Abstract
Background:
HPV associated oropharyngeal cancer is a clinically and biologically distinct disease from smoking related Head and Neck Squamous Cell Carcinoma (HNSCC). Despite its rapidly increasing incidence, the mutational landscape of HPV-positive oropharyngeal squamous cell carcinoma (OPSCC) remains under-studied.
Methods:
In this study, we present the first mutational analysis of the 46 HPV+ OPSCC tumors within the newly expanded 530 tumor HNSCC TCGA cohort. We also performed a separate exome sequencing analysis of 46 HPV+ oropharyngeal squamous cell carcinomas matched to their normal lymphocyte controls from the Johns Hopkins University (JHU) cohort.
Results:
There was a strikingly high 33% frequency of mutations within genes associated with chromatin regulation, including mutations in KMT2C, KMT2D, NSD1, CREBBP, EP300, and CTCF. In addition, the commonly altered genes PIK3CA and FGFR3 show distinct domain-specific hotspot mutations as compared to their HPV negative counterparts. PIK3CA shows a uniquely high rate of mutation within the helicase domain, and FGFR3 contains a predominance of hotspot S249C alterations that are not found in HPV-negative HNSCC.
Conclusion:
This analysis represents one of the largest studies to date of HPV+ OPSCC, and lends novel insight into the genetic landscape of this biologically distinct disease, including a high rate of mutation in histone and chromatin modifying genes, that may offer novel therapeutic targets.
Keywords: Head and neck squamous cell carcinoma, oropharyngeal squamous cell carcinoma, HPV, TCGA, exome sequencing, epigenetics
Precis
A high frequency of mutations within chromatin regulatory genes, as well as domain specific alterations within PIK3CA and FGFR3, are novel findings that distinguish the molecular signature of HPV positive oropharyngeal squamous cell carcinoma from its smoking-related counterpart.
Introduction
While smoking related Head and Neck Squamous Cell Carcinoma (HNSCC) has been on the decline, the incidence of Human Papillomavirus-associated oropharyngeal squamous cell carcinoma has rapidly increased and currently represents the most common form of HPV-associated cancer in the United States (1). However, only recently has it been recognized as a clinically distinct entity from HPV-negative HNSCC, reflected in both its improved prognosis and response to treatment (2). This distinction is further established in the newly released American Joint Committee on Cancer eighth edition staging criteria, which for the first time categorizes HPV-positive HNSCC tumors as separate from HPV-negative tumors (3).
Genomic mutational profiling has revealed HNSCC tumors to be remarkable for their high degree of inter-tumor heterogeneity, highlighting the challenge behind effective molecular targeting and the need for larger cohort analyses of these tumors. Whole exome sequencing has to date largely focused on HPV-negative tumors, and data on the HPV+ subpopulation remains limited. The Cancer Genome Atlas (TCGA) Network originally published on a cohort of 279 HNSCC tumors (4), with 36 of these identified as HPV-positive tumors, and only 22 of these being from the oropharynx. This analysis did not separately analyze oropharyngeal and non-oropharyngeal HPV-positive tumors however. The TCGA HNSCC census has since expanded to 530 total samples, and here we analyze the expanded Oropharyngeal HPV+ cohort, which now consists of 46 tumors. Non-oropharynx sites were excluded from analysis, as HPV positivity within these other subsites still has an unclear clinical role. The prior TCGA HPV+ analysis revealed 56% of tumors to contain PIK3CA somatic mutations and only one tumor of the cohort to contain a TP53 alteration, both hallmarks of HPV+ disease (4). Indeed, other WES studies by Agrawal, Stransky, and Pickering (5)(6)(7), also containing limited HPV+ data, have found only rare alterations in TP53, CDKN2A, EGFR, or NOTCH1, which are so commonly found in smoking related HNSCC. Liu et al found that within a subset of 15 HPV+ tumors, PIK3CA was the only mutated gene (6). These studies have also highlighted the comparatively low rate of driver mutations within HPV+ tumors; Agarwal noted a 4.8 Mut/Mb rate of HPV+ tumors as compared to a 20.6 Mut/Mb mutational rate for HPV-negative carcinoma (7). Further, recent analysis comparing HPV+ and HPV- oral cavity cancers has shown no distinct methylation clustering between the two cohorts (8), suggesting uncoupling of HPV infection from downstream epigenetic driver events reliant on promoter methylation, and further complicating our genetic understanding of HPV+ tumor progression.
Although HPV+ oropharynx tumors have clinically favorable outcomes compared to their HPV-negative counterparts, the low mutational rate and tumor heterogeneity suggests limited options for targeted therapeutic intervention, and a need for further focused genomic analysis of this unique oncologic entity. Here, we report whole exome analyses focused on HPV+ OPSCCs from an independent JHU cohort, as well as present an updated analysis of the expanded HPV+ TCGA cohort, to further elucidate signaling pathways that may potentially be targeted by novel molecular agents.
Materials and Methods
JHU OPSCC Cohort and Sequencing
Forty-six advanced stage primary HPV+ OPSCC tumor samples were collected under an approved Institutional Review Board protocol (#NA_00–36235) as previously described (9). Each sample was matched to their respective normal lymphocyte control derived from subjects’ blood. Full clinical characteristics of the cohort are presented in Supplementary Table S1. All tissue was submitted to the Johns Hopkins Tissue Core as part of the Head and Neck Cancer Specialized Program of Research Excellence (HNC-SPORE). Pathology of the primary tumors was confirmed by two independent pathologists and tumor tissue was microdissected to yield at least 80% tumor purity. Tumors were considered HPV positive if they were positive on p16 immunohistochemistry or if positive on in situ hybridization for high-risk subtypes, in accordance with the recent College of American Pathologist guidelines on HPV testing (10). In equivocal cases, HPV-16 E6 and E7 viral oncoproteins were detected via PCR for confirmation (Table S1).
Exome Sequencing, Filtering and Alignment
DNA was extracted using the DNeasy Blood and Tissue kit (Qiagen) for high-quality extraction per the manufacturer instructions. DNA samples from tumor and matched lymphocyte controls were quantified using a Qubit (ThermoFisher Scientific). Greater than 1ug of each sample was prepared using a sonication based library construction and enrichment method per the Beijing Genomics Institute (BGI) as previously described (11). The prepared DNA libraries were hybridized to Agilent SureSelect Human All Exon kit to capture the target exome and sequencing was executed with the Illumina HiSeq 4000 sequencing system (BGI) at a variable depth of 50–150X.
The exome sequencing pipeline was performed on 92 samples, which included the 46 tumor samples and 46 normal matched lymphocyte samples. To generate sequence alignment and variant calls, we implemented our exome analysis pipeline on the cfncluster v1.3.1 (12). Short reads were mapped to the human 1000 genomes v37 (13) by BWA-mem v.0.7.12. Subsequent processing was carried out with SAMtools v.1.1, Picard Tools v.1.96, Genome Analysis Toolkit (GATK) v2.4–9 (14) and, which consisted of the following steps: sorting and splitting of the BAM files, marking of duplicate reads, local realignment, indel realignment and recalibration of base quality scores. Somatic variants were called with Mutect. Oncotator was used to annotate variants which were then filtered to include exonic insertions and deletions, and nonsynonymous variants with ExAC and 1000 Genomes population allele frequency <0.05.
TCGA analysis
Data was downloaded from a recently expanded epigenomic analysis of 530 HNSCC tumor samples within TCGA using the TCGA data portal. Non-oropharynx sites were excluded from analysis, as HPV positivity within these subsites still has an unclear clinical role, and do not display the treatment sensitivity that is the hallmark of HPV+ OPSCC. Oropharynx samples were then selected for HPV positivity according to the previously described selection methods of the Comprehensive Genomic Characterization of Head and Neck Squamous Cell Carcinoma analysis (4). Samples were classified as HPV positive using an empiric definition of high coverage (>1000) mapped RNA-Seq reads to high risk HPV subtypes 16, 18, 33 and 35. Reads primarily aligned to E6 and E7 viral genes, and highly corresponded to known clinical demographics of HPV+ OPSCC, including younger age and white race. A total of 46 oropharyngeal tumors were deemed HPV+ per this criterion. Full clinical data is presented in Supplementary Table S2. Mutations were filtered as per above in order to include variants according to EXAC and human 1000 genomes v37. Genes were annotated via Oncotator and only variants with population allele frequency less than 0.05 included.
Results
Clinical Characteristics and Mutational Rates
Our cohort, gathered from the Johns Hopkins University (JHU) tissue core, contained 46 OPSCC HPV+ tumors and their paired normal lymphocytes. Clinical characteristics were consistent with national demographic data regarding this cancer population (Table 1). The average age of subjects was 55.7 years old, with the majority being male (89.1%, 41 of 46). There was a predominance of Caucasian subjects (95.7%, 44 of 46). Seventeen tumors were from never smokers (36.9%), 18 from former smokers (39.1%), and 11 from current smokers (23.9%). Sixteen of 46 (34.8%) had a significant smoking history, defined here as greater than 10 pack/years. The mutational rate of HPV associated carcinomas within our study is independent of smoking status. This holds true in subgroup analysis, whether comparing nonsmokers with current smokers (mutational rate 2.10 vs 2.56, p=0.69) or with former smokers (2.10 vs 2.82, p=0.36). There was no difference in mutagenic rates of smokers with a >10 pack/year smoking history versus <10 pack/years (3.25 vs 2.22, p=0.25), suggesting mutational effects are due to the HPV etiology of the disease and not smoking status. All patients presented with locoregional disease, with the majority classified as early stage disease per the AJCC 8th Edition. Five subjects (10.9%) had Stage I disease, 38 subjects (82.6%) had Stage II disease, and 3 subjects (6.5%) had Stage III disease. There were 3 recurrences within the cohort following primary treatment. Two of the recurrences occurred in Stage I disease and 1 recurrence occurred within a subject with Stage II carcinoma. There was no definable pattern of mutations within the tumors that had clinical recurrences. Two tumors contained PIK3CA mutations, and there were separate mutations of HRAS, FGFR3, KMT2D, CASP8, and FBXW7 within one sample each.
Table 1.
Clinical characteristics of subjects from the JHU cohort and the TCGA HPV+ OPSCC cohort
| Characteristic | JHU No. (%) (N=46) | TCGA No. (%) (N=46) |
|---|---|---|
| Mean Age (range) | 55.7 ± 9.0 (35–74) | 55.6 ± 9.0 (35–77) |
| Sex | ||
| Male | 41 (89.1) | 41 (89.1) |
| Female | 5 (10.9) | 5 (10.9) |
| Race | ||
| Caucasian | 44 (95.7) | 44 (95.7) |
| Other | 2 (4.4) | 2 (4.4) |
| Smoking Status | ||
| Never | 17 (36.9) | 20 (43.5) |
| <10 pack/years | 13 (28.3) | 4 (8.7) |
| >10 pack/years | 16 (34.8) | 22 (47.8) |
| Alcohol | ||
| No | 14 (30.4) | 8 (17.4) |
| Yes | 32 (69.6) | 38 (82.6) |
| TNM Stage (AJCC 8th Ed) | ||
| I | 5 (10.9) | 12 (26.0) |
| II | 38 (82.6) | 27 (58.7) |
| III | 3 (6.5) | 7 (15.2) |
| IV | 0 | 0 |
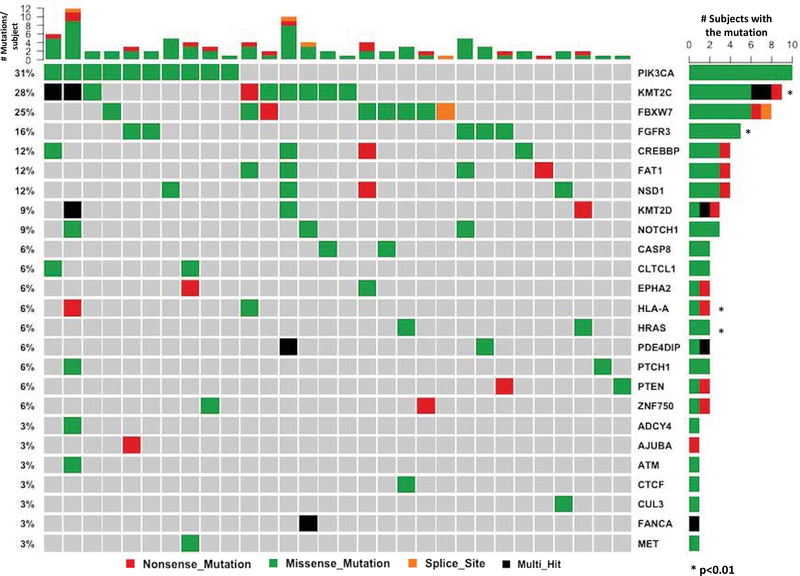
Exome sequencing of 46 tumors was compared to normal lymphocytes from the same subjects. A total of 18,862 genes were analyzed and the top 20 mutated genes along with variant classification shown in Figure 1. This analysis revealed 6 genes with p-value <0.01. The majority of variants contained missense mutations, with a high rate of enrichment (42%) of C>T transversions, followed by T>C transversions (25%) (Figure S1). This mirrors the HPV positive population within the prior TCGA analysis. In contrast, there is an elevated rate of G>T transversions within HPV negative HNSCC, with an increasing frequency associated with smoking status, as shown by Stransky et al and within the original TCGA analysis. There was an average rate of 2.45 mutations per tumor, similar to the low rate of mutagenesis within HPV+ tumors reported in other studies (15).
Figure 1.
Mutation events as determined by Exome Sequencing Analysis within the JHU cohort. Corresponding gene is listed on the right side, and frequency of the mutation listed on the left. Top histogram: number of mutations per subject.
We have also analyzed the oropharyngeal sub-population of the newly expanded TCGA cohort. 46 of these were HPV+, as determined by RNA-Seq high-risk subtype viral oncoprotein analysis. Mean age of these subjects is 55.6 years old, with 89.1% being male (Table 1). A majority of subjects had early stage cancers as per AJCC 8th Edition. Twelve subjects (26.0%) had Stage I disease, 27 subjects (58.7%) had Stage II disease, and 7 subjects (15.2%) had Stage III disease. There were 7 recurrences within the cohort, again with no unique pattern of mutations as compared to subjects without recurrences. Full Tumor mutations were determined by whole exome sequencing (4). There is an overall low mutational rate of 1.21 per Mb. This is lower than previous studies of HPV positive HNSCC and cervical squamous cell carcinomas. There is a particular predominance of TpC mutations (5.45 per Mb) compared to CpG mutations (3.94 per Mb). This phenomena is also seen in HPV related cervical cancer, whereas CpG dinucleotide alterations predominate in non-HPV related cancers such as colorectal, pancreatic, and glioblastoma multiforme (16). CpG transversions also correlate closely with degree of smoking in lung SCC and HPV-negative HNSCC (4)(17). The top 25 mutated genes along with variant classification are shown in Figure 2. There was a strong predominance of amplifications within 3q25–28 containing PIK3CA, and consistent with prior reports (18).
Figure 2.
TCGA top mutated genes. Genes are filtered to exclude common variants according to EXAC and 1000 Genomes Project (population allele frequency >0.05).
Chromatin regulators are significantly mutated within HPV+ OPSCC
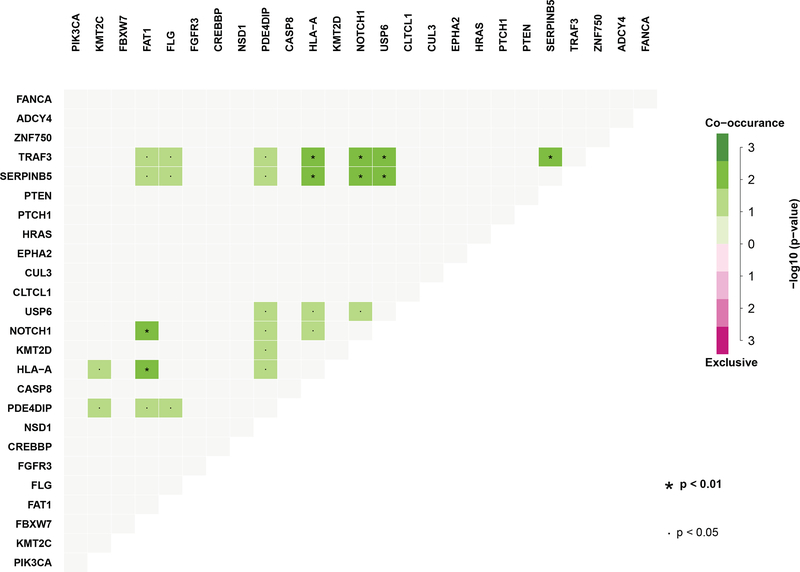
KMT2C, KMT2D, NSD1, CREBBP, EP300, and CTCF are all genes involved in histone-dependent epigenetic regulation, and in aggregate are altered in 33% (15 of 46 tumors) of our JHU cohort, and 24% (11 of 46 tumors) of the TCGA cohort. Interestingly, the majority of these within the JHU cohort are found concurrently with other mutations (Figure 3), perhaps reflecting a co-dependence on prior mutations for tumor propagation. KMT2C stands as one of the most highly altered genes within the JHU group (17.4%, 8 of 46 tumors). KMT2C is a histone methyltransferase, synonymous with the MLL class of proteins, which acts at transcription enhancer regions within cell growth pathways (19)(20). The closely related KMT2D (MLL2) mutation is also identified within 5 of 46 of the JHU tumors (10.8%).
Figure 3.
Mutational co-occurrences within exome sequencing of the JHU cohort
NSD1 is mutated within 9% (4 of 46 tumors) of the JHU cohort, and 2 tumors within the TCGA samples. NSD1 is a histone methyltransferase previously reported to be significantly altered within HPV negative HNSCC (4). This putative tumor suppressor gene is correlated with significant increased patient survival when the mutation is present, possibly related to a known strong association with genome-wide hypomethylation and corresponding platinum sensitivity (21)(22).
Mutations of the tumor suppressor CREBBP and its closely related paralogue EP300 have not been previously identified in genome-wide studies of HPV-positive HNSCC. Within the TCGA analysis, EP300 is the third most altered gene at 13% (6 of 46 tumors). There are 2 CREBBP mutations. Within our JHU cohort, CREBBP is mutated in 4 of 46 tumors (8.7%), and EP300 in 1 subject (2.2%). CREBBP/EP300 are acetyltransferases which modulate chromatin accessibility within enhancer networks, and inactivating mutations of these genes are highly recurrent within multiple types of lymphoma where they have been most studied (23). As global transcriptional coactivators they act on both proto-oncogenes and tumor suppressor genes, and loss of CREBBP/EP300 confers a clonal proliferative advantage within tumor cells (24).
CTCF was also found to be mutated in 1 JHU subject and 1 TCGA tumor. CTCF plays a role in establishing three-dimensional chromatin structure by delineating insulating boundary regions, preventing inappropriate interactions between enhancer and promoter loci (25).
Interestingly, Rb1 is mutated within 2 JHU samples (4.4%) and 4 TCGA samples (8.8%). The HPV+ E7 viral oncoprotein classically binds to Rb1, thereby disrupting the Rb-E2F complex, and prematurely pushing the cell cycle into S-phase (26). An Rb1 mutation may similarly act to disrupt binding to E2F independent of E7, but its dependence on E7 in this case remains unclear. Interestingly, one of the Rb1 mutated samples within the JHU cohort does contain expression of viral oncoproteins E245. Mechanistic and clinical studies have notably shown a clear role for Rb beyond its canonical control of E2F. Specifically, Rb plays an important role in histone modification and stabilization of the genome through epigenetic control (27), and may represent an alternate role for Rb mutation in HPV+ cancer progression.
Distinct PIK3CA and FGFR3 hotspot mutations predominate within HPV+ cancer
In our JHU cohort, the highest frequency of mutation was found within the PIK3CA gene, at a rate of 28% (13 of 46 subjects). Eight of these variants contain nonsynonymous SNVs located at the E545K hotspot position within the Helical PIK domain, and one within the E542K hotspot (Figure 4). Ten of the 13 subjects contain mutations within the helical domain, whereby only one mutation is located within the kinase domain at G1049R, immediately adjacent to the H1047R hotspot. TCGA analysis of HPV+ oropharynx tumors is markedly similar, with a likewise 28% frequency (13 of 46 tumors) of missense mutation of the PIK3CA gene. Notably, twelve of the 13 mutations were located within the E545K or E542K helical hotspot domain, and none within the kinase domain (Figure 4). In contrast, Liu et al showed that the majority of HPV-negative tumors contain mutations within the kinase domain of PIK3CA (6), highlighting a potentially distinct mechanistic difference despite alterations within the same gene.
Figure 4.
PIK3CA mutational gene mapping highlighting predominance of HPV+ OPSCC mutations within the helical domain. Top figure from our JHU cohort, and bottom figure from TCGA.
FGFR3 likewise shows a mutational signature distinct from HPV-negative tumors. The gene is mutated within 15% (6 of 46 tumors) of the JHU samples and 11% (5 of 46 tumors) of the TCGA cohort. 5 of 6 mutations within the JHU cohort occur at the S249C hotspot, and 4 of 5 mutations within TCGA occur at the same locus. FGFR3 mutational domain variants have not been previously reported within TCGA. In contrast to HPV+ tumors, TCGA HPV-negative tumors have only a 1.5% (7 of 466 samples) FGFR3 mutagenic rate, with only one of these located at the S249C hotspot.
PIK3CA and FGFR3 mutations predominate in HPV+ OPSCC, with a lesser role played by other mutations within the PIK3CA pathway. For example, only 2 tumors (4.4%, 2 of 46) contain HRAS mutations within the JHU samples. There was one HRAS mutation within TCGA HPV+ OPSCC. Two tumors contained PTEN mutations and two contained a PIK3R1 mutation in both our cohort and TCGA, respectively.
Notable Mutations within HPV+ OPSCC
We have identified potential driver mutations within HPV associated HNSCC that have only previously been identified within non-HPV associated carcinomas, suggesting a smoking-independent biological overlap between these anatomically related but distinct cancers. Three of the JHU tumors carried the CASP8 alteration. CASP8 mutations are a significant finding in HPV negative OSCC, yet to be identified in HPV+ HNSCC. The loss-of-function of the apoptosis gene CASP8 is notable for its strong correlation with an HRAS co-mutation in oral cavity cancer (4)(28), but there was no concurrent HRAS mutation within our study. Indeed, we found one recurrent G13D HRAS mutation in two subjects, and no KRAS or NRAS mutations within our entire cohort. The TCGA analysis revealed no CASP8, KRAS, or NRAS mutations within HPV+ OPSCC tumors.
SERPINB5 is mutated within 2 of the JHU samples, and reaches statistical significance based on its oncogenic clustering, suggesting its role as a driver. This gene has yet to be reported within HNSCC genome analyses. SERPINB5 is notable in our population for containing a high rate of co-occurring mutations (Figure 3), existing concurrently with NOTCH1, FAT1, USP6, PDE4DIP, TRAF3 and HLA-A. SERPINB5 is a serine protease inhibitor with tumor suppressive properties whose expression levels are correlated with poor response to chemoradiotherapy and worse overall survival in rectal squamous cell carcinoma (29).
We have identified mutations in FBXW7 within 8 of 46 tumors (17.4%) of the JHU cohort. This gene has previously been thought to represent a driver but has failed to reach statistical significance in prior exome sequencing of HPV+ HNSCCs (30). FBXW7 promotes the breakdown of Notch1 and Notch4, as well as degradation of the oncoproteins c-Myc and c-Jun (31). Clinically low expression of FBXW7 has been related to poor prognosis in colorectal cancer (32) as well as resistance to chemotherapy within lung cancer (33), highlighting its important role as a putative TSG. Importantly, within oral cavity cancer, FBXW7 may serve as a potential biomarker for clinical prognosis, as low histopathologic levels of the protein correlate with poor response to chemotherapy and lower overall survival (23). Notch1 itself is mutated within 6.5% (3 of 46) of patients within the JHU cohort and 6.5% (3 of 46) within TCGA.
Notably, CYLD is mutated within the TCGA cohort at a rate of 13% (6 of 46). CYLD inhibits NF-kB, a cellular proliferation switch in many cancers (34). PTCH1, a novel gene alteration, is mutated in 2 JHU tumor samples and one TCGA tumor. PTCH1 acts within the hedgehog signaling pathway, and its expression within certain tumor types has been shown to reliably predict response to Imatinib treatment (35). HLA-B (6.5%, 3 of 46), HLA-C (4.4%, 2 of 46), and HLA-J (2.2%, 1 of 46) are also all newly identified mutations, albeit altered at low rates.
Discussion
HPV-positive Oropharyngeal Carcinoma represents a distinct biological disease process from smoking related HNSCC. Here, we have defined the somatic mutational profile of a large cohort of HPV+ carcinomas, as well as analyzed the HPV+ OPSCC subpopulation of the newly expanded TCGA cohort, in order to further elucidate potential treatment strategies based on driver alterations.
Alterations of chromatin regulators have emerged as a distinct feature of cancer in recent years, and we show these genes to be highly mutated within HPV+ OPSCC. One-third of the top mutated genes found in our JHU cohort manage histone marks, or bind directly to chromatin, to exert transcriptional activation or silencing. Integration of viruses into the host genome is reliant on an open chromatin landscape (36), and may explain how dysregulation of these genes can assist in HPV integration. Just recently has HPV integration sites been shown to tightly correlate with specifically enriched histone marks (e.g. H3K27ac) in HPV+ oropharyngeal cancer (37). These specific histone marks are in turn associated with downstream genes known to be commonly altered in HNSCC. Our findings support the novel hypothesis that chromatin dysregulation paves the way for HPV integration.
CREBBP and its paralogue EP300 remodel chromatin via acetylation, and are altered in 15% of JHU tumors in this study. Likewise, within our updated TCGA analysis, EP300 is the third highest altered gene. CREBBP/EP300 is ubiquitously expressed, interacts with over 400 proteins, and is a global transcriptional coactivator, exerting a powerful control over the nucleosome (23)(38)(39). HPV oncoproteins E6 and E7 bind to the CREBBP/EP300 pair and inhibit their ability to acetylate p53 (36). Through this mechanism, p53 dependent gene activation is repressed, and consequently the cells ability to protect against DNA damage compromised. The binding of HPV oncoproteins to CREBBP/EP300 may explain why p53 remains notably non-mutated within HPV+ OPSCC, yet we see similar downstream epigenetic effects as in patients that do contain altered p53.
KMT2C, KMT2D, and NSD1 belong to an increasingly important group of enzymes known as protein methyltransferases (PMTs), which perform site-specific methylation of histone proteins. Only within the past decade have PMTs been implicated as drivers of carcinogenesis, with mutations strongly implicated in metastatic spread of squamous cell carcinoma (40). Within metastatic HPV+ anal cancer, KMT2C is the most commonly mutated gene at a rate of 39% (41). In squamous cell carcinoma of the esophagus, there is a higher rate of KMT2D mutations identified within metastases as compared to primary tumors (42). In our JHU study population, KMT2C is altered in a statistically significant 28% of subjects, and all subjects within this cohort presented with locoregional metastases. HPV+ OPSCC often presents clinically with large nodal disease, and it may be that alteration in chromatin regulation acts as a strong driver event in metastatic spread of HPV positive disease. In sum, the dysregulation of enhancer chromatin within HPV+ OPSCC oncogenic progression has not been well characterized previously, and here, we highlight its high rate of mutagenesis along with its potential mechanistic connection to HPV+ carcinoma.
There are currently no approved molecular therapies for HPV+ Head and Neck Cancer that are based on empiric genetic mutational data. Cetuximab, a monoclonal antibody against EGFR, was approved based on early studies within HNSCC showing an increase in copy number of this gene and its association with poor clinical prognosis (35). However, its use for HPV+ carcinoma is still being born out. Recent Phase III trials of pan-HER tyrosine kinase inhibitors have suggested a reduced efficacy of EGFR antagonists against HPV-positive tumors (43). Conversely, Bonner et al. did show an overall survival benefit with the addition of the EGFR antagonist Cetuximab to radiotherapy (44). While the majority of subjects within the study had OPSCC, HPV status was not known. Vermorken et al. also showed a survival benefit within their small HPV+ subpopulation (n=24) when Cetuximab was added to chemotherapy (45). Somewhat contradictory, within the JHU cohort there were no EGFR specific mutations, and only two tumors harbored an HRAS mutation, highlighting the need for more in-depth genomic investigations of driver events in order to bridge understanding with empiric clinical data.
Interventions upon genomic mutations downstream of EGFR, such as the PI3K signaling pathway, have only recently begun to be explored. We found PIK3CA to be the most frequently mutated gene within our JHU and TCGA study populations, at a rate of 31%. This is consistent with prior reports (18)(4), however here we demonstrate a difference in domain mutations between HPV positive and negative disease. Within our HPV+ cohort, mutations within the PIK3CA helical domain predominate, whereby kinase domain mutations are more prevalent within HPV negative tumors (6). This may have clinical impact. The BERIL-1 study has recently shown an improvement in disease-free survival with the addition of the pan-PI3K inhibitor Buparlisib to paclitaxel within metastatic Head and Neck Cancer (46). In subgroup analysis however, only subjects with HPV negative tumors derived survival benefit. Those with HPV positive HNSCC had no improvement in disease free survival or overall survival. This suggests that despite having a similar rate of PIK3CA mutations, HPV positive and negative disease may be specifically driven by domain alterations. In several other cancers, the specific PIK3CA domain alteration has been shown to be predictive of tumor response to treatment and survival. In urothelial carcinoma, mutations within the PIK3CA kinase domain correspond with high levels of AKT activation as compared to mutations within the helical domain (47)(48). And in an early phase clinical trial, patients with the kinase domain H1047R mutation had a significantly higher partial response rate to PI3K/AKT/mTOR inhibitors as compared to patients with helical domain E545K and E542K mutations (49). In breast cancer, patients with helical domain mutant tumors have been associated with poorer recurrence-free and overall survival, and conversely, those patients with kinase alterations had a better prognosis than those with even PIK3CA wild-type tumors (50). Improved clinical outcomes for HPV positive OPSCC may be dependent on isoform selective inhibition, with research ongoing. Recently, treatment of an HPV-positive PIK3CA-mutated HNSCC tumor-graft model showed significant growth inhibition following administration of the PI3K/mTOR inhibitor BEZ-235 (6).
Mechanistically, PIK3CA domain specific alterations have been linked with differential downstream pathway activations – mutations in the helical domain located on exon 9 corresponds strongly with features enabling cell migration and dissemination, while kinase alterations located on exon 20 are associated with aberrant proliferation (51). Furthermore, helical alterations are linked to oncogenesis by allowing the p110alpha catalytic subunit to escape the inhibitory effect of p85, releasing the brakes on growth factor signaling. Kinase mutants act via a different mechanism, directly activating the PI3K catalytic subunit, while still being acted upon by p85 inhibition (50). This may explain the difference in survival outcomes between HNSCC subjects with HPV positive versus HPV negative disease in the BERIL-1 trial. As the case with targetable BRAF V600E mutations, importance of selective inhibition against point mutations have become possible with deep genome-wide investigations. The PIK3CA pathway represents a possible avenue for successful intervention in HPV-positive tumors, requiring further investigation.
We likewise show a distinct FGFR3 domain mutation profile focused at the S249C hotspot, which is not found within HPV negative tumors. The S249C variant is prognostic, and correlates with significantly worse disease-free survival within HPV+ OPSCC (52). This is not true of the other FGFR3 domain variants. Moreover, targeted CHASM analysis of oncogenesis suggests this variant to have a high likelihood of being a driver (18). Within lung SCC, S249C mutations have been shown to drive cellular transformation in culture, and growth of cells expressing this mutation were inhibited by the FGFR kinase inhibitor ponatinib (53). This particular domain mutation has also been investigated within cervical and bladder carcinomas, showing a distinct profile from other FGFR3 domain variants. The S249C variant correlates with higher FGFR3 protein expression, non-16/18 HPV subtypes, and unfavorable clinical outcome (54)(55). Hotspot identification may become valuable as selective FGFR inhibitors have shown promising preclinical and clinical effects (55).
The main difference between the two cohorts was in the heterogeneous make-up of the top 25 genes from each cohort as compared to each other. Only 3 genes are found in common between the two cohorts – PIK3CA, FGFR3, and ZNF750. While this may possibly demonstrate the importance of these particular genes, it also highlights the dilemma of genome wide studies. Often, inter-study mutational findings are highly variable, blurring the lines between driver and passenger mutations, and highlighting an inherent limitation to the present study.
In conclusion, this study represents one of the largest exome analyses of HPV-positive oropharyngeal squamous cell carcinoma, including an updated analysis of the recently expanded TCGA HPV+ cohort, offering further insight into the genetic landscape of this biologically distinct disease from its smoking-related counterpart. Alterations of chromatin regulators have only recently emerged as a distinct and common feature of cancer, and here we highlight these genes to be altered in one-third of HPV+ OPSCC tumors. Particularly, KMT2C mutational burden reaches statistical significance, with other notably high mutagenic rates seen within CREBBP, EP300, and NSD1. Furthermore, while PIK3CA and FGFR3 mutations are commonly mutated within HNSCC, here we have shown a distinct domain mutation profile between HPV positive and negative tumors. Domain alterations have been suggested to be as clinically important as gene specific alterations, and their clinical relevance within HPV+ OPSCC requires further study as potential indicators of therapeutic response for targeted agents. Incorporating in-depth genome wide analyses will become ever more important in future clinical trials if we hope to improve survival outcomes and quality of life measures in HPV-positive oropharyngeal cancer.
Supplementary Material
Funding Source:
This publication was supported by the National Institute of Health (R01 DE023347 to J. Califano and S. Ren)
Footnotes
Disclosure of Potential Conflict of Interest: The authors declare no potential conflicts of interest
Bibliography
- 1.CDC [Internet]. [cited 2018. January 10]. Available from: https://www.cdc.gov/cancer/hpv/statistics/cases.htm
- 2.Ang KKP, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-tân PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2011;363(1):24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang SH, O’Sullivan B. Overview of the 8th Edition TNM Classification for Head and Neck Cancer. Curr Treat Options Oncol. 2017;18(7). [DOI] [PubMed] [Google Scholar]
- 4.Network TCGA. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science (80- ). 2012;333(6046):1157–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lui VWY, Hedberg M, Lu Y, Zeng Y, Zhang Q, Du Y, et al. Frequent mutation of the PI3K pathway iin head and neck cancer defines predictive biomarkers. Cancer Discov. 2014;3(7):761–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agrawal N, Frederick MJ, Pickering CR, Chang K, Li RJ, Fakhry C, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science (80- ). 2011;333(6046):1154–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basu B, Chakraborty J, Chandra A, Katarkar A, Ritesh J, Baldevbhai K, et al. Genome-wide DNA methylation profile identified a unique set of differentially methylated immune genes in oral squamous cell carcinoma patients in India. 2017;1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo T, Gaykalova DA, Considine M et al. Characterization of functionally active gene fusions in human papillomavirus related oropharyngeal squamous cell carcinoma. Int J Cancer. 2016;139(2):373–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fakhry C, Lacchetti C, Rooper LM, Jordan RC, Rischin D, Sturgis EM, et al. Human Papillomavirus Testing in Head and Neck Carcinomas : ASCO Clinical Practice Guideline Endorsement of the College of American Pathologists Guideline. J Clin Oncol. 2019;36(31). [DOI] [PubMed] [Google Scholar]
- 11.Patch A, Nones K, Kazakoff SH, Newell F, Wood S, Leonard C, et al. Germline and somatic variant identification using BGISEQ-500 and HiSeq X Ten whole genome sequencing. PLoS One. 2018;1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.cfncluster (https://github.com/awslabs/cfncluster).
- 13.Consortium GP. A global reference for human genetic variation. Nature. 2016;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mckenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit : A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mountzios G, Rampias T, Psyrri A. The mutational spectrum of squamous-cell carcinoma of the head and neck: targetable genetic events and clinical impact. Ann Oncol. 2014;25(10):1889–900. [DOI] [PubMed] [Google Scholar]
- 16.Rubin AF, Green P. Mutation patterns in cancer genomes. PNAS. 2009;106(51):21766–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cancer T, Atlas G. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;517(7536):576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seiwert TY, Zuo Z, Keck MK, Khattri A, Pedamallu CS, Stricker T, et al. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin cancer Res [Internet] 2015;21(3):632–41. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4305034&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ford DJ, Dingwall AK. The cancer COMPASS: Navigating the functions of MLL complexes in cancer. Cancer Genet [Internet] 2015;208(5):178–91. Available from: 10.1016/j.cancergen.2015.01.005 [DOI] [PubMed] [Google Scholar]
- 20.Chiappetta C, Mancini M, Lessi F, Aretini P, Gregorio V De, Puggioni C, et al. Whole-exome analysis in osteosarcoma to identify a personalized therapy. Oncotarget [Internet]. 2017;8(46):80416–28. Available from: http://www.oncotarget.com/fulltext/19010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peri S, Izumchenko E, Schubert AD, Slifker MJ, Ruth K, Serebriiskii IG, et al. NSD1 and NSD2 damaging mutations define a subset of laryngeal tumors with favorable prognosis. Nat Commun [Internet]. 2017;8(1):1772 Available from: 10.1038/s41467-017-01877-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bui N, Huang JK, Bojorquez-gomez A, Licon K, Sanchez KS. Disruption of NSD1 in head and neck cancer promotes favorable chemotherapeutic responses linked to hypomethylation. Mol Cancer Ther. 2018;Accepted,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Vlasevska S, Wells VA, Nataraj S, Holmes AB, Duval R, et al. The CREBBP Acetyltransferase Is a Haploinsuffi cient Tumor Suppressor in B-cell Lymphoma. Cancer Discov. 2017;7(3):322–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avantaggiati ML, Ogryzko V, Gardner K, Giordano A, Levine AS, Kelly K. Recruitment of p300 / CBP in p53-Dependent Signal Pathways. Nature. 1997;89:1175–84. [DOI] [PubMed] [Google Scholar]
- 25.Ausió J, Georgel PT. MeCP2 and CTCF : enhancing the cross-talk of silencers. Biochem Cell Biol. 2017;608(August):593–608. [DOI] [PubMed] [Google Scholar]
- 26.Dyson N The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12(617):2245–62. [DOI] [PubMed] [Google Scholar]
- 27.Dick FA, Goodrich DW, Sage J, Dyson NJ. Non-canonical functions of the RB protein in cancer. Nat Rev Cancer [Internet]. 2018; Available from: 10.1038/s41568-018-0008-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pickering CR, Zhang J, Yoo SY, Bengtsson L, Cortez E, Xie T, et al. Integrative genomic characterization of oral squamous cell carcinomaidentifies frequent somatic drivers. Cancer Discov. 2013;3(7):1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang I, Liu K, Ragunanan M, He H, Shiue Y. SERPINB5 Expression : Association with CCRT Response and Prognostic Value in Rectal Cancer. Int J Med Sci. 2018;15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayes DN, Van Waes C, Seiwert TY. Genetic landscape of human papillomavirus-associated head and neck cancer and comparison to tobacco-related tumors. J Clin Oncol. 2015;33(29):3227–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakayama KI, Nakayama K. Ubiquitin ligases : cell-cycle control and cancer. 2006;6(May):369–81. [DOI] [PubMed] [Google Scholar]
- 32.Iwatsuki M, Mimori K, Ishii H, Yokobori T, Takatsuno Y, Sato T, et al. Loss of FBXW7, a cell cycle regulating gene, in colorctal cancer : clinical significance. Int J Cancer. 2010;126(8):1828–37. [DOI] [PubMed] [Google Scholar]
- 33.Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, et al. SCF regulates cellular apoptosis by targeting Mcl-1 for ubiquitination and destruction. Nature. 2011;471(7336):104–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan C, Yarbrough WG, Issaeva N. Advances in biomarkers and treatment HPV-associated head and neck cancer strategies for. Oncoscience. 2018;5(May):140–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung CH, Ely K, Mcgavran L, Varella-garcia M, Parker J, Parker N, et al. J OURNAL OF C LINICAL O NCOLOGY Increased Epidermal Growth Factor Receptor Gene Copy Number Is Associated With Poor Prognosis in Head and Neck Squamous Cell Carcinomas. 2018;24(25). [DOI] [PubMed] [Google Scholar]
- 36.Durzynska J, Lesniewicz K, Poreba E. Mutation Research / Reviews in Mutation Research Human papillomaviruses in epigenetic regulations. Mutat Res Mutat Res [Internet]. 2017;772:36–50. Available from: 10.1016/j.mrrev.2016.09.006 [DOI] [PubMed] [Google Scholar]
- 37.Kelley DZ, Flam EL, Izumchenko E, Danilova LV, Wulf HA, Guo T, et al. Integrated Analysis of Whole-Genome ChIP-Seq and RNA-Seq Data of Primary Head and Neck Tumor Samples Associates HPV Integration Sites with Open Chromatin Marks. Cancer Res. 2017;77(23):6538–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gayther SA, Batley SJ, Linger L, Bannister A, Thorpe K, Chin S, et al. Mutations truncating the EP300 acetylase in human cancers. 2000;24(March):1–4. [DOI] [PubMed] [Google Scholar]
- 39.Asaduzzaman M, Constantinou S, Min H, Gallon J, Poonam ML, Selina S, et al. Tumour suppressor EP300 , a modulator of paclitaxel resistance and stemness , is downregulated in metaplastic breast cancer. 2017;461–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Copeland RA. Molecular Pathways : Protein Methyltransferases in Cancer. Clin Cancer Res. 2013;19(23):6344–51. [DOI] [PubMed] [Google Scholar]
- 41.Morris V, Rao X, Pickering C, Foo WC, Rashid A, Eterovic K, et al. Comprehensive Genomic Pro filing of Metastatic Squamous Cell Carcinoma of the Anal Canal. 2017;15(November):1542–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dai W, Ko JMY, Choi SSA, Yu Z, Ning L, Zheng H, et al. Whole-exome sequencing reveals critical genes underlying metastasis in oesophageal squamous cell carcinoma. J Pathol [Internet]. 2017;242(4):500–10. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28608921%0Ahttp://doi.wiley.com/10.1002/path.4925 [DOI] [PubMed] [Google Scholar]
- 43.Machiels JH, Haddad RI, Fayette J, Licitra LF, Tahara M, Vermorken JB, et al. Afatinib versus methotrexate as second-line treatment in patients with recurrent or metastatic squamous-cell carcinoma of the head and neck progressing on or after platinum-based therapy ( LUX-Head & Neck 1 ): an open-label , randomised phase 3 trial. 2015;583–94. [DOI] [PubMed] [Google Scholar]
- 44.Bonner JA, Harari PM, Giralt J, Cohen RB, Jones CU, Sur RK, et al. Radiotherapy plus cetuximab for locoregionally advanced head and neck cancer: 5-year survival data from a phase 3 randomised trial, and relation between cetuximab-induced rash and survival. Lancet Oncol. 2010;11(1):21–8. [DOI] [PubMed] [Google Scholar]
- 45.Vermorken JB, Psyrri A, Mesía R, Peyrade F, Beier F, Blas B De, et al. Impact of tumor HPV status on outcome in patients with recurrent and / or metastatic squamous cell carcinoma of the head and neck receiving chemotherapy with or without cetuximab : retrospective analysis of the phase III EXTREME trial. Ann Oncol. 2014;25(February):801–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soulières D, Faivre S, Mesía R, Remenár É, Li S, Karpenko A, et al. Buparlisib and paclitaxel in patients with platinum-pretreated recurrent or metastatic squamous cell carcinoma of the head and neck ( BERIL-1 ): a randomised ,. Lancet Oncol. 2017;18(March):323–35. [DOI] [PubMed] [Google Scholar]
- 47.Koncar RF, Feldman R, Bahassi EM, Hashemi Sadraei N. Comparative molecular profiling of HPV-induced squamous cell carcinomas. Cancer Med. 2017;6(7):1673–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ross RL, Askham JM, Knowles MA. PIK3CA mutation spectrum in urothelial carcinoma reflects cell context-dependent signaling and phenotypic outputs. Oncogene [Internet]. 2012;32(6):768–76. Available from: 10.1038/onc.2012.87 [DOI] [PubMed] [Google Scholar]
- 49.Janku F, Wheler JJ, Naing A, Falchook GS, Hong DS, Stepanek VM, et al. PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early phase clinical trials. Cancer Res. 2014;73(1):276–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barbareschi M, Buttitta F, Felicioni L, Cotrupi S, Barassi F, Grammastro M Del, et al. Different Prognostic Roles of Mutations in the Helical and Kinase Domains of the PIK3CA Gene in Breast Carcinomas. Clin Canc Res. 2007;13(20):6064–70. [DOI] [PubMed] [Google Scholar]
- 51.Mukohara T. PI3K mutations in breast cancer : prognostic and therapeutic implications. Breast Cancer Targets Ther. 2015;15(7):111–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bersani C, Sivars L, Haeggblom L, Dilorenzo S, Ährlund-richter A, Tertipis N, et al. Targeted sequencing of tonsillar and base of tongue cancer and human papillomavirus positive unknown primary of the head and neck reveals prognostic effects of mutated FGFR3. 2017;8(21):35339–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liao RG, Jung J, Tchaicha J, Wilkerson MD, Sivachenko A, Beauchamp EM, et al. Inhibitor-Sensitive FGFR2 and FGFR3 Mutations in Lung Squamous Cell Carcinoma. Cancer Res. 2013;73(16):5195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baldia PH, Maurer A, Heide T, Rose M, Stoehr R, Hartmann A, et al. Fibroblast growth factor receptor ( FGFR ) alterations in squamous differentiated bladder cancer : a putative therapeutic target for a small subgroup. 2016;7(44). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosty C, Aubriot M, Cappellen D, Bourdin J, Cartier I, Thiery JP, et al. Clinical and biological characteristics of cervical neoplasias with FGFR3 mutation. Mol Cancer. 2005;8:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.