OBJECTIVES:
Pancreatic ductal adenocarcinoma (PDAC) presents the lowest survival rate of all cancers because only 6% of patients reach five-year survival. Alterations in the expression of several microRNAs (miRNAs) occur in the tumor of PDAC and in preneoplastic lesions as the called intraductal papillary mucinous neoplasm (IPMN). Here, we aimed at identifying which miRNAs are significantly altered in liquid biopsies from patients with PDAC and IPMN to find new noninvasive biomarkers for early detection of PDAC.
METHODS:
We analyzed by real-time quantitative reverse transcription-PCR (qRT-PCR) the expression of 17 circulating miRNAs, previously found to be significantly overexpressed in tissue pancreatic neoplasms, in a set of 182 plasma samples (94 PDAC, 19 IPMN, 18 chronic pancreatitis, and 51 disease-free controls). Then, we analyzed CA19.9 levels in the same plasma set, and we assessed the diagnostic values of differentially expressed miRNAs, CA19.9, and all possible combinations.
RESULTS:
Of note, 16, 14, and 9 miRNAs were significantly increased in PDAC, IPMN, and chronic pancreatitis, respectively, compared with control plasmas. miR-21-5p, miR-33a-3p, miR-320a, and miR-93-5p showed the highest discriminating capacity for pancreatic neoplasia (PDAC or IPMN) with an area under the receiver operating characteristic curve (AUC) of 0.86, 0.85, 0.85, and 0.80, respectively. 2-miRNA combinations improved these performances reaching AUC = 0.90 for “miR-33a-3p+miR-320a.” Addition of CA19.9 increased the diagnostic potential of miRNA signatures even further achieving an AUC of 0.95 (93% sensitivity and 85% specificity) for the combination of “miR-33a-3p+miR-320a+CA19.9.”
CONCLUSIONS:
Novel signatures combining miRNAs and CA19.9 could be used as noninvasive biomarkers for early detection of PDAC.
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) presents the lowest survival rate of all cancers because only 6% of patients having this deadly malignancy survive 5 years or more after their diagnosis (1). Its poor prognosis has remained almost the same in the last decades, and PDAC has been predicted to become the second cause of cancer-related deaths in developed countries by 2030 (2). PDAC progresses with no noticeable symptoms in the early stages; thus, approximately 80% of cases are surgically unresectable at the time of diagnosis (3).
Although many biochemical markers have been examined in pancreatic cancer, none are useful for diagnosis in daily practice. Serum carbohydrate antigen 19.9 (CA19.9), used for monitoring response to therapy and predicting postoperative recurrence, has several limitations. It has limited sensitivity and specificity for PDAC diagnosis, it can be elevated in patients with nonmalignant obstructive jaundice resulting in impaired specificity, and it is often normal in precancerous masses (4,5). Moreover, this marker is unsuitable for 10% of the white population who are unable to express CA19.9 because it is Lewis genotype negative (6).
The dire prognosis of patients with PDAC and lack of sensitive and specific tumor markers urges us to find novel noninvasive biomarkers for an early effective detection of PDAC. To this end, our study focuses on identifying circulating microRNAs (miRNAs) altered in the onset of pancreatic cancer with relevant diagnostic characteristics.
Some miRNAs, short-length, noncoding RNAs that posttranscriptionally regulate gene expression, have been largely found deregulated in different stages of pancreatic cancer and in the preneoplastic cystic lesion, intraductal papillary mucinous neoplasm (IPMN) (7,8). This fact, along with the following intrinsic features of miRNAs, could confer these molecules the potential to be reliable biomarkers. MiRNAs, whose sequences are conserved across species, are very stable and resistant to RNAses among other harsh conditions such as extended storage or multiple freeze-thaw cycles. They are easily detectable in challenging samples by highly sensitive techniques and have been found in most body fluids (9,10).
In a previous study, we analyzed the miRNome of PDAC and IPMN tissues by next-generation sequencing. Subsequently, we validated a significant increased expression of 30 miRNAs by qRT-PCR in 2 independent sets of samples (11). MiRNA candidates extracted from these data have been used in the present multicenter study to identify new noninvasive biomarkers for early detection of pancreatic cancer.
METHODS
Study design and blood sample collection
A total of 190 subjects from 2 Spanish hospitals (168 from Hospital Clinic of Barcelona, Catalonia, and 22 from Hospital Donostia, Guipúzcua) were prospectively recruited from January 2009 to September 2013. A group of 96 patients were diagnosed with PDAC, 20 with IPMN, and 23 with chronic pancreatitis (CP), and 51 were control individuals (C). Controls were healthy subjects with no history of any cancer. From all these subjects, we obtained 10 mL of peripheral venous blood in sterile ethylenediaminetetraacetic acid–treated anticoagulant (K2EDTA) tubes (BD Vacutainer, Toronto, Canada) before resection or any treatment. Blood samples were kept on ice during transportation and immediately processed for plasma separation.
Eight subjects were finally discarded for the study owing to low miRNA extraction efficiency as detected by spiked-in cel-miR-39 levels (n = 6) and to the presence of hemolysis (n = 2). Clinicopathologic characteristics of participants in this study are summarized in Table S1 (see Supplementary Digital Content 2, http://links.lww.com/CTG/A26). The Institutional Ethics Committee of Hospital Clinic of Barcelona approved the study (March 27, 2008), and written informed consent was obtained from all patients in accordance with the Declaration of Helsinki.
From our previous study (11), we selected 17 candidate miRNAs based on the following criteria: significantly overexpressed in both PDAC and IPMN vs healthy pancreatic tissues, high discriminative capacity between PDAC or IPMN and healthy (considering areas under the receiver operating characteristic (ROC) curves for PDAC (AUC) > 0.82), and high expression levels in tissue assuring detectable expression levels in the circulation (with mean cycle threshold (Ct) values ≤ 28, with the exception of miR-548d-3p that presented mean Ct > 30 but a high AUC = 0.93).
Plasma processing
Blood samples were centrifuged at 1,600g for 10 minutes at 4 °C. The superior phase, which contains the plasma, was transferred into new tubes and centrifuged at 16,000g for 10 minutes at 4 °C. The resultant supernatant of plasma poor in platelets was collected and kept at −80 °C until RNA extraction.
RNA extraction
Total RNA was isolated from 500 μL plasma by using the miRNeasy Mini Kit (Qiagen, Hilden, Germany), following the manufacturers protocol with some variations in the initial steps. Denaturation of plasma was achieved by TRIzol LS Reagent (Thermo Fisher Scientific, Foster City, CA), and during its incubation, we added 5 μL of spike-in cel-miR-39 (5 nM) as a reference for normalization of technical variations between samples. The elution volume with RNAse-free water was 30 μL.
Analysis of miRNA expression by real-time qRT-PCR
MiRNA expression was assessed by singleplex qRT-PCR using TaqMan miRNA Assays (Thermo Fisher Scientific) according to the manufacturer protocol. Considering the general low levels of miRNAs in plasma, the amount of input RNA per reaction was standardized by extracting RNA from exactly the same plasma volume. PCR reactions were prepared in a 384-well optical plate and run on the ViiA7 Real-Time PCR System (Thermo Fisher Scientific) with the following program: polymerase activation at 95 °C for 10 minutes and a PCR of 50 cycles at 95 °C for 15 seconds and 60 °C for 1 minute. Negative controls did not show any amplification. All samples were amplified per duplicate. Data were normalized against cel-miR-39 as the technical control. Currently, there is no consensus on a suitable endogenous reference miRNA (12). Ct values were calculated from automatic threshold using QuantStudio Real Time PCR Software. Preamplification reaction was only performed for miR-548d-3p because its expression levels in plasma were very low or undetectable by qRT-PCR. This reaction consisted of 14 cycles of preamplification using TaqMan PreAmp Master Mix (Thermo Fisher Scientific). The preamplified product was diluted 1/2 and analyzed by qRT-PCR as indicated earlier.
Hemolysis assessment
Sample hemolysis can affect plasma miRNA levels because of miRNA contamination from red blood cells (13,14). Therefore, hemolyzed samples were discarded before RNA isolation, and confirmation was done by qRT-PCR quantification of miR-451-5p. Samples found with high miR-451 levels were classified as hemolyzed and also discarded.
Detection of plasma CA19.9
CA19.9 levels in plasma were determined by an electrochemiluminescent technique in a Cobas 4000 analyzer (Roche Diagnostics, Indianapolis, IN), considering as normal those concentrations equal to or below 37 U/mL.
Data analysis
Relative expression levels of miRNAs were calculated as (−∆Ct = −(Ct of specific miRNA – Ct of spike-in control cel-miR-39)). Fold change values were transformed to a log2 scale, and a t test was applied. The false discovery rate (FDR) was computed using the Benjamini & Hochberg procedure (FDR < 0.05 is considered as significant). Heatmaps or scatter plots, 3D, were done by using made4 R-package. Evaluation of predictability of miRNAs was performed using logistic regression adjusted by age and sex (generalized linear model binomial distribution). ROC analysis plots and derived cut-points, as well as overall discriminative accuracy parameters, were computed using pROC R-package considering each miRNA expression as a continuous variable. Sensitivity and specificity were calculated from the optimum cut-point associated with the minimum distance between the ROC curve and upper left corner. All possible combinations of miRNAs were computed with multivariate logistic regression models, and ROC AUCs of miRNA signatures were calculated using samples with no missing values and sorted with the Brier score. From multivariate analysis, we can see whether miRNAs in the model are correlated or are independent variables. To compute the maximum number of variables to be used in the models, we followed the rule of “10 events per variable considering the n from the smallest group (n = 51)” to prevent overfitting. Taking into account that models were adjusted by age and sex, we considered that the maximum number of miRNAs and/or CA19.9 to put in the models was 3. For cross-validation, we computed Harrell optimism using 5-fold cross-validation and 1,000 resamples. Predicted risk score probability for each sample was computed from multivariate logistic regression model, and these values were used to derive their respective box plots.
RESULTS
qRT-PCR confirms overexpression of a group of selected miRNAs in the circulation of patients harboring PDAC or IPMN
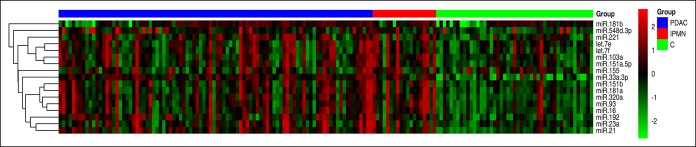
The qRT-PCR expression analysis of 17 candidate miRNAs in plasma samples clearly showed a differential expression profile between patients with PDAC or IPMN and controls (Figure 1), with a remarkable overlap between patients with IPMN and PDAC.
Figure 1.
Circulating miRNA expression profiles distinguish between patients with PDAC or IPMN and control individuals. Heatmap showing expression profiling by qRT-PCR of 17 candidate miRNAs in plasma samples from PDAC (n = 94), IPMN (n = 19), and C (n = 51). Red pixels correspond to an increased abundance of miRNA in the indicated sample, whereas green pixels indicate decreased miRNA levels. C, control; IPMN, intraductal papillary mucinous neoplasm; PDAC, pancreatic ductal adenocarcinoma.
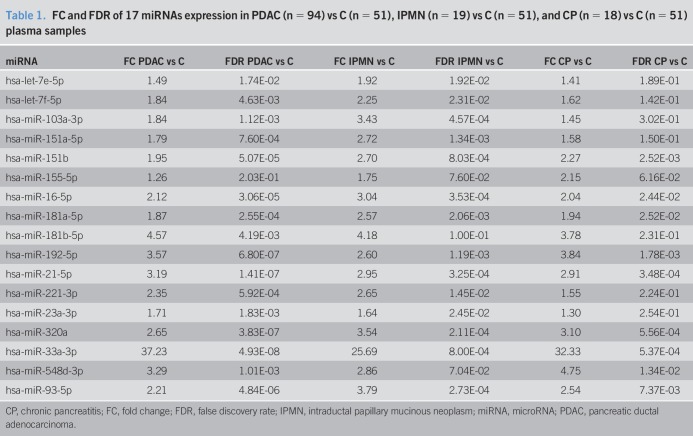
From the 17 circulating miRNAs analyzed in the final 182 plasma samples, the following 16 were significantly upregulated in PDAC compared with the control group: let-7e-5p, let-7f-5p, miR-103a-3p, miR-151a-5p, miR-151b, miR-16-5p, miR-181a-5p, miR-181b-5p, miR-192-5p, miR-21-5p, miR-221-3p, miR-23a-3p, miR-320a, miR-33a-3p, miR-548d-3p, and miR-93-5p. Regarding the comparative analysis of miRNA expression between IPMN and control plasmas, 14 of 17 miRNAs were also significantly overexpressed: let-7e-5p, let-7f-5p, miR-103a-3p, miR-151a-5p, miR-151b, miR-16-5p, miR-181a-5p, miR-192-5p, miR-21-5p, miR-221-3p, miR-23a-3p, miR-320a, miR-33a-3p, and miR-93-5p. Among them, there were 9 miRNAs (miR-151b, miR-16-5p, miR-181a-5p, miR-192-5p, miR-21-5p, miR-320a, miR-33a-3p, miR-548d-3p, and miR-93-5p) with also significant higher levels in CP than in C. Fold change and FDR values are specified in Table 1, and box plots of all miRNAs analyzed are depicted in Figure S1 (see Supplementary Digital Content 1, http://links.lww.com/CTG/A25).
Table 1.
FC and FDR of 17 miRNAs expression in PDAC (n = 94) vs C (n = 51), IPMN (n = 19) vs C (n = 51), and CP (n = 18) vs C (n = 51) plasma samples
Validated miRNAs in plasma samples show high accuracy to discriminate between patients with PDAC or IPMN and controls
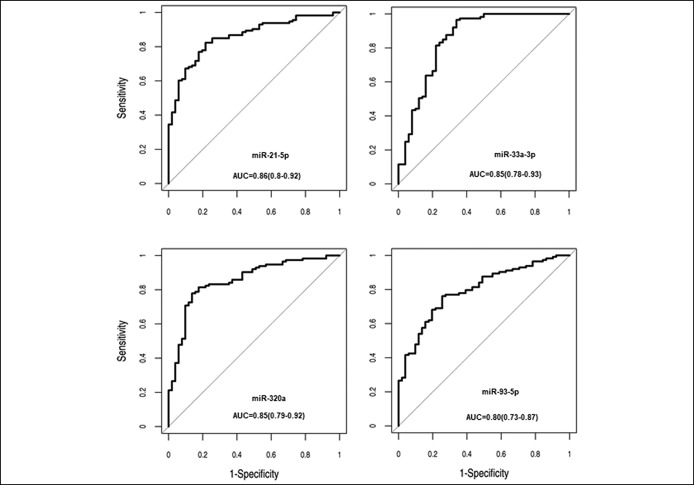
The diagnostic potential of all miRNAs validated as significantly upregulated in our plasma set was assessed by ROC analysis. Regarding the capacity to discriminate patients with pancreatic neoplasia (including PDAC or IPMN) from controls, these miRNAs presented individually AUC ranging from 0.64 to 0.86. miR-21-5p, miR-33a-3p, miR-320a, and miR-93-5p were the ones showing the highest discriminating accuracy (Figure 2 and Table 2). In addition, the diagnostic capacity to distinguish PDAC vs controls and IPMN vs controls, separately, is detailed in Table S2 (see Supplementary Digital Content 3, http://links.lww.com/CTG/A27). The performances of these combinations for PDAC or IPMN separately are shown in Table S3 (see Supplementary Digital Content 4, http://links.lww.com/CTG/A28). It is important to highlight that the best 4 combinations contained miR-33a-3p.
Figure 2.
Plasma miR-21-5p, miR-33a-3p, miR-320a, and miR-93-5p present a high capacity to distinguish patients with pancreatic neoplasia from control individuals. ROC curves of the top 4 miRNAs illustrating high discriminatory power to detect pancreatic neoplasia (PDAC or IPMN). Results are obtained from miRNA analysis by qRT-PCR in PDAC or IPMN (n = 113) and C (n = 51) plasma samples. AUC, area under the curve.
Table 2.
Summary data from ROC curve analysis of all miRNAs analyzed in plasma samples from patients with pancreatic neoplasia (PDAC or IPMN, n = 113) and control individuals (n = 51)
Signatures combining 2 miRNAs with CA19.9 further improve the accuracy for diagnosing pancreatic neoplasia
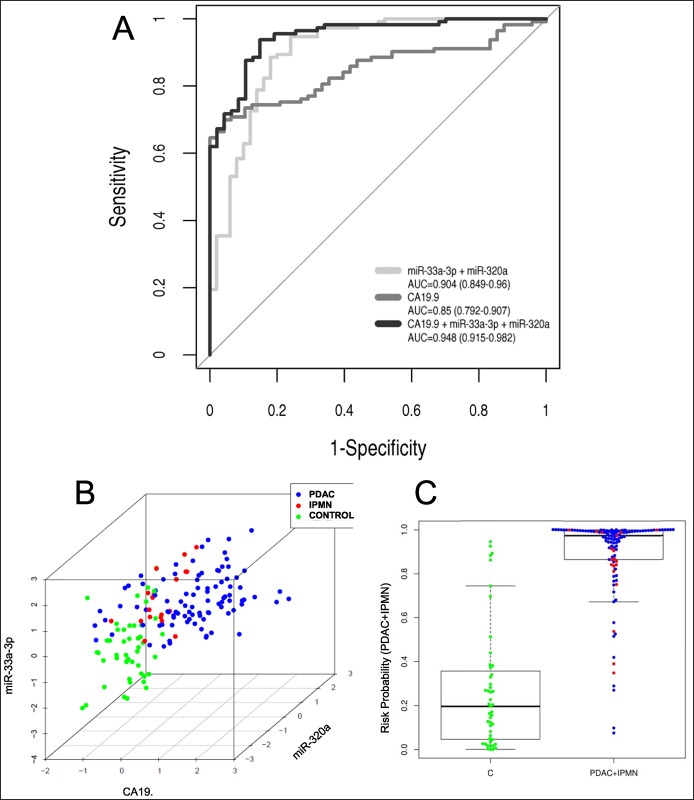
We next examined CA19.9 levels on our set of plasma samples to see whether adding CA19.9 to these miRNA signatures could improve diagnostic performance. CA19.9 showed an AUC of 0.85 (95% confidence interval: 0.91–0.79) to discriminate patients with pancreatic neoplasia from controls.
Addition of CA19.9 to the above-mentioned 2-miRNA signatures clearly improved diagnostic performances reaching AUC values of 0.95 for the signatures: “miR-33a-3p+miR-320a+CA19.9,” “miR-33a-3p+miR-21+CA19.9,” and “miR-33a-3p+miR-548-3p+CA19.9” and 0.94 for the signature “miR-33a-3p+miR-93+CA19.9” (Table 3). As we can see, all of them showed very similar performances with high sensitivity and specificity to discriminate between patients with PDAC or IPMN and controls. Among them, we chose to plot the ROC curve from the signature combining miR-33a-3p and miR-320a plasma expression levels with CA19.9, for being the one with the highest AUC, the lowest Brier score (0.079 vs > 0.081), and very high sensitivities and specificities to detect patients with pancreatic neoplasia (Figure 3a). Cross-validation for this signature reached a corrected AUC of 0.939 (0.921–0.952). As we can see in Figure 3b, plasma samples from control individuals appeared quite differentiated from PDAC or IPMN samples, depending on the scaled values of miR-33a-3p, miR-320a, and CA19.9. It is worth to highlight that in the corresponding multivariate model, the 3 variables (miR-33a-3p, miR-320a, and CA19.9) are identified as independent (see Table S4, Supplementary Digital Content 5, http://links.lww.com/CTG/A29). According to this signature, only 8.5% of the plasma samples showed a probability risk higher than 0.5 of being misclassified (Figure 3c). Considering the group of PDAC independently, we observed that the diagnostic power of 2-miRNA signatures for PDAC improved with the use of CA19.9 (see Table S3, Supplementary Digital Content 4, http://links.lww.com/CTG/A28). By contrast, the value of CA19.9 to detect IPMN cases from controls is lower than all individual miRNAs analyzed with a poor AUC of 0.62 (see Table S2, Supplementary Digital Content 3, http://links.lww.com/CTG/A27), and its combination with the best miRNA signatures does not improve at all the diagnostic performances of miRNAs (data not shown).
Table 3.
Summary data from ROC curve analysis of the 4 top 2-miRNA signatures for detecting pancreatic neoplasia (PDAC or IPMN) from plasma samples and their values when combined with CA19.9 (adjusted by age and sex)
Figure 3.
Performance of the best plasma miRNA signature to discriminate between patients with pancreatic neoplasia and control individuals in our plasma set. (a) Receiver operating characteristic curve of the top miRNA signature “miR-33a-3p+miR-320a” to detect pancreatic neoplasia (PDAC or IPMN). Results are obtained from miRNA analysis by qRT-PCR in PDAC or IPMN (n = 113) and C (n = 51) plasma samples. CA19.9 diagnostic performance for patients with pancreatic neoplasia and its combination with the above-mentioned miRNA signature are also shown. (b) Scatter graph according to the scaled values of miR-33a-3p, miR-320a, and CA19.9 for PDAC, IPMN, and C plasma set (n = 182). (c) Probabilistic sensitivity analysis according to the plasma signature “miR-33a-3p+miR-320a+CA19.9.” AUC, area under the curve; C, control; IPMN, intraductal papillary mucinous neoplasm; PDAC, pancreatic ductal adenocarcinoma.
DISCUSSION
In this study, we have shown that analysis of a panel of circulating miRNAs in plasma from a multicenter cohort results in a significant differential expression profile between PDAC or IPMN and healthy individuals. All of them had been previously found overexpressed in pancreatic tissue from IPMN and PDAC patients as we had previously published (11). These results confirm that there is a very similar miRNA expression pattern between tissue and plasma for the miRNAs tested. Whether this coincidental trend of alteration is due to the fact that these circulating cancer-associated miRNAs are actively secreted from their originating cells, passively released from necrotic tumor cells, or are mere byproducts of other responses mediated by the carcinogenesis would require further investigation. There are a few studies which have already analyzed the relationship between circulating and tissue miRNAs demonstrating different sources of circulating miRNAs (15–17).
Several studies have already reported altered expression profiles of circulating miRNAs in pancreatic cancer (18–21). Early on, Wang et al. (22) identified miR-21, miR-210, miR-155, and miR-196a as being upregulated in plasma of PDAC compared with healthy individuals. Carlsen et al. (23) described a group of 7 deregulated miRNAs in PDAC plasmas vs disease controls, and Liu et al. (24) demonstrated elevated expression of numerous miRNAs in the serum of pancreatic cancer. However, our study, apart from confirming already reported aberrantly expressed miRNAs in PDAC (such as miR-155-5p, miR-16-5p, miR-181a-5p, miR-181b, miR-192-5p, miR-21-5p, and miR-221-3p), also describes new altered circulating miRNAs in PDAC that have never been reported before (let-7e-5p, let-7f-5p, miR-103a-3p, miR-151a-5p, miR-151b, miR-23-3p, miR-320a, miR-33a-3p, miR-548d-3p, and miR-93). Besides, most of them have never been reported as altered in IPMN cases (25,26).
IPMNs, the only radiographically identifiable precursor lesions of pancreatic cancer, have the potential to progress into invasive carcinoma. Approximately 30% of IPMNs that are surgically resected ultimately demonstrate invasive disease (27). Detecting them before its malignant transformation is crucial for patient survival. However, the identification of high-grade dysplasia and early invasive carcinoma and the timing and frequency of malignant progression remain a critical issue in the management of these patients (28). Permuth-wey et al. (29) defined a 5-miRNA signature that discriminated between malignant and benign IPMNs, supporting miRNA relevance in the progression of IPMNs to malignancy. In the clinical practice, only carcinoembryonic antigen and CA.19.9 are used regardless of their very low sensitivities concerning malignant IPMNs. Better molecular knowledge of these lesions and new biomarkers are required to identify those individuals who will benefit from an early surgery and those who will benefit from surveillance (30).
Herein, among the 17 miRNAs analyzed, we have identified a panel of 14 circulating miRNAs significantly overexpressed in the plasma of both patients with PDAC and IPMN relative to control individuals, suggesting that they could serve as potential noninvasive biomarkers indicative of a neoplastic process in the pancreas. Two other miRNAs, miR-181b-5p and miR-548d-3p, were significantly upregulated in PDAC plasmas but not in premalignant IPMNs, suggesting that their late increase in levels could be related to the malignant progression of IPMNs. Whether these 2 miRNAs could be specific of invasive or high-risk IPMNs and could serve to distinguish between low- and high-risk IPMNs should be confirmed in subsequent studies designed for this purpose. Moreover, these results have to be taken with a cautious note because the group of IPMN samples is small, and further studies also including malignant IPMN would be required to confirm these molecules as involved in malignant transformation. Regarding the CP group, 6 miRNAs significantly altered in IPMN and PDAC (let-7e-5p, let-7f-5p, miR-103a-3p, miR-151a-5p, miR-221-3p, and miR-23a-3p) had normal levels in CP plasmas, suggesting that these 6 miRNAs could be more specific of pancreatic neoplasia. However, these results should be validated in larger cohorts of patients.
Several studies have reported that levels of circulating miRNAs in combination with CA19.9 work efficiently for screening pancreatic cancer. Liu et al. (26) concluded that the combination of miR-16, miR-196a, and CA19.9 was more effective among various combinations in discriminating patients with PDAC from patients without PDAC. Schultz et al. (31) observed that a miRNA panel for diagnosing PDAC, tested in whole blood, could be more useful in combination with serum CA19.9. In accordance, we have demonstrated that addition of CA19.9 to the best miRNA signatures in our plasma set improves the overall diagnostic performance. Combination of the miRNAs found here and other new blood-based biomarkers as circulating tumor DNA or circulating tumor cells could be additionally studied to try to further enhance surveillance for invasive PDAC.
We describe here, for the first time, the signature “miR-33a-3p+miR-320a” able to discriminate patients harboring malignant pancreatic cancer or premalignant IPMNs from healthy individuals with high accuracy (88% sensitivity and 82% specificity), and addition of CA19.9 increases this performance (93% sensitivity and 85% specificity), suggesting that this combination could be a promising biomarker signature for noninvasive early detection of pancreatic neoplasia.
Nevertheless, before we can translate miRNA expression evaluation into a clinical routine, it is necessary to standardize the methodologies used. A comprehensive review of circulating miRNAs by Jarry et al. (32) unveils great contradictions in the results and low levels of reproducibility across different laboratories that have used different methods.
Nonetheless, it is noteworthy that the results presented here could help detecting pancreatic malignant lesions in asymptomatic stages, even before malignancy, helping to improve the fatal prognosis and life expectancy of this disease. We can also consider that a test like this could be useful for early diagnosis likely on suspicion of pathology; for that purpose, the behavior of these biomarkers in other pancreatic pathologies and tumor types such as solid pseudopapillary neoplasm and neuroendocrine tumors should be further studied, given that they have a different clinical management. Determining the functional role of miRNAs in circulation is critical to their advancement as suitable biomarkers (33). In further studies, we aim to elucidate the biological implication of these miRNAs in PDAC. It is important to note that most of the plasmatic miRNAs that we have found deregulated in pancreatic cancer are already described to be associated with tumor biology (34), and others have not been studied yet.
CONCLUSIONS
We have found a panel of altered circulating miRNAs that hold great promise as a proof of principle for PDAC noninvasive early detection. In addition, we have defined a biomarker signature consisting of 2 miRNAs combined with CA19.9 to detect pancreatic neoplasia with the highest diagnostic values reported to date. However, before these novel biomarkers can be routinely used in a clinical setting, it is essential to validate them in larger cohorts of patients by using standardized approaches to the isolation, quantification, and miRNA normalization to ensure reproducibility.
CONFLICTS OF INTEREST
Guarantor of the article: Meritxell Gironella, PhD.
Specific author contributions: Conception and design: M.G. Provision of study materials or patients: L.M., À.G., M.C., L.B., and A.C. Collection and assembly of data: E.V.N., S.D.S., and M.G. Data analysis and interpretation: E.V.-N., S.D.S., M.V-.C., J.J.L., A.C., and M.G. Manuscript writing: E.V.-N., A.C., and M.G. All authors gave final approval of the version to be published.
Financial support: This work was supported by grants from Instituto de Salud Carlos III (PI13/02192 and PI17/01003), Fundación Científica de la Asociación Española contra el Cáncer (GCB13131592CAST), and Ministerio de Economía y Competitividad (RTC-2015-3850-1 and SAF2014-54453-R) and cofunded by FEDER-European Union. CIBEREHD is funded by the Instituto de Salud Carlos III. We also acknowledge the support of CERCA Programme and SGR 653/Generalitat de Catalunya. This work was developed at the Centro Esther Koplowitz, Barcelona, Spain. We are indebted to the Biobank core facilities of the IDIBAPS. Work supported by the Xarxa de Bancs de Tumors de Catalunya (XBTC). SDS is funded by Ministerio de Economia y Competitividad (FPI). M.V-.C. is funded by Ministerio de Educación Cultura y Deporte (FPU).
Potential competing interests: Part of the results presented in the article pertain to a filed patent application.
Study Highlights.
WHAT IS KNOWN
✓ Feasibility of detecting and analyzing miRNAs from plasma and serum.
✓ Tissue miRNA expression profiles can distinguish between patients with PDAC or IPMN and healthy individuals.
✓ Some plasma signatures combining several miRNAs to discriminate PDAC vs controls have already been described.
WHAT IS NEW HERE
✓ Discovery of a new group of miRNAs commonly upregulated in the plasma of both patients with IPMN and PDAC.
✓ Validation of significant overexpression of 16 and 14 miRNAs in PDAC and IPMN, respectively, in a new plasma set of patients. Confirming similar miRNA expression pattern between plasma and pancreatic tissue for this malignancy.
✓ Discovery of new biomarker plasma signatures, combining miRNAs and CA19.9, with high capacity to discriminate pancreatic neoplasia from healthy individuals.
TRANSLATIONAL IMPACT
✓ Description of a potential noninvasive strategy for early detection of pancreatic cancer.
Supplementary Material
ACKNOWLEDGEMENTS
We are sincerely grateful to the patients for their collaboration and to Dr. Salvador Navarro and Dr. Laureano Fernandez-Cruz for helping us in the inclusion of patients for the study. We also appreciate the collaboration of nurses from Hospital Clinic of Barcelona and Hospital Universitario Donostia who help us to get the samples.
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at http://links.lww.com/CTG/A25, http://links.lww.com/CTG/A26, http://links.lww.com/CTG/A27, http://links.lww.com/CTG/A28, http://links.lww.com/CTG/A29
REFERENCES
- 1.De Angelis R, Sant M, Coleman MP, et al. Cancer survival in Europe 1999–2007 by country and age: Results of EUROCARE-5—A population-based study. Lancet Oncol 2014;15:23–34. [DOI] [PubMed] [Google Scholar]
- 2.Rahib L, Smith BD, Aizenberg R, et al. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014;74:2913–21. [DOI] [PubMed] [Google Scholar]
- 3.Stathis A, Moore MJ. Advanced pancreatic carcinoma: Current treatment and future challenges. Nat Rev Clin Oncol 2010;7:163–72. [DOI] [PubMed] [Google Scholar]
- 4.Goggins M, Koopmann J, Yang D, et al. NACB: Practice Guidelines and Recommendations for use of tumor markers in the Clinic National Academy of Clinical Biochemistry (NACB) guidelines for the use of tumor markers in pancreatic ductal adenocarcinoma. Clin Chem 2005;3:1–38. [Google Scholar]
- 5.Ballehaninna UK, Chamberlain RS. The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: An evidence based appraisal. J Gastrointest Oncol 2012;3:105–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winter JM, Yeo CJ, Brody JR. Diagnostic, prognostic, and predictive biomarkers in pancreatic cancer. J Surg Oncol 2013;107:15–22. [DOI] [PubMed] [Google Scholar]
- 7.Li A, Yu J, Kim H, et al. MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls. Clin Cancer Res 2013;19:3600–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ali S, Almhanna K, Chen W, et al. Differentially expressed miRNAs in the plasma may provide a molecular signature for aggressive pancreatic cancer. Am J Transl Res 2010;3:28–47. [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 2008;105:10513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18:997–1006. [DOI] [PubMed] [Google Scholar]
- 11.Vila-Navarro E, Vila-Casadesús M, Moreira L, et al. MicroRNAs for detection of pancreatic neoplasia: Biomarker discovery by next-generation sequencing and validation in 2 independent cohorts. Ann Surg 2017;265:1226–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kroh EM, Parkin RK, Mitchell PS, et al. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 2010;50:298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirschner MB, Edelman JJ, Kao SC, et al. The impact of hemolysis on cell-free microRNA biomarkers. Front Genet 2013;4:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirschner MB, Kao SC, Edelman JJ, et al. Haemolysis during sample preparation alters microRNA content of plasma. PLoS One 2011;6:e24145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma R, Jiang T, Kang X. Circulating microRNAs in cancer: Origin, function and application. J Exp Clin Cancer Res 2012;31:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: A potential marker for colorectal cancer screening. Gut 2009;58:1375–81. [DOI] [PubMed] [Google Scholar]
- 17.Pigati L, Yaddanapudi SCS, Iyengar R, et al. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One 2010;5:e13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong X, Du Y, Wang G, et al. Detection of differentially expressed microRNAs in serum of pancreatic ductal adenocarcinoma patients: miR-196a could be a potential marker for poor prognosis. Dig Dis Sci 2011;56:602–9. [DOI] [PubMed] [Google Scholar]
- 19.Xu J, Cao Z, Liu W, et al. Plasma miRNAs effectively distinguish patients with pancreatic cancer from controls: A multicenter study. Ann Surg 2016;263:1173–9. [DOI] [PubMed] [Google Scholar]
- 20.Bauer AS, Keller A, Costello E, et al. Diagnosis of pancreatic ductal adenocarcinoma and chronic pancreatitis by measurement of microRNA abundance in blood and tissue. PLoS One 2012;7:e34151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawaguchi T, Komatsu S, Ichikawa D, et al. Clinical impact of circulating miR-221 in plasma of patients with pancreatic cancer. Br J Cancer 2013;108:361–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J, Chen J, Chang P, et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila) 2009;2:807–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlsen AL, Joergensen MT, Knudsen S, et al. Cell-free plasma microRNA in pancreatic ductal adenocarcinoma and disease controls. Pancreas 2013;42:1107–13. [DOI] [PubMed] [Google Scholar]
- 24.Liu R, Chen X, Du Y, et al. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem 2012;58:610–8. [DOI] [PubMed] [Google Scholar]
- 25.Wei L, Yao K, Gan S, et al. Clinical utilization of serum- or plasma-based miRNAs as early detection biomarkers for pancreatic cancer. Medicine 2018;97:e12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Gao J, Du Y, et al. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int J Cancer 2012;131:683–91. [DOI] [PubMed] [Google Scholar]
- 27.Strauss A, Birdsey M, Fritz S, et al. Intraductal papillary mucinous neoplasms of the pancreas: Radiological predictors of malignant transformation and the introduction of bile duct dilation to current guidelines. Br J Radiol 2016;89:20150853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allen PJ. The management of intraductal papillary mucinous neoplasms of the pancreas. Surg Oncol Clin N Am 2010;19:297–310. [DOI] [PubMed] [Google Scholar]
- 29.Permuth-Wey J, Chen D-T, Fulp WJ, et al. Plasma microRNAs as novel biomarkers for patients with intraductal papillary mucinous neoplasms of the pancreas. Cancer Prev Res 2015;8:826–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moris D, Damaskos C, Spartalis E, et al. Updates and critical evaluation on novel biomarkers for the malignant progression of intraductal papillary mucinous neoplasms of the pancreas. Anticancer Res 2017;37:2185–94. [DOI] [PubMed] [Google Scholar]
- 31.Schultz NA, Dehlendorff C, Jensen BV, et al. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA 2014;311:392–404. [DOI] [PubMed] [Google Scholar]
- 32.Jarry J, Schadendorf D, Greenwood C, et al. The validity of circulating microRNAs in oncology: Five years of challenges and contradictions. Mol Oncol 2014;8:819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parikh A, Lee C, Joseph P, et al. microRNA-181a has a critical role in ovarian cancer progression through the regulation of the epithelial-mesenchymal transition. Nat Commun 2014;5:2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giovannetti E, Funel N, Peters GJ, et al. MicroRNA-21 in pancreatic cancer: Correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res 2010;70:4528–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.