Abstract
Understanding the evolutionary background of a bacterial isolate has applications for a wide range of research. However generating an accurate species phylogeny remains challenging. Reliance on 16S rDNA for species identification currently remains popular. Unfortunately, this widespread method suffers from low resolution at the species level due to high sequence conservation. Currently, there is now a wealth of genomic data that can be used to yield more accurate species designations via modern phylogenetic methods and multiple genetic loci. However, these often require extensive expertise and time. The Automated Multi-Locus Species Tree (autoMLST) was thus developed to provide a rapid ‘one-click’ pipeline to simplify this workflow at: https://automlst.ziemertlab.com. This server utilizes Multi-Locus Sequence Analysis (MLSA) to produce high-resolution species trees; this does not preform multi-locus sequence typing (MLST), a related classification method. The resulting phylogenetic tree also includes helpful annotations, such as species clade designations and secondary metabolite counts to aid natural product prospecting. Distinct from currently available web-interfaces, autoMLST can automate selection of reference genomes and out-group organisms based on one or more query genomes. This enables a wide range of researchers to perform rigorous phylogenetic analyses more rapidly compared to manual MLSA workflows.
INTRODUCTION
Identifying an unknown bacterial isolate is not only a necessity for academic classification but is an integral piece of data for a variety of research. This information helps guide growth requirements, downstream comparative analysis, and understanding a specific phenotype in context. For drug discovery efforts, this is especially useful, as secondary metabolite potentials are enriched in certain phyla, with differences seen down to the species level (1). Species delineation remains a challenge however due to factors such as horizontal gene transfer (HGT), homologous recombination, and incomplete lineage sorting. Genome based methods have historically served as a powerful tool to discriminate species with the use of DNA-DNA hybridization methods (DDH). Currently this method has been largely supplanted by genomic sequencing of conserved areas, such as the 16S ribosomal DNA sequences present in all bacteria (2). Thanks to cheap sequencing and rapid processing using tools such as BLAST, 16S sequence analysis has been the workhorse of identifying bacterial isolates (3–6). Unfortunately, complications such as using partial 16S sequences (7) or multiple variants (8) can be a source of misleading designations. This highly conserved sequence may also result in ambiguous designations due to similar sequence similarity within genera (9). In light of this, additional similarity methods using whole genome data, such as Average Nucleotide Identity (ANI) (10) or in silico DDH (11), have helped to delineate species. These both provide a summary score for the degree and extent of homology between two genomes. Additionally, morphological and chemical data remains an important step in defining a type strain - an isolate that represents a particular species; however this solution is unsuitable for high-throughput classification.
One issue with similarity-based approaches is that it is difficult to interpret when no close relative exists in current databases. A solution to this problem is to model evolutionary history using phylogenetic methods. Initial implementations include similarity based tree construction using Neighbor-Joining (NJ) (12) or rapid k-mer approaches such as CVTree3 (13,14); however these do not take into account parameters of evolution such as higher rates of transitions compared with transversions. Computationally rigorous character-based approaches, e.g. maximum-likelihood, are alternatives that include these evolutionary parameters and often yield more accurate results over similarity based approaches (15). Unfortunately, the variety of processing techniques discourages widespread use as best practices are not immediately apparent to non-specialists. Recently this barrier to use is being reduced through accessible web interfaces that utilize the computationally expensive maximum-likelihood approaches, such as IQ-TREE (16,17) and RaxML (18). Additional measures such as model finding, included in IQ-TREE (19), ensure higher confidence in evolutionary reconstruction as the choice of model can give varied results (20). These advancements provide a more rigorous analysis over similarity methods, however this process can often be insufficient in delineating species splits using 16S data alone due to limited phylogenetic signal in the highly conserved sequence.
A solution to this issue is the use of Multi-Locus Sequence Analysis (MLSA)—a technique that integrates many genomic loci to increase phylogenetic signal. By analyzing many conserved genes, often including 16S data, a higher resolution species tree can be inferred (21). The choice of genomic loci is important however, as many considerations can impair accurate estimation (22). For example, limiting to genes unlikely to be horizontally transferred is an important consideration. Criteria such as using single copy ubiquitous housekeeping genes and low evolutionary selection pressures have shown to help focus on those with low phylogenetic noise (23). Another option is the use of whole genome phylogenies, which carries the risk of including genes with conflicting phylogenetic signal. Unfortunately, these advantages come with the cost of computationally expensive workflows with an esoteric set of options and processing steps. Even the seemingly trivial selection of appropriate genomes to include can be a source of error; for example, selecting an inappropriate out-group organism will lead to misleading ancestral splits (24). The choice of genomes will also impair gene selection, which may require timely curation to identify appropriate single copy genes. Other important downstream analyses, such as proper partitioning of alignments before tree inference (25), may also lead to conflicting results and tree topologies.
To help remove these issues we created The Automated Multi-Locus Species Tree (autoMLST), a free to use webserver for generating high-resolution species trees. Unlike currently available pipelines: EDGAR (26), Phylogeny.fr (27) and GTDB (28), autoMLST automates all steps in the process including organism and gene selection, offers de novo construction of maximum-likelihood trees, and includes useful features such as model finding and tree annotations. Average Nucleotide Identity (ANI) estimates are also provided and overlaid on the resulting tree using MASH (29) to help delineate species boundaries and final tree interpretation. To aid in the important application of natural product drug discovery, autoMLST includes additional visualizations of secondary metabolite potential so a quick assessment can be made on which isolates to focus on. Other options such as bootstrap analysis and gene tree consistency filtering are also included. One such option is the use of coalescent theory to infer species trees. In addition to helping to corroborate an evolutionary hypothesis, this can be beneficial for recent or rapidly diverging lineages (30,31). In short, the server aims for an accessible ‘BLAST-like’ workflow to obtain a rapid high-resolution species tree and to identify closely related reference genomes.
METHODS AND IMPLEMENTATION
Workflow and inputs
Two provided pipelines for phylogenetic inference in autoMLST are available: ‘placement mode’, which leverages pre-analyzed gene trees, and ‘de novo mode’, which automates Maximum-likelihood tree generation from scratch (Figure 1). Up to 20 simultaneous genomes in Fasta, EMBL or Genbank formats are used as input to the server; alternatively NCBI accession numbers can be submitted. Each step is automated by default but can also be manually curated for organism and gene selection. All options and interpretation of output results can be seen from the help section at: https://automlst.ziemertlab.com/help
Figure 1.
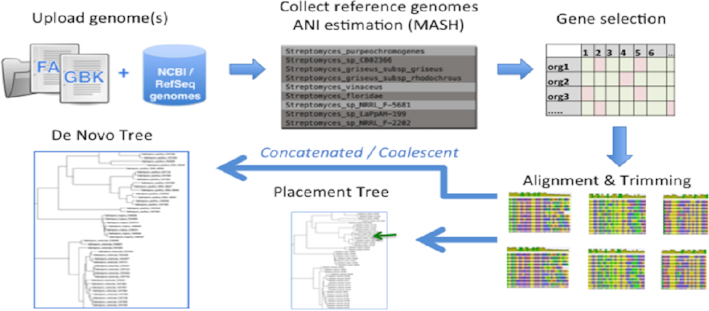
autoMLST workflow depicting placement and de novo mode. Estimated ANI values with reference genomes are found which is used for organism selection. This set is then screened for single copy genes present in every genome and prioritized based on MLSA criteria. Multiple sequence alignments are then obtained and trimmed. Final maximum-likelihood inference is calculated depending on the options and mode used.
Reference and genome selection
Reference genomes were obtained from NCBI Refseq (32) in September 2017 and incorporated into a SQL database including taxon metadata. To reduce redundant strains the top ten highest quality genomes were retained for genomes of the same species. This was determined using the most complete ‘assembly level’ and ‘taxid’ metadata. Genomes marked as type strain or reference genome were added and those with ambiguous genus designations were removed.
Using the MASH ANI estimator all query genomes are compared to the collected database such that a total of 50 reference and query organisms are used. Reference genomes are then selected by allotting half of the open positions to genomes with the nearest average to the entire query set with the other half devoted to references nearest to individual queries. This results in a tree balanced with informative taxa spanning evolutionary gaps in queries. Type strains are given priority by allowing for higher distances (∼5% ANI) over non-type strains. For the placement mode workflow, some of these genomes were used to produce reference alignments and gene trees. A total of 128 families were found to have over 10 type strain genomes, ranging from 11 to 313 members, which were then used to build each of the family specific reference sets.
Gene selection
Searches for gene homologs are preformed using HMMER (33) and essential gene models. These models were collected from Pfam (34) and ‘equivologs’, orthologous genes with confirmed conserved functions, from TIGRFAM (35). A list of these models can be found in Supplemental S1. These searches are added to a matrix of pre-identified homologs present in reference organisms, which is then screened to identify all single copy homologs; Genes that pass bit-score trusted cutoffs of each model and show over 50% coverage of both model and query are added. This list is further prioritized to focus on genes with stronger purifying selection using pre-calculated dN/dS values and a maximum of 100 genes are selected for downstream analysis. The Dn/Ds values are averaged from codon alignments of reference organisms using Pal2Nal (36) and the PAML (37) application ‘yn00’. An optional filtering step is also provided, which discriminates genes with larger median pairwise Robinson–Foulds (RF) distances to all guide trees before preforming species inference.
Alignment and tree construction
Placement mode leverages pre-built DNA alignments of all selected single copy genes and their subsequent trees which are combined using ASTRAL-III (38) to infer species trees. Gene tree placement is done via the evolutionary placement algorithm (EPA) in RAxML (39) using alignments that have query organisms added with MAFFT (40). By default the rapid ‘FFT-NS-2’ alignment is used by both placement and de novo modes; this can optionally run in local iterative mode for improved accuracy. All alignments are then trimmed using trimAl (41) using the ‘automated1’ setting. DNA alignments are likewise produced using MAFFT for de novo mode and extra options for bootstrap analysis and model finding are provided via IQ-TREE (16,42); this is also used to infer the final species tree via a partitioned concatenated alignment of selected genes. Alternatively, the coalescent pipeline can be applied in de novo mode which will construct all gene trees with IQ-TREE before inferring a final species tree with ASTRAL-III.
Additional tree annotations
The Biosynthetic Gene Cluster (BGC) coloring scheme illustrates conservative counts of secondary metabolite potential taken form an antiSMASH v4 (43) analysis of all reference genomes in the database. BGCs found on contig edges were given a count of 0.5 to avoid overestimation due to those found on separated contigs. Five bins were then defined for all counts with respect to various BGC types. These were centered on the mean of non-zero counts from all reference organisms with one standard deviation as the width. Annotations for genome size and percent GC were also added. These are taken from NCBI’s prokaryotic summary files and eight bins were selected to produce a histogram of relatively even amplitudes.
ANI clans and validation
Groups of organisms with closely matching ANI values, ‘clans’, were based on pairwise MASH distances of all reference genomes. All distances at various thresholds were used as input for Markov clustering using the MCL application (44) to assign unique clan IDs. These were done at 97%, 95% and 90% ANI similarity thresholds such that groups above these values were clustered. These groupings were also used to validate generated trees by checking if related genomes clade together on tree branches; this was done by using the Environment for Tree Exploration (ETE3) python library (45) to identify the largest monophyletic group (strictly homogeneous) for each ANI clan. The proportion of maximum monophyletic members to the total was then used to assess tree clades; a score of 100% would be given if all members appear in one branch with no other genomes included. This is done for every non-singleton ANI clan and the average is reported for each tree tested at various ANI clan definitions. Two additional validations were also performed as detailed in the supplemental. Finally a comparison to a manual high-resolution phylogeny (46) was performed using the default de novo mode.
RESULTS
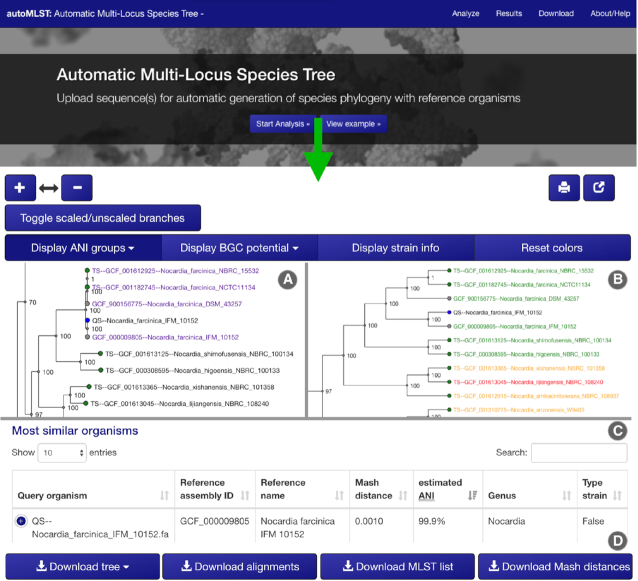
Here we introduce autoMLST a user-friendly, rapid web tool to delineate bacterial species based on genomic data from multiple loci (Figure 2). The server is publicly available at automlst.ziemertlab.com with no login requirement. From the start page you can easily reach the intuitive analysis panel and begin by simply uploading up to 20 bacterial genomes; Each genome is represented by exactly one file in single or multi-record FASTA/EMBL/GenBank format. The pipeline is fully automated by default but can optionally guide users through custom organism or gene selections before processing the MLSA by selecting the appropriate options.
Figure 2.
Tree visualization provides options to toggle branch lengths, zoom, search and color the final tree. (A) ANI group coloring. (B) Secondary metabolite coloring. (C) Sortable table of ANI values with search function. (C) Export functions to download trees, alignments, and supporting information.
Depending on chosen options, performance results showed manageable runtimes between 4 and 5 min for the default de novo workflow allowing for approximately 500 daily submissions on one server. Roughly, 4× more time is needed when using model finding and bootstrap analysis, whereas placement mode showed average runtimes of less than a minute.
After processing, the generated species trees are presented with a set of useful annotation and export functions to help explore the results (Figure 2). For example, type strains and query organisms are highlighted and ANI ‘clans’ are directly labeled on the tree to identify species boundaries. A special application for the natural product community includes the estimation of BGC diversity from antiSMASH analysis. Additionally, a reanalyze button in the final results allows for manual curation options with greater ease, e.g. for removal of organisms in the set that might be problematic. All code for the webserver and workflow scripts are open source and available at: https://bitbucket.org/ziemertlab/automlst if extra throughput is required.
Tree validation
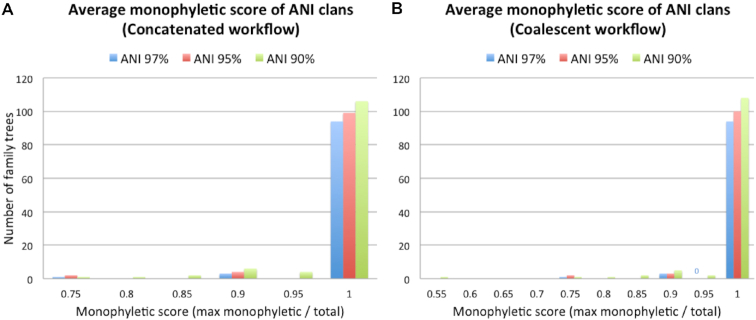
Multiple validation steps were taken in order to assess the quality and accuracy of generated phylogenetic trees. First, scoring of family trees via ANI clan definitions showed the vast majority of trees, over 90%, had perfect grouping of ANI clans into monophyletic clades for all grouping thresholds (Figure 3). Similar results were seen for the coalescent workflow with the exception of one tree showing an average score <0.75. Some trees could not be scored as they only formed singleton ANI clans, which are not considered as this inflates average scoring; therefore further validation was performed using bootstrap analysis (Supplemental S2) and branch length to ANI distance correlation (Supplemental S3).
Figure 3.
Histograms of monophyletic scoring of ANI clans at three thresholds: 97%, 95% and 90% ANI. (A) Concatenated workflow. (B) Coalescent workflow.
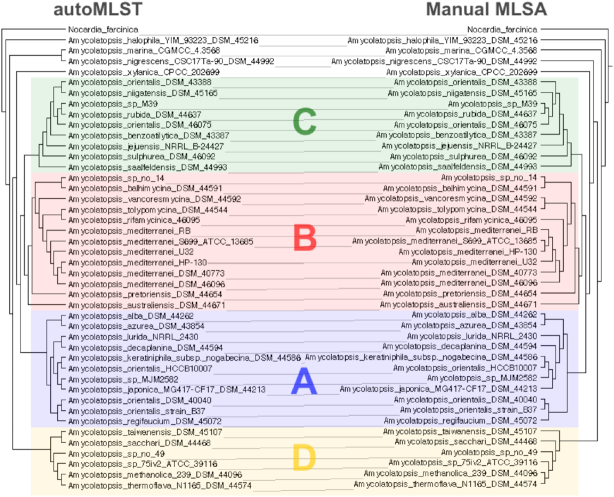
Furthermore, we compared an automated Amycolatopsis tree with a previously defined MLSA from Adamek et al. (46). This was compared to one generated with autoMLST using default parameters in de novo mode and was found to contain all major clade definitions with subtle differences in topology (Figure 4). Of these differences, variations in deep ancestry were seen in addition to strain level ambiguities; Mainly these occur in areas of lower bootstrap support and indicate uncertainty using either method (Supplementary Figures S4 and S5). These differences are likely the result of autoMLST using 85 genes compared to seven selected in the manual procedure. Notably, the automated gene selection overlapped with five of the manually selected genes as well as 19 genes commonly used in the pubMLST database (47)—a resource for sequence typing that uses well characterized marker genes. A consequence of the larger gene selection is fewer polytomies (unresolved bifurcation) were seen in the autoMLST tree, e.g. A. mediterranei clade (Supplemental S6). Despite these minor difference in difficult to resolve evolutionary splits, autoMLST was able to highlight all major sub-clades in a fraction of the hands on time of the Manual workflow.
Figure 4.
Comparison of trees generated automatically with autoMLST (left) a manual MLSA (right) provided by Dr Adamek (46). Groups defined in this study are indicated using the same color scheme and labels as in Adamek et al. Comparison was made using the tanglegram algorithm in dendroscope (51). Further details can be seen in the Supplementary Figures S4 and S5.
DISCUSSION AND CONCLUSIONS
As bacterial species definitions remain a challenge, with known misnomers and ambiguous assignment due to human error (48), it is important to maintain a rigorous procedure for processing newly sequenced genomes. With the expected rise of data, and eventual maturation of meta-genome assembled genomes (MAGs) into high-quality draft genomes, it is equally important to have rapid and accessible procedures to process them. While 16S classification has largely been a practical solution to taxonomic profiling it can have the disadvantage of low resolution for closely related species. Classification via ANI is becoming a popular proposal to solve the taxonomy difficulties for prokaryotes (49), however these similarity measures alone may have trouble resolving closely related strains compared to character-based methods. A viable alternative is the use of MLSA methods that can leverage several evolutionary markers from a simple draft genome.
One of the main motivations for designing this tool is to not only make these methods more accessible but also reduce the hand on time so that many alternate approaches and datasets can be explored. We also aimed to provide helpful annotations for specific applications, one of which is an active use case in our lab for natural product prospecting. These methods are especially important when intra-genus or intra-species differences are under consideration, e.g. distinguishing promising organisms within a genus or species for drug discovery (46). Thus, we have incorporated counts of various BGC types of interest as an initial heuristic to assess query organism potential by adding this coloring scheme directly on the resulting tree. Future efforts aim to expand on these visualizations by illustrating overlap of secondary metabolite potential using gene cluster networking approaches such as BiG-SCAPE (50) so that product diversity can also be estimated. This can potentially highlight clades with high diversity of clusters despite low absolute counts. We have added other additional properties of interest such as genome size and GC content to help show differences between clades. In addition to prioritizing query genomes, the server aims to provide a rapid collection of related species for downstream comparative analysis or heterologous host selection.
autoMLST is shown to be a quick solution to performing these MLSA methods with the ease of current 16S analysis. While having an automated solution is beneficial we also stress the importance of using high quality genomes and performing manual confirmation of an evolutionary hypothesis. Ensuring alignments are free of artifacts via the export functions and comparing various organism and gene sets is an important step, e.g. adding alternate organisms and confirming little impact on original tree topology is seen. Examining branch length variation and provided ANI distance scores against tree topology is another important quality control. This process of retesting is also encouraged via the reanalyze function to allow researchers to test several methods, organisms or gene sets if needed; this can help to eliminate problematic data, such as poor quality draft genomes that may reduce the number of informative genes selected. In short, this server has greatly improved the hands-on time in generating high-resolution species trees and provides several optional processing steps to obtain a more rigorous taxonomic classification.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Daniel Huson, Marnix Medema, Martina Adamek, Daniel Maennle, and Athina Gavriilidou for valued discussions, testing and feedback on autoMLST.
Contributor Information
Mohammad Alanjary, Interfaculty Institute of Microbiology and Infection Medicine Tübingen, University of Tübingen, Tübingen, Germany; German Centre for Infection Research (DZIF), Partner Site Tübingen, Tübingen, Germany.
Kat Steinke, Interfaculty Institute of Microbiology and Infection Medicine Tübingen, University of Tübingen, Tübingen, Germany; German Centre for Infection Research (DZIF), Partner Site Tübingen, Tübingen, Germany.
Nadine Ziemert, Interfaculty Institute of Microbiology and Infection Medicine Tübingen, University of Tübingen, Tübingen, Germany; German Centre for Infection Research (DZIF), Partner Site Tübingen, Tübingen, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Center for Infectious Biology [DZIF 9.704 to N.Z.]. Funding for open access charge: German Center for Infectious Biology [DZIF 9.704].
Conflict of interest statement. None declared.
REFERENCES
- 1. Jensen P.R., Williams P.G., Oh D.-C., Zeigler L., Fenical W. Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl. Environ. Microbiol. 2007; 73:1146–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Woese C.R., Kandler O., Wheelis M.L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. U.S.A. 1990; 87:4576–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 4. Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013; 41:590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006; 72:5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cole J.R., Wang Q., Fish J.A., Chai B., McGarrell D.M., Sun Y., Brown C.T., Porras-Alfaro A., Kuske C.R., Tiedje J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014; 42:633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Peplies J., Kottmann R., Ludwig W., Glöckner F.O. A standard operating procedure for phylogenetic inference (SOPPI) using (rRNA) marker genes. Syst. Appl. Microbiol. 2008; 31:251–257. [DOI] [PubMed] [Google Scholar]
- 8. Louca S., Doebeli M., Parfrey L.W. Correcting for 16S rRNA gene copy numbers in microbiome surveys remains an unsolved problem. Microbiome. 2018; 6:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Conville P.S., Witebsky F.G. Analysis of multiple differing copies of the 16S rRNA Gene in five clinical isolates and three type strains of nocardia species and implications for species assignment. J. Clin. Microbiol. 2007; 45:1146–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Konstantinidis K.T., Tiedje J.M. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:2567–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meier-Kolthoff J.P., Auch A.F., Klenk H.-P., Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013; 14:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987; 4:406–425. [DOI] [PubMed] [Google Scholar]
- 13. Qi J., Luo H., Hao B. CVTree: a phylogenetic tree reconstruction tool based on whole genomes. Nucleic Acids Res. 2004; 32:W45–W47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zuo G., Hao B. CVTree3 Web Server for Whole-genome-based and Alignment-free Prokaryotic Phylogeny and Taxonomy. Genomics Proteomics Bioinformatics. 2015; 13:321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang Z., Rannala B. Molecular phylogenetics: principles and practice. Nat. Rev. 2012; 13:303–314. [DOI] [PubMed] [Google Scholar]
- 16. Nguyen L.T., Schmidt H.A., Von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015; 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Trifinopoulos J., Nguyen L.T., von Haeseler A., Minh B.Q. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016; 44:W232–W235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006; 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- 19. Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., Von Haeseler A., Jermiin L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017; 14:587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ripplinger J., Sullivan J. Does choice in model selection affect maximum likelihood analysis. Syst. Biol. 2008; 57:76–85. [DOI] [PubMed] [Google Scholar]
- 21. Guo Y.P., Zheng W., Rong X.Y., Huang Y. A multilocus phylogeny of the Streptomyces griseus 16S rRNA gene clade: Use of multilocus sequence analysis for streptomycete systematics. Int. J. Syst. Evol. Microbiol. 2008; 58:149–159. [DOI] [PubMed] [Google Scholar]
- 22. Doroghazi J.R., Buckley D.H. Widespread homologous recombination within and between Streptomyces species. ISME J. 2010; 4:1136–1143. [DOI] [PubMed] [Google Scholar]
- 23. Glaeser S.P., Kämpfer P. Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst. Appl. Microbiol. 2015; 38:237–245. [DOI] [PubMed] [Google Scholar]
- 24. de la Torre-Bárcena J.E., Kolokotronis S.-O., Lee E.K., Stevenson D.W., Brenner E.D., Katari M.S., Coruzzi G.M., DeSalle R. The impact of outgroup choice and missing data on major seed plant phylogenetics using genome-wide est data. PLoS One. 2009; 4:e5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kainer D., Lanfear R. The Effects of Partitioning on Phylogenetic Inference. Mol. Biol. Evol. 2015; 32:1611–1627. [DOI] [PubMed] [Google Scholar]
- 26. Blom J., Kreis J., Spanig S., Juhre T., Bertelli C., Ernst C., Goesmann A. EDGAR 2.0: an enhanced software platform for comparative gene content analyses. Nucleic Acids Res. 2016; 44:W22–W28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.-F., Guindon S., Lefort V., Lescot M. et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008; 36:W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Parks D.H., Chuvochina M., Waite D.W., Rinke C., Skarshewski A., Chaumeil P.A., Hugenholtz P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018; 36:996. [DOI] [PubMed] [Google Scholar]
- 29. Ondov B.D., Treangen T.J., Melsted P., Mallonee A.B., Bergman N.H., Koren S., Phillippy A.M. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016; 17:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Simmons M.P., Gatesy J. Coalescence vs. concatenation: Sophisticated analyses vs. first principles applied to rooting the angiosperms. Mol. Phylogenet. Evol. 2015; 91:98–122. [DOI] [PubMed] [Google Scholar]
- 31. Liu L., Yu L., Kubatko L., Pearl D.K., Edwards S. V Coalescent methods for estimating phylogenetic trees. Mol. Phylogenet. Evol. 2009; 53:320–328. [DOI] [PubMed] [Google Scholar]
- 32. Pruitt K.D., Brown G.R., Hiatt S.M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C.M., Hart J., Landrum M.J., McGarvey K.M. et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014; 42:D756–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Finn R.D., Clements J., Eddy S.R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011; 39:W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A. et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016; 44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Haft D.H., Selengut J.D., White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003; 31:371–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Suyama M., Torrents D., Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006; 34:W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007; 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- 38. Zhang C., Rabiee M., Sayyari E., Mirarab S. ASTRAL-III: Polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics. 2018; 19:15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kozlov A.M., Zhang J., Yilmaz P., Glöckner F.O., Stamatakis A. Phylogeny-aware identification and correction of taxonomically mislabeled sequences. Nucleic Acids Res. 2016; 44:5022–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013; 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009; 25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hoang D.T., Chernomor O., Von Haeseler A., Minh B.Q., Vinh L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018; 35:518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Blin K., Wolf T., Chevrette M.G., Lu X., Schwalen C.J., Kautsar S.A., Suarez Duran H.G., de Los Santos E.L.C., Kim H.U., Nave M. et al. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017; 45:W36–W41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. van Dongen S., Abreu-Goodger C. Using MCL to extract clusters from networks. Methods Mol. Biol. 2012; 804:281–295. [DOI] [PubMed] [Google Scholar]
- 45. Huerta-Cepas J., Serra F., Bork P. ETE 3: reconstruction, analysis, and visualization of phylogenomic Data. Mol. Biol. Evol. 2016; 33:1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Adamek M., Alanjary M., Sales-Ortells H., Goodfellow M., Bull A.T., Winkler A., Wibberg D., Kalinowski J., Ziemert N. Comparative genomics reveals phylogenetic distribution patterns of secondary metabolites in Amycolatopsis species. BMC Genomics. 2018; 19:426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jolley K.A., Maiden M.C.J. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010; 11:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Van Belkum A., Welker M., Dunne W.M., Girard V. The infallible microbial identification test: Does it exist. J. Clin. Microbiol. 2015; 53:1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Garrity G.M. A New Genomics-Driven Taxonomy of Bacteria and Archaea: Are We There Yet. J. Clin. Microbiol. 2016; 54:1956–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Navarro-Muñoz J., Selem-Mojica N., Mullowney M., Kautsar S., Tryon J., Parkinson E., De Los Santos E., Yeong M., Cruz-Morales P., Abubucker S. et al. A computational framework for systematic exploration of biosynthetic diversity from large-scale genomic data. 2018; bioRxiv doi: 10.1101/445270, 17 October 2018, preprint: not peer reviewed. [DOI]
- 51. Huson D.H., Scornavacca C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 2012; 61:1061–1067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.