Abstract
The Interactive Tree Of Life (https://itol.embl.de) is an online tool for the display, manipulation and annotation of phylogenetic and other trees. It is freely available and open to everyone. The current version introduces four new dataset types, together with numerous new features. Annotation options have been expanded and new control options added for many display elements. An interactive spreadsheet-like editor has been implemented, providing dataset creation and editing directly in the web interface. Font support has been rewritten with full support for UTF-8 character encoding throughout the user interface. Google Web Fonts are now fully supported in the tree text labels. iTOL v4 is the first tool which supports direct visualization of Qiime 2 trees and associated annotations. The user account system has been streamlined and expanded with new navigation options, and currently handles >700 000 trees from more than 40 000 individual users. Full batch access has been implemented allowing programmatic upload and export of trees and annotations.
INTRODUCTION
Phylogenetic trees are important tools in a wide variety of biological and other scientific fields, also serving as contextualization for various data types (1). This is illustrated by the heavy usage of tools to create such trees (e.g. MEGA, (2)). The visualization of such trees has been covered by various software tools for many years (3,4), including iTOL (5), which introduced the annotation of trees with various types of additional data. Nowadays, various other software packages offer additional tree annotation features, both online and as standalone packages, for example ETE toolkit (6), Dendroscope (7) or Evolview (8). Here, we report on recent developments of iTOL, which have expanded and streamlined its functionality, making it more powerful and easier to use.
BASIC FEATURES
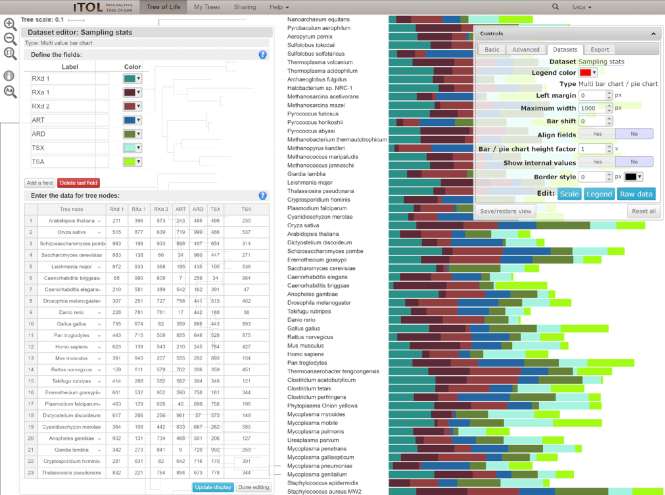
iTOL is an online tool, accessible with any modern web browser (Figure 1). The tree display engine is implemented in pure Javascript and uses the HTML5 Canvas element for visualization.
Figure 1.
iTOL’s user interface. Phylogenetic tree annotated with a multi-value bar chart is displayed, with the interactive dataset editor open. Data is entered through a spreadsheet-like interface. Tree nodes can be easily selected with an autocomplete search interface. Clicking the nodes in the editor zooms and pans the tree to show their exact location. Dataset legends and scales can be similarly edited.
Input types and basic functions
iTOL supports commonly used phylogenetic tree formats: Newick, Nexus (9) and phyloXML (10). Phylogenetic placements files created by EPA (11) and pplacer (12) are also supported. Current version introduces support for QIIME 2 (13) trees and annotation files. QIIME 2 QZA trees (Phylogeny[Rooted] or Phylogeny[Unrooted]) can be uploaded directly, and annotated with the following types of data:
FeatureData[Taxonomy]: Leaf labels will be automatically assigned, and confidence values will appear as a bar chart dataset.
FeatureTable[Frequency]: A multi value bar chart will be created with sample frequencies in different tree leaves.
FeatureData[AlignedSequence]: Multiple sequence alignment dataset will be created automatically.
All additional data used for various types of tree annotation are provided in plain text files, and simply dragged and dropped onto the trees visualized in the user's web browser.
iTOL provides most common functions available in any phylogenetic tree viewer. In addition to standard display formats (rectangular, circular and unrooted), iTOL v4 supports the slanted phylogram display mode. Trees can be manipulated in various ways, and basic editing functions allow users to interactively delete or move single nodes or whole clades. Clades can also be pruned or collapsed, either manually or automatically, based on various parameters (such as associated bootstrap values or average branch length distances). Trees can be re-rooted manually on any node, or automatically using the midpoint rooting method. Tree leaves can be sorted in various ways, either manually or automatically.
A complete overview of features added since the last publication (14) can be accessed through iTOL’s version history page (https://itol.embl.de/version_history.cgi).
ANNOTATING TREES
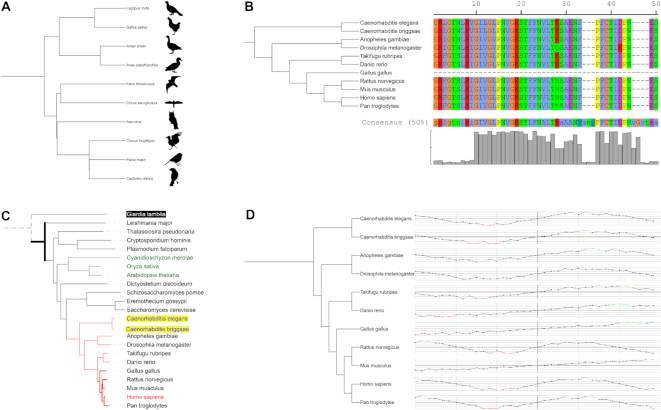
The current version introduces several new annotation features, which expand the user control over the individual display elements, as well as four new dataset types (Figure 2).
Figure 2.
New dataset types in iTOL v4. Each tree in iTOL can be annotated with an unlimited number of datasets. Dataset files are drag-dropped onto the trees displayed in the browser and visualized automatically. (A) Images: External bitmap and SVG images can be linked to any tree node, any individually scaled, rotated and positioned. (B) Multiple sequence alignments: Raw FASTA multiple sequence alignments are supported. Several standard color schemes are available, and custom colors can be defined for each residue. Consensus sequence and the residue conservation graph are calculated automatically. (C) Branch and label styles: colors and styles for tree nodes and labels can now be specified through a separate dataset type. There can be multiple different styles datasets present, allowing users to simply switch between various tree colors and styles. (D) Line charts: Visualized as a series of points, optionally connected with a line. Vertical and horizontal axis lines are supported, as well as automatic coloring of the dots/lines based on user specified value thresholds. Vertical and horizontal display styles are available, both on rectangular and circular trees.
Tree style and colors
iTOL supports individual style and colors for each individual node and label in the tree. The current version greatly expands the support for different fonts and font styles, with full UTF-8 character set support throughout the user interface, and the ability to use any font from the Google Web Fonts list. In addition, background color of any text label can be changed independently now.
Displaying branch length, support and metadata values
iTOL v4 expands the bootstrap visualization options with full support for MRBAYES (15) and The New Hampshire X (NHX) formatted metadata in the tree nodes, as well as multi-value node support values, e.g. as provided by IQ-TREE (16). These metadata values are parsed and available as selectable sources for iTOL’s existing bootstrap display options.
Display of branch lengths and metadata sourced text labels has been improved with additional options for specifying their exact location on the branches, as well as their size and format. Branch lengths and internal tree scales can now be reversed (displayed as ages), i.e. starting with zero at the farthest leaf, and increasing toward the root of the tree.
Datasets and the interactive editor
iTOL currently supports 18 different dataset types, with 4 new types introduced since version 3 (Figure 2). All datasets can be uploaded as plain text files and use simple predefined templates which are available through iTOL’s help pages. A major new feature in iTOL v4 is the interactive editor, which allows users to create and edit the datasets directly in the web user interface (Figure 1). Raw data as well as dataset legends, scales and other information can be edited directly through a spreadsheet-like interface, with dynamically updating tree visualization.
iTOL can now display several dataset types (binary datasets, protein domain architectures, connections, branch symbols and internal pie-charts) in the unrooted tree display mode as well. Even though the individual dataset entries are not aligned in this mode, they still provide users with valuable annotation options.
Many graphical elements in various datasets and tree colored ranges now support user defined borders, with individually changeable colors and widths.
USER ACCOUNTS
In addition to the anonymous direct tree upload and annotation, iTOL provides a freely available personal account system, which currently has more than 40 000 registered users, managing more than 700 000 uploaded trees. The current version brings various updates to the account pages, making them easier to use. Creation and modification times for all trees are tracked and can be used to sort various tree tables. Additional navigation options are now available in the tree display pages, providing shortcuts to other trees in the same user project. Tree information popups have been extended with information on user workspaces and projects, as well as currently used tree views (user saved tree display states).
EXPORT
One of iTOL’s strengths is the creation of high-quality figures for publication or inclusion into other documents. The current version extends the export functions with automatically generated legends for colored ranges, and several dataset types, like heatmaps. Backend server has been extended to make the export of bitmap formats faster.
BATCH ACCESS
iTOL offers programmatic access to both its tree upload and export interfaces. All iTOL features available interactively through the web interface can also be accessed through the batch interface. Complete redesign of iTOL in version 3 introduced ‘What-You-See-Is-What-You-Get’ export capabilities, where the user's browser performs the initial creation of the SVG (Scalable Vector Format) image, making our old batch access system incompatible. The current version solves this problem by using scriptable headless browsers on the server side (phantomjs, http://phantomjs.org). Example upload/download scripts are provided in several programming languages.
DATA AVAILABILITY
The Interactive Tree Of Life is freely available at https://itol.embl.de.
FUNDING
This work has been partially supported by the German Federal Ministry of Education and Research with the project number 031A537B. This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No EC/H2020/ES/ERC-AdG-669830). Funding for open access charge: European Molecular Biology Laboratory.
Conflict of interest statement. None declared.
REFERENCES
- 1. McTavish E.J., Drew B.T., Redelings B., Cranston K.A.. How and why to build a unified tree of life. Bioessays. 2017; 39:1700114–1700122. [DOI] [PubMed] [Google Scholar]
- 2. Kumar S., Stecher G., Tamura K.. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016; 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Page R.D. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996; 12:357–358. [DOI] [PubMed] [Google Scholar]
- 4. Zmasek C.M., Eddy S.R.. ATV: display and manipulation of annotated phylogenetic trees. Bioinformatics. 2001; 17:383–384. [DOI] [PubMed] [Google Scholar]
- 5. Letunic I., Bork P.. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007; 23:127–128. [DOI] [PubMed] [Google Scholar]
- 6. Huerta-Cepas J., Serra F., Bork P.. ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 2016; 33:1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huson D.H., Scornavacca C.. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 2012; 61:1061–1067. [DOI] [PubMed] [Google Scholar]
- 8. He Z., Zhang H., Gao S., Lercher M.J., Chen W.H., Hu S.. Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016; 44:W236–W241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wilgenbusch J.C., Swofford D.. Inferring evolutionary trees with PAUP*. Curr. Protoc. Bioinformatics. 2003; doi:10.1002/0471250953.bi0604s00. [DOI] [PubMed] [Google Scholar]
- 10. Han M.V., Zmasek C.M.. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics. 2009; 10:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Berger S.A., Krompass D., Stamatakis A.. Performance, accuracy, and Web server for evolutionary placement of short sequence reads under maximum likelihood. Syst. Biol. 2011; 60:291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matsen F.A., Kodner R.B., Armbrust E.V.. pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics. 2010; 11:538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lawley B., Tannock G.W.. Analysis of 16S rRNA gene amplicon sequences using the QIIME software package. Methods Mol. Biol. 2017; 1537:153–163. [DOI] [PubMed] [Google Scholar]
- 14. Letunic I., Bork P.. Interactive Tree of Life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016; 44:W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ronquist F., Teslenko M., van der Mark P., Ayres DL., Darling A., Höhna S., Larget B., Liu L., Suchard MA., Huelsenbeck JP.. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012; 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q.. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015; 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Interactive Tree Of Life is freely available at https://itol.embl.de.