Abstract
The liver kinase B1-AMP-activated protein kinase (LKB1-AMPK) pathway has been identified as a new target for cancer therapy, because it controls the glucose and lipid metabolism in response to alterations in nutrients and intracellular energy levels. In the present study, we aimed to identify genetic variants of the LKB1-AMPK pathway genes and their associations with pancreatic cancer (PanC) risk using 15,418 participants of European ancestry from two previously published PanC genome-wide association studies. We found that six novel tagging single-nucleotide polymorphisms (SNPs) (i.e., MAP2 rs35075084 T>deletion, PRKAG2 rs2727572 C>T and rs34852782 A>deletion, TP53 rs9895829 A>G, and RPTOR rs62068300 G>A and rs3751936 G>C) were significantly associated with an increased PanC risk. The multivariate logistic regression model incorporating the number of unfavorable genotypes (NUGs) with adjustment for age and sex showed that carriers with 5-6 NUGs had an increased PanC risk (odds ratio=1.24, 95% confidence interval=1.16-1.32 and P<0.0001), compared to those with 0-4 NUGs. Subsequent expression quantitative trait loci (eQTL) analysis further revealed that these SNPs were associated with significantly altered mRNA expression levels either in 373 normal lymphoblastoid cell lines (TP53 SNP rs9895829, P<0.05) or in whole blood cells of 369 normal donors from the genotype-tissue expression project (GTEx) database [RPTOR SNP rs60268947 and rs28434589, both in high linkage disequilibrium (r2>0.9) with RPTOR rs62068300, P<0.001]. Collectively, our findings suggest that these novel SNPs in the LKB1-AMPK pathway genes may modify susceptibility to PanC, possibly by influencing gene expression.
Keywords: single-nucleotide polymorphism, genome-wide association study, pancreatic cancer risk
Introduction
Pancreatic cancer (PanC) is one of the most lethal human malignancies, with an overall five-year survival rate lower than 10% and a median survival of six months, and PanC is also the fourth leading cause of cancer-related death and is expected to become the second within the next decade in the United States 1,2. The exact cause of PanC is not yet well understood, though several risk factors have been identified, including smoking, morbid obesity, having a family history of PanC or pancreatitis, and having certain hereditary conditions 3,4. More importantly, patients with PanC rarely exhibit symptoms at the early stages, until the disease reaches an advanced stage, which is one of the main reasons for the observed, generally poor survival rates 4,5. Therefore, prevention and early diagnosis at a curable stage are desperately needed to reduce PanC mortality.
Since Otto Warburg first proposed a connection between cellular metabolism and tumorigenesis nearly 100 years ago, pointing out a new direction for cancer research 6,7, numerous studies have been reported on molecular mechanisms that link the signaling pathways controlling the metabolism to cell growth, in which the metabolism reprogramming was found to be necessary for cancer initiation and progression, a hallmark of cancer 8,9. PanC is characterized by a severely hypoxic and nutrient-deprived microenvironment, with specific metabolically adaptive mechanisms, such as the Warburg effect, glutamine addiction and autophagy, that all contribute to PanC development and progression, in addition to both oncogenes/tumor suppressors and tumor microenvironment 10,11. Therefore, targeting any of specific metabolic adaptions becomes an emerging strategy for PanC diagnosis and treatment 12,13.
LKB1 (liver kinase B1, also known as STK11) directly activates the AMPK (AMP-activated protein kinase), which is responsible for nutrient sensing and metabolism reprogramming, and LKB1 is inactivated by mutations found in PanC, and the loss of LKB1 is thought to drive tumorigenesis 14-16.
AMPK is a master regulator of metabolic homeostasis by sensing cellular energy status, the AMP:ATP ratio. When there is an increase in the cellular AMP:ATP ratio, which reflects a decrease in energy supply, AMPK is phosphorylated and activated, promoting catabolic processes and inhibiting anabolic processes to increase the energy level 17.
Recent studies found that meisoindigo can preferentially kill cancer stem cells by interfering cell metabolism via inhibition of LKB1 and activation of AMPK in PanC 18. Similar results have also been drawn from metformin, which influences PanC progression by activating the LKB1-AMPK pathway, including inhibition of cell division, promotion of apoptosis and autophagy, down-regulation of circulating insulin, and activation of the immune system 19.
Therefore, we hypothesize that genetic variants of the LKB1-AMPK pathway genes are associated with PanC risk. To test such a hypothesis, we conducted a comprehensive meta-analysis of genetic variants in the LKB1-AMPK pathway genes using two previously published genome-wide association study (GWAS) datasets from the PanScan (i.e., the Pancreatic Cancer Cohort Consortium) and Pancreatic Cancer Case-Control Association Study. We also focused our analysis on the identified single-nucleotide polymorphisms (SNPs) that may change the mRNA expression levels of the genes and thus are likely have functional consequences.
Methods
Study Population and GWAS Data
The analysis used two previously published PanC GWASs: Pancreatic Cancer Cohort Consortium (PanScan, phs000206.v5.p3) and the Pancreatic Cancer Case-Control Association Study (dbGaP#:phs000648.v1.p1), which included 15,418 participants (8,474 cases and 6,944 controls). The PanScan GWAS has three phases, including PanScan I, II and III (1,760 cases and 1,780 controls in PanScan I; 1,457 cases and 1,666 controls in PanScan II, 1,538 cases and 0 controls in PanScan III) 20-23. We then merged the PanScan II and PanScan III into one dataset, PanScan II/III, because PanScan III was a case-only study 24. Another GWAS dataset was from the Pancreatic Cancer Case-Control Consortium (PanC4) that consisted of 3,719 cases and 3,498 controls from the United States, Europe and Australia (Supplementary Fig. S1). All the participants in these GWASs were of European ancestry, and a written informed consent was obtained from all study participants. All methods were performed in accordance with the relevant guidelines and regulations for each of the participating institutions, and the present study followed the protocols approved by Duke University Health System Institutional Review Board.
Gene selection, genotyping, and imputation
The keywords “LKB1-AMPK” was searched in Molecular Signatures Database (MSigDB) (http://www.broadinstitute.org/gsea/msigdb/index.jsp), and all the 58 related genes located on autosomal chromosomes were selected from REACTOME and PID (details presented in Supplementary Table S1). The GWAS genotyping was performed using Illumina HumanHap550v3.0, Human 610-Quadv1_B, HumanOmniExpress-12v1.0 and HumanOmniExpressExome-8v1.0 25-27. The SNPs located both in these genes and their ± 500-kb flanking regions were extracted for further imputation by the IMPUTE2 software, using the 1000 Genomes (Phase 1, Release 3) Project as the reference dataset 28,29.
For quality control, the imputed SNPs with an information/accuracy score > 0.4 were qualified for further analysis (with details presented in Table 1 and Supplemental Fig. S2). As a result, there were 14,557 SNPs, 15,866 SNPs and 14,263 SNPs within 5.0 kb up- and down-streams of the 58 LKB1-AMPK pathway genes from populations of the PanScan I, PanScan II/III and panC4 studies, respectively. The final meta-analysis for all three dataset contained 12,777 SNPs that meets the inclusion criteria: a call rate > 95%, MAF > 1% and Hardy-Weinberg equilibrium (HWE) test P value > 1 × 10−5.
Table 1.
Associations between 16 SNPs in the AMPK-LKB1 pathway genes and pancreatic cancer risk with an FPRP ≤ 0.02.
| SNP rs ID# | Locus | Gene | Allelea | IS1b | IS2b | IS3b | EAF1c | EAF2c | EAF3c | OR (95% CI)d | Pe | FPRP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs35075084 | 2q34 | MAP2 | CT>C | 0.540 | 0.602 | 0.545 | 0.019 | 0.023 | 0.023 | 0.76 (0.64-0.90) | 0.0010 | 0.020 |
| rs2727572 | 7q36.1 | PRKAG2 | C>T | 0.982 | 0.979 | 0.982 | 0.473 | 0.454 | 0.445 | 1.08 (1.03-1.13) | 0.0011 | 0.019 |
| rs12668489 | 7q36.1 | PRKAG2 | C>T | 0.985 | 0.982 | 0.986 | 0.473 | 0.456 | 0.448 | 1.08 (1.03-1.13) | 0.0010 | 0.018 |
| rs2538046 | 7q36.1 | PRKAG2 | G>A | 0.995 | 0.993 | 0.991 | 0.505 | 0.486 | 0.474 | 1.09 (1.04-1.14) | 0.0003 | 0.005 |
| rs34852782 | 7q36.1 | PRKAG2 | TA>T | 0.886 | 0.860 | 0.791 | 0.283 | 0.272 | 0.275 | 1.10 (1.05-1.16) | 0.0002 | 0.003 |
| rs17884306 | 17p13.1 | TP53 | C>T | 0.979 | 0.973 | 0.971 | 0.060 | 0.058 | 0.057 | 0.83 (0.75-0.93) | 0.0004 | 0.007 |
| rs17879377 | 17p13.1 | TP53 | C>T | 0.972 | 0.969 | 0.966 | 0.051 | 0.053 | 0.050 | 0.84 (0.74-0.94) | 0.0011 | 0.020 |
| rs9891744 | 17p13.1 | TP53 | C>T | 0.980 | 0.975 | 0.976 | 0.060 | 0.059 | 0.058 | 0.82 (0.73-0.91) | 0.0001 | 0.002 |
| rs75732100 | 17p13.1 | TP53 | C>T | 0.985 | 0.980 | 0.981 | 0.060 | 0.058 | 0.058 | 0.82 (0.74-0.92) | 0.0002 | 0.004 |
| rs35850753 | 17p13.1 | TP53 | C>T | 0.963 | 0.948 | 0.952 | 0.021 | 0.022 | 0.023 | 0.74 (0.62-0.88) | 0.0004 | 0.008 |
| rs9895829 | 17p13.1 | TP53 | A>G | 0.991 | 0.987 | 0.991 | 0.061 | 0.058 | 0.058 | 0.82 (0.74-0.92) | 0.0002 | 0.003 |
| rs17883323 | 17p13.1 | TP53 | G>T | 0.989 | 0.987 | 0.991 | 0.061 | 0.058 | 0.057 | 0.83 (0.74-0.92) | 0.0002 | 0.004 |
| rs8079544 | 17p13.1 | TP53 | G>T | 1.000 | 1.000 | 1.000 | 0.060 | 0.058 | 0.058 | 0.82 (0.74-0.92) | 0.0002 | 0.003 |
| rs62068300 | 17q25.3 | RPTOR | G>A | 0.973 | 0.974 | 0.978 | 0.308 | 0.320 | 0.330 | 0.92 (0.87-0.97) | 0.0008 | 0.015 |
| rs17848685 | 17q25.3 | RPTOR | C>G | 0.729 | 0.701 | 0.822 | 0.213 | 0.228 | 0.234 | 0.91 (0.86-0.97) | 0.0011 | 0.019 |
| rs3751936 | 17q25.3 | RPTOR | G>C | 0.897 | 0.883 | 0.906 | 0.264 | 0.268 | 0.258 | 0.91 (0.86-0.97) | 0.0008 | 0.014 |
Abbreviations: SNP: single nucleotide polymorphism; FPRR: false positive report probability; IS: information score; EAF: effect allele frequency; OR: odds ratio; CI: confidence interval.
Referring to “common allele/effect allele.”
IS1 was EAF in PanScan I controls; IS2 was EAF in PanScan II/III controls; IS3 was EAF in PanC4 controls.
EAF1 was EAF in PanScan I controls; EAF2 was EAF in PanScan II/III controls; EAF3 was EAF in PanC4 controls.
Fixed effects models were used when no heterogeneity was found between studies (Q test P > 0.10 and I2 < 50.0%); otherwise, random effects models were used.
Obtained from the meta-analysis of the three studies.
Association analysis
Unconditional multivariable logistic regression including sex, age and top five principal components was performed using PLINK (version 1.90), assuming an additive genetic model, with assessment of genomic data to control for potential population stratification. The principal components were computed by genome-wide complex trait analysis and the top five principal components with P value less than 0.001 were selected from the logistic regression analysis from all the three studies (Supplementary Table S2), and an odds ratio and its 95% CI were estimated for each SNP with PLINK 30. A meta-analysis was further employed on the results of a log-additive model of 12,777 SNPs using the fixed-effects inverse-variance method based on β estimates and standard errors with Stata software (v 12, State College, Texas, US). Cochran’s Q statistics and I2 were used to assess the heterogeneity 31. If the Cochran’s Q-test P value > 0.100 and the heterogeneity statistic I2 < 50%, a fixed-effects model was applied. Otherwise, a random-effects model was employed.
False positive report probability (FPRP) is the probability of no true association between a genetic variant and disease, given a statistically significant finding. We chose FPRP to correct for multiple testing, because more than 90% of SNPs (12,379 out of 12,777 under investigation) included in the present study were imputed. The FPRP approach with a prior probability of 0.01 and a hazards ratio (HR) of 2.0 was assigned for an association with genotypes and alleles of each SNP to reduce the probability of false-positive findings. The association between each SNP and PanC risk was evaluated with an additive genetic model in which a cut-off FPRP value ≤ 0.02 was considered as a significant association. The multivariable stepwise logistic regression model was also employed to identify independent SNPs. The number of unfavorable genotypes (NUGs) of SNPs with independent effects was calculated to assess the classification performance of the model. All individuals were further dichotomized into low-risk group (0-4 NUGs) and high-risk group (5-6 NUGs) for additional analysis. Besides, Haploview v4.2 32 was used to produce the Manhattan plot and Linkage disequilibrium (LD) plot, and LocusZoom 33 was employed to construct the regional association plots by using the dataset from the 1000 Genomes Project. Linear regression was used to analyze the correlations between SNPs and corresponding mRNA expression levels. All statistical analyses were performed with SAS software (version 9.4; SAS Institute, Cary, NC, USA), if not specified otherwise.
In silico functional prediction and validation
To predict potential functions of the significant SNPs, we used two in silico tools: RegulomeDB (http://regulomedb.org/) 34 and HaploReg (http://www.broadinstitute.org/mammals/haploreg/haploreg.php) 35. We performed the expression quantitative trait loci (eQTL) analysis to estimate the associations between SNPs and mRNA expression levels of the corresponding genes by using the mRNA expression data from the lymphoblastoid cell lines of 373 Europeans available in the 1000 Genomes Project 36 and 127 tumor tissues in The Cancer Genome Atlas (TCGA) 37 as well as the eQTL results from the genotype-tissue expression project (GTEx) database for the whole blood (n=369) and normal pancreatic tissues (n=220) (http://www.gtexportal.org/home/) 38. In addition, we also compared the mRNA expression levels of targeted gene between tumor and adjacent normal tissues available in the Oncomine™ database (https://www.oncomine.org/) 39.
Results
Single locus analysis
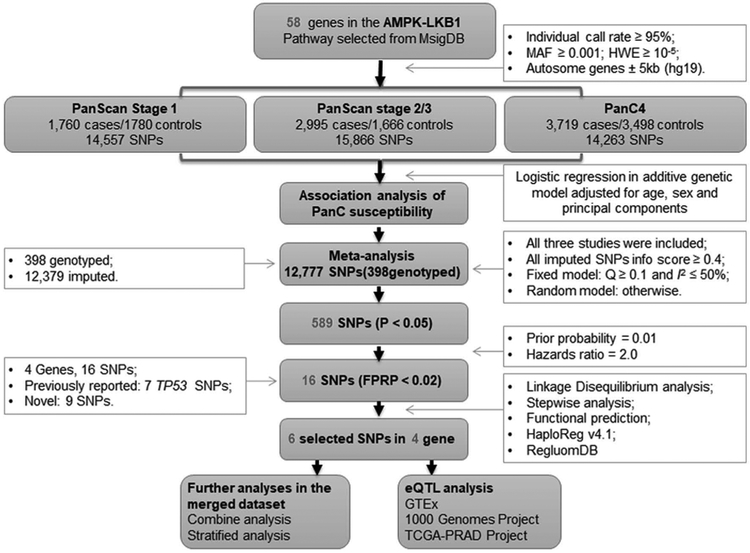
The research workflow of the present study design is shown in Fig. 1. We first estimated the associations between selected SNPs [with a minor allele frequency (MAF) ≥ 0.01] and PanC risk with the unconditional logistic regression analysis for each of the three populations of European ancestry with 14,557 SNPs, 15,866 SNPs and 14,263 SNPs for PanScan I, PanScan II/III and PanC4, respectively; the single locus analysis revealed that these three study populations had 623, 1713 and 911 SNPs with a nominal P < 0.05, respectively (Supplemental Fig. S3). Then, we included a total of 12,777 SNPs in a meta‐analysis of the three populations and found that 589 SNPs remained to be associated with PanC risk at P < 0.05 in an additive genetic model, of which 16 SNPs on MAP2, PRKAG2, TP53 and RPTOR passed multiple testing corrections with an FPRP ≤ 0.02 (Supplemental Fig. S4a; Table 1).
Fig. 1. The workflow of the analysis.
MAF: minor allele frequency; HWE: Hardy-Weinberg equilibrium; SNP: single nucleotide polymorphism; PanC: pancreatic cancer; FPRP: false positive report probability; eQTL: expression quantitative trait loci.
Although seven SNPs of TP53 (i.e., rs17884306, rs17879377, rs9891744, rs75732100, rs9895829, rs17883323 and rs8079544) located at 17p13 have been previously reported by the AURORA pathway-based analyses 40, the TP53 rs35850753 and other eight SNPs (i.e., MAP2 rs35075084, PRKAG2 rs2727572, rs12668489, rs2538046 and rs34852782 and RPTOR rs62068300 and rs17848685, rs3751936) located at 2q34, 7q36.1 and 17q25.3, respectively, are novel findings, for which we performed additional in silico analysis for their functional relevance. The results of these SNPs in each of GWAS datasets and the final meta‐analysis are summarized in Table 2. All these SNPs showed a low heterogeneity among the three GWAS datasets (all Q‐test P > 0.100 and I2 < 50.0).
Table 2.
The results of these 16 SNPs in each of GWAS datasets and the final meta-analysis.
| SNP rs ID# | Allele | Position | Gene | PanScan_stage1 | PanScan_stage2 | PanC4 | All GWAS combined | Heterogeneity | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | Q | I2 | ||||
| rs35075084 | CT>C | 210473015 | MAP2 | 0.78 (0.54-1.13) | 0.188 | 0.88 (0.66-1.18) | 0.400 | 0.68 (0.54-0.86) | 0.001 | 0.76 (0.64-0.90) | 0.0010 | 0.398 | 0 |
| rs2727572 | C>T | 151298246 | PRKAG2 | 1.03 (0.94-1.13) | 0.509 | 1.08 (1.00-1.18) | 0.064 | 1.10 (1.03-1.18) | 0.005 | 1.08 (1.03-1.13) | 0.0011 | 0.541 | 0 |
| rs34852782 | TA>T | 151566171 | PRKAG2 | 1.08 (0.97-1.20) | 0.144 | 1.11 (1.01-1.22) | 0.036 | 1.11 (1.03-1.20) | 0.005 | 1.10 (1.05-1.16) | 0.0002 | 0.902 | 0 |
| rs12668489 | C>T | 151301725 | PRKAG2 | 1.04 (0.95-1.15) | 0.362 | 1.08 (0.99-1.17) | 0.087 | 1.10 (1.03-1.17) | 0.005 | 1.08 (1.03-1.13) | 0.0010 | 0.675 | 0 |
| rs2538046 | G>A | 151314162 | PRKAG2 | 0.94 (0.85-1.03) | 0.161 | 1.07 (0.98-1.16) | 0.132 | 1.11 (1.04-1.19) | 0.002 | 1.09 (1.04-1.14) | 0.0003 | 0.715 | 0 |
| rs17884306 | C>T | 7572101 | TP53 | 0.81 (0.65-0.99) | 0.044 | 0.82 (0.68-0.99) | 0.039 | 0.85 (0.73-0.99) | 0.032 | 0.83 (0.75-0.93) | 0.0004 | 0.912 | 0 |
| rs17879377 | C>T | 7574721 | TP53 | 0.88 (0.70-1.10) | 0.256 | 0.78 (0.64-0.95) | 0.015 | 0.85 (0.73-1.00) | 0.044 | 0.84 (0.74-0.94) | 0.0011 | 0.706 | 0 |
| rs9891744 | C>T | 7574864 | TP53 | 0.79 (0.64-0.98) | 0.032 | 0.81 (0.67-0.98) | 0.028 | 0.83 (0.72-0.97) | 0.016 | 0.82 (0.73-0.91) | 0.0001 | 0.817 | 0 |
| rs75732100 | C>T | 7576348 | TP53 | 0.80 (0.65-0.99) | 0.037 | 0.83 (0.69-1.00) | 0.046 | 0.84 (0.72-0.97) | 0.018 | 0.82 (0.74-0.92) | 0.0002 | 0.947 | 0 |
| rs35850753 | C>T | 7578671 | TP53 | 0.94 (0.67-1.31) | 0.712 | 0.60 (0.43-0.83) | 0.002 | 0.73 (0.58-0.93) | 0.010 | 0.74 (0.62-0.88) | 0.0004 | 0.168 | 43.97 |
| rs9895829 | A>G | 7578679 | TP53 | 0.79 (0.64-0.98) | 0.029 | 0.83 (0.69-1.00) | 0.046 | 0.84 (0.72-0.97) | 0.020 | 0.82 (0.74-0.92) | 0.0002 | 0.913 | 0 |
| rs17883323 | G>T | 7579619 | TP53 | 0.79 (0.64-0.98) | 0.029 | 0.82 (0.68-0.99) | 0.042 | 0.84 (0.73-0.98) | 0.024 | 0.83 (0.74-0.92) | 0.0002 | 0.896 | 0 |
| rs8079544 | G>T | 7580052 | TP53 | 0.80 (0.65-0.99) | 0.036 | 0.82 (0.68-0.99) | 0.039 | 0.83 (0.72-0.97) | 0.017 | 0.82 (0.74-0.92) | 0.0002 | 0.948 | 0 |
| rs62068300 | G>A | 78574727 | RPTOR | 0.96 (0.87-1.07) | 0.479 | 0.92 (0.84-1.01) | 0.083 | 0.90 (0.84-0.96) | 0.003 | 0.92 (0.87-0.97) | 0.0008 | 0.537 | 0 |
| rs17848685 | C>G | 78599562 | RPTOR | 1.00 (0.89-1.12) | 0.998 | 0.92 (0.83-1.02) | 0.099 | 0.87 (0.80-0.94) | 0.001 | 0.91 (0.86-0.97) | 0.0011 | 0.150 | 47.36 |
| rs3751936 | G>C | 78938204 | RPTOR | 1.00 (0.90-1.11) | 0.960 | 0.89 (0.81-0.98) | 0.017 | 0.89 (0.82-0.96) | 0.003 | 0.91 (0.86-0.97) | 0.0008 | 0.178 | 41.98 |
Abbreviations: SNP: single nucleotide polymorphism; GWAS: genome-wide association study; OR: odds ratio; CI: confidence interval.
LD analysis and stepwise analysis
For the LD analysis, three (i.e., rs2727572, rs12668489 and rs2538046) of the four PRKAG2 SNPs shared a high LD (r2 ≥ 0.80, Supplemental Fig. S4b and Supplemental Fig. S5a); seven (i.e., rs17884306, rs17879377, rs9891744, rs75732100, rs9895829, rs17883323 and rs8079544) of the eight TP53 SNPs shared a high LD (r2 ≥ 0.80, Supplemental Fig. S4d and Supplemental Fig. S5c); and three RPTOR SNPs (i.e., rs62068300, rs17848685 and rs3751936) showed a low LD (r2 ≤ 0.80, Supplemental Fig. S4c and Supplemental Fig. S5b). Having considered functional prediction and LD, we selected final eight SNPs, including MAP2 rs35075084, two proxy PRKAG2 SNPs (i.e., rs2727572 and rs34852782), two proxy TP53 SNPs (i.e., rs9895829 and rs35850753) and all three SNPs of RPTOR (i.e., rs62068300, rs17848685 and rs3751936), for further analysis. Next, we assessed these eight representative SNPs in the presence of age, sex and top five principal components in a multivariate stepwise logistic regression model. As a result, the genotypes of six SNPs remained significantly and independently associated with PanC risk (Supplemental Table S3).
Genotype effect and the joint‐effect of the six significant SNPs
After stepwise analysis, we found that the genotypes of MAP2 rs35075084 T>deletion, TP53 rs9895829 A>G, PRKAG2 rs2727572 C>T and rs34852782 A>deletion, and RPTOR rs62068300 G>A and rs3751936 G>C were significantly associated with PanC risk in both additive and dominant genetic models.
In the additive model, the association between each of these six novel SNPs and PanC risk had a linear trend as the frequency of the minor allele increased (trend test: P = 0.0008, P = 0.0004, P = 0.0007, P = 0.0002, P = 0.0009 and P = 0.0009, respectively, Table 3). Consistent with previous results of the single locus analysis, the rs2727572 T allele was associated with an increased PanC risk [odds ratio (OR) = 1.11, 95% confidence interval (CI) = 1.04-1.19 and P = 0.0034], the rs34852782 deleted allele was associated with an increased PanC risk (OR = 1.09, 95% CI = 1.02-1.16 and P = 0.0076), while the rs35075084 deleted allele, rs9895829 G, rs62068300 A and rs3751936 C alleles were associated with a reduced risk (OR = 0.75, 95% CI = 0.64-0.89, P = 0.0007, OR = 0.83, 95% CI = 0.75-0.92, P = 0.0004, OR = 0.91, 95% CI = 0.85-0.97, P = 0.0026 and OR = 0.91, 95% CI = 0.86-0.97, P = 0.0051, respectively), compared with their corresponding wild-type allele (Table 3).
Table 3.
Analysis of associations between PanC risk and the six SNPs in the dataset of PanScan and PanC4 studies.
| SNP rs ID# & genetic model |
Group | OR (95% CI)a | Pb | ||
|---|---|---|---|---|---|
| Genotype | Case (%) | Control (%) | |||
| MAP2 rs35075084 CT>Cb | |||||
| Additive | TT | 8221 (96.49) | 6798 (95.60) | 1.00 | -- |
| T- | 295 (3.46) | 310 (4.36) | 0.75 (0.64-0.89) | 0.0006 | |
| -- | 4 (0.05) | 3 (0.04) | 0.99 (0.22-4.44) | 0.9871 | |
| Trend test | 0.0008 | ||||
| Dominant | T-+-- | 299 (3.51) | 313 (4.40) | 0.75 (0.64-0.89) | 0.0007 |
| TP53 rs9895829 A>G | |||||
| Additive | AA | 7729 (90.56) | 6315 (88.62) | 1.00 | -- |
| AG | 788 (9.23) | 790 (11.09) | 0.83 (0.75-0.93) | 0.0006 | |
| GG | 18 (0.21) | 21 (0.29) | 0.73 (0.39-1.37) | 0.3285 | |
| Trend test | 0.0004 | ||||
| Dominant | AG+GG | 806 (9.44) | 811 (11.38) | 0.83 (0.75-0.92) | 0.0004 |
| PRKAG2 rs2727572 C>T | |||||
| Additive | CC | 2432 (28.50) | 2146 (30.00) | 1.00 | -- |
| CT | 4170 (48.87) | 3485 (48.72) | 1.09 (1.01-1.17) | 0.0283 | |
| TT | 1930 (22.62) | 1522 (21.28) | 1.17 (1.07-1.28) | 0.0008 | |
| Trend test | 0.0007 | ||||
| Dominant | CT+TT | 6100 (71.50) | 4980 (69.62) | 1.11 (1.04-1.19) | 0.0034 |
| PRKAG2 s34852782 TA>Tb | |||||
| Additive | AA | 4237 (49.98) | 3687 (51.98) | ||
| A- | 3492 (41.49) | 2893 (40.79) | 1.06 (0.99-1.13) | 0.1161 | |
| -- | 749 (8.83) | 513 (7.23) | 1.29 (1.15-1.46) | <.0001 | |
| Trend test | 0.0002 | ||||
| Dominant | A-+-- | 4241 (50.02) | 3406 (48.02) | 1.09 (1.02-1.16) | 0.0076 |
| RPTOR rs62068300 G>A | |||||
| Additive | GG | 4093 (47.97) | 3278 (46.00) | ||
| GA | 3639 (42.65) | 3116 (43.73) | 0.92 (0.86-0.98) | 0.0149 | |
| AA | 801 (9.39) | 732 (10.27) | 0.85 (0.76-0.95) | 0.0043 | |
| Trend test | 0.0009 | ||||
| Dominant | GA+AA | 4440 (52.03) | 3848 (54.00) | 0.91 (0.85-0.97) | 0.0026 |
| RPTOR rs3751936 G>C | |||||
| Additive | GG | 4887 (25.28) | 3886 (27.01) | 1.00 | -- |
| GC | 3119 (49.55) | 2710 (49.40) | 0.93 (0.87-1.00) | 0.0347 | |
| CC | 513 (25.17) | 512 (23.59) | 0.82 (0.72-0.93) | 0.0025 | |
| Trend test | 0.0009 | ||||
| Dominant | GC+CC | 3632 (74.72) | 3222 (72.99) | 0.91 (0.86-0.97) | 0.0051 |
Abbreviations: PanC: pancreatic cancer; SNP: single nucleotide polymorphism; OR: odds ratio; CI: confidence interval.
Obtained from Logistic regression models with adjustment for age, sex, and top five significant principal components.
Base deletion.
To better estimate the joint association between the six SNPs and PanC risk, we combined risk genotypes of rs35075084 TT, rs2727572 CT/TT, rs34852782 A−/−-, rs9895829 AA, rs62068300 AA and rs3751936 GG into a single genetic score as the NUGs in a dominant model. The significant association between an increased NUGs and an increased PanC risk remained with a significant trend (Ptrend < 0.0001, Table 4). We then dichotomized all individuals into low-risk group (0-4 NUGs) or high-risk group (5-6 NUGs). As shown in Table 4, PanC risk in high-risk group was significantly greater than the low-risk group (OR = 1.24, 95% CI = 1.16-1.32, P < 0.0001).
Table 4.
Combined risk genotypes of the six SNPs and the risk of pancreatic cancer.
| NUG | Group | OR (95% CI)a | Pb | |
|---|---|---|---|---|
| Case (%) | Control (%) | |||
| 0 | 1 (0.01) | 1 (0.01) | 1.00 | -- |
| 1 | 34 (0.40) | 50 (0.71) | 1.75 (0.11-29.08) | 0.6963 |
| 2 | 459 (5.44) | 467 (6.61) | 1.48 (0.94-2.34) | 0.0911 |
| 3 | 1738 (20.59) | 1571 (22.25) | 1.66 (1.07-2.59) | 0.0253 |
| 4 | 3014 (35.70) | 2626 (37.20) | 1.75 (1.12-2.72) | 0.0133 |
| 5 | 2450 (29.02) | 1836 (26.01) | 2.04 (1.31-3.18) | 0.0016 |
| 6 | 746 (8.84) | 509 (7.21) | 2.23 (1.42-3.51) | 0.0005 |
| Trend test | <.0001 | |||
| 0-4 | 5246 (62.14) | 4715 (66.78) | 1.00 | -- |
| 5-6 | 3196 (37.86) | 2345 (33.22) | 1.24 (1.16-1.32) | <.0001 |
Abbreviations: SNP: single nucleotide polymorphism; PanC: pancreatic cancer; NUG: number of unfavorable genotype; OR: odds ratio; CI: confidence interval.
Risk genotypes were rs35075084 TT, rs2727572 CT+TT, rs34852782 A-+--, rs9895829 AA, rs62068300 GG and rs3751936 GG.
Logistic regression analyses were adjusted for age, sex and the top five principal components.
Since the difference in the distribution of age existed in all the dataset, and age is a known risk factor for PanC, we performed subgroup analysis by age group (i.e., < 60y, 60 – 70y and > 70y) and sex to assess any potential interaction. We found that the risk associated with high-risk NUGs was more evident in the >70 year group (OR = 1.27, 95% CI = 1.13-1.41, P < 0.0001) and males (OR = 1.25, 95% CI = 1.15-1.37, P < 0.0001); however, there was no evidence for an interaction among and between these strata (P > 0.05 for all, Supplementary Table S4).
Genotype and phenotype correlation analysis
To explore the potential function of these six SNPs, we used online prediction tools and performed eQTL analysis. We found that four SNPs are located in the intronic regions, one SNP in 5’-UTR and one SNP in 3’-UTR (Supplemental Table S5). All SNPs are located in the enhancer region of histone H3 mono acetyl K27, which is associated with the higher activation of transcription and defined as an active enhancer marker. Besides, SNP of TP53 and two SNPs of RPTOR also located in DNase I hypersensitive sites where chromatins are sensitive to cleavage by the DNase I and lost its condensed structure, functionally related to transcriptional activity (Supplementary Fig. S6).
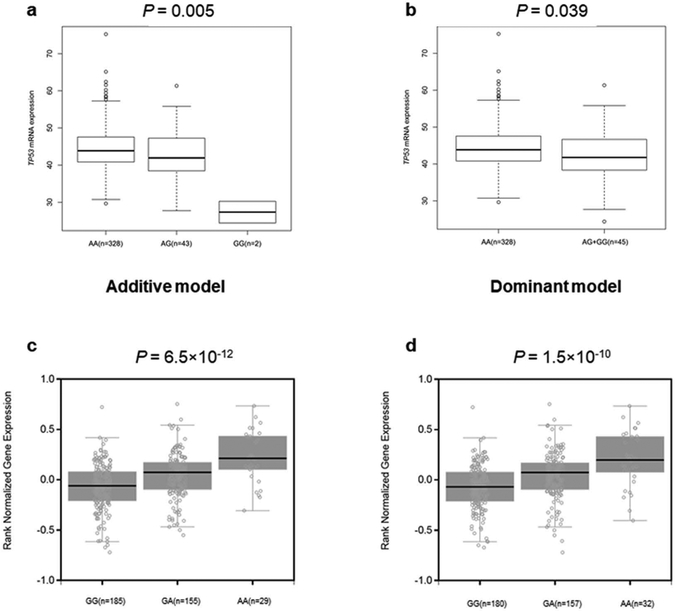
We evaluated correlations between SNPs and corresponding mRNA levels in 373 normal lymphoblastoid cell lines from the 1000 Genomes Project. We found rs9895829 G allele was significantly correlated with decreased levels of TP53 mRNA expression in both additive and dominent model (P = 0.005 and 0.039) by using Student’s t test or linear regression analysis of the logarithm transformed expression values (log2) (Fig. 2a-2b). No other allele was significantly correlated with increased/decreased levels of mRNA expression. However, the rs2727572 T allele of PRKAG2, rs62068300 A allele of RPTOR and rs3751936 C allele of RPTOR were correlated with an observable increased/decreased mRNA expression level in both additive and dominant models, though the differences did not reach the statistical level (P = 0.363 and 0.625, P = 0.299 and 0.092 and P = 0.163 and 0.177, respectively. Supplementary Fig. S7). For SNP rs35075084 and rs34852782, the eQTL results are not available. Next, we used the data from 127 Europeans in the TCGA-PAAD Project to query the eQTL results and assess the correlations. However, we failed to impute the genotype of six SNPs based on the current quality control.
Fig. 2. The eQTL analyses of the functional SNPs.
The eQTL results of the SNP rs9895829 of TP53 in 373 normal lymphoblastoid cell lines from the 1000 Genomes Project in (a) additive and (b) dominant model and (c-d) GTEx results of the two SNPs in high LD with SNP rs62068300 of RPTOR in whole blood cells of 369 normal participants. eQTL: expression quantitative trait loci; SNP: single nucleotide polymorphism; GTEx: genotype-tissue expression; LD: Linkage disequilibrium.
In addition, we used the data from the GTEx database and found that SNP rs60268947 and rs28434589, located in the intron of RPTOR and in high LD (r2 = 0.91 and r2 = 0.96, respectively) with SNP rs62068300, had a significant correlation with an increased level of RPTOR mRNA expression in whole blood cells of 369 donors from GTEx (P = 6.5 × 10−12 and P = 1.5 × 10−10, respectively, Fig. 2c-2d). For other SNPs, there are no significant correlations from GTEx (Supplemental Table S6).
We also assessed the differences in mRNA expression levels of MAP2, PRKAG2, TP53 and RPTOR between adjacent normal pancreatic tissues and pancreatic tumor tissues (n=220) from the Oncomine database. We found that MAP2 and TP53 mRNA expression levels were statistically significantly higher (P = 0.002 and P = 1.38 × 10−6, respectively) in tumor tissues than in normal pancreas, and the mRNA expression levels of PRKAG2 and RPTOR were also higher in tumor tissues than in normal pancreas, though the differences were not statistically significant (P = 0.061 and P = 0.196) (Supplementary Fig. S8).
Discussion
In the present study, we evaluated the associations between genetic variants in the 58 LKB1-AMPK pathway genes and PanC risk, using the two existing GWAS datasets: PanScan I, II/III from PanScan study and PanC4 from Pancreatic Cancer Case-Control Association Study. Through the meta-analysis, we identified six novel potential susceptibility loci of MAP2, PRKAG2, TP53 and RPTOR for PanC risk, located at 2q34, 7q36.1, 17p13.1 and 17q25.3, respectively. We further showed that these variants were independently or jointly associated with an increased PanC risk. Further eQTL analysis revealed that those six novel SNPs might influence the mRNA expression levels of corresponding genes, particularly true for the MAP2 rs35075084 and TP53 rs9895829.
LKB1 was initially identified as a tumor suppressor gene responsible for the familial Peutz-Jeghers syndrome and associated with increased risk for gastrointestinal tract cancers, including PanC 41. The AMPK, which is highly conserved in all eukaryotic cells and exists as a trimeric complex consisting of a catalytic subunit (α subunit) and two regulatory subunits (β and γ subunits), plays a role in the regulation of cellular energy homeostasis by maintaining cellular energy homeostasis in response to an increased AMP:ATP ratio and restores energy balance by inhibiting anabolic processes that consume ATP, while promoting catabolic processes that generate ATP 12,13,17.
Together, the LKB1-AMPK pathway genes serve as a metabolic checkpoint and a central metabolic switch that governs glucose and lipid metabolisms in response to alterations in nutrients and intracellular energy levels 42,43. Besides, LKB1-AMPK also controls for cell growth in response to environmental nutrient changes. For example, one central down-stream pathway suppressed by the LKB1-AMPK pathway is the mammalian target of rapamycin (mTOR) pathway, which controls cell growth in all eukaryotes; this signaling pathway is inhibited through the AMPK phosphorylation of tuberous sclerosis complex 2 (TSC2) and regulatory associated protein of mTOR (RPTOR) in conditions of low intracellular ATP levels 44-47.
Microtubule associated protein 2 (MAP2) is localized primarily in dendrites of neurons and involved in microtubule assembly 48,49. It has been reported that MAP2 participates in the outgrowth of neuronal processes and synaptic plasticity and controls for selective axonal cargo sorting by regulating kinesin activity 50. In addition, MAP2 is also involved in the protein kinase A-induced decrease in the invasiveness of glioma cells 51. In the present study, we found that the MAP2 rs34852782 deleted allele was associated with PanC risk, likely due to the resultant increase in the mRNA expression.
The protein kinase AMP-activated non-catalytic subunit γ 2 (PRKAG2), as a member of AMPK γ subunit family, mutations in which can cause inappropriate AMPK activation under resting conditions and lead to hypertrophic cardiomyopathy associated with the Wolff-Parkinson-White syndrome 52. Previous studies revealed a nominal association of PRKAG2 SNPs with diabetes incidence 53 and suggested that PRKAG2 variants were involved in feed efficiency traits in beef steers 54. The present study suggests that SNPs rs2727572 and rs34852782 located in the intron of PRKAG2 were associated with PanC risk, and the variant-associated gene expression may be the mechanism underlying the observed association, but additional studies are needed to validate this speculation.
Tumor protein p53 (TP53) has many mechanisms of anticancer function and plays a role in apoptosis, genomic stability, and inhibition of angiogenesis 55. TP53 somatically mutated in 50-80% of PanC 2,56-58. As we mentioned before, SNPs rs9895829 of TP53 has been previously reported by the AURORA pathway-based analyses. In our present study we found that SNPs rs9895829 had a significant correlation with a decreased TP53 mRNA expression and was associated with PanC risk.
RPTOR is a crucial component of MTORC1 and negatively regulates mTOR 59. When the intracellular energy level is low, AMPK directly phosphorylates RPTOR at Ser722 and Ser792, reducing mTOR kinase activities 60,61. One study suggested that SNP rs11868112 of RPTOR exhibits a strong association with temperature variables that contribute to climate adaptations 62. The present study found that SNPs rs62068300 and rs3751936 of RPTOR were associated with PanC risk. Because two SNPs (rs60268947 and rs28434589) in high LD (r2 > 0.90) with rs62068300 had a significant correlation with an increased RPTOR mRNA expression in whole blood cells from GTEx, and genetic variants in RPTOR are likely to play a role in carcinogenesis of PanC.
Although the present study observed associations between six novel genetic variants in the LKB1-AMPK pathway genes and PanC risk, it has also several limitations. First, we had no access to family history in the publically available datasets, which might have an impact on PanC risk. Second, since we only used the available online tools and eQTL analysis to evaluate function of a particular SNP, further functional investigations are required. Third, we are still not sure which SNP in the LKB1-AMPK pathway genes may have played a major role in or how jointly they may have an impact on PanC risk. Finally, because all selected subjects in two GWAS studies were from Caucasian populations, the results may not be generalizable to the general populations.
In summary, we report some significant associations between genetic variants in 58 LKB1-AMPK pathway genes and PanC risk in European populations. Six SNPs (i.e., MAP2 rs35075084 T > deletion, PRKAG2 rs2727572 C > T and rs34852782 A > deletion, TP53 rs9895829, RPTOR rs62068300 G > A and rs3751936 G > C) were found to be significantly associated with an increased PanC risk, possibly by influencing their gene expression. More population validations and additional functional studies are needed to explore possible molecular mechanisms in the etiology of PanC.
Supplementary Material
Acknowledgements
The authors would like to thank Wenjun Yang and Wei Dai for their technical support. We thank all the participants of the PanScan GWAS Study and Pancreatic Cancer Case Control Association Study and acknowledge dbGaP repository for their help in providing the cancer genotyping datasets described as follows.
PanScan
The PanScan project was funded in whole or in part with federal funds from the National Cancer Institute (NCI), US National Institutes of Health (NIH) under contract number HHSN261200800001E. This work was also supported by NIH/NCI K07 CA140790, the American Society of Clinical Oncology Conquer Cancer Foundation, the Howard Hughes Medical Institute, the Lustgarten Foundation, the Robert T. and Judith B. Hale Fund for Pancreatic Cancer Research and Promises for Purple. A full list of acknowledgments for each participating study is provided in the Supplementary Note of the manuscript with PubMed ID: 25086665. The dbGaP accession number for the present study is phs000206.v5.p3.
PanC4
The patients and controls for this study were derived from the following PANC4 studies: Johns Hopkins National Familial Pancreas Tumor Registry, Mayo Clinic Biospecimen Resource for Pancreas Research, Ontario Pancreas Cancer Study (OPCS), Yale University, MD Anderson Case Control Study, Queensland Pancreatic Cancer Study, University of California San Francisco Molecular Epidemiology of Pancreatic Cancer Study, International Agency of Cancer Research and Memorial Sloan Kettering Cancer Center. This work is supported by NCI R01CA154823 Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN2682011000111. The dbGaP accession number for this study used in this manuscript is phs000648.v1.p1.
TCGA
The results published here are in whole or part based upon data generated by The Cancer Genome Atlas pilot project established by the NCI and NHGRI. Information about TCGA and the investigators and institutions that constitute The Cancer Genome Atlas (TCGA) Research Network can be found at “http://cancergenome.nih.gov”. The TCGA SNP data analyzed here are requested through dbGAP (accession#: phs000178.v1.p1).
Funding sources
Qingyi Wei was partly supported by the Duke Cancer Institute as part of the P30 Cancer Center Support Grant (Grant ID: NIH CA014236). Xinyuan Xu was supported by National Scholarship Council of China.
Abbreviations:
- PanC
pancreatic cancer
- SNP
single nucleotide polymorphism
- NUGs
number of unfavorable genotypes
- OR
odds ratio
- CI
confidence interval
- GWAS
genome-wide association studies
- FPRP
false positive report probability
Footnotes
Conflicts of Interest: None declared.
References
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer research. 2014;74(11):2913–2921. [DOI] [PubMed] [Google Scholar]
- 2.Kleeff J, Korc M, Apte M, et al. Pancreatic cancer. Nature reviews Disease primers. 2016;2:16022. [DOI] [PubMed] [Google Scholar]
- 3.Raimondi S, Maisonneuve P, Lowenfels AB. Epidemiology of pancreatic cancer: an overview. Nature reviews Gastroenterology & hepatology. 2009;6(12):699–708. [DOI] [PubMed] [Google Scholar]
- 4.Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet (London, England). 2011;378(9791):607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet (London, England). 2016;388(10039):73–85. [DOI] [PubMed] [Google Scholar]
- 6.Warburg O On the origin of cancer cells. Science (New York, NY). 1956;123(3191):309–314. [DOI] [PubMed] [Google Scholar]
- 7.Vander Heiden MG, DeBerardinis RJ. Understanding the Intersections between Metabolism and Cancer Biology. Cell. 2017;168(4):657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. [DOI] [PubMed] [Google Scholar]
- 9.Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23(1):27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen R, Neuzillet C, Tijeras-Raballand A, Faivre S, de Gramont A, Raymond E. Targeting cancer cell metabolism in pancreatic adenocarcinoma. Oncotarget. 2015;6(19):16832–16847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer cell. 2012;21(3):309–322. [DOI] [PubMed] [Google Scholar]
- 12.Faubert B, Boily G, Izreig S, et al. AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell Metab. 2013;17(1):113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia D, Shaw RJ. AMPK: Mechanisms of Cellular Energy Sensing and Restoration of Metabolic Balance. Molecular cell. 2017;66(6):789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su GH, Hruban RH, Bansal RK, et al. Germline and Somatic Mutations of the STK11/LKB1 Peutz-Jeghers Gene in Pancreatic and Biliary Cancers. The American Journal of Pathology. 1999;154(6):1835–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waddell N, Pajic M, Patch AM, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540):495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kottakis F, Nicolay BN, Roumane A, et al. LKB1 loss links serine metabolism to DNA methylation and tumorigenesis. Nature. 2016;539(7629):390–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin SC, Hardie DG. AMPK: Sensing Glucose as well as Cellular Energy Status. Cell metabolism. 2018;27(2):299–313. [DOI] [PubMed] [Google Scholar]
- 18.Cheng X, Kim JY, Ghafoory S, et al. Methylisoindigo preferentially kills cancer stem cells by interfering cell metabolism via inhibition of LKB1 and activation of AMPK in PDACs. Molecular oncology. 2016;10(6):806–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Souza A, Khawaja KI, Masud F, Saif MW. Metformin and pancreatic cancer: Is there a role? Cancer chemotherapy and pharmacology. 2016;77(2):235–242. [DOI] [PubMed] [Google Scholar]
- 20.Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nature genetics. 2009;41(9):986–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petersen GM, Amundadottir L, Fuchs CS, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nature genetics. 2010;42(3):224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolpin BM, Rizzato C, Kraft P, et al. Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nature genetics. 2014;46(9):994–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Childs EJ, Mocci E, Campa D, et al. Common variation at 2p13.3, 3q29, 7p13 and 17q25.1 associated with susceptibility to pancreatic cancer. Nature genetics. 2015;47(8):911–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duan B, Hu J, Liu H, et al. Genetic variants in the platelet-derived growth factor subunit B gene associated with pancreatic cancer risk. International journal of cancer. 2018;142(7):1322–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu C, Miao X, Huang L, et al. Genome-wide association study identifies five loci associated with susceptibility to pancreatic cancer in Chinese populations. Nat Genet. 2011;44(1):62–66. [DOI] [PubMed] [Google Scholar]
- 26.Wolpin BM, Rizzato C, Kraft P, et al. Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat Genet. 2014;46(9):994–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009;41(9):986–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abecasis GR, Altshuler D, Auton A, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. [DOI] [PubMed] [Google Scholar]
- 33.Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database issue):D930–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lappalainen T, Sammeth M, Friedländer MR, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501(7468):506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Network CGAR. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Consortium G. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng Y, Liu H, Duan B, et al. Potential Functional Variants in SMC2 and TP53 in the AURORA Pathway Genes and Risk of Pancreatic Cancer. Carcinogenesis. 2019. February 22. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Lier MG, Westerman AM, Wagner A, et al. High cancer risk and increased mortality in patients with Peutz-Jeghers syndrome. Gut. 2011;60(2):141–147. [DOI] [PubMed] [Google Scholar]
- 42.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nature reviews Cancer. 2009;9(8):563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang YL, Guo H, Zhang CS, et al. AMP as a low-energy charge signal autonomously initiates assembly of AXIN-AMPK-LKB1 complex for AMPK activation. Cell metabolism. 2013;18(4):546–555. [DOI] [PubMed] [Google Scholar]
- 44.Green AS, Chapuis N, Lacombe C, Mayeux P, Bouscary D, Tamburini J. LKB1/AMPK/mTOR signaling pathway in hematological malignancies: from metabolism to cancer cell biology. Cell cycle. 2011;10(13):2115–2120. [DOI] [PubMed] [Google Scholar]
- 45.Kottakis F, Bardeesy N. LKB1-AMPK axis revisited. Cell research. 2012;22(12):1617–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kishton RJ, Barnes CE, Nichols AG, et al. AMPK Is Essential to Balance Glycolysis and Mitochondrial Metabolism to Control T-ALL Cell Stress and Survival. Cell metabolism. 2016;23(4):649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer cell. 2007;12(1):9–22. [DOI] [PubMed] [Google Scholar]
- 48.Sa nchez C D-N J, Avila J Phosphorylation of microtubule-associated protein 2 (MAP2) and its relevance for the regulation of the neuronal cytoskeleton function.pdf. Progress in Neurobiology. 2000. [DOI] [PubMed] [Google Scholar]
- 49.Halpain LDaS. The MAP2 Tau family of microtubule-associated proteins.pdf. Genome Biology. 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gumy LF, Katrukha EA, Grigoriev I, et al. MAP2 Defines a Pre-axonal Filtering Zone to Regulate KIF1- versus KIF5-Dependent Cargo Transport in Sensory Neurons. Neuron. 2017;94(2):347–362 e347. [DOI] [PubMed] [Google Scholar]
- 51.Zhou Y, Wu S, Liang C, et al. Transcriptional upregulation of microtubule-associated protein 2 is involved in the protein kinase A-induced decrease in the invasiveness of glioma cells. Neuro-oncology. 2015;17(12):1578–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arad M, Seidman CE, Seidman JG. AMP-activated protein kinase in the heart: role during health and disease. Circulation research. 2007;100(4):474–488. [DOI] [PubMed] [Google Scholar]
- 53.Jablonski KA, McAteer JB, de Bakker PI, et al. Common variants in 40 genes assessed for diabetes incidence and response to metformin and lifestyle intervention in the diabetes prevention program. Diabetes. 2010;59(10):2672–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lindholm-Perry AK, Kuehn LA, Oliver WT, et al. DNA polymorphisms and transcript abundance of PRKAG2 and phosphorylated AMP-activated protein kinase in the rumen are associated with gain and feed intake in beef steers. Animal genetics. 2014;45(4):461–472. [DOI] [PubMed] [Google Scholar]
- 55.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000. [DOI] [PubMed] [Google Scholar]
- 56.Yachida S, Iacobuzio-Donahue CA. Evolution and dynamics of pancreatic cancer progression. Oncogene. 2013;32(45):5253–5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patel SJ, Sanjana NE, Kishton RJ, et al. Identification of essential genes for cancer immunotherapy. Nature. 2017;548(7669):537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yachida S, White CM, Naito Y, et al. Clinical significance of the genetic landscape of pancreatic cancer and implications for identification of potential long-term survivors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18(22):6339–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim Do-Hyung DDS, Ali Siraj M., King Jessie E. RRL, Erdjument-Bromage Hediye PT, and Sabatini David M.. mTOR Interacts with Raptor to Form a Nutrient-Sensitive Complex that Signals to the Cell Growth Machinery. Cell. 2002. [DOI] [PubMed] [Google Scholar]
- 60.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk. Nature cell biology. 2011;13(2):132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feng Y, Yao Z, Klionsky DJ. How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends in cell biology. 2015;25(6):354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun C, Southard C, Witonsky DB, Kittler R, Di Rienzo A. Allele-specific down-regulation of RPTOR expression induced by retinoids contributes to climate adaptations. PLoS genetics. 2010;6(10):e1001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.