Abstract
RNA has become one of the major research topics in molecular biology. As a central player in key processes regulating gene expression, RNA is in the focus of many efforts to decipher the pathways that govern the transition of genetic information to a fully functional cell. As more and more researchers join this endeavour, there is a rapidly growing demand for comprehensive collections of tools that cover the diverse layers of RNA-related research. However, increasing amounts of data, from diverse types of experiments, addressing different aspects of biological questions need to be consolidated and integrated into a single framework. Only then is it possible to connect findings from e.g. RNA-Seq experiments and methods for e.g. target predictions. To address these needs, we present the RNA Workbench 2.0 , an updated online resource for RNA related analysis. With the RNA Workbench we created a comprehensive set of analysis tools and workflows that enables researchers to analyze their data without the need for sophisticated command-line skills. This update takes the established framework to the next level, providing not only a containerized infrastructure for analysis, but also a ready-to-use platform for hands-on training, analysis, data exploration, and visualization. The new framework is available at https://rna.usegalaxy.eu , and login is free and open to all users. The containerized version can be found at https://github.com/bgruening/galaxy-rna-workbench.
INTRODUCTION
Together with the focus on RNA as regulatory key player, the number and complexity of datasets ready for analysis is steadily increasing. Although many tools for the analysis of such data exist, they are often tailored to specific experiments and not always easy to install, adapt, and run appropriately. The challenge for the individual researcher remains to chain them into useful workflows and pipelines. Often this task is further complicated, as many tools are only available for the command line, limiting their user base to computer-savvy biologists and bioinformaticians.
Although pitfalls during the installation process of tools can be circumvented with package managers like condahttps://conda.io and its BioConda (1) channel, or Docker containers, it remains with the user to set up the appropriate computational environment. Many of these needs were already addressed with the release of the RNA Workbench (2). Based on the framework (3), containerized in a Docker instance, the workbench guarantees simple access, easy extension and flexible adaption to personal and security needs. This enables users to run sophisticated analyses that are independent of command-line knowledge while utilizing's integrated and powerful workflow manager. With the current release of the RNA Workbench 2.0 we now additionally provide the user with a pre-configured, ready-to-use compute environment, running on dedicated hardware, available at https://rna.usegalaxy.eu.
The RNA Workbench 2.0 is developed and maintained by a community consisting of experts in RNA bioinformatics and, as well as a growing number of users, and tool developers. Our commitment to keep the workbench fit for future standards and needs is one of the reasons for the release of this update. We aim to provide researchers with an up-to-date reliable and robust framework for RNA data analysis. In this release, we integrated many new RNA-related tools, and updated well established suites, such as the ViennaRNA (4) package, covering a broad variety of use-cases.
Currently, we provide more than 100 bioinformatics tools that are dedicated to different research areas of RNA biology including RNA structure analysis, RNA alignment, RNA annotation, RNA-protein interaction, ribosome profiling, RNA-Seq pre-processing and analysis, as well as RNA target prediction. The complete list of tools can be found at https://rna.usegalaxy.eu or https://github.com/bgruening/galaxy-rna-workbench.
Taking advantage of Galaxy ’s powerful workflow manager allows users to easily connect single tools into computational pipelines. For common RNA related tasks we provide >25 ready-to-use workflows combining, e.g. established tools for RNA-Seq processing and analysis. For each workflow we provide a dedicated training to guide researchers through the analysis. Training is a key aspect of our effort in bringing high-quality RNA bioinformatics to researchers. Thus, each training accompanying a workflow comes with a test dataset, allowing interested users to get hands-on experience with their tools and workflows of interest. Keeping such trainings up-to-date and functional is a cooperative endeavour together with the Galaxy Training Network , which hosts Galaxy Training Material (5), a collection of tutorials developed and maintained by the worldwide Galaxy community. In case a user requires a novel workflow to answer a research question that is not covered by existing ones or to incorporate specific tools, we encourage users to share these workflows and if possible adequate training data and material. This directly enables all users to benefit from contributions to our community, which distributes shared knowledge and in return helps to maintain and enhance workflows and trainings where possible.
GOALS
A main intention behind the development of the original RNA Workbench was the creation of an easy-to-use and deploy environment for training and self-empowerment of biologists in RNA bioinformatics. The RNA Workbench was downloaded >2000 times, used for research, training courses (e.g. within de.NBI (6)), and has even been integrated into the B3Africa toolset (7). The ongoing need for such a comprehensive collection of RNA bioinformatics tools, workflows and resources led to the development of RNA Workbench 2.0. Although the provision of RNA Workbench as in a Docker made it easy to maintain, deploy and use, we became aware that there is additional need for an instance with freely available compute resources. Our target audience, mainly RNA biologists, requested an even more easy-to-use and ready-to-go way of accessing this collection. With the realization of the European server (https://usegalaxy.eu), we gained access to an infrastructure that would allow exactly that. Thus, with RNA Workbench 2.0 we provide an updated and ready-to-use webserver, satisfying user requests and enabling even more scientists to participate in RNA research.
TOOLS AND IMPROVEMENTS
In addition to providing the RNA Workbench 2.0 as a portable Docker container (https://github.com/bgruening/galaxy-rna-workbench), users can now directly use integrated tools, workflows and tutorials at a free online instance of Galaxy. This makes the use of the RNA Workbench 2.0 even easier, and allows users to train and run data analysis workflows without the need to set up hardware, software environments, or even Docker. New workflows and tutorials ease introduction to the environment, and guide users through analysis tasks step by step. Continuous exchange of workflows, tours and training material with the Galaxy Training Network ensures that the RNA Workbench 2.0 remains a state-of-the-art training and research resource. New and updated tools and workflows are continuously integrated and made available in close cooperation between the user and developer community. This includes also updates to the underlying packages in BioConda. Updated tools are for example LocaRNA (8), RNAz (9), (10), AREsite2 (11) and Infernal (12). In addition new tools like edgeR (13), CMV (14), RNAlien (15), MultiQC (16) and scPipe (17) have been added. A complete list of available tools can be found at https://rna.usegalaxy.eu.
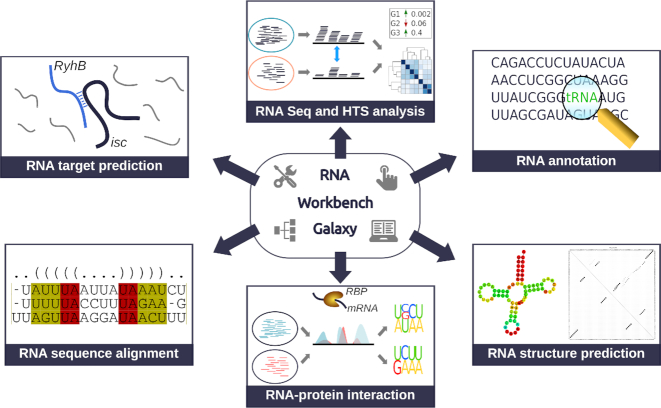
Figure 1 provides an overview of tools and workflows dedicated to specific topics of RNA research in version 2 of the RNA Workbench.
Figure 1.
Overview of RNA research topics, dedicated tools and example workflows in RNA Workbench 2.0 RNA target prediction enables to analyze potential interaction partners of RNA molecules. Included annotation tools allow the discovery of homologous sequences in genomes. The secondary structure of input RNA sequences can be predicted and visualized or for example used to create sequence-structure alignments. High-throughput and RNA sequencing data analysis can be performed with available tools and results directly intersected with e.g. databases for RNA-protein interactions.
TRAINING
A key aspect behind the development of the original and now updated RNA Workbench was to provide an accessible platform, easing the process of gaining expertise in and applying bioinformatics. To this end, considerable effort went into extensive documentation and a large set of training material, empowering beginners and non-bioinformaticians to use, adapt, and apply workflows based on their needs and standards. The recently published Galaxy Training Material provides users with a collection of hands-on training material and data on many topics of (not exclusively high-throughput sequencing (HTS)-related) life-science research. This collection is constantly improved and extended in an international community effort, including de.NBI, ELIXIR and EMBL. We tightly integrate Galaxy Training Material into the RNA Workbench 2.0 , exchanging workflows and training material on various RNA related topics. As an example, for RNA-Seq data analyses we provide training instances as specific introduction to the topic. These consist of self-explanatory presentation slides, hands-on training documentation and a Galaxy Interactive Tour guiding through the analysis workflow with all required input files ready-to-use, hosted by Zenodo.
WORKFLOWS
One of the strengths of the framework is that users can easily create, customize and share their workflows with other users of the same or other instances. A workflow is not only a chain of tools applied to a fixed dataset, Galaxy workflows also save tool versions, required data formats and other metadata ensuring a maximum of reproducibility. The built-in graphical workflow editor facilitates repurposing or adaptation of workflows.
A set of >25 workflows dedicated to specific analysis goals is included in RNA Workbench 2.0. We provide for example a set of workflows for the analysis of non-coding RNA and cover a range of analysis tasks, from structure conservation and coding potential of homologous RNAs, based on Locarna (8) and RNAz (9), as well as automatic construction of RNA family models, based on RNAlien (14). The workbench features workflows for processing, analyzing and visualizing data from RNA-Seq, CLIP-Seq, RNA folding, network analysis, sRNA-Seq, RNA family model construction and more.
Datasets for analysis can be imported from a local source, from dedicated databases or via link, easing the integration of data from different sources. Training datasets can be imported directly from Zenodo.
TOURS
Another training aspect is provided via Galaxy Interactive Tours. These guide users through an entire analysis in an interactive and explorative way. In contrast to training videos, a Galaxy Interactive Tour can be easily created, updated and improved to guide the Galaxy user step-by-step, e.g. through a whole HTS analysis starting from uploading the data to using complex analysis tools. The RNA workbench currently integrates more than 15 Galaxy Interactive Tours. These range from general tours introducing new users to the Galaxy interface and its usage, with RNA-seq example datasets, to specialized tours, e.g. illustrating secondary structure prediction of RNA molecules using parts of the ViennaRNA package.
INPUTS AND OUTPUTS
Users of the RNA Workbench 2.0 have access to a diverse set of Galaxy implemented data formats and format conversion tools. Common formats for sequence and/or structure information are readily accepted as input, generic data can be imported and converted to fit tool specifications, guaranteeing reproducibility and interoperability. Output data follows the same principle, defined by the analysis tool, but can be converted to a range of standard and specific formats, including plots and figures. For the latter, the RNA Workbench 2.0 contains tools for visualizations of RNA-Seq related data (e.g. mQC (18), MultiQC (16), sRNAPipe (19)), RNA structure datasets, such as dot-bracket strings RNA 2D or 3D structures or RNA family models and alignments (cmv (15)).
COMMUNITY CONTRIBUTIONS
The RNA Workbench 2.0 is hosted on GitHub (https://github.com/bgruening/galaxy-rna-workbench) and users are welcome to suggest new tools, workflows and tours to be made available through GitHub and the workbench Docker container. Tools should be published to the Galaxy Tool Shed (20) via https://github.com/bgruening/galaxytools followed by a pull request at GitHub. After passing continuous integration tests and approval after manual review, new tools will be integrated into the RNA Workbench. More information about tool development can be found on the Galaxycommunity page. Workflows can easily be contributed by running them at https://rna.usegalaxy.eu and sharing them, ideally accompanied by test datasets and a shared history of the workflow run. A pull request adding them to the workflow folder of https://github.com/bgruening/galaxy-rna-workbench, will allow us to merge the workflow into the workbench. When contributing workflows, users should make sure that all tools needed for the workflow are integrated into the RNA Workbench 2.0. If not, please add these tools beforehand following above steps, or request them to be added by opening an appropriate issue at GitHub. Galaxy interactive tours can be contributed similarly, by opening a pull request and including tours in the tours folder of https://github.com/bgruening/galaxy-rna-workbench after approval.
USE CASES
de.NBI
The ‘German Network for Bioinformatics Infrastructure–(de.NBI )’ is an academic and non-profit infrastructure supported by the German Federal Ministry of Education and Research. As German partner of ELIXIR (https://www.elixir-europe.org) it provides bioinformatics services to users in life science research and biomedicine in Europe (6). The partners organize training events, courses and summer schools on tools, standards and compute services provided by de.NBI and ELIXIR to assist researchers to more effectively exploit their data. The RNA Workbench and also RNA Workbench 2.0 have in part been developed by researchers funded by de.NBI with the aim to generate a free and easy to use platform for training and education. As such, the RNA Workbench is ready for and has been used in de.NBI training courses. With the publication of RNA Workbench 2.0 this will become even easier, as trainers and trainees have access to a ready-to-use instance, including dedicated hardware, simply connecting via a web browser.
B3Africa
The Bridging Biomolecular Researcher and Biobanking in Africa (B3Africa) created the eB3Kit, an informatics platform for comprehensive management of samples and associated data (21) to support the establishment of research integrated biobanks (22). A key priority of the project is to strengthen the research capacity in resource constrained areas. As bioinformatics is a rapidly advancing field leading to constant changes in the demand for tools and procedures, the bioinformatics module has been designed to integrate a pre-existing platform satisfying the following key requirements. (i) An active community providing access to new tools, algorithms and training through a standardized interface, (ii) an accessible API enabling the B3Africa project to interact with the software without changing the supported codebase and (iii) the ability to download tools and databases for access without internet connection. Fulfilling these requirements, the RNA Workbench has been implemented in the BIBBOX appstore (7) and is used as the preferred solution to showcase both the eB3Kit and the Galaksio interface for simplified workflow management (23). Throughout the project successful showcases of the eB3Kit using the RNA Workbench have been conducted, e.g. in Lyon (France), Banjul (Gambia) and at Lake Naivasha in the Rift Valley region of Kenya.
DISCUSSION
An active community developing and applying the RNA workbench in training (e.g. within de.NBI, ELIXIR and B3Africa) and research, the RNA Workbench has become an important resource for best practices in RNA and high-throughput sequencing bioinformatics in Galaxy.
In this work, we present an update to this resource with the creation of the ready-to-use webserver instance. Users benefit from this setup as they can now directly browse to https://rna.usegalaxy.eu and use a pre-configured instance of RNA Workbench , without needing to have any software installed on their own system except for a browser. This enables researchers not only to become familiar with a set of RNA-related bioinformatics tasks, running one of the provided tutorials and/or accompanying workflows, but also to compute and analyze data on dedicated hardware. For users concerned with data regulations, e.g. when working on patient data, or users with their own dedicated hardware, we also provide an updated Docker container, similar to the first version of RNA Workbench. A RNA Workbench instance started with this container provides the same tools, workflows, trainings and tours as the online instance and can easily be extended with additional tools via the Galaxy Tool Shed. As for the first version of RNA Workbench , each tool in the workbench is also available as a BioConda package as well as a Docker/rkt container (BioContainers). The Docker container offers a comprehensive virtualized RNA workbench that can be deployed on every standard Linux, Windows and OSX computer, but can at the same time employ high-performance- or cloud-computing infrastructure.
Similar to the first version, this release is developed and maintained by a constantly growing RNA and Galaxy community. This community approach helps to keep the workbench up-to-date and valuable for research. Moreover, all components such as tools, workflows, visualizations, interactive tours and training material can be easily integrated into any available Galaxy instance for teaching, learning or exploratory purposes. Every user is encouraged to contribute and add to this collection, which is tightly integrated into the Galaxy Training Material , providing state-of-the-art learning material.
To our knowledge, the RNA workbench is a unique suite without direct competitors. Existing workbenches, such as miARma-Seq (24), the UEA Small RNA Workbench (25) or the NCBI genome workbench, are all tailored to specific analysis tasks. In addition, our focus on accessibly, flexibility in workflow assembly and application, training and the interaction with the community are all major benefits of RNA Workbench 2.0.
ACKNOWLEDGEMENTS
We thank de.NBI and ELIXIR for supporting bioinformatics infrastructure. Thanks also to the Galaxy community, especially to the Freiburg Galaxy Team, for developing, maintaining and supporting this great framework. We also like to acknowledge the BioConda and BioContainers community for setting new standards in reproducible software deployments. Furthermore, the authors acknowledge the support of many upstream developers like Chao Zhang, Steven Verbruggen, the sRNAPipe team Pierre Pouchin, Silke Jensen and Emilie Brasset that helped us to integrate their tools into the RNA Workbench 2.0 and accepted patches and to whom we wish a wonderful day, every day.
FUNDING
German Federal Ministry of Education and Research [BMBF grants 031 A538A/A538C de.NBI-RBC awarded to P.F.S. and R.B., 031L0101C de.NBI-epi awarded to B.G., 031L0106 de.STAIR (de.NBI)]; German Research Foundation for the Collaborative Research Center 992 Medical Epigenetics [SFB 992/1 2012 and SFB 992/2 2016 awarded to R.B.]. Funding for open access charge: German Federal Ministry of Education and Research.
Conflict of interest statement. None declared.
REFERENCES
- 1. Grüning B., Dale R., Sjödin A., Chapman B.A., Rowe J., Tomkins-Tinch C.H., Valieris R., Köster J., Bioconda T.. Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat. Methods. 2018; 15:475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Grüning B.A., Fallmann J., Yusuf D., Will S., Erxleben A., Eggenhofer F., Houwaart T., Batut B., Videm P., Bagnacani A. et al.. The RNA workbench: best practices for RNA and high-throughput sequencing bioinformatics in Galaxy. Nucleic Acids Res. 2017; 45:W560–W566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Afgan E., Baker D., Batut B., Van Den Beek M., Bouvier D., Ech M., Chilton J., Clements D., Coraor N., Grüning B. et al.. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018; 46:W537–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lorenz R., Bernhart S.H., Zu Siederdissen C.H., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Alg. Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Batut B., Hiltemann S., Bagnacani A., Baker D., Bhardwaj V., Blank C., Bretaudeau A., Brillet-Gueguen L., čech M., Chilton J. et al.. Community-Driven Data Analysis Training for Biology. Cell Syst. 2018; 6:752–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tauch A., Al-Dilaimi A.. Bioinformatics in Germany: toward a national-level infrastructure. Brief. Bioinform. 2019; 20:370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Müller H., Malservet N., Quinlan P., Reihs R., Penicaud M., Chami A., Zatloukal K., Dagher G.. From the evaluation of existing solutions to an all-inclusive package for biobanks. Health Technol. 2017; 7:89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Will S., Joshi T., Hofacker I.L., Stadler P.F., Backofen R.. LocARNA-P: accurate boundary prediction and improved detection of structural RNAs. RNA. 2012; 18:900–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gruber A.R., Findeiß S., Washietl S., Hofacker I.L., Stadler P.F.. Rnaz 2.0: improved noncoding RNA detection. Pac. Symp. Biocomput. 2010; 2010:69–79. [PubMed] [Google Scholar]
- 10. Videm P., Rose D., Costa F., Backofen R.. BlockClust: Efficient Clustering and Classification of Non-Coding RNAs from Short Read RNA-Seq Profiles. Bioinform. 2014; 30:i274–i282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fallmann J., Sedlyarov V., Tanzer A., Kovarik P., Hofacker I.L.. AREsite2: an enhanced database for the comprehensive investigation of AU/GU/U-rich elements. Nucleic Acids Res. 2016; 44:D90–D95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nawrocki E.P., Eddy S.R.. Infernal 1.1: 100-fold faster RNA homology searches. Bioinform. 2013; 29:2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinform. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eggenhofer F., Hofacker I.L., zuSiederdissen C.H.. RNAlien-unsupervised RNA family model construction. Nucleic Acids Res. 2016; 44:8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eggenhofer F., Hofacker I.L., Backofen R.. CMVVisualization for RNA and protein family models and their comparisons. Bioinform. 2018; 1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ewels P., Magnusson M., Lundin S., Kaller M.. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinform. 2016; 32:3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tian L., Su S., Dong X., Amann-Zalcenstein D., Biben C., Seidi A., Hilton D.J., Naik S.H., Ritchie M.E.. scPipe: a flexible R/bioconductor preprocessing pipeline for single-cell RNA-sequencing data. PLoS Comput. Biol. 2018; 14:e1006361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Verbruggen S., Menschaert G.. mQC: a post-mapping data exploration tool for ribosome profiling. Comput. Methods Programs Biomed. 2018; doi:10.1016/j.cmpb.2018.10.018. [DOI] [PubMed] [Google Scholar]
- 19. Pogorelcnik R., Vaury C., Pouchin P., Jensen S., Brasset E.. sRNAPipe: a Galaxy-based pipeline for bioinformatic in-depth exploration of small RNAseq data. Mob DNA. 2018; 9:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blankenberg D., Von Kuster G., Bouvier E., Baker D., Afgan E., Stoler N., Taylor J., Nekrutenko A.. Dissemination of scientific software with Galaxy ToolShed. Genome Biol. 2014; 15:403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Klingström T., Mendy M., Meunier D., Berger A., Reichel J., Christoffels A., Bendou H., Swanepoel C., Smit L., Mckellar-Basset C. et al.. Supporting the development of biobanks in low and medium income countries. IST-Africa Week Conference. 2016; Durban: IEEE; 1–10. [Google Scholar]
- 22. Slokenberga S., Reichel J., Niringiye R., Croxton T., Swanepoel C., Okal J.. EU data transfer rules and African legal realities: is data exchange for biobank research realistic?. Data Privacy Law Int. 2018; 9:30–48. [Google Scholar]
- 23. Klingström T., Hernández-deDiego R., Collard T., Bongcam-Rudloff E.. Galaksio, a user friendly workflow-centric front end for Galaxy. EMBnet. J. 2017; 23:e897. [Google Scholar]
- 24. Andres-Leon E., Nunez-Torres R., Rojas A.M.. miARma-Seq: a comprehensive tool for miRNA, mRNA and circRNA analysis. Sci. Rep. 2016; 6:25749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stocks M.B., Mohorianu I., Beckers M., Paicu C., Moxon S., Thody J., Dalmay T., Moulton V.. The UEA sRNA Workbench (version 4.4): a comprehensive suite of tools for analyzing miRNAs and sRNAs. Bioinform. 2018; 34:3382–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]