Abstract
Phylogeny.fr, created in 2008, has been designed to facilitate the execution of phylogenetic workflows, and is nowadays widely used. However, since its development, user needs have evolved, new tools and workflows have been published, and the number of jobs has increased dramatically, thus promoting new practices, which motivated its refactoring. We developed NGPhylogeny.fr to be more flexible in terms of tools and workflows, easily installable, and more scalable. It integrates numerous tools in their latest version (e.g. TNT, FastME, MrBayes, etc.) as well as new ones designed in the last ten years (e.g. PhyML, SMS, FastTree, trimAl, BOOSTER, etc.). These tools cover a large range of usage (sequence searching, multiple sequence alignment, model selection, tree inference and tree drawing) and a large panel of standard methods (distance, parsimony, maximum likelihood and Bayesian). They are integrated in workflows, which have been already configured (‘One click’), can be customized (‘Advanced’), or are built from scratch (‘A la carte’). Workflows are managed and run by an underlying Galaxy workflow system, which makes workflows more scalable in terms of number of jobs and size of data. NGPhylogeny.fr is deployable on any server or personal computer, and is freely accessible at https://ngphylogeny.fr.
INTRODUCTION
Inference and interpretation of phylogenetic trees are required in a large number of studies covering a large spectrum of biological areas (comparative genomics, functional prediction, metagenomics, species identification, taxonomy, molecular epidemiology, population genetics, etc.).
Phylogeny.fr (1) had originally been designed to facilitate phylogenetic analyses by implementing workflows based on the following steps: (i) BLAST-based sequence searching; (ii) multiple sequence alignment; (iii) alignment curation; (iv) phylogenetic tree inference; (v) tree visualization. It has been widely used in several contexts, some we did not expect when designing Phylogeny.fr, such as very large teaching classes where hundreds of jobs were (still are) submitted simultaneously, or large scale genome annotation studies, where phylogenies were built for thousands of gene families using custom submission scripts. Since its launch in 2008, Phylogeny.fr has been cited >3000 times and currently runs >200 workflows per day.
In the past decade, several kinds of solutions to support phylogenetic analyses have been developed.
First are online services dedicated to one specific phylogenetic tool that generally comes with a key publication (e.g. MAFFT (2), PhyML (3), FastME (4), BOOSTER (5)). The number of such web services is increasing with the publication of new tools, offering a large number of options, while increasing the difficulty to correctly select them. Most importantly, performing a phylogenetic analysis implies chaining such tools and managing their inputs and outputs, that is storing them and reformatting them between many formats such as Fasta, Nexus, Newick and Phylip.
Integrative web services have thus emerged to answer part of the difficulties listed above, by allowing users to chain and execute several tools online. Phylogeny.fr (1) is widely used and cited, and CIPRES (6), TRex (7) and Phylemon (8) also belong to this category. In the same spirit, SeaView (9) and MEGA (10) offer integrative solutions for phylogenetic analysis, while providing a standalone software to be installed locally. These integrative solutions usually consider preselected tools and/or analyses, and may have difficulties to evolve in terms of tool updating and chaining. Moreover, while such integrative solutions were particularly interesting ten years ago, the analyses that run nowadays have drastically changed in terms of number and size of sequences and CPU requirements.
In parallel, scientific workflow systems (Galaxy (11,12)) have reached a level of maturity that makes them convenient for scheduling the execution of complex and large-scale analyses, while properly managing data by tracking consumed and produced data. A third kind of solution has then been based on such systems. This is the case of Osiris (13) that offers access to several phylogenetic tools through Galaxy, or Armadillo (14) that implements its own workflow manager dedicated to phylogenetics. Such solutions are highly flexible as they provide numerous tools and a way to combine them easily, and thus make them close to the unified framework described by Guang et al. (15). However, they remain difficult to use for end-users, as they are expected to select and parameterize all tools using the workflow system graphical user interface.
NGPhylogeny.fr, the Next Generation Phylogeny.fr web service introduced in this paper, has been built to (i) have a general scope, offering a large panel of phylogenetic tools to fit anyone needs; (ii) be flexible, allowing to easily add, update or remove tools; (iii) be scalable, able to support large-scale analyses by integrating simple and fast methods, and relying on a workflow system that enables the distribution of parallel computations on large clusters; (iv) be turnkey, avoiding users to manage installation on their own computers while ensuring reproducibility; and (v) be user-adaptable, providing several usage levels from pure end-users to bioinformaticians with technical skills who may prefer to use NGPhylogeny.fr on their own servers rather than on the public one.
To do so, NGPhylogeny.fr is built upon two components: (i) the Galaxy workflow system that deals with the management of tool executions and (ii) a graphical user interface making the use of the Galaxy workflow system transparent to users. In the next sections we first focus on how NGPhylogeny.fr can be used by end-users, while the last section describes how advanced users with more technical skills can exploit additional aspects of it.
PHYLOGENETIC WORKFLOWS
All NGphylogeny.fr workflows are based on the tools listed in Table 1. The choice of tools will mainly depend on the size of the dataset and the application.
Table 1.
List of tools currently integrated in NGPhylogeny.fr
| Step | Tool name | New | Version | Dataset size ability | ‘One click’ | ‘Advanced’ | ‘A la carte’ | ‘Stand-alone’ |
|---|---|---|---|---|---|---|---|---|
| MSA | Clustal Ω (16) | Yes | 1.2.4.1 | Very large | ✓ | ✓ | ||
| MSA | MAFFT (2) | Yes | 7.407 | Large | ✓ | ✓ | ✓ | ✓ |
| MSA | MUSCLE (17) | Up | 3.8.37 | Medium | ✓ | ✓ | ||
| AC | Gblocks (18) | - | 0.91b | Very large | ✓ | ✓ | ||
| AC | trimAl (21) | Yes | 1.4.1 | Very large | ✓ | ✓ | ||
| AC | BMGE (19) | Yes | 1.12 | Medium | ✓ | ✓ | ✓ | ✓ |
| AC | Noisy (20) | Yes | 1.5.12.1 | Small | ✓ | ✓ | ||
| TI (Fast max-likelihood) | FastTree (22) | Yes | 2.1.10 | Very large | ✓ | ✓ | ✓ | ✓ |
| TI (Distance) | FastME (4) | Yes | 2.1.6.1 | Large | ✓ | ✓ | ✓ | ✓ |
| TI (Parsimony) | TNT (23) | Yes | 1.5.0a | Large | ✓ | ✓ | ||
| TI (Max-likelihood) | PhyML (3) | Up | 3.1 | Medium | ✓ | ✓ | ✓ | ✓ |
| TI (PhyML+MS) | PhyML (3)+SMS (24) | Yes | 1.8.1 | Medium | ✓ | ✓ | ✓ | ✓ |
| TI (Bayesian) | MrBayes (25) | Yes | 3.2.6 | Small | ✓ | ✓ | ||
| TV | Newick Utilities (26) | Yes | 1.6 | Large | ✓ | ✓ | ✓ | ✓ |
| BS | BOOSTER (5) | Yes | 0.2.4 | Large | ✓ | ✓ | ✓ |
Step: MSA for multiple sequence alignment, AC for alignment curation, TI for tree inference, TV for tree visualization, BS for branch support, and MS for model selection. New: Yes for new tools, Up for updated tools and - for tools already present in Phylogeny.fr. Dataset size ability: dataset dimension able to be analyzed by each tool, very large (typically >10 000 sequences), large (>5000), medium (>1000), small (≤1000). ‘One click’, ‘Advanced’, ‘A la carte’ and ‘Stand-alone’: tools that are available in each run mode.
For multiple sequence alignment, very large datasets will preferably be run with Clustal Ω (16); medium to large datasets can be run with MAFFT (2); and small to medium datasets can be computed using Muscle (17).
Regarding alignment curation, Gblocks (18) and trimAl (21) are the methods of choice for very large datasets; BMGE (19) will mainly be used for medium datasets to large datasets, while Noisy (20), though very accurate (27), will be dedicated to small datasets.
Lastly, for tree inference, with very large datasets (>5000 sequences) users can choose FastTree (fast combination of distance and likelihood); with large datasets (in the order of several thousand sequences) users will typically select FastME (distance) or TNT (parsimony), while with small to medium datasets they will prefer PhyML+SMS (likelihood based plus model selection). MrBayes will be a method of choice for relatively small datasets, when users are interested in the posterior distribution of phylogenetic trees induced by their data.
All the workflows take a FASTA file as input, preferably unaligned, and produce multiple sequence alignment files (FASTA or PHYLIP) and phylogenetic tree files (Newick format). For each type of results, NGPhylogeny.fr proposes a dedicated viewer: Multiple sequence alignments are visualized dynamically using the BioJS MSAViewer plugin (28); Phylogenetic trees are visualized dynamically using PRESTO (http://www.atgc-montpellier.fr/presto) built on the phylotree.js plugin (29) or via upload to iTOL (30); Other formats such as images, text, or html are displayed in the browser.
Several flavors of workflows are available, depending on user’s level of expertise. These workflows differ mainly by the tools that are executed at each step and their parameters (see Table 1 for the list of available tools).
The first kind of workflows, called ‘One click’, is dedicated to users wanting to execute fully automatic workflows with default tools and parameters that we estimate to be adapted to most cases. The four ‘One click’ workflows differ only at the tree inference step, which can be performed by FastTree (22), FastME (4), PhyML (3) or PhyML+SMS (24).
The second kind of workflows, called ‘Advanced’, is directed to users wanting to execute already structured but customized workflows, with default tools and specific parameters. These workflows have the exact same structure as ‘One click’ ones, that is with the same steps and available tools, but users can specify the parameter values of these tools. It is worth noticing that we integrated Felsenstein Bootstrap Proportions (FBP) and Transfer Bootstrap Expectation (TBE) (5) for branch support computation to several tree inference tools, which can be configured at this step and was not available in Phylogeny.fr.
The last kind of workflows, called ‘A la carte’, provides the users with a workflow maker, which enables the construction of fully customized workflows, made of any available tools and parameter values. Workflows built this way are composed of any combination of steps, and users just have to select the tools they want to run. The workflow so constructed is parameterized just as ‘Advanced’ workflows.
Lastly, all tools can be executed individually without being integrated in a workflow. All workflow results can be reused as input of individual tools and be further analyzed without being downloaded and re-uploaded.
BLAST-SEARCH
Beyond the needs associated with execution and configuration of phylogenetic analyses, there is also a need to guide users in selecting sequences on which the analysis will be performed. The Blast-Search module, provided by NGPhylogeny.fr, implements such a sequence search interface. Blast-Search is a successor of BlastExplorer (31), and uses BLAST (32) to retrieve and compare sequences that are similar enough to a user input sequence. To do so, Blast-Search runs ‘blastn’, ‘blastp’, ‘tblastn’ or ‘blastx’ either by querying databases installed on the Institut Pasteur Galaxy server (33), or by querying the public NCBI BLAST databases (the latter is only available on standalone mode).
The use of Blast-Search can be summarized as follows: First, the user pastes an input sequence of interest (in FASTA format) and submits the form. Once the BLAST job is finished, only sequences passing the e.value and query coverage thresholds (given by the user) are considered. A fast multiple-alignment is then built by using the query sequence as reference, ignoring insertions on matching sequences, merging potential multiple High Scoring Pairs (HSP), and filling the holes with gaps. This fast alignment is then used to compute a distance matrix and a distance based tree, which is visualized dynamically to enable the deletion of unwanted sequences or groups of sequences, hence building a clean dataset. The final dataset, constituted of the user input and its matching sequences, can be downloaded in FASTA format or used as input of any of the NGPhylogeny.fr workflows.
USE CASES
We now provide two use cases illustrating the benefit of using NGPhylogeny.fr.
Blast-Search and ‘One click’ tree building
In this use case, we take as reference the human Tripartite motif-containing protein 5 isoform α (gene TRIM5, Uniprot id: Q9C035), a retrovirus restriction factor notably involved in inhibiting some strains of retroviruses.
The aim of the analysis is to place this protein in its close evolutionary context. This task, involving many tools, is largely facilitated by NGPhylogeny.fr and its ability to connect the different steps of the analysis, that is sequence selection with Blast-Search, multiple sequence alignment, model selection and tree inference.
To launch the analysis, we execute a Blast-Search run, with the sequence of TRIM5α protein as input, using ‘blastp’ on ‘nrprot’ hosted by the Institut Pasteur Galaxy Server. We select the first 100 best matches having an e.value lower than 10−5 and covering the query on at least 80% of its length. Once the run is finished, in ∼20 min, we obtain 100 sequences having a length of ∼500 amino acids. Using the tree visualizer, we select sequences from the ape clade (hominoidae), that is, orangutans (pongo), chimpanzees (pan), gorillas, human and gibbons (hylobatidae), and delete all other sequences. We obtain a dataset made of 38 sequences that we give as input of the PhyML+SMS ‘One click’ workflow, as it is very accurate and fast with dataset of such size.
Results are obtained in less than 5 min, and are shown in Figure 1, displaying the workflow monitoring page, the curated alignment, as well as the final phylogenetic tree (displayed also with SH-like supports in Supplementary Figure S1). The input sequence is well-placed among other known Human sequences in the tree and the taxonomy of apes is globally well-structured and well supported (SH-like), with human sequences closest to chimpanzees, then gorillas, orangutans and finally gibbon sequences. We also built a MrBayes workflow (‘A la carte’) with default options, which ran in ∼5 min, and gave the same topology and high branch supports (Supplementary Figure S2).
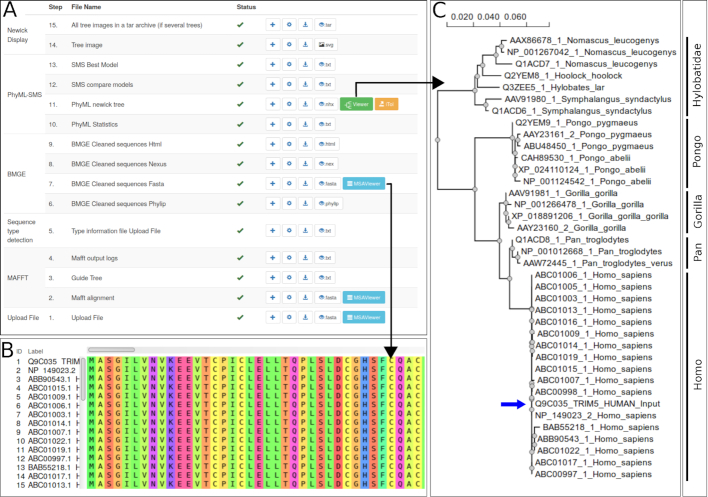
Figure 1.
Analysis of human TRIM5α protein with Blast-Search and PhyML+SMS ‘One click’ workflow. (A) NGPhylogeny.fr workflow monitoring page. All workflow steps are listed and their status indicated as pending, running, finished successfully (green check mark) or with error. For each result, a link to a dedicated viewer is provided. (B) Multiple sequence alignment visualization. The multiple sequence alignment resulting of the cleaning step can be visualized dynamically in the browser. (C) Phylogenetic tree visualization. Human, Chimpanzee, Gorilla, Orangutan and Gibbon sequences are branched as expected. The blue arrow indicates the user input sequence.
Analysing large viral sequence dataset (‘A la carte’)
In this case study, we analyze a very large viral sequence dataset of several thousands of sequences for which we want to build a phylogenetic tree with branch supports. Such an analysis is particularly CPU-intensive and cannot be executed in other phylogenetic analysis solutions, including the former Phylogeny.fr. Here, thanks to the integration of FastTree and bootstrap support in NGPhylogeny.fr, results can be computed in ∼8 h on Institut Pasteur web server.
To run this use case, we downloaded the HIV data sequence file located at https://ngphylogeny.fr/static/hiv_pol.fa.zip, which contains 9,147 HIV pol gene DNA sequences of length ∼1,050 nucleotides (5). Using the workflow maker, we then build a workflow including the following steps: (i) sequence alignment with MAFFT; (ii) tree inference with FastTree and (iii) tree rendering with Newick Display. The workflow is configured such that MAFFT has default options, and FastTree considers sequences as nucleotidic and has bootstrap support turned ON with 100 replicates. The resulting tree shows the HIV subtypes, which are well grouped and supported with TBE (Supplementary Figure S3).
DISCUSSION
This paper introduces NGPhylogeny.fr, the new version of Phylogeny.fr. NGPhylogeny.fr is based on modern Web technologies and relies on the Galaxy workflow system, to provide flexible, modular and scalable analyses, via a user-friendly graphical interface.
Thanks to this new architecture and the new integrated tools, NGPhylogeny.fr (i) allows any user to easily perform complete analyses (as shown in our first use case) and (ii) pushes the limits of what is possible with Phylogeny.fr and other solutions in terms of number and size of sequences, such as large viral datasets (as shown in our second use case). Last but not least, NGPhylogeny.fr is easy to maintain and update, and straightforward to install and deploy.
Currently NGPhylogeny.fr is limited to single gene analyses. We plan to extend NGPhylogeny.fr to multi-gene, phylogenomics studies, to make it possible to analyze several multiple-alignments of gene sequences with the same workflow, combine the results into a species tree, and reconcile the gene trees with the species tree.
ARCHITECTURE AND IMPLEMENTATION
The architecture of NGPhylogeny.fr consists of two main components working together: (i) a Galaxy workflow system on which tools and workflows are stored and executed and (ii) a user interface implemented in Python/Django allowing end-users to run their workflows without having to know how to use the Galaxy system.
The Galaxy instance stores workflows and tools, and is responsible for running the jobs, and monitoring their execution until completion. We wrapped phylogenetic tools in Galaxy XML wrappers, and built the workflows upon them. To facilitate tool interoperability and workflow development, we implemented the Galaxy wrappers such that all alignment and tree inference tools take FASTA and PHYLIP formats as input. Moreover, to prevent sequence name errors, wrappers first clean sequence names conflicting with the Newick format or temporarily rename sequences with automatically generated names. The user interface takes care of presenting available workflows, assembling tools, formatting input forms, storing previous runs, checking job runs, and visualizing output files. The two components communicate via the Galaxy API using the bioblend python library (34).
For advanced users interested in deploying their own instance of NGphylogeny.fr, and be able to add other tools and modify the workflows, we additionally provide an easy way to install, update and deploy NGPhylogeny.fr via Docker (https://www.docker.com/). The web interface as well as the custom Galaxy instance containing all phylogenetic tools and workflows are packed into two Docker images that are automatically configured to run a local NGPhylogeny.fr instance (well suited for teaching classes for example).
DATA AVAILABILITY
NGPhylogeny.fr is freely available at https://ngphylogeny.fr. Source codes of the web interface, wrappers and worflows are available on GitHub at C3BI-pasteur-fr/ngphylogeny-django and C3BI-pasteur-fr/ngphylogeny-galaxy. The two Docker images are stored on Docker Hub at evolbioinfo/ngphylogeny-galaxy and evolbioinfo/ngphylogeny.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the ‘Genome Informatics and Phylogenetics’ (GIPhy) expertise group of Institut Pasteur for helpful discussions and testing, as well as the IT System Department of Institut Pasteur, in particular Eric Deveaud who manages installation and update of tools. We also thank Jean-Michel Claverie and his team, who initiated the Phylogeny.fr project ten years ago.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Institut Français de Bioinformatique [ANR-11-INBS-0013]; INCEPTION project [PIA/ANR-16-CONV-0005]. Funding for open access charge: Institut Pasteur.
Conflict of interest statement. None declared.
REFERENCES
- 1. Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.-F., Guindon S., Lefort V., Lescot M. et al.. Phylogeny. fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008; 36:W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Katoh K., Standley D.M.. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013; 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O.. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010; 59:307–321. [DOI] [PubMed] [Google Scholar]
- 4. Lefort V., Desper R., Gascuel O.. FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 2015; 32:2798–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lemoine F., Domelevo Entfellner J.-B., Wilkinson E., Correia D., Dàvila Felipe M., De Oliveira T., Gascuel O.. Renewing Felsenstein’s phylogenetic bootstrap in the era of big data. Nature. 2018; 556:452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Miller M.A., Pfeiffer W., Schwartz T.. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE), 2010. 2010; IEEE; 1–8. [Google Scholar]
- 7. Boc A., Diallo A.B., Makarenkov V.. T-REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 2012; 40:W573–W579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sánchez R., Serra F., Tárraga J., Medina I., Carbonell J., Pulido L., de María A., Capella-Gutíerrez S., Huerta-Cepas J., Gabaldón T. et al.. Phylemon 2.0: a suite of web-tools for molecular evolution, phylogenetics, phylogenomics and hypotheses testing. Nucleic Acids Res. 2011; 39:W470–W474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gouy M., Guindon S., Gascuel O.. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2009; 27:221–224. [DOI] [PubMed] [Google Scholar]
- 10. Kumar S., Stecher G., Li M., Knyaz C., Tamura K.. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018; 35:1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goecks J., Nekrutenko A., Taylor J.. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010; 11:R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Afgan E., Baker D., Batut B., van den Beek M., Bouvier D., Čech M., Chilton J., Clements D., Coraor N., Grüning B.A. et al.. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018; 46:W537–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oakley T.H., Alexandrou M.A., Ngo R., Pankey M.S., Churchill C.K., Chen W., Lopker K.B.. Osiris: accessible and reproducible phylogenetic and phylogenomic analyses within the Galaxy workflow management system. BMC Bioinformatics. 2014; 15:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lord E., Leclercq M., Boc A., Diallo A.B., Makarenkov V.. Armadillo 1.1: an original workflow platform for designing and conducting phylogenetic analysis and simulations. PLoS One. 2012; 7:e29903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guang A., Zapata F., Howison M., Lawrence C.E., Dunn C.W.. An integrated perspective on phylogenetic workflows. Trends Ecol. Evol. 2016; 31:116–126. [DOI] [PubMed] [Google Scholar]
- 16. Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J. et al.. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011; 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000; 17:540–552. [DOI] [PubMed] [Google Scholar]
- 19. Criscuolo A., Gribaldo S.. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 2010; 10:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dress A.W., Flamm C., Fritzsch G., Grünewald S., Kruspe M., Prohaska S.J., Stadler P.F.. Noisy: identification of problematic columns in multiple sequence alignments. Algorith. Mol. Biol. 2008; 3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T.. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009; 25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Price M.N., Dehal P.S., Arkin A.P.. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010; 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goloboff P.A., Farris J.S., Nixon K.C.. TNT, a free program for phylogenetic analysis. Cladistics. 2008; 24:774–786. [Google Scholar]
- 24. Lefort V., Longueville J.-E., Gascuel O.. SMS: smart model selection in PhyML. Mol. Biol. Evol. 2017; 34:2422–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ronquist F., Teslenko M., Van Der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P.. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012; 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Junier T., Zdobnov E.M.. The Newick utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics. 2010; 26:1669–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tan G., Muffato M., Ledergerber C., Herrero J., Goldman N., Gil M., Dessimoz C.. Current methods for automated filtering of multiple sequence alignments frequently worsen single-gene phylogenetic inference. Syst. Biol. 2015; 64:778–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yachdav G., Wilzbach S., Rauscher B., Sheridan R., Sillitoe I., Procter J., Lewis S.E., Rost B., Goldberg T.. MSAViewer: interactive JavaScript visualization of multiple sequence alignments. Bioinformatics. 2016; 32:3501–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shank S.D., Weaver S., Pond S. L.K.. phylotree.js—a JavaScript library for application development and interactive data visualization in phylogenetics. BMC Bioinformatics. 2018; 19:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Letunic I., Bork P.. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016; 44:W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dereeper A., Audic S., Claverie J.-M., Blanc G.. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 2010; 10:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 33. Mareuil F., Doppelt-Azeroual O., Ménager H.. A public Galaxy platform at Pasteur used as an execution engine for web services [version 1; not peer reviewed]. F1000Research. 2017; 6:1030. [Google Scholar]
- 34. Sloggett C., Goonasekera N., Afgan E.. BioBlend: automating pipeline analyses within Galaxy and CloudMan. Bioinformatics. 2013; 29:1685–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NGPhylogeny.fr is freely available at https://ngphylogeny.fr. Source codes of the web interface, wrappers and worflows are available on GitHub at C3BI-pasteur-fr/ngphylogeny-django and C3BI-pasteur-fr/ngphylogeny-galaxy. The two Docker images are stored on Docker Hub at evolbioinfo/ngphylogeny-galaxy and evolbioinfo/ngphylogeny.