Abstract
PubTator Central (https://www.ncbi.nlm.nih.gov/research/pubtator/) is a web service for viewing and retrieving bioconcept annotations in full text biomedical articles. PubTator Central (PTC) provides automated annotations from state-of-the-art text mining systems for genes/proteins, genetic variants, diseases, chemicals, species and cell lines, all available for immediate download. PTC annotates PubMed (29 million abstracts) and the PMC Text Mining subset (3 million full text articles). The new PTC web interface allows users to build full text document collections and visualize concept annotations in each document. Annotations are downloadable in multiple formats (XML, JSON and tab delimited) via the online interface, a RESTful web service and bulk FTP. Improved concept identification systems and a new disambiguation module based on deep learning increase annotation accuracy, and the new server-side architecture is significantly faster. PTC is synchronized with PubMed and PubMed Central, with new articles added daily. The original PubTator service has served annotated abstracts for ∼300 million requests, enabling third-party research in use cases such as biocuration support, gene prioritization, genetic disease analysis, and literature-based knowledge discovery. We demonstrate the full text results in PTC significantly increase biomedical concept coverage and anticipate this expansion will both enhance existing downstream applications and enable new use cases.
INTRODUCTION
Automated text mining is becoming increasingly important for accessing and extracting knowledge within the biomedical literature (1). Web-based tools simplify distributing results from state-of-the-art text mining systems, due to their platform independence and the lack of installation, maintenance, and infrastructure requirements (2). As such, several web-based tools have been recently developed to provide automated concept annotations to support downstream text mining tasks in the biomedical domain (3–9). PubTator (10) was one of the first such systems to provide automated concept annotations of several important biomedical concept types – genes/proteins, genetic variants, diseases, chemicals, and species – across all PubMed article abstracts. PubTator features a PubMed-like web interface for ease of use, with a RESTful web service (API) added in April 2015 (2). PubTator is updated with new PubMed articles daily, and the API has currently served annotated abstracts for ∼300 million requests. Third-party researchers have used PubTator in a variety of use cases, including biocuration support (11–13), gene prioritization (14,15), genetic disease analysis (16), literature-based knowledge discovery (17,18) and downstream text mining (7,19–24).
An important limitation of PubTator has been the lack of results from full text articles. Human indexers and biocurators both require access to full text (25,26), and studies have shown that text mining efforts limited to abstracts lack important knowledge present in the full text (25,27,28). Full text articles are more complex than abstracts and ∼40 times longer, making them more difficult for text mining. Moreover, in the past full text has been significantly less available than abstracts. In recent years, however, the availability of full text articles for text mining has increased dramatically, with the percentage of articles in PubMed Central available for text mining approaching ∼80% (29).
In this work, we describe PubTator Central (PTC), a new implementation of the PubTator service expanding automated concept annotation to full text articles and including a web interface designed for full text, higher throughput architecture, updated concept annotation methods, and an expanded set of concept types. PTC annotates PubMed abstracts and the full text documents in PubMed Central Text Mining (PMC-TM) subset, including both the Open Access Subset and the Author Manuscript Collection. PTC thus contains over 29 million abstracts and ∼3 million full text documents, which expand the total number of annotations nearly four-fold over abstracts alone. PTC includes a completely new web interface, featuring semantic search and improved navigation in full text. The server-side architecture is significantly redesigned, exploiting nonrelational data to increase throughput despite the increased load. PTC annotates the concept types supported by the original PubTator system (genes/proteins, genetic variants, diseases, chemicals, and species) and expands the annotated concept types to include cell lines. Updated concept identification methods and a new disambiguation module based on cutting-edge deep learning techniques provide increased accuracy. Annotations in PTC are available in multiple formats (XML, JSON and tab delimited) via the online interface, a RESTful web service or FTP download. PTC is synchronized with PubMed and PMC-TM, with new articles added daily.
SYSTEM OVERVIEW
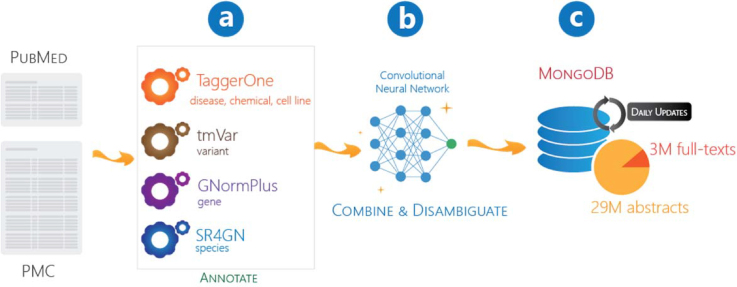
New articles in PubMed or PMC-TM are first processed through a series of concept taggers (Figure 1A) to obtain annotations for each bioconcept type. In this manuscript, an annotation consists of a contiguous text span, a concept type, and an accession identifier. The disambiguation module (Figure 1B) then resolves annotation conflicts (overlapping annotations). Annotated articles are subsequently stored in a MongoDB database (Figure 1C), and made available to users via the new PTC web interface and the RESTful API for programmatic access. To ensure consistency, the input/output text files for each step in the PTC processing pipeline are handled by BioC, a community-driven biomedical text processing data format for improved interoperability (30).
Figure 1.
PTC processing pipeline. PubMed abstracts and PMC-TM full text articles are annotated by multiple concept taggers (A), conflicts/overlapping annotations handled by the disambiguation module (B) and results stored in the database (C).
Concept annotation improvements
While the concept taggers used in PubTator (10) demonstrated high performance in their respective benchmarks (31–34), the content of full text articles is more complex than abstracts, making them more difficult to annotate accurately. Hence, our concept taggers are either modified and/or re-trained (if a corpus of full-text article is available) for improved performance. Specifically, genes/proteins are annotated in PTC by GNormPlus (35) and normalized to NCBI Gene identifiers. GNormPlus integrates several text mining approaches (e.g. AB3P (36) for abbreviation resolution and SimConcept (37) for composite mentions) for improved accuracy. Genetic variants are annotated by tmVar 2.0 (38), an improved version of tmVar (31) that maps recognized variant mentions to dbSNP RS identifiers. For PTC, tmVar 2.0 was re-trained using both abstracts and full text to improve performance. Species continue to be tagged by SR4GN (33), which provides NCBI taxonomy identifiers. Diseases, chemicals and cell lines are all annotated by TaggerOne (39), using separate models. Diseases and chemical names are normalized to MeSH identifiers while cell lines are normalized to Cellosaurus (40). We improved the performance of TaggerOne for chemicals by re-training the model using a combination of the BioCreative V CDR corpus (41) and the CHEMDNER corpus (42).
Table 1 lists the different taggers and their performance in PubTator vs. PTC. Detailed evaluation results (i.e. recall and precision) are reported in the Supplementary material (Supplementary Table S1). The taggers for two concept types were evaluated on full text corpora (genetic variants (24) and species (43)), while the tagger for cell lines was evaluated on a corpus derived from figure captions (44). The taggers for the remaining concept types were evaluated on corpora derived from abstracts (genes (45), diseases (46) and chemicals (41)).
Table 1.
Improvements in concept tagger performance from PubTator to PTC for each concept type
| Performance | ||||
|---|---|---|---|---|
| Type | Training/evaluation corpus | Doc type | PubTator | PTC |
| Gene | BioCreative II GN (45) | Abstract | GenNorm (34) 80.10% | GNormPlus (35) 86.70% |
| Variant | BRONCO (24) | Full text | tmVar (31) N/A | tmVar 2.0 (38) 86.24% |
| Disease | NCBI Disease (46) | Abstract | DNorm (32) 80.60% | TaggerOne (39) 83.70% |
| Chemical | BioCreative V CDR (41) | Abstract | Dictionary 53.82% | TaggerOne 89.50% |
| Species | Linnaeus (43) | Full text | SR4GN (33) 85.42% | SR4GN (33) 85.42% |
| Cell Line | BioCreative VI BioID corpus (44) | Full text (caption) | N/A | TaggerOne 83.10% |
Performance listed is the F1 score for concept identification (normalization). The previous version of tmVar does not provide accession identifiers (dbSNP RS numbers) for variants located within the text. Cell line annotations are new in PTC.
Concept disambiguation module
Recognizing multiple concept types simultaneously occasionally results in text spans being annotated as more than one bioconcept type. For example, ‘CO2’ may be recognized as both the chemical ‘carbon dioxide’ (MESH: D002245) and the gene ‘complement C2’ (EntrezGene:717). Overlapping annotations were originally disambiguated in PubTator using a rule-based approach: a priority ordering was applied based on the precision of each concept tagger (from highest to lowest), specifically mutation > species > gene > chemical > disease. While this method is straightforward and useful, incorrect disambiguation remained a significant source of error. For example, if a cell line (e.g. ‘A2780S’ in PMID:25026335) or chemical (e.g. ‘C3368-A’ in PMID:7767952) were erroneously also marked as a genetic variant, then the rule-based disambiguation would always consider the mention to be a genetic variant.
Accurately disambiguating the bioconcept type is difficult without considering the surrounding context. We therefore approached this task as a classification problem and developed a novel convolutional neural network (CNN) (47) based method, which identifies the most likely bioconcept type using the syntax and semantics of both the span being classified and the surrounding words. Our disambiguation model consists of one CNN for syntactic features and a second for semantics, concatenated to a fully connected layer then output with a softmax layer.
Due to the high cost of annotating human ground truth, we trained our disambiguation model using a dataset constructed by cross-referencing annotations in multiple human-curated databases (e.g. CTDbase, gene2pubmed) with ambiguous annotations in PubTator. First, we examined the PubTator results to identify overlapping annotations to more than one bioconcept type. We then identified the likely correct type for each set of overlapping annotations by determining if one matched a manual annotation within the human-curated database for that type. Using a holdout portion of this dataset, the rule-based approach demonstrated an accuracy of 55.7%, while the new disambiguation model demonstrated a significantly higher accuracy of 85.2%. The disambiguation module is available as an open source download: (https://www.ncbi.nlm.nih.gov/CBBresearch/Lu/Demo/tmTools/download/BioConceptDisambiguation.zip).
Improved web interface for interactive access
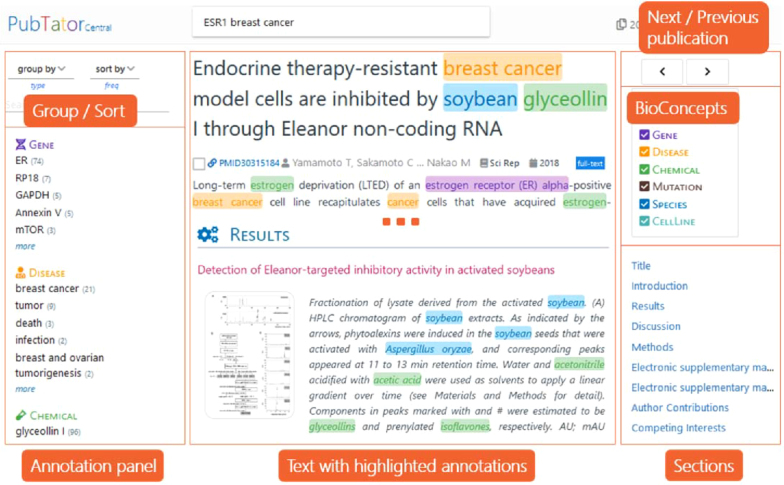
PubTator Central (PTC) features a completely new web interface, designed specifically for use with full text articles and implemented using the popular web framework AngularJS. The publication view page, shown in Figure 2, allows users to view and download concept annotations in individual articles. The left column of the page provides a quick summary of all concepts annotated within the article, grouped by concept type or section. Users may quickly navigate to concepts of interest within the annotation summary, then click to locate them in the article text. The annotation summary can be sorted by either annotation frequency or the position of the annotation within the text, and may be grouped by article section or concept type. The filter search box limits the concepts shown in the summary to those matching the input text. The center column of the publication view page shows the full text of the article with the annotated bioconcepts highlighted. Clicking on any annotation will display a tooltip showing the concept type for the annotation, its accession identifier, and a link to an external website with a description. The tooltip also contains three buttons: the magnifying glass button starts a new search for articles annotated with this concept, the RSS button subscribes the user to the RSS feed for this concept (to notify the user of new articles annotated with this concept), and the report button allows users to report a misannotated concept. The right column of the publication view page provides additional navigation and visualization tools. The ‘Next/Previous publication’ panel allows users to quickly scroll through search results. The ‘BioConcepts’ panel allows users to toggle the text highlighting for each concept type. The ‘Sections’ panel allows the user to quickly navigate to any desired section in the article.
Figure 2.
Displaying the abstract or full-text of a publication and related tools.
Users arrive at the publication view page by first locating articles of interest using either keyword or semantic searches. Keyword search uses the PubMed e-utils API to retrieve the list of relevant PMIDs for the query, which are then retrieved from our database and displayed in reverse date order. Semantic search returns articles annotated with a specific bioconcept, which may be of any of the six types supported by PTC (genes, diseases, chemicals, mutations, species and cell lines). Semantic searches utilize a specialized query format: @[ConceptType]@[ConceptID], for example: @gene@2099. Users may also limit results only full text articles using the ‘full-text only’ filter.
PTC allows users to organize articles into collections, which may then be viewed or downloaded together. Articles may be added to a collection via the publication view page or on the collection definition window using either a list of PMIDs entered manually, a file containing the list of PMIDs, or via query, which will add all matching articles. Articles can be removed from a collection by editing the list of PMIDs.
API for programmatic access
The PTC RESTful web service provides programmatic access to PTC results in a straightforward tab-delimited format (PubTator format), and two BioC-based formats: BioC-XML and BioC-JSON (48). PMC-TM full text articles require either BioC-XML or BioC-JSON, but PubMed abstracts are supported in all three formats. Table 2 presents examples of the web queries used to access the web service. Full details and code samples are provided at the online tutorial (https://www.ncbi.nlm.nih.gov/research/pubtator/api.html).
Table 2.
Usage for PTC RESTful web service API
| Description | URL |
|---|---|
| Abstract example | https://www.ncbi.nlm.nih.gov/research/pubtator-api/publications/export/pubtator?pmids=26739349,28483577 |
| Full text example | https://www.ncbi.nlm.nih.gov/research/pubtator-api/publications/export/biocxml?pmcids=PMC4743391 |
Server-side architecture improvements
Articles are preprocessed and stored in a MongoDB database. A Django web server handles requests from both the web application and RESTful API clients. Articles and their annotations are stored in each output format directly, allowing the web server to return requested data without conversion. Due to the enhancements in the back-end database, the PTC API is ∼70 times faster than the PubTator API. The maximum number of articles that may be requested per query has also increased from 100 for PubTator to 1000 for PTC.
A detailed point-by-point comparison between PubTator and PTC can be found in the Supplementary material (Supplementary Table S2).
USE CASES
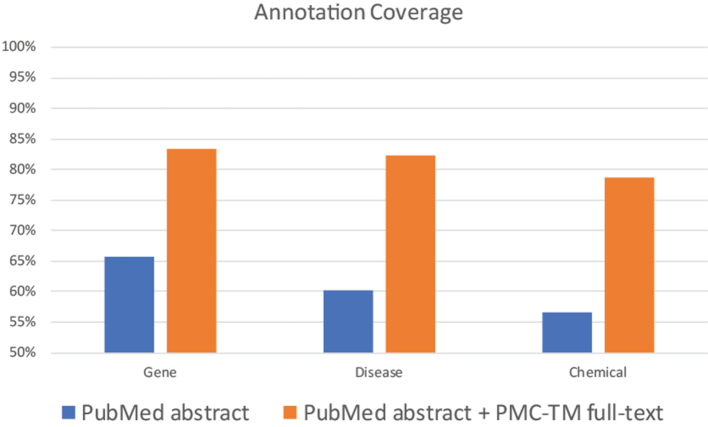
As an expansion of PubTator functionality, PTC provides increased accuracy and speed to existing PubTator use cases; in the past these have included a wide variety of third-party research (3,7,11–24). PTC also significantly increases the amount of knowledge that may be extracted from each article by mining its full text. To demonstrate this increase, we compared the results of our concept taggers in abstracts and full text articles against the annotations in human-curated databases. We considered three concept types: genes (using GeneRIF), diseases and chemicals (both using MeSH), and limited our analysis to articles present in the PMC-TM subset. As shown in Figure 3: our concept taggers extract between 56% and 65% of the manual annotations when applied to abstracts, but extract between 78% and 83% when applied to full text (an increase of 18–22%). Analysis of a sample of the concepts missed in both the abstracts and full text indicates that the remaining issues can be categorized into three groups. First, some diseases and chemicals are not easily annotated since the corresponding concept is not present in MeSH; this causes difficulties in both automated and manual annotation. Second, some errors (∼20%) are due to either boundary errors or type mismatches by the concept taggers or the disambiguation module. Third, the source text for the manual annotation may only be present in the supplementary material or figures, which are currently out of scope.
Figure 3.
Comparison of annotation coverage between processing PubMed abstracts and processing both abstracts and full text articles from PMC-TM.
We next describe two use cases demonstrating the advantages of the full text concept annotations provided by PTC.
Case 1: Enhancing downstream text mining applications
Several text mining groups using PubTator annotations (7,19,27) have sought full text annotations to provide more extensive automated literature analyses for a variety of downstream applications. Previous work has shown that the content of the full article text is substantially different from the abstracts (49). This is demonstrated by LitVar (28), a semantic search engine for genomic variants in PubMed and PMC, where mining the PMC-TM set (containing full text articles) increased the number of variants extracted approximately 2.6 times compared to only mining PubMed abstracts. Information retrieval is also more effective when performed on paragraphs from full text articles rather than only on abstracts (50). We therefore anticipate PTC enhancing a variety of downstream text mining applications.
Case 2: Improving biocuration support
Support for full text articles has been identified as a top priority for text mining workflows supporting biocuration (51). The full article text is an important source of information at multiple stages in the curation process, including document triage, where the curator must determine whether an article should be accepted for curation. Previous work integrating PubTator with the UniProtKB/Swiss-Prot curation workflow demonstrated that ranking the articles by the number of automatically-identified protein mentions is a highly effective method for identifying curatable articles on protein function (12). Variations of this approach may be applicable for other biocuration goals, such as identifying mutations associated with genetic diseases by simultaneously considering the counts of genes, mutations and diseases. Text mining may also reduce the amount of work the curator must perform during curation by providing preannotations: automated annotations of at least part of the curation task (13). Automated concept annotations in full text thus have the potential to improve the scalability of manual curation both by helping identify curatable articles and reducing the manual effort required.
CONCLUSION
We have described PubTator Central (PTC), a web-based system for automated concept annotations in PubMed abstracts and PMC-TM full text articles. PTC supports six concept types: genes/proteins, genetic variants, diseases, chemicals and cell lines. The new PTC web interface is designed for ease of use with full text articles and the server-side architecture supports significantly higher throughput. PTC includes updated concept identification methods and a new disambiguation model for increased accuracy. Annotations in PTC are available in multiple formats (XML, JSON and tab delimited) via the online interface, a RESTful web service or FTP download. PTC is updated daily, currently containing approximately 3 million full text articles and 29 million abstracts. The full text articles expand the amount of annotated text ∼4-fold over abstracts alone, enhancing existing downstream applications and enable new use cases. We anticipate PTC to become an important resource for future studies using knowledge extracted from the biomedical literature.
In future work, we intend to improve the search function to allow keyword and semantic queries to be combined. We also intend to include additional concept types (e.g. anatomical entities such as cellular components, cell types and tissues) and to improve the accuracies of the concept taggers in full text, especially in areas with minimal textual context, such as tables and supplementary material (52).
DATA AVAILABILITY
PubTator Central (PTC) is publicly available at https://www.ncbi.nlm.nih.gov/research/pubtator/.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH Intramural Research Program, National Library of Medicine, National Institutes of Health. Funding for open access charge: NIH Intramural Research Program, National Library of Medicine, National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Singhal A., Leaman R., Catlett N., Lemberger T., McEntyre J., Polson S., Xenarios I., Arighi C., Lu Z.. Pressing needs of biomedical text mining in biocuration and beyond: opportunities and challenges. Database. 2016; 2016:baw161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wei C.-H., Peng Y., Leaman R., Davis A.P., Mattingly C.J., Li J., Wiegers T.C., Lu Z.. Assessing the state of the art in biomedical relation extraction: overview of the BioCreative V chemical-disease relation (CDR) task. Database. 2016; 2016:baw032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Garcia-Pelaez J., Rodriguez D., Medina-Molina R., Garcia-Rivas G., Jerjes-Sánchez C., Trevino V.. PubTerm: a web tool for organizing, annotating and curating genes, diseases, molecules and other concepts from PubMed records. Database. 2019; 2019:bay137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Soto A.J., Przybyła P., Ananiadou S.. Thalia: Semantic search engine for biomedical abstracts. Bioinformatics. 2018; bty871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Matos S. Configurable web-services for biomedical document annotation. J. Cheminform. 2018; 2018:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Venkatesan A., Kim J.-H., Talo F., Ide-Smith M., Gobeill J., Carter J., Batista-Navarro R., Ananiadou S., Ruch P., McEntyre J.. SciLite: a platform for displaying text-mined annotations as a means to link research articles with biological data. Wellcome Open Res. 2016; 1:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee S., Kim D., Lee K., Choi J., Kim S., Jeon M., Lim S., Choi D., Kim S., Tan A.-C. et al.. BEST: next-generation biomedical entity search tool for knowledge discovery from biomedical literature. PLoS One. 2016; 11:e0164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thomas P., Starlinger J., Vowinkel A., Arzt S., Leser U.. GeneView: a comprehensive semantic search engine for PubMed. Nucleic Acids Res. 2012; 40:W585–W591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rak R., Rowley A., Black W., Ananiadou S.. Argo: an integrative, interactive, text mining-based workbench supporting curation. Database. 2012; 2012:bas010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wei C.-H., Kao H.-Y., Lu Z.. PubTator: a Web-based text mining tool for assisting Biocuration. Nucleic Acids Res. 2013; 41:W518–W522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee K., Famiglietti M.L., McMahon A., Wei C.-H., MacArthur J.A.L., Poux S., Breuza L., Bridge A., Cunningham F., Xenarios I. et al.. Scaling up data curation using deep learning: An application to literature triage in genomic variation resources. PLoS Comput. Biol. 2018; 14:e1006390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Poux S., Arighi C.N., Magrane M., Bateman A., Wei C.-H., Lu Z., Boutet E., Bye-A-Jee H., Famiglietti M.L., Roechert B. et al.. On expert curation and scalability: UniProtKB/Swiss-Prot as a case study. Bioinformatics. 2017; 33:3454–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Burger J.D., Doughty E., Khare R., Wei C.-H., Mishra R., Aberdeen J., Tresner-Kirsch D., Wellner B., Kann M.G., Lu Z. et al.. Hybrid curation of gene–mutation relations combining automated extraction and crowdsourcing. Database. 2014; 2014:bau094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu J.-L., Zhao M.. A PubMed-wide study of endometriosis. Genomics. 2016; 108:151–157. [DOI] [PubMed] [Google Scholar]
- 15. Shao Q., Byrum S.D., Moreland L.E., Mackintosh S.G., Kannan A., Lin Z., Morgan M., Brendan C., Stack J., Cornelius L.A., Tackett A.J. et al.. A proteomic study of human Merkel cell carcinoma. J. Proteomics Bioinform. 2013; 6:275–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang L.-C., Ross K.E., Baffi T.R., Drabkin H., Kochut K.J., Ruan Z., D’Eustachio P., McSkimming D., Arighi C., Chen C. et al.. Integrative annotation and knowledge discovery of kinase post-translational modifications and cancer-associated mutations through federated protein ontologies and resources. Sci. Rep. 2018; 8:6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qin X., Wang S., Wu Y., Xia J.. Evaluation of the performance of BioNLP tools for discovering causal genes in terms with pathway enrichment. J. Phys. Conf. Ser. 2018; 1069:012037. [Google Scholar]
- 18. Lee K., Shin W., Kim B., Lee S., Choi Y., Kim S., Jeon M., Tan A.C., Kang J.. HiPub: translating PubMed and PMC texts to networks for knowledge discovery. Bioinformatics. 2016; 32:2886–2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pyysalo S., Baker S., Ali I., Haselwimmer S., Shah T., Young A., Guo Y., Högberg J., Stenius U., Narita M. et al.. LION LBD: a literature-based discovery system for cancer biology. Bioinformatics. 2018; bty845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Percha B., Altman R.B.. A global network of biomedical relationships derived from text. Bioinformatics. 2018; 34:2614–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nentidis A., Bougiatiotis K., Krithara A., Paliouras G., Kakadiaris I.. Results of the fifth edition of the BioASQ Challenge. BioNLP. 2017; 48–57. [Google Scholar]
- 22. Singhal A., Simmons M., Lu Z.. Text mining genotype-phenotype relationships from biomedical literature for database curation and precision medicine. PLoS Comput Biol. 2016; 12:e1005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mahmood A.S.M.A., Wu T.-J., Mazumder R., Vijay-Shanker K.. DiMeX: a text mining system for mutation-disease association extraction. PLoS One. 2016; 11:e0152725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee K., Lee S., Park S., Kim S., Kim S., Choi K., Tan A.C., Kang J.. BRONCO: Biomedical entity Relation ONcology COrpus for extracting gene-variant-disease-drug relations. Database. 2016; 2016:baw043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mork J., Aronson A., Demner-Fushman D.. 12 years on - Is the NLM medical text indexer still useful and relevant. J. Biomed. Semantics. 2017; 8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lu Z., Hirschman L.. Biocuration workflows and text mining: overview of the BioCreative 2012 Workshop Track II. Database. 2012; 2012:bas043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Westergaard D., Stærfeldt H.-H., Tønsberg C., Jensen L.J., Brunak S.. A comprehensive and quantitative comparison of text-mining in 15 million full-text articles versus their corresponding abstracts. PLoS Comput. Biol. 2018; 14:e1005962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Allot A., Peng Y., Wei C.-H., Lee K., Phan L., Lu Z.. LitVar: a semantic search engine for linking genomic variant data in PubMed and PMC. Nucleic. Acids. Res. 2018; 46:W530–W536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Comeau D.C., Wei C.-H., Doğan R.I., Lu Z.. PMC text mining subset in BioC: about 3 million full text articles and growing. Bioinformatics. 2019; btz070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peng Y., Tudor C.O., Torii M., Wu C.H., Vijay-Shanker K.. iSimp in BioC standard format: enhancing the interoperability of a sentence simplification system. Database. 2014; 2014:bau038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wei C.-H., Harris B.R., Kao H.-Y., Lu Z.. tmVar: A text mining approach for extracting sequence variants in biomedical literature. Bioinformatics. 2013; 29:1433–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Leaman R., Doğan R.I., Lu Z.. DNorm: disease name normalization with pairwise learning to rank. Bioinformatics. 2013; 29:2909–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wei C.-H., Kao H.-Y., Lu Z.. SR4GN: a species recognition software tool for gene normalization. PLoS One. 2012; 7:e38460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wei C.-H., Kao H.-Y.. Cross-species gene normalization by species inference. BMC Bioinformatics. 2011; 12:S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wei C.-H., Kao H.-Y., Lu Z.. GNormPlus: An integrative approach for tagging genes, gene families, and protein domains. Biomed Res Int. 2015; 2015:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sohn S., Comeau D.C., Kim W., Wilbur W.J.. Abbreviation definition identification based on automatic precision estimates. BMC Bioinformatics. 2008; 9:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wei C.-H., Leaman R., Lu Z.. SimConcept: a hybrid approach for simplifying composite named entities in biomedical text. IEEE J. Biomed. Health Inform. 2015; 19:1385–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wei C.-H., Phan L., Feltz J., Maiti R., Hefferon T., Lu Z.. tmVar 2.0: integrating genomic variant information from literature with dbSNP and ClinVar for precision medicine. Bioinformatics. 2017; 34:80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Leaman R., Lu Z.. TaggerOne: joint named entity recognition and normalization with semi-Markov Model. Bioinformatics. 2016; 32:2839–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bairoch A. The Cellosaurus, a Cell-Line Knowledge Resource. J. Biomol. Tech. 2018; 29:25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li J., Sun Y., Johnson R.J., Sciaky D., Wei C.-H., Leaman R., Davis A.P., Mattingly C.J., Wiegers T.C., Lu Z.. BioCreative V CDR task corpus: a resource for chemical disease relation extraction. Database. 2016; 2016:baw068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Krallinger M., Rabal O., Leitner F., Vazquez M., Salgado D., Lu Z., Leaman R., Lu Y., Ji D., Lowe D.M. et al.. The CHEMDNER corpus of chemicals and drugs and its annotation principles. J. Cheminform. 2015; 7:S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gerner M., Nenadic G., Bergman C.M.. LINNAEUS: a species name identification system for biomedical literature. BMC Bioinformatics. 2010; 11:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Arighi C., Hirschman L., Lemberger T., Bayer S., Liechti R., Comeau D., Wu C.. Bio-ID track overview. Proc. BioCreative Workshop. 2017; 482:376. [Google Scholar]
- 45. Morgan A.A., Lu Z., Wang X., Cohen A.M., Fluck J., Ruch P., Divoli A., Fundel K., Leaman R., Hakenberg J. et al.. Overview of BioCreative II gene normalization. Genome Biol. 2008; 9:S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Doğan R.I., Leaman R., Lu Z.. NCBI disease corpus: a resource for disease name recognition and concept normalization. J. Biomed. Inform. 2014; 47:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kim Y. Convolutional neural networks for sentence classification. EMNLP. 2014; 1746–1751. [Google Scholar]
- 48. Comeau D.C., Doğan R.I., Ciccarese P., Cohen K.B., Krallinger M., Leitner F., Lu Z., Peng Y., Rinaldi F., Torii M. et al.. BioC: a minimalist approach to interoperability for biomedical text processing. Database. 2013; 2013:bat064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cohen K.B., Johnson H.L., Verspoor K., Roeder C., Hunter L.E.. The structural and content aspects of abstracts versus bodies of full text journal articles are different. BMC Bioinformatics. 2010; 11:492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lin J. Is searching full text more effective than searching abstracts. BMC Bioinformatics. 2009; 10:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hirschman L., Burns G.A., Krallinger M., Arighi C., Cohen K.B., Valencia A., Wu C.H., Chatr-Aryamontri A., Dowell K.G., Huala E. et al.. Text mining for the biocuration workflow. Database. 2012; 2012:bas020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yepes A.J., Verspoor K.. Literature mining of genetic variants for curation: quantifying the importance of supplementary material. Database. 2014; 2014:bau003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
PubTator Central (PTC) is publicly available at https://www.ncbi.nlm.nih.gov/research/pubtator/.