Abstract
The prevalence of infection in chronic wounds is well documented in the literature, but not optimally studied due to the drawbacks of current methodologies. Here, we describe a tractable and simplified ex vivo human skin model of infection that addresses the critical drawbacks of high costs and limited translatability. Wounds were generated from excised abdominal skin from cosmetic procedures and cultured, inoculated with Staphylococcus aureus strain UAMS-1, or under aseptic conditions. After three days, the infected wounds exhibited biofilm formation and significantly impaired re-epithelialization compared to the control. Additionally, pro-migratory and pro-reparative genes were significantly downregulated while pro-inflammatory genes were significantly upregulated, demonstrating molecular characterizations of impaired healing as in chronic wounds. This model allows for a simplified and versatile tool for the study of wound infection and subsequent development of novel therapies.
Introduction
Chronic wounds continue to be a major clinical problem, affecting an estimated 6.5 million patients with a cost of $25 billion each year in the US alone.1 Many of the underlying pathogenic diseases remain difficult to manage, yet share a unifying contributing factor of persistent bacterial infection.2 A plethora of animal models have been designed to model chronic wounds, including infected wounds3, however these in vivo models are costly, labor-intensive, and remain limited in their translation to the clinic.4 Although ex vivo models using human skin samples have improved our understanding of wound infections, they are limited by equipment cost and availability, need for injecting bacteria directly into the dermis (rather than topical inoculation), lack of context for critical inflammatory innate immune responses, and absence of pro-reparative marker analysis.5, 6 Thus, a new and more universally accessible model is needed to better characterize both the innate inflammatory and pro-reparative responses to skin wound infection and rapidly extrapolate these findings toward development of novel therapies.
We herein present a standardized and more readily tractable human skin explant model for wound infection to serve as an effective tool for testing potential wound therapies targeting infected wounds. S. aureus clinical isolate strain UAMS-1 was chosen as the infectious agent because it is among the most frequently cultured and/or genomically identified organisms in chronic wounds and for its ability to form biofilm.7,8 Wound biofilms pose a significant clinical challenge, as the aggregation of microbial cells covered by a self-initiated extracellular matrix both limits access to, and confers resistance to, antibiotics and the underlying immune response. Even after surgical debridement, complete removal remains highly unlikely and may result in the dissemination of infection.9 Thus, the recreation of biofilm in an ex vivo human skin wound model would allow for study of one of the most significant challenges to chronic wound therapy.
Materials and Methods
Skin collection, inoculum preparation, wound induction, and culture
Skin discarded at the time of elective panniculectomy or lower body lift procedures were obtained under an IRB-approved human subjects protocol. For this model, adipose tissue was removed, and skin was washed with 3x ABAM (Life Technologies, Carlsbad CA) and PBS. Circular wounds through the epidermis and superficial dermis were created using 3 mm diameter biopsy punch tools. Then, the wound and rim of tissue was excised using 8mm punch tools, washed twice in culture medium, Dulbecco’s Modified Eagle Medium (DMEM) (Thermo Fisher Scientific, Waltham MA) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals, Flowery Branch GA). The clinical isolate UAMS-1 (gift of Mark Smeltzer), was incubated for 16 hours at 37°C in tryptic soy broth normalized to a concentration of 106 colony forming units (CFU)ml−1 in DMEM. Either 10μl of culture medium or bacterial inoculum (10,000 CFUs) in culture medium in exponential growth phase was applied to the wound surfaces. Skin explants were cultured individually in 1ml culture medium for 72 hours, with the epithelium maintained at the air-liquid interface, and with medium changes every 24 hours to select for adherent bacteria and facilitate biofilm formation. Skin wounds were collected after 72 hours.
Histological analysis and quantitation of wound re-epithelialization
Skin explants fixed overnight in 4% paraformaldehyde were transferred to 70% ethanol, paraffin-processed via the Sakura Tissue-Tek VIP processor 6 (Sakura Finetek, Torrance CA). Wound tissue was bisected and 5μm sections taken from the center (widest diameter of the wound) were stained with Gill’s Hematoxylin III (American MasterTech, Lodi CA) and Eosin (VWR, Radnor PA) stains, then imaged and analyzed for re-epithelialization using a BioRevo BZ-9000 Fluorescence Microscope and accompanying software (Keyence, Osaka Japan). Wound closure was expressed as the percentage of epithelial migration from both wound edges relative to the original length of the wound bed, using the following equation:
Scanning electron microscopy
Skin explants fixed overnight in Karnovsky’s fixative were washed in PBS, dehydrated with serial ethanol washes followed by critical point drying via the Tousimis 931 GL Super Critical Autosamdri (Tousimis Research Corp, Rockville MD). The explants were gold-sputter coated via Pelco Auto Sputter Coater SC-7 (Ted Pella, Redding CA) and imaged with the Philips XL30 (FEI Company, Hillsboro OR) using 20kV voltage.
Bacterial enumeration in skin
A quarter of each wound was placed in sterile PBS, homogenized using the Tissue-Tearor (Biospec, Bartlesville, OK) for 25 seconds at maximum speed on ice, then serially diluted 10−1 to 10−5 in triplicates, plated onto tryptic soy agar plates and incubated at 37°C for 24 hours. Colonies were counted and back calculated to quantify colony forming units (CFU).
Assessment of tissue viability
After 72 hours of ex vivo incubation, wound tissues were minced with surgical scissors and incubated for 30 minutes in dispase II solution (Roche, Basel Switzerland)(2.4μg/ml) at 37°C. After this, tissue samples were incubated at 37°C for 3 hours with 2mg/ml Collagenase D (Roche, Basel Switzerland) under constant agitation. The resulting single cell suspensions were washed with DMEM (Thermo Fisher Scientific, Waltham MA) supplemented with 10% FBS, 2mM L-glutamine, 0.15% hydrogencarbonate, 1mM sodium pyruvate, nonessential amino acids, 100μg/ml gentamycin and labeled with LIVE/DEAD Fixable Yellow Dead Cell Stain kit (Invitrogen, Carlsbad CA). Approximately 200,000 events were acquired from each sample on the BD Fortessa flow cytometer (BD Biosciences, San Jose CA) equipped with 405 nm, 488 nm, 642 nm, and 785 nm lasers to determine the percentage of live cells.
Relative mRNA expression
Snap-frozen samples were pulverized using the Biopulverizer (Biospec, Bartlesville OK) and RNA extraction was carried out according to Qiagen RNeasy kit protocol. Once purified, cDNA was generated using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City CA) and SuperScript III Reverse Transcriptase kit (Invitrogen, Carlsbad CA). Changes in gene expression were measured by qPCR based on the CFZX384 Touch Real-Time PCR Detection System (Bio-Rad, Hercules CA).
Protein quantitation
Skin samples were placed in 250μl NP40 Cell Lysis Buffer (Invitrogen, Carlsbad CA) supplemented with 0.1M PMSF (Sigma, St. Louis MO) and Protease Inhibitor Cocktail P-2714 (Sigma, St. Louis MO), vortexed, then centrifuged at 12,000rpm for 10 minutes at 4°C. Protein concentration in the lysate was determined using the Bradford assay, and latent TGFβ was quantitated using the TGFβ Singleplex kit (EMD Millipore, Burlington MA). TNFα levels were determined using the TNFα Duoset Kit (R&D Systems, Minneapolis MN) on 700μl of culture media collected at the 72-hour endpoint.
Statistics
Six skin donors were used for this study. Each assay was conducted with at least three different donors with technical triplicates. For the ELISA and Singleplex assays, duplicate samples from three different donors were used. qPCR data were analyzed using CFX Maestro software (Bio-Rad, Hercules CA). For all other statistical analyses, two-tailed Student’s t tests were used. The error bars are represented as mean ± SEM in the figures, with p values reported according to the following: *p<0.05, ** p<0.01. ***p<0.001.
Results
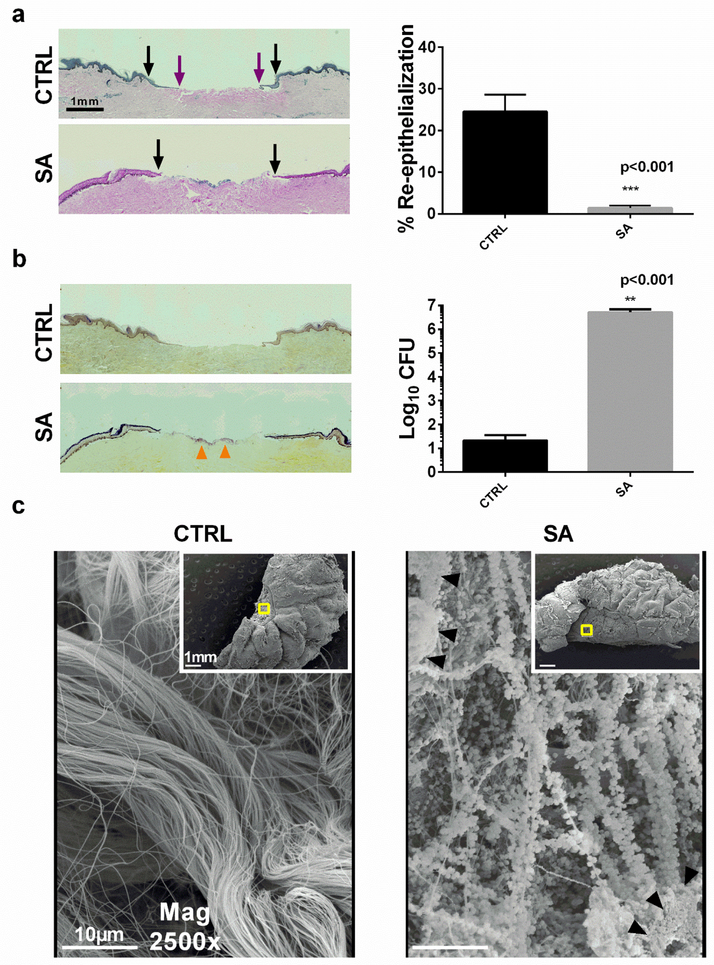
When the explanted skin wounds are cultured under aseptic conditions for 72 hours, they demonstrate early wound re-epithelialization, which is completely lost when S. aureus infection is localized within the wound (Figure 1a). Bacterial infection is confirmed both by Gram stain of the wound (and detection of greater than 106 organisms/gram wound tissue homogenate) (Figure 1b). The localization of infection to the wound bed was confirmed by scanning electron microscopy. Interestingly, biofilm on the wound bed can be observed as early as 72-hours as noted by the presence of the characteristic extracellular matrix (Figure 1c).
Figure 1. Wound re-epithelialization and wound infection in human skin explants.
Hematoxylin and Eosin stain of wounds 72 hours post infection. Re-epithelialization from the wound border (black arrows) across the wound bed (purple arrows), and percentage re-epithelialization of non-infected (CTRL) and S. aureus infected (SA) wounds. (b) Wounds stained with modified Gram stain17. Bacterial aggregation is indicated by orange arrows. Corresponding CFU counts of skin homogenates, normalized to gram of tissue. (c) Scanning electron microscopy of wound (inset) and wound bed. Biofilm aggregates (black triangles) with characteristic extracellular matrix production are present in wound beds infected with SA. (data are represented as mean ± SEM, n= 6 skin donors).
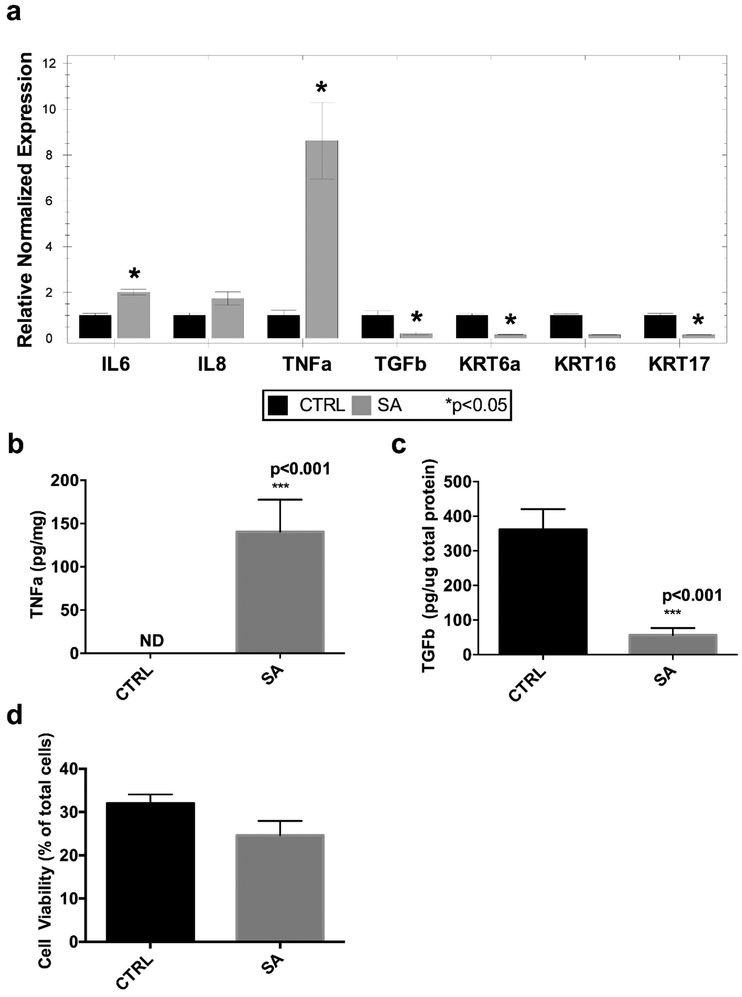
The infection produces a statistically significant upregulation of pro-inflammatory cytokines TNFα and IL-6, as well as a non-statistically significant increase in IL-8 (Figure 2a,b), demonstrating the induction of a skin innate inflammatory response to infection. On the other hand, to examine how skin wound infection impairs healing in this model, we found a decreased expression of the pro-reparative cytokine, TGFβ, a cytokine that is vital to the wound healing process by modulating extracellular matrix production and wound re-epithelialization10,11 (Figure 2a,c). Keratins (Krt) 6a, 16, and 17, reported to be upregulated in response to injury and in turn act to remodel and facilitate keratinocyte migration10,12 are likewise downregulated in the infected wounds (Figure 2a). The observed genetic and biochemical changes are not due to increased cell death in the infected ex vivo explant, as cell viability was similar in the infected and non-infected control explants (Figure 2d). Since most of the epidermis is differentiated by the 72 hour incubation time point, and the epidermis contributes to the majority of the tissue cellularity, the low percentage (about 30%) of total viable cells reflects the contribution of non-viable differentiated keratinocytes to the total cell count.
Figure 2. Characterization of inflammatory and pro-reparative responses.
(a) Gene expression of pro-inflammatory and pro-reparative cytokines, and pro-migratory keratins using RT-qPCR. (b) TNFα protein quantitation in culture media, normalized to gram tissue. (c) TGFβ protein quantitation in tissue lysate, normalized to total protein. (d) Cell viability of skin tissue via flow cytometry. (data represented as mean ± SEM, n= 3–4 skin donors); IL-6 = interleukin-6, TNFα = tumor necrosis factor alpha, TGFβ = transforming growth factor beta, KRT = keratin.
Discussion
This ex vivo model uses human skin and demonstrates how wound infection impairs healing, by the upregulation of innate skin inflammatory responses, and downregulation of expression of genes involved in the repair process. The decrease in K6a is of particular interest in view of the recent discovery of antimicrobial peptides (AMP) derived from cytoplasmic proteasome processing of K6a in the corneal epithelium in response to exposure to microbially-derived ligands.13 The observed infection-induced reduction of the parental K6a in this model, may thus result in decrease in AMP production and persistence of infection.
Of interest is the noted separation at the dermal-epidermal junction in the infected wounds (Figure 1). This was previously noted in ex vivo human skin wounds infected with S. aureus14 and may be due to release of Staphopain A, virulence factors and/or other extracellular proteases with collagenolytic activity during invasion of the wound bed.15 This provides yet another interesting prospect for future investigation.
We note that the model is limited by the absence of a systemic inflammatory response, and cellular recruitment, as well as the use of a single infecting microorganism, when in vivo wound infections are often polymicrobial. However, the lack of a systemic immune response allows for the focused study of the local innate immune and host reparative responses to injury and infection. Additionally, the mono-microbial model could be modified to included mixed infection if desired. Despite these limitations, this model allows for an easily tractable, ex vivo method for investigation of potential therapeutics for human wound infections.
mRNA primers
| Target | Forward | Reverse |
|---|---|---|
| TNF α | GAAAGCATGATCCGGGACGTG | GATGGCAGAGAGGAGGTTGAC |
| TGF-β | CTAATGGTGGAAACCCACAACG | TATCGCCAGGAATTGTTGCTG |
| IL6 | GGTACATCCTCGACGGCATCT | GTGCCTCTTTGCTGCTTTCAC |
| Krt6a | CTAAAGTGCGTCTGCTA | TGGGTGCTCAGATGGTATA |
| Krt16 | GACCGGCGGAGATGTGAAC | CTGCTCGTACTGGTCACGC |
| Krt17 | AAGATCCGTGACTGGTACCAGAGG | GATGTCGGCCTCCACACTCAGG |
| GAPDH | ATGGGGAAGGTGAAGGTCG | GGGGTCATTGATGGCAACAATA |
| 18S | CCCAACTTCTTAGAGGGACAAG | GCTTATGACCCGCACTTACT |
| TBP | GTGAACATCATGGATCAGAACAACA | AAGATAGGGATTCCGGGAGTCAT |
Acknowledgements
We thank Dr. Michael Wong and Anthony Gallegos for facilitating collection of discarded human skin. We also thank Jennifer Tran and Gloria Nguyen for support in preparation of bacterial cultures and CFU enumeration. This work was supported in part by NIH/NINR R01NR015649 (MTC and IP) and NIH/NIAID R21AI08064 and California Institute for Regenerative Medicine Grant PC1–08118 to RRI. The authors have no conflicts of interest to disclose.
References
- 1.Sen CK, Gordillo GM, Roy S, Kirsner R, Lambert L, Hunt TK et al. Human skin wounds: a major and snowballing threat to public health and the economy. Wound Repair Regen 2009; 17:763–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mustoe T Understanding chronic wounds: a unifying hypothesis on their pathogenesis and implications for therapy. Am J Surg 2004; 187:65S–70S. [DOI] [PubMed] [Google Scholar]
- 3.Grada A, Mervis J, Falanga V. Research techniques made simple: Animal models of wound healing. J Invest Dermatol 2018; 138:2095–2105. [DOI] [PubMed] [Google Scholar]
- 4.Elliot S, Wikramanayake TC, Jozic I, Tomic-Canic M. A modeling conundrum: murine models for cutaneous wound healing. J Invest Dermatol 2018;138:736–40. [DOI] [PubMed] [Google Scholar]
- 5.Steinstraesser L, Sorkin M, Niederbichler AD, Becerikli M, Stupka J, Daigeler A, et al. A novel human skin chamber model to study wound infection ex vivo. Arch Dermatol Res 2010; 302: 357–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schaudinn C, Dittmann C, Jurisch J, Laue M, Günday-Türeli N, Blume-Peytavi U, et al. Development, standardization and testing of a bacterial wound infection model based on ex vivo human skin. PLoS ONE 2017; 12: e0186946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loesche M, Gardner SE, Kalan L, Horwinski J, Zheng Q, Hodkinson BP, et al. Temporal stability in chronic wound microbiota is associated with poor healing. J Invest Dermatol 2017;137:237–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin JM, Zenilman JM, Lazarus GS. Molecular microbiology: new dimensions for cutaneous biology and wound healing. J Invest Dermatol 2010;130:38–48. [DOI] [PubMed] [Google Scholar]
- 9.Attinger C, Wolcott R. Clinically Addressing Biofilm in Chronic Wounds. Adv Wound Care 2012;1:127–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pastar I, Stojadinovic O, Yin NC, Ramirez H, Nusbaum AG, Sawaya A, et al. Epithelialization in wound healing: a comprehensive review. Adv Wound Care 2014;3:445–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schultz GS, Davidson JM, Kirsner RS, Bornstein P, Herman IM. Dynamic reciprocity in the wound microenvironment. Wound Repair Regen 2011;19:134–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazzalupo S, Wong P, Martin P, Coulombe PA. Role for keratins 6 and 17 during wound closure in embryonic mouse skin. Dev Dyn 2003;226:356–65. [DOI] [PubMed] [Google Scholar]
- 13.Chan JKL, Yuen D, Too PH, Sun Y, Willard B, Man D, Tam C. Keratin 6a reorganization for ubiquitin-proteasomal processing is a direct antimicrobial response. J Cell Biol 2018;217:731–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramirez HA, Pastar I, Jozic I, Stojadinovic O, Stone RC, Ojeh N, et al. Staphylococcus aureus Triggers Induction of miR-15B-5P to Diminish DNA Repair and Deregulate Inflammatory Response in Diabetic Foot Ulcers. J Invest Dermatol 2018;138:1187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohbayashi T, Irie A, Murakami Y, Murakami Y, Nowak M, Potempa J, et al. Degradation of fibrinogen and collagen by staphopains, cysteine proteases released from Staphylococcus aureus. Microbiology 2011;157:786–92. [DOI] [PubMed] [Google Scholar]
- 16.Wolcott RD, Hanson JD, Rees EJ, Koenig LD, Phillips CD, Wolcott RA, et al. Analysis of the chronic wound microbiota of 2,963 patients by 16S rDNA pyrosequencing. Wound Repair Regen 2015;24:163–174. [DOI] [PubMed] [Google Scholar]
- 17.Becerra SC, Roy DC, Sanchez CJ, Christy RJ, Burmeister DM. An optimized staining technique for the detection of Gram positive and Gram negative bacteria within tissue. BMC Res Notes 2016;9:216. [DOI] [PMC free article] [PubMed] [Google Scholar]