Abstract
MyDGR is a web server providing integrated prediction and visualization of Diversity-Generating Retroelements (DGR) systems in query nucleotide sequences. It is built upon an enhanced version of DGRscan, a tool we previously developed for identification of DGR systems. DGR systems are remarkable genetic elements that use error-prone reverse transcriptases to generate vast sequence variants in specific target genes, which have been shown to benefit their hosts (bacteria, archaea or phages). As the first web server for annotation of DGR systems, myDGR is freely available on the web at http://omics.informatics.indiana.edu/myDGR with all major browsers supported. MyDGR accepts query nucleotide sequences in FASTA format, and outputs all the important features of a predicted DGR system, including a reverse transcriptase, a template repeat and one (or more) variable repeats and their alignment featuring A-to-N (N can be C, T or G) substitutions, and VR-containing target gene(s). In addition to providing the results as text files for download, myDGR generates a visual summary of the results for users to explore the predicted DGR systems. Users can also directly access pre-calculated, putative DGR systems identified in currently available reference bacterial genomes and a few other collections of sequences (including human microbiomes).
INTRODUCTION
Mutations are among the important driving forces of the evolution of all organisms and viruses and their adaptation to new niches. The diversity-generating retroelements (DGRs) are genetic elements that can produce targeted, massive variations in the genomes that carry these elements (1). The DGR systems rely on error-prone reverse transcriptases to produce mutagenized cDNA (containing A-to-N mutations) from a template region (TR), to replace a segment called variable region (VR) that is similar to the TR region—this process is called mutagenic retrohoming. The DGR system was first found in Bordetella phage (BPP-1) (1), which was shown to contribute to its host tropsim specificity; specifically, the VR is part of a gene (called target gene) encoding for the phage’s receptor-binding protein (Mtd). It was projected that the DGR system could potentially generate massive variations (e.g. >1018 Mtd variants) in Bordetella phage (2,3) as a result of the targeted mutagenesis mechanism.
Studies have revealed sequence and structural features important for the mutagenic retrohoming mechanism (4). The DGR-specific reverse transcriptases belong to a large family of RT genes, which also include RT genes associated with group II introns, retrons, phage infection retroelements (Abi), and some CRISPR-Cas defense systems (5). A gene called avd that encodes accessory variability determinant (Avd) protein is often found with other core DGR elements. The tertiary structure of Avd and mutational analysis revealed a strict correspondence between retrohoming and the interaction of Avd with RT, suggesting that the RT-Avd complex is important for DGR retrohoming (6). Handa et al (7) recently showed that a complex of the RT and Avd protein along with DGR RNA were necessary and sufficient for synthesis of template-primed, covalently linked RNA-cDNA molecules. Additional sequence features of DGR systems include the IMH (initiation of mutagenic retrohoming) site (at the end of the VR) and the IMH* site found in the TR segment: IMH marks the 3’ boundary of A-to-N mutagenesis in the VR (8) and is often followed by a GC-rich inverted repeat required for efficient mutagenic retrohoming (9); and the IMH* in the TR segment differs from IMH and is not followed by an inverted repeat, thereby distinguishing the TR donor sequence from the recipient target DNA sequence (10). Although GC-rich inverted repeats in the downstream of IMH sites were found to be essential for efficient mutagenic retrohoming for the Bordetella and Legionella DGRs (9,10), these repeats are not considered universal features of DGR systems (3).
DGR systems have evolved to confer important functions to their hosts. The DGR system in Bordetella phage mediates the phage tropism specificity (1). Legionella pneumophila contains a DGR system that diversifies a gene encoding for a lipoprotein that is anchored in the outer leaflet of the outer membrane (9). Paul et al reported intact DGRs in two distinct intraterrestrial archaeal systems, including a novel virus that appears to infect archaea in the marine subsurface and two uncultivated nanoarchaea from the terrestrial subsurface (11). A recent survey revealed a large number of DGR systems in temperate phages, leading to a hypothesis that DGR may be a ubiquitous mechanism underlying phage-bacteria interaction in the human microbiome (12). Cornuault et al. found that a large fraction of Faecalibacterium prausnitzii phages (10 of the 18 phages) contain DGR systems, and they hypothesized that these DGR systems contribute to the phages’ adaptation to the digestive tract environment, considering that F. prausnitzii is found depleted in inflammatory bowel disease (IBD) patients (13). Lastly, Paul et al. found prominent DGR systems in genomically reduced organisms from the bacterial candidate phyla radiation (CPR) and uncultivated phyla belonging to the archaeal superphylum called DPANN (14). The great variability of target genes revealed in all these studies, including our own study of the DGR systems in human microbiomes (15), implies important roles of DGRs in many undiscovered biological processes.
While DGR target proteins share low sequence identity, the structures of several such proteins have revealed the C-type lectin (CLec) fold as a conserved scaffold for accommodating massive sequence variation (16–18). A recent example is the target protein encoded by a prophage of the thermophile Thermus aquaticus: its variable region is nearly identical in structure to those of several other DGR variable proteins containing the CLec fold despite the low sequence identity among them (17). Wu et al classified the VR-encoding domains of the variable proteins into several classes based on their sequence alignments, including three C-type lectin folds (CLec1, CLec2 and CLec3), two Ig fold classes (named Ig1 and Ig2), and several additional classes of unknown VR domains (3).
To the best of our knowledge, currently there are only two publicly available computational tools for automated prediction of DGR systems, DiGReF (19) and our own tool DGRscan (15). Considering the biological importance of DGR systems, and their potential applications in molecular display (20), there is a clear need to develop a web server for automated annotation of DGR with friendly user interface, providing annotations of DGR systems in individual bacterial genomes and metagenomes.
MATERIALS AND METHODS
Detection of core components of DGR system using DGRscan
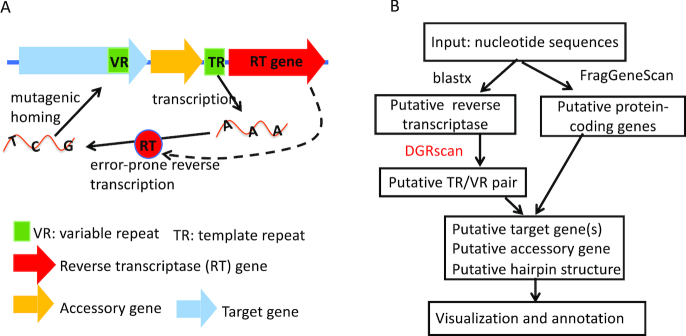
MyDGR is built upon an improved version of DGRscan we previously developed (15). As shown in Figure 1A, a minimal DGR system consists of a RT gene and a TR-VR pair, and DGRscan was devised based on finding these core components. In particular, MyDGR uses the de novo search function in DGRscan (see Figure 1B). Given an input nucleotide sequence, MyDGR first identifies putative RT genes by searching the translated nucleotide sequence against a protein database of 155 RT proteins (21) (using blastx). If it finds putative RT genes, it then scans in the neighborhood of each of these putative RT genes (10 kb in both ends), searching for segments that potentially form a TR-VR pair: two repeats that are similar to each other spanning at least 60 bp with seven or more substitutions involving adenines in one of the repeats (i.e. the TR), allowing only a small fraction (≤30%) of the substitutions to be involved in non-As in the putative TR. Although rare, TR and VR may be on opposite strands in some genomes (3,19)—myDGR does not limit its search for TR–VR pairs on the same strand. A dynamic programming algorithm is used for aligning the candidate TR-VR pairs; however, to speed up the alignment process, a full dynamic programming is called only when a seed match of at least 60 bp (without indels) is found between two candidate segments. We also note that using putative RT as the constraint not only significantly reduces the search space of TR-VR pairs, but also helps eliminate potential false DGRs.
Figure 1.
Illustration of DGR systems and their prediction using myDGR. (A) The main components in DGR sytsems; (B) the diagram of myDGR pipeline, which uses DGRscan to search for potential TR-VR pairs, enhanced with other functions including identification of target genes and accessory genes.
Prediction of remote target genes using DGRscan-remote
MyDGR uses an added option of DGRscan (DGRscan-remote), which enables searching for remote target genes once a target gene close to the RT gene is predicted. First, the predicted TR sequence is used to search (using blastn (22)) for similar segments in the same sequence. These segments will be further examined by DGRscan-remote, which searches for A-to-N substitutions, the hallmark feature of the VR regions found in target genes.
To predict domains in putative target proteins (encoded by the core target gene and remote ones), myDGR searches (using hmmscan) them against curated Hidden Markov Models (HMM) of known target-protein-associated domains (including CLec1, CLec2, CLec3, Ig1 and Ig2) (3), Pfam-A domains (23), and CDD domains (24). We apply an e-value cutoff of 0.001 for the domain prediction. When putative domains overlap, the one with lower e-value is chosen over the others. Also as CDD consists of domains from Pfam and other sources, myDGR only considers CDD domains that do not overlap with Pfam-A domains to avoid redundancy.
Prediction of GC-rich inverted repeats (hairpin structures)
MyDGR looks for potential GC-rich inverted repeats (which form hairpin structures) in the downstream of putative VR regions by searching for short segments that can form a GC-rich stem of 7–10 bp in length, connecting a loop of 4 nt with the motif of GRNA (R = A or G, N can be any), that is 4–34 bp away from the VRs; these features were shown to be important for target site recognition (10).
Prediction of putative accessory genes
Accessory genes are often found in the immediate neighborhood of the core DGR components (i.e. the RT, TR and the nearby VR-containing target gene). MyDGR extracts adjacent two genes in both directions of identified RT gene, and compare their protein sequences against the HMMs of domains built from previously identified accessory genes (3) using hmmscan search (25). The protein predicted to contain one of these domains is reported as the putative accessory gene. However, accessory genes are poorly conserved, and thus some of them might be missed by the similarity searches. If that happens, the visual summary of the DGR systems produced by myDGR will provide a convenient way for users to manually check potential accessory genes.
Datasets
We tested myDGR on three collections of data sets. The first collection is composed of 372 genomes predicted to contain DGR systems (3), including 246 of reference genomes (‘core set’) and 126 genomically-reduced genomes (‘CPR’ set) (11,14,26) (we refer to this data set as Zimmerly collection). The second dataset contains 29 contigs assembled from the human virome (27), and the third dataset contains 559 assembly scaffolds from the human microbiome (HMP) dataset (28,29). We note the latter two datasets were used to test DGRscan and the results were reported in our previous publication (15). We applied myDGR to these two datasets and made the results (putative DGR systems and their annotations) available on myDGR server.
UTILITY AND WEB INTERFACE
Input
MyDGR takes query nucleotide sequences in FASTA format. As an optional input, a user may upload gene predictions (in gff format) of the query nucleotide sequence; otherwise, myDGR calls FragGeneScan (30) to predict open reading frames (ORFs) in the query nucleotide sequences. An example of input is provided on the Prediction page at the myDGR website.
Details on the web server usage can be found on the website Help page. Once a job is submitted, the user is redirected to a webpage reporting the status of the job, which automatically refreshes every 10s until the job is completed. This page can be bookmarked for later uses. Results also can be retrieved by providing job IDs to the web site. If the user provides an email address, a notification containing a link to the result page will be sent when the job finishes.
Output
MyDGR provides fast annotation of DGR systems: annotation of an average size bacterial genome takes several minutes. The output results are presented to the user through both an interactive viewer and downloadable files. The interactive viewer provides visualization of annotated DGR systems, including a global view showing the location of the systems in the input sequence, and views of the different components. Further information including location and annotation of each component will be shown by simply hovering the mouse over on each locus. Links are provided to show additional information, including the target gene, alignment of the TR and VR, and putative hairpin in the downstream of the VR region. Text files of the results are also available for download.
Pre-calculated putative DGR systems in reference genomes and other collections of sequences
MyDGR provides the results of putative DGR systems predicted in reference bacterial genomes (downloaded from the NCBI ftp site) through their accession numbers and/or species names. In addition, we made available myDGR predictions for the three datasets we used.
EVALUATION AND DEMONSTRATION EXAMPLES
Our previous publication (15) reported the evaluation of DGRscan using the human virome dataset, which showed accurate and fast prediction of DGR systems. Here we further tested the integrated pipeline myDGR using the Zimmerly collection (3) (see details on myDGR web server under the Collection page). For comparison, we focused on RT genes, target genes, and the TR/VR pairs. For most of the DGR systems, myDGR’s predictions are consistent with previous annotations, with the exceptions of six cases. We believe that these six cases either contain atypical DGR systems (e.g. CALI01000035.1 involves only three A-to-N substitutions in its reported TR-VR pair), or are likely false positives (e.g. the reported TR-VR pair in LCHU01000008.1 involves only four A-to-N substitutions but nine other mutations; and the putative TR-VR pair in LCDE01000016.1 involves seven A-to-N substitutions where N is G in all case and the TR and VR are very short). Among the 366 DGRs predicted by myDGR, in 246 (67.2%) cases the TRs exactly match with those previously reported (3), and the number increases to 354 (96.7%) when ≥80% overlap between the TR regions is required.
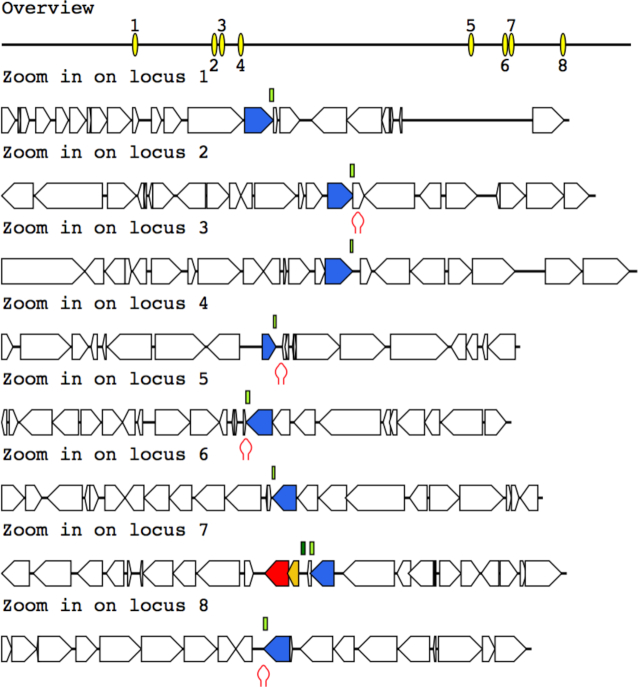
To demonstrate the functionalities of myDGR, we show the DGR systems identified from three genomes (users can explore these DGR systems on myDGR web server under Demo page). The first one is the DGR system in Bordetella phage. Just as expected, the predicted DGR system contains a RT gene and a target gene, and an accessory gene in between. MyDGR was also able to predict a hairpin structure following the VR region in this genome. The target protein is found to contain the CLec1 domain at its C-terminal (the VR part) and the Mtd domain. The second case is the DGR system in the T. denticola genome, which was predicted to contain a large number of target genes (including one that is close to the RT gene, and seven that scatter across the genome, as shown clearly in the graphical output from myDGR) (Figure 2). Manual examination of the TR-VR alignments and domain composition of the proteins encoded by the target genes suggest that all eight genes appear to be typical target genes. All eight of the target proteins contain the FGE-sulfatase domain, and seven contain the CLec1 domain in their VR-coding regions. The putative accessory gene contains a HRDC domain, a domain often found in accessory genes.
Figure 2.
The DGR system found in T. denticola. This DGR system contains eight target genes, one close to the RT gene, and the rest scatter across the genome. Note that the core component of the DGR system (shown as locus 7 in the graph) includes the RT gene (red arrow), a target gene (blue), and a putative accessory gene (orange). Putative hairpin structures (shown as red hairpins in the figure) are found in the downstream of some target genes. Other protein-coding genes are shown as open arrows in the figure.
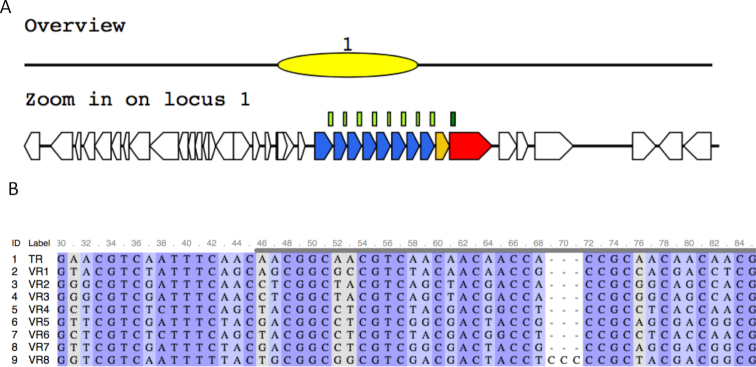
The third example is the DGR system found in a contig assembled from Stenotrophomonas sp. SKA14 (GenBank ID: ACDV01000044.1; contig id: ctg_1108481805216). Interestingly, in this case, all eight target genes are located close in tandem (see Figure 3A). The farthest target gene to the core DGR is longer than the other seven with about the same length. All target proteins contain the typical CLec2 domain. Alignment of the VR regions from all target genes and the TR region clearly shows the typical A-to-N substitutions (Figure 3B).
Figure 3.
The DGR system found in Stenotrophomonas sp. SKA14. The DGR system contains eight target genes, all in one cluster (A). The core DGR system contains a RT gene (red arrow), a target gene (blue), and a putative accessory gene (orange), which contains a AVD domain. (B) The alignment between the TR and VR regions, highlighting A-to-N substitutions.
We note that myDGR can predict DGR systems in which the TR and VRs are found on the opposite strands of the input sequence, or TR and VRs are on the same strand but are on the opposite strand of the RT gene (although those cases are rare). If applicable, MyDGR can also predict multiple DGR systems (each with its own RT and TR) in a single genome. We provide cases showing these different scenarios on myDGR server (Demo page).
FUNDING
Division of Biological Infrastructure, National Science Foundation [DBI-1262588]; National Institute of Allergy and Infectious Diseases, National Institute of Health [1R01AI108888]. Funding for open access charge: National Institute of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Liu M., Deora R., Doulatov S.R., Gingery M., Eiserling F.A., Preston A., Maskell D.J., Simons R.W., Cotter P.A., Parkhill J. et al.. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science. 2002; 295:2091–2094. [DOI] [PubMed] [Google Scholar]
- 2. Bikard D., Marraffini L.A.. Innate and adaptive immunity in bacteria: mechanisms of programmed genetic variation to fight bacteriophages. Curr. Opin. Immunol. 2012; 24:15–20. [DOI] [PubMed] [Google Scholar]
- 3. Wu L., Gingery M., Abebe M., Arambula D., Czornyj E., Handa S., Khan H., Liu M., Pohlschroder M., Shaw K.L. et al.. Diversity-generating retroelements: natural variation, classification and evolution inferred from a large-scale genomic survey. Nucleic Acids Res. 2018; 46:11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. LaRoche-Johnston F., Monat C., Coulombe S., Cousineau B.. Bacterial group II introns generate genetic diversity by circularization and trans-splicing from a population of intron-invaded mRNAs. PLoS Genet. 2018; 14:e1007792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Toro N., Nisa-Martinez R.. Comprehensive phylogenetic analysis of bacterial reverse transcriptases. PLoS ONE. 2014; 9:e114083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alayyoubi M., Guo H., Dey S., Golnazarian T., Brooks G.A., Rong A., Miller J.F., Ghosh P.. Structure of the essential diversity-generating retroelement protein bAvd and its functionally important interaction with reverse transcriptase. Structure. 2013; 21:266–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Handa S., Jiang Y., Tao S., Foreman R., Schinazi R.F., Miller J.F., Ghosh P.. Template-assisted synthesis of adenine-mutagenized cDNA by a retroelement protein complex. Nucleic Acids Res. 2018; 46:9711–9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Doulatov S., Hodes A., Dai L., Mandhana N., Liu M., Deora R., Simons R.W., Zimmerly S., Miller J.F.. Tropism switching in Bordetella bacteriophage defines a family of diversity-generating retroelements. Nature. 2004; 431:476–481. [DOI] [PubMed] [Google Scholar]
- 9. Arambula D., Wong W., Medhekar B.A., Guo H., Gingery M., Czornyj E., Liu M., Dey S., Ghosh P., Miller J.F.. Surface display of a massively variable lipoprotein by a Legionella diversity-generating retroelement. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:8212–8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guo H., Tse L.V., Nieh A.W., Czornyj E., Williams S., Oukil S., Liu V.B., Miller J.F.. Target site recognition by a diversity-generating retroelement. PLoS Genet. 2011; 7:e1002414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Paul B.G., Bagby S.C., Czornyj E., Arambula D., Handa S., Sczyrba A., Ghosh P., Miller J.F., Valentine D.L.. Targeted diversity generation by intraterrestrial archaea and archaeal viruses. Nat. Commun. 2015; 6:6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Benler S., Cobian-Guemes A.G., McNair K., Hung S.H., Levi K., Edwards R., Rohwer F.. A diversity-generating retroelement encoded by a globally ubiquitous Bacteroides phage. Microbiome. 2018; 6:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cornuault J.K., Petit M.A., Mariadassou M., Benevides L., Moncaut E., Langella P., Sokol H., De Paepe M.. Phages infecting Faecalibacterium prausnitzii belong to novel viral genera that help to decipher intestinal viromes. Microbiome. 2018; 6:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Paul B.G., Burstein D., Castelle C.J., Handa S., Arambula D., Czornyj E., Thomas B.C., Ghosh P., Miller J.F., Banfield J.F. et al.. Retroelement-guided protein diversification abounds in vast lineages of Bacteria and Archaea. Nat. Microbiol. 2017; 2:17045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ye Y. Identification of diversity-generating retroelements in human microbiomes. Int. J. Mol. Sci. 2014; 15:14234–14246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Le Coq J., Ghosh P.. Conservation of the C-type lectin fold for massive sequence variation in a Treponema diversity-generating retroelement. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:14649–14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Handa S., Shaw K.L., Ghosh P.. Crystal structure of a Thermus aquaticus diversity-generating retroelement variable protein. PLoS ONE. 2019; 14:e0205618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dai W., Hodes A., Hui W.H., Gingery M., Miller J.F., Zhou Z.H.. Three-dimensional structure of tropism-switching Bordetella bacteriophage. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:4347–4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schillinger T., Lisfi M., Chi J., Cullum J., Zingler N.. Analysis of a comprehensive dataset of diversity generating retroelements generated by the program DiGReF. BMC Genomics. 2012; 13:430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Overstreet C.M., Yuan T.Z., Levin A.M., Kong C., Coroneus J.G., Weiss G.A.. Self-made phage libraries with heterologous inserts in the Mtd of Bordetella bronchiseptica. Protein Eng. Des. Sel. 2012; 25:145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schillinger T., Zingler N.. The low incidence of diversity-generating retroelements in sequenced genomes. Mob. Genet. Elem. 2012; 2:287–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L.. BLAST+: architecture and applications. BMC Bioinformatics. 2009; 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sonnhammer E.L., Eddy S.R., Durbin R.. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins. 1997; 28:405–420. [DOI] [PubMed] [Google Scholar]
- 24. Marchler-Bauer A., Lu S., Anderson J.B., Chitsaz F., Derbyshire M.K., DeWeese-Scott C., Fong J.H., Geer L.Y., Geer R.C., Gonzales N.R. et al.. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011; 39:D225–D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eddy S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011; 7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rinke C., Schwientek P., Sczyrba A., Ivanova N.N., Anderson I.J., Cheng J.F., Darling A., Malfatti S., Swan B.K., Gies E.A. et al.. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013; 499:431–437. [DOI] [PubMed] [Google Scholar]
- 27. Minot S., Bryson A., Chehoud C., Wu G.D., Lewis J.D., Bushman F.D.. Rapid evolution of the human gut virome. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:12450–12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huttenhower C., Gevers D., Knight R., Abubucker S., Badger J.H., Chinwalla A.T., Creasy H.H., Earl A.M., FitzGerald M.G., Fulton R.S. et al.. Structure, function and diversity of the healthy human microbiome. Nature. 2012; 486:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Human Microbiome Project Consortium. A framework for human microbiome research. Nature. 2012; 486:215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rho M., Tang H., Ye Y.. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res. 2010; 38:e191. [DOI] [PMC free article] [PubMed] [Google Scholar]