:1003–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tang J., Wennerberg K., Aittokallio T.. What is synergy? The saariselka agreement revisited. Front. Pharmacol. 2015; 6:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. He L., Kulesskiy E., Saarela J., Turunen L., Wennerberg K., Aittokallio T., Tang J.. Methods for High-throughput drug combination screening and synergy scoring. Methods Mol. Biol. 2018; 1711:351–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Holbeck S.L., Camalier R., Crowell J.A., Govindharajulu J.P., Hollingshead M., Anderson L.W., Polley E., Rubinstein L., Srivastava A., Wilsker D. et al.. The national cancer institute ALMANAC: a comprehensive screening resource for the detection of anticancer drug pairs with enhanced therapeutic activity. Cancer Res. 2017; 77:3564–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. O’Neil J., Benita Y., Feldman I., Chenard M., Roberts B., Liu Y., Li J., Kral A., Lejnine S., Loboda A. et al.. An unbiased oncology compound screen to identify novel combination strategies. Mol. Cancer Ther. 2016; 15:1155–1162. [DOI] [PubMed] [Google Scholar]
- 28. Forcina G.C., Conlon M., Wells A., Cao J.Y., Dixon S.J.. Systematic quantification of population cell death kinetics in mammalian cells. Cell Syst. 2017; 4:600–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Licciardello M.P., Ringler A., Markt P., Klepsch F., Lardeau C.H., Sdelci S., Schirghuber E., Muller A.C., Caldera M., Wagner A. et al.. A combinatorial screen of the CLOUD uncovers a synergy targeting the androgen receptor. Nat. Chem. Biol. 2017; 13:771–778. [DOI] [PubMed] [Google Scholar]
- 30. Ritz C., Baty F., Streibig J.C., Gerhard D.. Dose-Response analysis using R. PLoS One. 2015; 10:e0146021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Han K., Jeng E.E., Hess G.T., Morgens D.W., Li A., Bassik M.C.. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat. Biotechnol. 2017; 35:463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boettcher M., Tian R., Blau J.A., Markegard E., Wagner R.T., Wu D., Mo X., Biton A., Zaitlen N., Fu H. et al.. Dual gene activation and knockout screen reveals directional dependencies in genetic networks. Nat. Biotechnol. 2018; 36:170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z. et al.. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018; 46:D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kanehisa M., Sato Y., Furumichi M., Morishima K., Tanabe M.. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019; 47:D590–D595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chisamore M.J., Gentile M.A., Dillon G.M., Baran M., Gambone C., Riley S., Schmidt A., Flores O., Wilkinson H., Alves S.E.. A novel selective androgen receptor modulator (SARM) MK-4541 exerts anti-androgenic activity in the prostate cancer xenograft R-3327G and anabolic activity on skeletal muscle mass & function in castrated mice. J. Steroid Biochem. Mol. Biol. 2016; 163:88–97. [DOI] [PubMed] [Google Scholar]
- 36. Tang J., Tanoli Z.U., Ravikumar B., Alam Z., Rebane A., Vaha-Koskela M., Peddinti G., van Adrichem A.J., Wakkinen J., Jaiswal A. et al.. Drug target commons: A community effort to build a consensus knowledge base for Drug-Target interactions. Cell Chem. Biol. 2018; 25:224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bairoch A. The cellosaurus, a cell-line knowledge resource. J. Biomol. Tech. 2018; 29:25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hatzis C., Bedard P.L., Birkbak N.J., Beck A.H., Aerts H.J., Stem D.F., Shi L., Clarke R., Quackenbush J., Haibe-Kains B.. Enhancing reproducibility in cancer drug screening: how do we move forward. Cancer Res. 2014; 74:4016–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mason D.J., Stott I., Ashenden S., Weinstein Z.B., Karakoc I., Meral S., Kuru N., Bender A., Cokol M.. Prediction of antibiotic interactions using descriptors derived from molecular structure. J. Med. Chem. 2017; 60:3902–3912. [DOI] [PubMed] [Google Scholar]
- 40. Preuer K., Lewis R.P.I., Hochreiter S., Bender A., Bulusu K.C., Klambauer G.. DeepSynergy: predicting anti-cancer drug synergy with Deep Learning. Bioinformatics. 2018; 34:1538–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ianevski A., He L., Aittokallio T., Tang J.. SynergyFinder: a web application for analyzing drug combination dose–response matrix data. Bioinformatics. 2017; 33:2413–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Luo H., Zhang P., Huang H., Huang J., Kao E., Shi L., He L., Yang L.. DDI-CPI, a server that predicts drug-drug interactions through implementing the chemical-protein interactome. Nucleic Acids Res. 2014; 42:W46–W52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cheng F., Kovacs I.A., Barabasi A.L.. Network-based prediction of drug combinations. Nat. Commun. 2019; 10:1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. van der Meer D., Barthorpe S., Yang W., Lightfoot H., Hall C., Gilbert J., Francies H.E., Garnett M.J.. Cell model Passports—a hub for clinical, genetic and functional datasets of preclinical cancer models. Nucleic Acids Res. 2019; 47:D923–D929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.