Abstract
Objective:
Targeted biopsy validation of magnetic resonance fingerprinting (MRF) and diffusion mapping for characterizing peripheral zone (PZ) prostate cancer and non-cancers.
Materials and Methods:
104 PZ lesions in 85 patients who underwent MRI were retrospectively analyzed with apparent diffusion coefficient (ADC) mapping, MRF and targeted biopsy (cognitive or in-gantry). A radiologist blinded to pathology drew regions-of-interest on targeted lesions and visually normal peripheral zone (NPZ) on MRF and ADC maps. Mean T1, T2 and ADC were analyzed using linear mixed models. Generalized estimating equations logistic regression analyses were used to evaluate T1 and T2 relaxometry combined with ADC in differentiating pathologic groups.
Results:
Targeted biopsy revealed 63 cancers (low-grade cancer/Gleason score 6=10, clinically significant cancer/Gleason score ≥ 7=53), 15 prostatitis and 26 negative biopsies. Prostate cancer T1, T2 and ADC (mean±SD, 1660±270 ms, 56±20 ms, 0.70×10−3±0.24×10−3 mm2/s) were significantly lower than prostatitis (mean±SD, 1730±350 ms 77±36 ms, 1.00×10−3±0.30×10−3 mm2/s) and negative biopsies (mean±SD, 1810±250 ms, 71±37 ms, 1.00×10−3 ±0.33×10−3 mm2/s). For cancer versus prostatitis, ADC was sensitive and T2 specific with comparable area under curve (AUC). (AUCT2=0.71, AUCADC=0.79, difference between AUCs not significant p=0.37). T1+ADC (AUCT1+ADC=0.83) provided best separation between cancer and negative biopsies. Low-grade cancer T2 and ADC (mean±SD, 75±29 ms, 0.96×10−3 ±0.34×10−3 mm2/s) were significantly higher than clinically significant cancers (mean±SD, 52±16 ms, 0.65± 0.18×10−3 mm2/s) and T2+ADC (AUCT2+ADC=0.91) provided best separation.
Conclusion:
T1 and T2 relaxometry combined with ADC mapping may be useful for quantitative characterization of prostate cancer grades and differentiating cancer from non-cancers for PZ lesions seen on T2w images.
Keywords: Magnetic Resonance Fingerprinting, Prostate Cancer, Peripheral Zone, Relaxometry, Quantitative MRI
Introduction
Interpretation of prostate mpMRI sequences as guided by Prostate Imaging, Reporting and Data System version 2 (PIRADS v2) is currently qualitative1. However, there is increasing interest in quantitative evaluation for more objective lesion assessment2–4. Prior studies have shown that the histological differences between normal prostate tissue, prostate cancers and inflammation are associated with measurable differences in T2 and T2* relaxation times and apparent diffusion coefficient (ADC) 5–13. In clinical practice, ADC mapping is the only technique used quantitatively in prostate MRI and has been shown to reflect cancer aggressiveness14–19 and partially separate cancer from prostatitis 20,21. Magnetic Resonance Fingerprinting (MRF) represents another framework for performing relaxometry and allows simultaneous measurement of T1 and T2 relaxation times in a clinically feasible time22,23. In MRF, user controllable system parameters such as flip angle, time of echo (TE), time of repetition (TR), etc. are allowed to vary in a pseudo-random manner such that unique signal evolutions are produced for each combination of tissue properties (T1, T2, etc.) and a dictionary of all possible signal evolutions is computed for that sequence. Obtained signal evolutions are matched against a best entry in the dictionary on a pixel-by-pixel basis, with relaxation properties used to generate the matched entry assigned to that pixel as the measured T1 and T2. This yields simultaneous, rapid and co-registered T1 and T2 maps that provide combined quantitative information24, with several potential advantages over traditional mapping methods that typically measure either T1 or T2 relaxation times per acquisition7,8,13,25. While relaxation property measurements will necessarily vary slightly based on the system imperfections and confounders that are accounted for in the dictionary26–29, MRF-based relaxometry has been found to be repeatable and reproducible in both phantom and in-vivo assessment30,31. Initial application to prostate imaging showed excellent separation between normal peripheral zone, and cancer or prostatitis using a combined quantitative protocol comprising of MRF-relaxometry and echo planar imaging (EPI) based DWI 32. That study also showed moderate accuracy for separating low-grade (Gleason score 6) from intermediate-high grade prostate (Gleason score 7 and above) cancers using quantitative criteria32. However, these results were based on transrectal ultrasound (TRUS) guided biopsy as a pathology reference and a small dataset with cognitive targeting. TRUS-guided biopsy is prone to sampling errors and can either underestimate the grade of cancer or miss cancer altogether 33 while targeted biopsy methods can produce better correlation with the actual pathology34,35. The purpose of this study was to provide targeted biopsy validation of combined MRF-based relaxometry and diffusion mapping for characterizing prostate cancer grades and differentiating prostate cancer from prostatitis and negative biopsies in the peripheral zone of prostate.
Materials and Methods
Patients
This Institutional Review Board approved and Health Insurance Portability and Accountability Act compliant study is a retrospective evaluation of MRF data collected prospectively between September 2014 and April 2018, from patients with suspected prostate cancer who had MRI followed by targeted biopsy (either cognitive or in-gantry biopsy). Written informed consent was obtained from all participants. Exclusion criteria included previous history of prostatectomy, pelvic radiation, chemotherapy or hormonal therapy.
Diagnostic MRI scans and in-gantry biopsies were performed at 3T (Verio or Skyra; Siemens, Erlangen, Germany) using a body array coil and no endorectal coil. The diagnostic MRI protocol is given in Table 1 and in-gantry biopsy protocol in a supplementary table (Supplementary Digital Content 1). MRF acquisitions and b-values for diffusion were kept constant to ensure consistency in quantitative MRI evaluation.
Table 1:
Imaging Parameters for Diagnostic MRI
| Sequence | TR (ms) / TE) (ms) |
Field of) View) (mm) |
Resolution) (mm) |
Matrix | Flip) angle) (degrees) |
Slice) thickness) (mm) |
b Value (s/mm2) |
Sequence Duration (minutes) |
|---|---|---|---|---|---|---|---|---|
| Localizer- 3 plane | 2000/95 | 305×285 | 1.2×1.2 | 320×240 | 150 | 5 | 0.02 | |
| Three plane single-shot fast spin echo | 2000/92 | 305×244 | 1.2×1.2 | 384×308 | 150 | 5 | 0.32 | |
| Transverse turbo spin-echo T2w | 8600/103 | 160×160 | 0.6×0.6 | 320×320 | 150 | 3 | 3:30 | |
| Diffusion weighted imaging | 7900/88 | 240×240 | 1.2×1.2 | 198×198 | 3 | 50, 600, 1000, 1400 | 4:46 | |
| MR fingerprinting | 13-15 | 400×400 | 1×1 | 400×400 | 5 –75 | 5 | 0.39 per slice | |
| Pre-contrast T1w imaging with DCE perfusion* | 3.34/1.02 | 240×240 | 1.9×1.9 | 128× 128 | 15 | 3 | 4:31 | |
| Post contrast T1w* | 3.63/1.33 | 240×240 | 1.0×1.0 | 128×128 | 9 | 2 | 0.23 |
Abbreviations: TR: Time of Repetition, TE: Time of Echo, DCE: Dynamic Contrast Enhanced.
The patients in cognitive biopsy group underwent a non-contrast MRI protocol
Cognitive biopsies of cancer suspicious lesions were performed in combination with 12-core TRUS biopsies. Targeted lesions were localized based on MRI reads and visualized on TRUS using a prostate sector map and internal landmarks for reference. In-gantry biopsies were performed with a dedicated MR-compatible biopsy device (DynaTRIM, In Vivo, Gainesville, FL) using the assisted planning software (DynaLOC; Invivo) for guiding biopsy needle placement. For in-gantry biopsies, needle placement in the lesion was confirmed with a scan prior to taking biopsy samples. The median interval between MRF and cognitive biopsy was 21 days (range 6–133 days). For in-gantry biopsy, MRF with ADC mapping were performed at the time of biopsy.
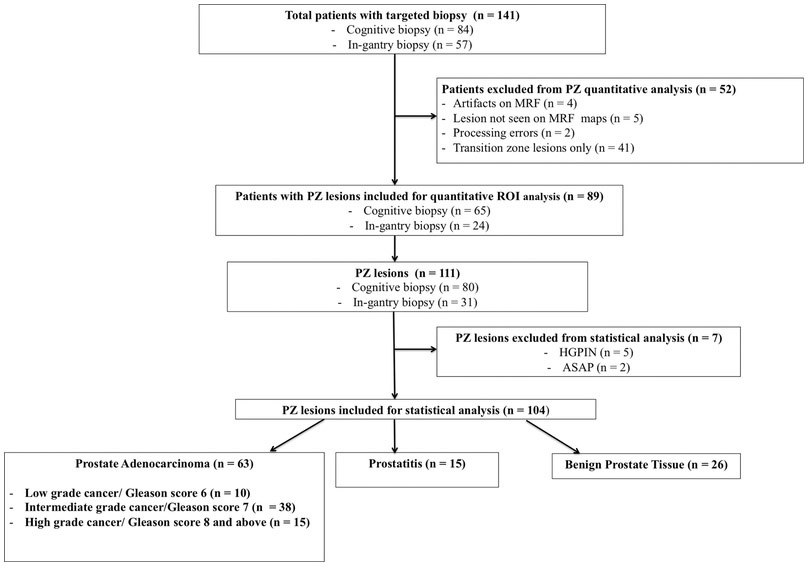
141 patients (median 64 years, range 42–81 years) underwent clinical MRI with MRF and targeted biopsy (84 cognitive and 57 in-gantry biopsy). All cognitive biopsy patients were biopsy naïve while 35/57 in-gantry patients had previous TRUS biopsies. The median time interval between prior TRUS and in-gantry biopsy was 16.5 months (2–132 months). Eleven patients were excluded from quantitative analysis due to technical limitations [artifacts on MRF maps, (n=4), lesion not visualized on MRF maps (n=5) and failed reconstruction of MRF maps (n=2)] and if they had only transition zone lesions (n=41). Lesions with histopathologic diagnosis other than cancer, prostatitis or benign prostatic tissue were further excluded from quantitative ROI analysis (Fig.1). None of the targeted lesions had visible post-biopsy hemorrhage to preclude analysis. Part of the dataset (37 patients with 27 prostate cancer lesions) used in this study was also used in a previous publication (reference withheld for blinded review). However, that study did not evaluate lesions with negative biopsies and the results of TRUS biopsy was used as final reference standard for MRF values in cases of discordance.
Figure 1: Flow diagram of patient and lesion selection.
PZ = peripheral zone, HGPIN = high-grade intraepithelial neoplasia ASAP = atypical small acinar proliferation, ROI = region of interest.
MRF Acquisition and Post-Processing
MRF with fast imaging with steady-state precession (MRF-FISP)23 was utilized and the whole prostate was covered. Acquisition time was 39 seconds per slice, and total scan time 5–10 minutes, depending on prostate size. A dictionary containing expected MRF signal evolutions was calculated with T1 20–2950ms and T2 9–500ms, and MRF maps obtained by template matching the signal timecourse in each pixel, as described previously23. For patients recruited between September 2014 and September 2017, the raw MRF data were processed offline on Matlab (Matlab 2014a; MathWorks, Natick, Mass) with offline reconstruction time of 190 seconds per slice. For patients recruited after October 2017, a Gadgetron-based framework was used for rapid online reconstruction of MRF data36 and quantitative T1 and T2 maps in DICOM format were directly available real-time on the MR scanner. A prior comparison of offline and online reconstruction methods showed that MRF T1 and T2 values were similar for both reconstruction methods37.
Clinical Interpretation and Quantitative ROI analysis
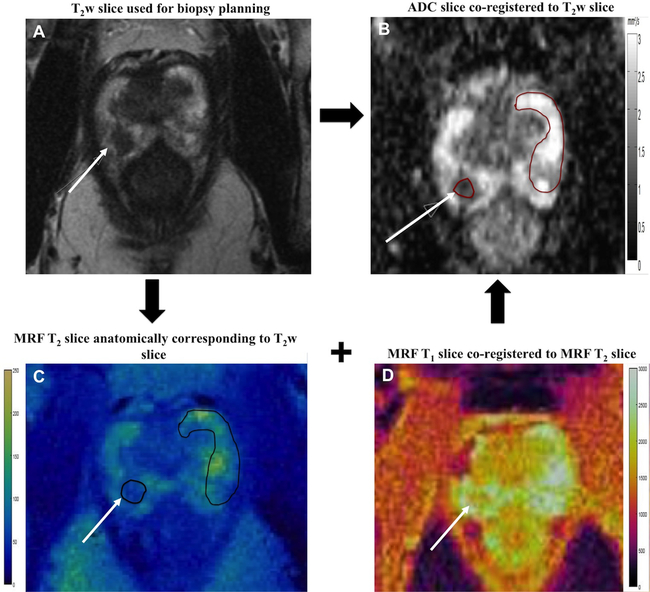
Targeted biopsy lesions were evaluated based on PIRADSv2 by a fellowship-trained body radiologist (18 years radiology experience) who also performed all in-gantry targeted biopsies with 1–6 cores obtained per lesion (median 3 cores). Another radiologist (8 years experience) who was blinded to the clinical information and pathology diagnosis but aware of the locations of the targeted lesions retrospectively drew regions-of-interest (ROIs) on suspicious peripheral zone lesions and on the contralateral visually normal peripheral zone (NPZ) on both MRF and ADC maps. As a part of acquisition scheme, both T2w and ADC slices were anatomically co-registered while MRF T1 and T2 maps were anatomically co-registered. The T2w slice with the largest lesion area and used for biopsy planning, was taken as the reference slice and the T2 MRF slice anatomically corresponding to this T2w slice was selected. Lesions and NPZ ROIs were drawn on the selected T2 MRF slice and, both T1 and T2 were obtained simultaneously from these ROIs . Again using T2w slice and T2 map as the reference, lesion and NPZ ROIs were replicated independently at the corresponding locations on the ADC maps. Fig. 2 depicts the image analysis workflow. The lesion ROI sizes ranged from 6–442 mm2 (median 55 mm2). For each targeted lesion and NPZ, the mean T1, T2, and ADC were recorded. Based on targeted-core biopsy reports, final pathologic diagnosis for each targeted lesion was recorded. For cancers, Gleason scores were recorded. For targeted lesions for which more than one Gleason score was given, the highest score was recorded as the final pathological diagnosis.
Figure 2: Regions of Interest (ROI) analysis.
Cancer suspicious lesions (solid arrow) were identified based on axial T2w slice (A) and ADC map (B). The anatomically corresponding MRF slices (C, D) were aligned with T2w slice and lesions ROIs were drawn on MRF map (black oval). As MRF maps were co-registered, both T1 and T2 values were simultaneously obtained from single MRF ROI. Independent ROIs were drawn on ADC map (red oval) co-registered to the T2w slice. ROIs were also drawn on the visually normal peripheral zone (NPZ) covering whole contralateral NPZ.
Statistical Analysis
Lesions diagnosed as cancer, prostatitis and negative on biopsy were included for analysis. Mean T1, T2, and ADC were compared between individual biopsy groups and with NPZ using linear mixed models. Generalized estimating equations logistic regression analysis was used to assess the utility of MR fingerprinting–derived T1, T2, and ADC in the differentiation of 1) All prostate cancers from (a) prostatitis (b) negative biopsies and (c) all non-cancers (prostatitis + negative biopsies) and 2) Clinically significant cancers from (a) low-grade cancers (b) all non-cancers (prostatitis + negative biopsies) and (c) all clinically insignificant lesions (low-grade cancers + prostatitis + negative biopsies).
Low-grade cancer was defined as Gleason 3+3=6, clinically significant cancer was defined as Gleason score≥7, as Gleason 6 cancers are considered for active surveillance at our institution. Low-grade cancers were grouped with non-cancers and compared them with clinically significant cancers to see if quantitative mapping could be used to differentiate lesions that do not need intervention (low-grade cancers, prostatitis, benign prostatic tissue) versus lesions that are clinically significant.
Receiver operating characteristic curves and areas under the receiver operating characteristic curve (AUC) (C- statistics) were obtained from logistic regressions by using the linear predictors obtained from the generalized estimating equations regressions. For significant univariate models with best AUCs, the cut-off points for maximum sensitivity and specificity were obtained using Youden’s J statistics. Statistical analyses were performed using SAS 9.4 (SAS Institute, Cary, NC).
Results
In 89 patients with peripheral zone lesions, 111 lesions were targeted (80 cognitive sampling, 31 in-gantry sampling). 63 lesions were prostate cancer (10 Low Grade (Gleason score 6), 38 Intermediate Grade (Gleason score 7), 15 High Grade (Gleason score ≥ 8)), 15 prostatitis, 26 negative with biopsy showing normal prostatic tissue and 7 had another diagnosis (5 high-grade prostatic intraepithelial neoplasia and 2 atypical small acinar proliferation). These 7 lesions (4 patients) were excluded and the remaining 104 lesions (85 patients) were analyzed (Fig. 1). T1 and T2 numbers were available for all 104 targeted lesions included in final analysis, and ADC measurement was not available for one lesion due to distorted ADC map. NPZ ROIs T1, T2 measurements were available for 82 patients for comparison with the measurements in the different histologic groups and were not drawn for 3 patients due to lack of visually normal peripheral zone on T2w images.
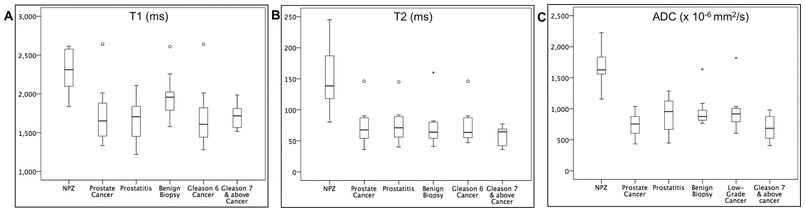
Mean T1, T2 and ADC for NPZ, histologically proven prostate cancer including low-grade cancer and clinically significant cancers, prostatitis and negative biopsies are summarized in Table 2 and the distributions depicted as box-and-whisker plots in Fig. 3. Table 3 summarizes the AUCs for regression models. The best diagnostic performance cut-off points are summarized in Table 4.
Table 2:
Summary of Means of T1, T2 and ADC of normal peripheral zone and different histopathologic groups
| Group (Number of samples) |
T1 (ms) Mean ± SD |
T2 (ms) Mean ± SD |
ADC (×10−3 mm2/s) Mean ± SD |
|---|---|---|---|
| Normal Peripheral Zone (n=82) | 2240±360 | 146±61 | 1.68±0.31 |
| Prostate Cancer (n=63) | 1660±270 | 56±20 | 0.70±0.24 |
| Prostatitis (n=15) | 1760±350 | 77±36 | 1.00±0.30 |
| Biopsy-proven Benign Prostatic Tissue (n=26) | 1810±250 | 71±37 | 1.00±0.33 |
| Low-grade cancer/Gleason score = 6 (n=10) | 1690±400 | 75±29 | 0.96±0.34 |
| Clinically significant cancers/Gleason score≥7 (n=53) | 1650±240 | 52±16 | 0.65±0.18 |
| Non-Cancers (Prostatitis + Benign Prostatic tissue) (n=41) | 1790±290 | 73 ±37 | 1.00±0.32 |
Figure 3: Box and whisker plots of (A) T1 (B) T2 and (C) ADC measurements for normal peripheral zone (NPZ) and different histologic groups.
The boxes represent the interquartile (IQ) range between 25-75th percentiles, the lines within boxes represent medians and the whiskers represent measurements 1.5 times interquartile range. The circles and crosses represent outliers beyond 1.5 times and beyond 3 times the IQ ranges respectively.
Table 3:
Differentiation of various groups with T1, T2 and ADC and their combinations.
| Groups Compared |
T1 AUC | T2 AUC | ADC AUC |
T1+T2 AUC |
T1+ ADC AUC |
T2+ ADC AUC |
T1+ T2+ ADC AUC |
Highest AUC# |
|---|---|---|---|---|---|---|---|---|
| All Prostate cancers versus Non-Cancers | ||||||||
| Prostate Cancer (n=63) vs. Prostatitis (n=15) | 0.60 (0.41-0.78) | 0.71* (0.55-0.88) | 0.79* (0.65-0.93) | 0.71 (0.54-0.88) | 0.76 (0.59-0.92) | 0.79 (0.64-0.94) | 0.79 (0.65-0.95) | ADC (0.79) comparable to T2 (0.71) Difference between two AUCs not significant (p=0.37) |
| Prostate Cancer (n=63) vs. Negative Biopsies (n=26) | 0.67* (0.55-0.79) | 0.62 (0.49-0.75) | 0.80* (0.69-0.90) | 0.67 (0.55-0.79) | 0.83* (0.74-0.93) | 0.80 (0.70-0.93) | 0.83 (0.74-0.93) | T1+ADC (0.83) |
| Prostate Cancer (n=63) vs. Non-cancers (n=41) | 0.64* (0.53-0.75) | 0.66* (0.55-0.77) | 0.80* (0.71-0.89) | 0.68 (0.57-0.78) | 0.80* (0.71-0.89) | 0.80 (0.71-0.89) | 0.80 (0.71-0.89) | T1+ADC (0.80) ADC (0.80) |
| Clinically-significant (CS) cancers versus other histologic groups | ||||||||
| CS Cancer (n=53) vs. Low-grade cancers (n=10) | 0.48 (0.25-0.71) [0.553] | 0.77* (0.61-0.92) [0.012] | 0.84* (0.71-0.97) [0.002] | 0.76 (0.61-0.92) | 0.85 (0.74-0.96) | 0.91* (0.82-0.99) | 0.90 (0.79-1.00) | T2+ADC (0.91) |
| CS Cancer (n=53) vs. Non-cancers (n=41) | 0.64* (0.53-0.76) [0.028] | 0.70* (0.59-0.81) [0.029] | 0.84* (0.76-0.92) [<0.0001] | 0.70 (0.60-0.81) | 0.85 (0.77-0.93) | 0.86* (0.78-0.93) | 0.86 (0.74-0.94) | T2+ADC (0.86) |
| CS Cancer (n=53) vs. Non-cancers + Low-grade cancers (n=51) | 0.61 (0.50-0.72) [0.064] | 0.71* (0.61-0.81) [0.0002] | 0.84* (0.76-0.92) [<0.0001] | 0.70 (0.60-0.80) | 0.85 (0.77-0.92) | 0.86* (0.79-0.93) | 0.86 (0.80-0.93) | T2+ADC (0.86) |
Indicate models with significant variables (p < 0.05) obtained from generalized estimating equation (GEE) logistic regression analysis. The numbers in parenthesis indicate 95% confidence intervals and numbers in brackets indicate P-value for the variables in the univariate models.
The highest AUC represents model(s) with significant variables after GEE logistic regression analysis.
Clinically-significant cancers included all cancers with Gleason score ≥ 7 while low-grade cancers was denoted by cancers with Gleason score =6
Negative biopsy =Targeted lesions with biopsy report of benign prostatic tissue
Table 4:
Best-performance cut-off values for T1, T2 and ADC based on regression models. For multivariate regression models, the individual cut-off values contributed independently to overall performance. The numbers in parenthesis indicate sensitivity and specificity for respective cut-off values.
| Groups Compared | T1
(Sensitivity/ Specificity) |
T2 (Sensitivity/ Specificity) |
ADC (Sensitivity/ Specificity) |
|---|---|---|---|
| All Prostate cancers versus Non-Cancers | |||
| Prostate Cancer (n=63) vs. Prostatitis (n=15) | Regression model not significant | 68 ms (79%67%) | 1.04×10−3 mm2/s (98%/53%) |
| Prostate Cancer (n=63) vs. Negative Biopsies (n=26) | 1720 ms (68%/62%) | Regression model not significant | 0.75×10−3 mm2/s (62%/92%) |
| Prostate Cancer (n=63) vs. Non-cancers (n=41) | 1720 ms (67%/59%) | 67 ms (79%/46%) | 0.75×10−3 mm2/s (62%/87.5%) |
| Clinically-significant (CS) cancers versus other histologic groups | |||
| CS Cancer (n=53) vs. Low-grade cancers (n=10) | Regression model not significant | 52 ms (62%/90%) | 0.78×10−3 mm2/s (73.5%/80%) |
| CS Cancer (n=53) vs. Non-cancers (n=41) | 1720 ms (68%/58/5%) | 52 ms (62%/71%) | 0.75×10−3 mm2/s (70%87.5%) |
| CS Cancer (n=53) vs. Clinically Insignificant lesions (Non-cancers + Low-grade cancers) (n=51) | 1730 ms (68%/55%) | 60 ms (62%/74.5%) | 0.75×10−3 mm2/s (70%/86%) |
All Prostate Cancers versus Non- cancers
Prostate Cancer versus Prostatitis:
Means of T1, T2 and ADC differed significantly between prostate cancer and prostatitis (p=0.039 for T1, p=0.015 for T2, p<0.0001 for ADC). Both T2 and ADC were significant predictors in logistic regression models with both having moderate diagnostic performance for separation (Table 3). AUCT2 was 0.71 while AUCADC was 0.79 with no significant difference between the two AUCs (p=0.37).
Prostate Cancer versus Negative Biopsies:
Means of T1, T2 and ADC differed significantly between prostate cancer and negative biopsies (p=0.0029 for T1, p=0.0058 for T2, p<0.0001 for ADC) Best separation was provided by T1+ADC (AUCT1+ADC=0.83) and was significantly higher than AUCADC (p=0.028) (Table 3).
Prostate Cancer versus Non-Cancers (Prostatitis and Negative Biopsies):
Means of T1, T2 and ADC differed significantly between prostate cancer and all non-cancers (p=0.0009 for T1, p=0.0004 for T2, p<0.0001 for ADC). Both ADC and T1+ADC had comparable diagnostic performances for separation (AUCADC=0.797, AUCADC+T1= 0.801) (Table 3) (Figure 4b).
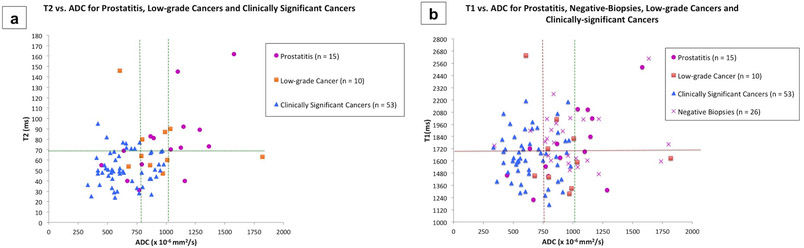
Figure 4: Quantitative characterization with combined MRF-relaxometry and ADC mapping.
(a) Scatterplot of T2 versus ADC for prostatitis (n = 15), low-grade cancers (n =10) and clinically significant cancers (n = 53). ADC value of 1.04 × 10−3 mm2/s is sensitive but not specific for differentiating all cancers from prostatitis (right vertical line). ADC value of 0.78 × 10−3 mm2/s (left vertical line) is the best cut-off for differentiating clinically significant cancers from low-grade cancers and prostatitis. In the ADC overlap zone (between two vertical lines), a T2 ≤ 68 ms is additionally helpful in differentiating cancers from prostatitis (horizontal line).
(b) Scatterplot of T1 versus ADC for non-cancers including prostatitis (n = 15), negative biopsies (n = 26), low-grade cancers (n =10) and clinically significant cancers (n = 53). ADC values of 0.75 × 10−3 mm2/s followed by T1 of 1720 ms are the best cut-offs for differentiating cancers from non-cancers (horizontal line). In the ADC overlap zone (between vertical lines), while five clinically significant cancers had T1 > 1720 ms, they also had T2 ≤ 68 ms.
Clinically Significant Prostate Cancers versus Low-grade cancers and Non-Cancers
Clinically significant cancer versus low-grade cancers:
Means of T2 and ADC differed between low-grade and high/intermediate grade cancer (p< 0.0031 for T2 and p<0.0001 for ADC) and both were significant univariable predictors with similar diagnostic performances for differentiating cancer grades (AUCT2=0.77, AUCADC=0.84, difference between two AUCs not significant, p=0.48). The best separation was obtained with T2+ADC (AUCT2+ADC=0.91) (Table 3).
Clinically significant cancer versus all Non-cancers (Prostatitis and Negative Biopsies)
Means of T1, T2 and ADC differed between clinically significant prostate cancer and all non-cancers (p=0.0003 for T1, p=0.0004 for T2, p<0.0001 for ADC). Best separation was provided by T2+ADC (AUCT2+ADC=0.86) and was significantly higher than AUCADC (p=0.04) (Table 3).
Clinically significant cancer versus Clinically insignificant lesions (Low-grade cancers and non-cancers)
Mean T1, T2 and ADC differed between clinically significant prostate cancer and low-grade cancers + non-cancers (p=0.0027 for T1, p=0.0003 for T2, p0.0001 for ADC). Best separation was provided by T2+ADC (AUCT2+ADC=0.86), and was significantly higher than AUCADC (p=0.005) (Table 3).
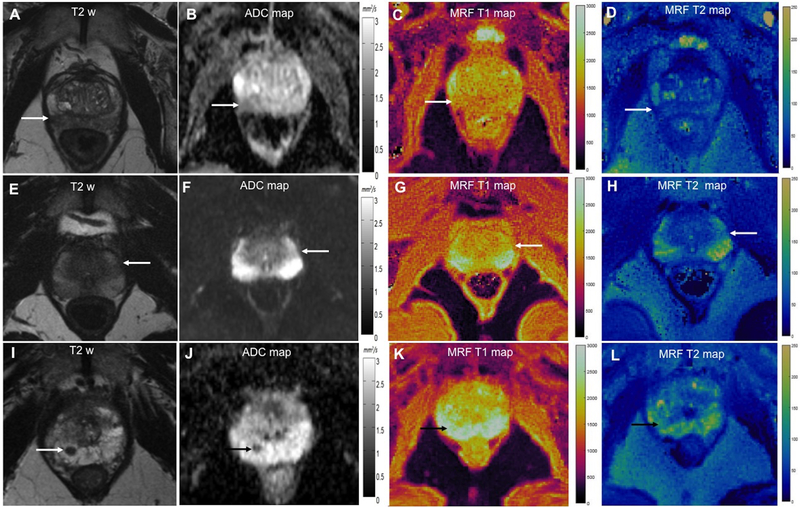
Figure 5 shows representative cases from our dataset.
Figure 5: Comparison of ADC, T1 and T2 values for targeted biopsy-proven prostate cancer (A-D), prostatitis (E-H) and benign prostatic tissue (I-L).
Prostate cancer: T2w image (A) shows focal dark lesion against diffuse dark background signal in right peripheral zone with ADC of 0.87 × 10−3 mm2/s (B). T1 and T2 values were 1560 ms and 42 ms respectively.
Prostatitis: T2w (E) shows a wedge-shaped mildly dark lesion in left peripheral zone with ADC of 0.87 × 10−3 mm2/s (F). T1 and T2 values were higher than cancer at 1770 ms and 83 ms respectively.
Benign prostatic tissue: T2w (I) shows a focal lesion in right apical peripheral zone with ADC of 0.82 × 10−3 mm2/s. Based on suspicious morphology on clinical MRI, biopsy was performed which revealed benign prostatic tissue. T1 and T2 values were higher than cancer at 2310 ms and 73 ms respectively.
Discussion
This study provides targeted biopsy validation of MRF-based relaxometry and ADC mapping for prostate imaging and adds to previous work on the demonstration of a combined quantitative exam using MRF and ADC mapping. Using targeted biopsy as a pathology reference allowed better exploration of the differences in relaxation times and ADC between grades of prostate cancer, prostatitis and negative biopsies and quantitative comparison of these histologic groups with visually NPZ. As reported previously 32 and expected due to the choice of ROIs, mean T1, T2 and ADC in visually NPZ were higher than prostate cancer and prostatitis (Table 2). Histologically, the long T2 and high ADC in NPZ have been attributed to the larger volume of glandular lumen which has “water-like” T2 relaxation times and shows increased diffusivity within the lumen38,39. The longer T1 in NPZ may relate to the proteinaceous contents of the glandular sections within the lumen12. The destruction of glandular architecture in cancers is also associated with decreased secretory function40, which may potentially account for the difference in T1 relaxation times between NPZ and cancer. More interestingly, targeted lesions diagnosed as normal prostatic tissue on biopsy, despite confirmed intra-lesional needle positions, had T1, T2 and ADC lower than visually NPZ, but higher than prostate cancer (Table 2). While the exact histological basis for these changes in negative biopsies is not known, these may represent non-specific changes in peripheral zone as prior ischemic/biopsy/inflammatory sequelae or may be attributed to the proposed existence of two populations of water protons in normal prostate tissue, one with characteristic long T2 and ADC within the glandular lumen and the other with shorter T2 and ADC due to increased stromal content12,38,39.
There were significant differences in T1 and T2 between prostate cancer and non-cancers (prostatitis and negative biopsies), which have not been reported previously 32. T1 and T2 were found to be complementary to ADC for differentiating prostate cancers from negative biopsies and prostatitis, respectively (Table 3). Previous studies have shown an overlap in ADC values between prostatitis, negative biopsies and prostate cancer. ADC values are dependent on the b-values used and the MR system gradient performance; thus no absolute ADC cut-off value can be recommended for diagnosis20,21,41. In practice, ADC values between 0.75–0.95×10−3 mm2/s, are the usual recommended thresholds for diagnosing malignancy 1. In this study too, an ADC value of less than 0.75×10−3 mm2/s was specific for differentiating a) prostate cancers from non-cancers and b) clinically significant cancers from both non-cancers and low-grade cancers, but missed cancers with higher ADC values (Table 4, Fig. 4). Vice-versa, a higher ADC cutoff of 1.04×10−3 mm2/s was sensitive for separating prostate cancer from prostatitis but had lower specificity due to a considerable overlap in ADC values between low-grade cancers, clinically significant cancers and prostatitis (Table 4, Fig. 4). However using T2 values below 68 ms may be additionally useful in differentiating prostatitis from prostate cancers for lesions with overlapping ADC values between 0.75–1.0×10−3 mm2/s (as shown in Fig. 4a). Similarly, T1 values below 1720 ms may be useful in separating cancers from non-cancers in the ADC overlap zone (Fig. 4b). Such additional measures of quantification may potentially improve pre-biopsy characterization of indeterminate or equivocal lesions seen on mpMRI, subject to future prospective validation.
For separation of cancers and non-cancers, AUCT2 and AUCADC were higher than the previously reported AUCT2 of 0.52–0.74 and AUCADC of 0.66–0.697,32 which may be due to better pathologic correlation provided by targeted biopsy while the T1 differences between prostate cancers and non-cancers is an additional finding in this study. The MRF-T2 values for different histopathologic groups are lower compared to values previously reported elsewhere 5,7,10,25,42–44 and may relate to differences from multiple spin-echo mapping7,43,45, such as noise floor effects at long echo times.
Both T2 and ADC had comparable performance for differentiating low-grade from clinically significant cancers, with the combination of T2 and ADC being additive (Table 3). Again, the AUCT2 from targeted biopsy validation is higher than the AUCT2 of 0.67–0.77 reported previously using TRUS biopsy7,32 while the AUCADC for differentiating grades of cancers is comparable to the AUCADC of 0.70–0.82 reported previously 4,16,17,46–49. At the microstructural level, higher Gleason grades are correlated with increased nuclear count and area, increased epithelial and decreased luminal and stromal volume fractions50. While ADC was previously shown to correlate better with tissue composition changes and increased cellularity metrics as compared to T26,50,51, both tissue properties had similar performance for predicting cancer aggressiveness in this study. Due to the FISP acquisition scheme utilized22,23, MRF as implemented is less adversely affected by rectal gas than echo planar imaging based diffusion acquisitions (Fig. 6). Subject to future validation, relaxation time mapping obtained in this manner could potentially have quantitative utility as an alternative to ADC mapping in situations when DWI is distorted due to susceptibility artifacts. Mean T2 and ADC for low-grade cancers were similar to those of prostatitis and benign biopsies (Table 2). This is concordant with previous results7 and the knowledge that low-grade cancers often have a have a low fraction of tumor cells intermixed with normal prostatic tissue51 and have lower epithelial and higher luminal fraction compared to higher grade cancers12.
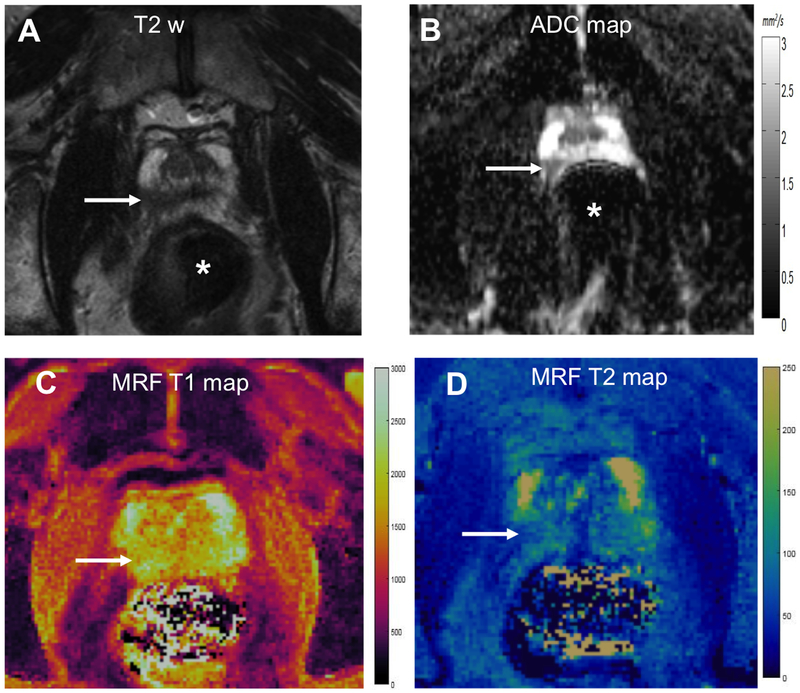
Figure 6: MRF with susceptibility artifacts on ADC mapping in a biopsy-proven case of 4 + 3 = 7 cancer.
T2w image (A) shows ill-defined dark lesion in right apical peripheral zone (arrow) with low ADC value of 0.60 × 10−3 mm2/s (B). However due to gas in rectum (*), there is susceptibility artifact on ADC map with anteroposterior deformation of the gland. MRF T1 (A) and T2 (B) color maps are relatively unaffected by rectal gas and corresponding lesion T1 and T2 values were 1600 ms and 52 ms respectively.
This study had several limitations. First, only peripheral zone lesions were analyzed in this study. This is because both peripheral and transition zones have different histological characteristics and are evaluated differently on conventional MRI, with ADC being the primary sequence for peripheral zone lesions and T2w imaging being the primary sequence for transition zone lesions. Separate analysis evaluating transition zone lesions will add further insight on the utility of this approach in prostate imaging. Second, the utilities of relaxometry and ADC mapping were utilized for lesion characterization and not for detection. Third, since the resolution of the technique is not comparable yet to T2w imaging, volumetric analysis was not performed and this remains a limitation of the work at this time. Efforts are underway at multiple institutions to develop and implement MRF examinations with higher spatial resolutions that would be better suited for detection and volumetric analysis in the future. Fourth as targeted biopsy correlation was used instead of whole-mount prostatectomy specimens for a more practical and clinically feasible histologic validation, our dataset contained of a larger number clinically significant cancers versus low-grade cancers and prostatitis. This introduces a potential selection bias because targeted biopsy is known to detect a higher number of clinically significant cancers as compared to TRUS biopsy or prostatectomy33. In the future, a prospective analysis accompanied by prostatectomy correlations may also allow analysis of larger subject/lesion populations. Fifth, cognitive biopsy was the predominant biopsy method in our study because in our institution, in-gantry biopsy was performed more often for anterior transition zone lesions and in patients with prior negative biopsies and this may have introduced an element of sampling bias. Finally, this was a single-center retrospective study with a single-reader analysis. Thus, the findings described need future prospective validation with larger datasets obtained from multi-institutional studies.
Conclusions
This work shows that the combination of T1 and T2 relaxometry can be complementary to ADC in predicting prostate cancer aggressiveness and may help in additional separation of cancers from prostatitis and negative biopsies for lesions on T2w images in the peripheral zone.
Supplementary Material
Acknowledgments
Conflicts of interest and Sources of funding:
Authors Ananya Panda, Wei-Ching Lo, Yun, Jiang, Mark Griswold and Vikas Gulani, received research support from Siemens Healthineers as part of a research grant to the University. The MR Fingerprinting technology has also been licensed by Siemens. Royalty payments have not yet started but are expected to start over the next 2–3 months. Other authors, namely Gregory O’Connor, Seunghee Margevicius, Mark Schluchter and Lee Ponsky do not have industry grant support to report. Other funding sources included NIH grants 1R01CA208236, 1R01EB016728, 1R01DK098503, 1R01EB017219.
Abbreviations:
- mpMRI
Multiparametric Magnetic Resonance Imaging
- DWI
Diffusion weighted Imaging
- MRF
Magnetic Resonance Fingerprinting
- NPZ
Normal Peripheral Zone
- ADC
Apparent Diffusion Coefficient
- TRUS
Transrectal Ultrasound
References
- 1.Barentsz JO, Weinreb JC, Verma S, et al. Synopsis of the PI-RADS v2 Guidelines for Multiparametric Prostate Magnetic Resonance Imaging and Recommendations for Use. Eur. Urol. 2016;69(1):41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Metzger GJ, Kalavagunta C, Spilseth B, et al. Detection of Prostate Cancer: Quantitative Multiparametric MR Imaging Models Developed Using Registered Correlative Histopathology. Radiology. 2016;279(3):805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peng Y, Jiang Y, Yang C, et al. Quantitative analysis of multiparametric prostate MR images: differentiation between prostate cancer and normal tissue and correlation with Gleason score--a computer-aided diagnosis development study. Radiology. 2013;267(3):787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donati OF, Afaq A, Vargas HA, et al. Prostate MRI: Evaluating Tumor Volume and Apparent Diffusion Coefficient as Surrogate Biomarkers for Predicting Tumor Gleason Score. Clin. Cancer Res 2014;20(14):3705–3711. [DOI] [PubMed] [Google Scholar]
- 5.Gibbs P, Tozer DJ, Liney GP, et al. Comparison of quantitative T2 mapping and diffusion-weighted imaging in the normal and pathologic prostate. Magn. Reson. Med 2001;46(6):1054–1058. [DOI] [PubMed] [Google Scholar]
- 6.Gibbs P, Liney GP, Pickles MD, et al. Correlation of ADC and T2 measurements with cell density in prostate cancer at 3.0 Tesla. Invest. Radiol 2009;44(9):572–576. [DOI] [PubMed] [Google Scholar]
- 7.Hoang Dinh A, Souchon R, Melodelima C, et al. Characterization of prostate cancer using T2 mapping at 3T: A multi-scanner study. Diagn. Interv. Imaging 2015;96(4):365–372. [DOI] [PubMed] [Google Scholar]
- 8.Hoang Dinh A, Melodelima C, Souchon R, et al. Quantitative Analysis of Prostate Multiparametric MR Images for Detection of Aggressive Prostate Cancer in the Peripheral Zone: A Multiple Imager Study. Radiology. 2016;280(1):117–127. [DOI] [PubMed] [Google Scholar]
- 9.Wu L-M, Chen X-X, Xuan H-Q, et al. Feasibility and preliminary experience of quantitative T2* mapping at 3.0 T for detection and assessment of aggressiveness of prostate cancer. Acad. Radiol 2014;21(8):1020–1026. [DOI] [PubMed] [Google Scholar]
- 10.Simpkin CJ, Morgan VA, Giles SL, et al. Relationship between T2 relaxation and apparent diffusion coefficient in malignant and non-malignant prostate regions and the effect of peripheral zone fractional volume. Br. J. Radiol 2013;86(1024):20120469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langer DL, van der Kwast TH, Evans AJ, et al. Prostate Tissue Composition and MR Measurements: Investigating the Relationships between ADC, T2, Ktrans, ve, and Corresponding Histologic Features. Radiology. 2010;255(2):485–494. [DOI] [PubMed] [Google Scholar]
- 12.Chatterjee A, Bourne RM, Wang S, et al. Diagnosis of Prostate Cancer with Noninvasive Estimation of Prostate Tissue Composition by Using Hybrid Multidimensional MR Imaging: A Feasibility Study. Radiology. 2018;287(3):864–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mai J, Abubrig M, Lehmann T, et al. T2 Mapping in Prostate Cancer. Invest. Radiol 2018;Publish Ahead of Print. Available at: https://journals.lww.com/investigativeradiology/Abstract/publishahead/T2_Mapping_in_Prostate_Cancer.98961.aspx. Accessed December 29, 2018. [Google Scholar]
- 14.Anwar SSM, Anwar Khan Z, Shoaib Hamid R, et al. Assessment of Apparent Diffusion Coefficient Values as Predictor of Aggressiveness in Peripheral Zone Prostate Cancer: Comparison with Gleason Score. Int. Sch. Res. Not 2014. Available at: https://www.hindawi.com/journals/isrn/2014/263417/. Accessed May 7, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.deSouza NM, Riches SF, Vanas NJ, et al. Diffusion-weighted magnetic resonance imaging: a potential non-invasive marker of tumour aggressiveness in localized prostate cancer. Clin. Radiol 2008;63(7):774–782. [DOI] [PubMed] [Google Scholar]
- 16.Hambrock T, Somford DM, Huisman HJ, et al. Relationship between Apparent Diffusion Coefficients at 3.0-T MR Imaging and Gleason Grade in Peripheral Zone Prostate Cancer. Radiology. 2011;259(2):453–461. [DOI] [PubMed] [Google Scholar]
- 17.Turkbey B, Shah VP, Pang Y, et al. Is Apparent Diffusion Coefficient Associated with Clinical Risk Scores for Prostate Cancers that Are Visible on 3-T MR Images? Radiology. 2011;258(2):488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bittencourt LK, Barentsz JO, de Miranda LCD, et al. Prostate MRI: diffusion-weighted imaging at 1.5T correlates better with prostatectomy Gleason grades than TRUS-guided biopsies in peripheral zone tumours. Eur. Radiol 2012;22(2):468–475. [DOI] [PubMed] [Google Scholar]
- 19.Glazer DI, Hassanzadeh E, Fedorov A, et al. Diffusion-weighted endorectal MR imaging at 3T for prostate cancer: correlation with tumor cell density and percentage Gleason pattern on whole mount pathology. Abdom. Radiol 2017;42(3):918–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagel KNA, Schouten MG, Hambrock T, et al. Differentiation of prostatitis and prostate cancer by using diffusion-weighted MR imaging and MR-guided biopsy at 3 T. Radiology. 2013;267(1):164–172. [DOI] [PubMed] [Google Scholar]
- 21.Esen M, Onur MR, Akpolat N, et al. Utility of ADC measurement on diffusion-weighted MRI in differentiation of prostate cancer, normal prostate and prostatitis. Quant. Imaging Med. Surg 2013;3(4):210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma D, Gulani V, Seiberlich N, et al. Magnetic resonance fingerprinting. Nature. 2013;495(7440):187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Y, Ma D, Seiberlich N, et al. MR Fingerprinting Using Fast Imaging with Steady State Precession (FISP) with Spiral Readout. Magn. Reson. Med. 2015;74(6):1621–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panda A, Mehta BB, Coppo S, et al. Magnetic Resonance Fingerprinting-An Overview. Curr. Opin. Biomed. Eng 2017;3:56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chatterjee A, Devaraj A, Mathew M, et al. Performance of T2 Maps in the Detection of Prostate Cancer. Acad. Radiol. Available at: https://www.sciencedirect.com/science/article/pii/S1076633218301880. Accessed May 7, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buonincontri G, Sawiak SJ. MR fingerprinting with simultaneous B1 estimation. Magn. Reson. Med 2016;76(4):1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buonincontri G, Schulte RF, Cosottini M, et al. Spiral MR fingerprinting at 7 T with simultaneous B1 estimation. Magn. Reson. Imaging. 2017;41:1–6. [DOI] [PubMed] [Google Scholar]
- 28.Hamilton JI, Jiang Y, Ma D, et al. Investigating and reducing the effects of confounding factors for robust T1 and T2 mapping with cardiac MR fingerprinting. Magn. Reson. Imaging. 2018;53:40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cloos MA, Knoll F, Zhao T, et al. Multiparametric imaging with heterogeneous radiofrequency fields. Nat. Commun 2016;7:ncomms12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang Y, Ma D, Keenan KE, et al. Repeatability of magnetic resonance fingerprinting T1 and T2 estimates assessed using the ISMRM/NIST MRI system phantom. Magn. Reson. Med 2017;78(4):1452–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lo WC, Jiang Y, Bittencourt LK, et al. Multicenter repeatability and reproducibility of MR Fingerprinting In: Intl Soc Magn Reson Med (26)2018. Vol 26 Paris, France; 2018. Available at: http://cds.ismrm.org/protected/18MPresentations/abstracts/4503.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu AC, Badve C, Ponsky LE, et al. Development of a Combined MR Fingerprinting and Diffusion Examination for Prostate Cancer. Radiology. 2017;283(3):729–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siddiqui MM, George AK, Rubin R, et al. Efficiency of Prostate Cancer Diagnosis by MR/Ultrasound Fusion-Guided Biopsy vs Standard Extended-Sextant Biopsy for MR-Visible Lesions. J. Natl. Cancer Inst. 2016;108(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bjurlin MA, Mendhiratta N, Wysock JS, et al. Multiparametric MRI and targeted prostate biopsy: Improvements in cancer detection, localization, and risk assessment. Cent. Eur. J. Urol 2016;69(1):9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wegelin O, van Melick HHE, Hooft L, et al. Comparing Three Different Techniques for Magnetic Resonance Imaging-targeted Prostate Biopsies: A Systematic Review of In-bore versus Magnetic Resonance Imaging-transrectal Ultrasound fusion versus Cognitive Registration. Is There a Preferred Technique? Eur. Urol 2017;71(4):517–531. [DOI] [PubMed] [Google Scholar]
- 36.Hansen MS, Sørensen TS. Gadgetron: an open source framework for medical image reconstruction. Magn. Reson. Med 2013;69(6):1768–1776. [DOI] [PubMed] [Google Scholar]
- 37.Lo W-C, Jiang Yun FD, Griswold Mark, et al. MR Fingerprinting using a Gadgetron-based reconstruction In: Intl Soc Magn Reson Med (26)2018. Vol 26 Paris, France; 2018. Available at: https://cds.ismrm.org/protected/18MPresentations/abstracts/3525.html. [Google Scholar]
- 38.Wang S, Peng Y, Medved M, et al. Hybrid Multidimensional T2 and Diffusion-Weighted MRI for Prostate Cancer Detection. J. Magn. Reson. Imaging JMRI 2014;39(4):781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Storås TH, Gjesdal K-I, Gadmar ØB, et al. Prostate magnetic resonance imaging: Multiexponential T2 decay in prostate tissue. J. Magn. Reson. Imaging. 2008;28(5):1166–1172. [DOI] [PubMed] [Google Scholar]
- 40.Liney GP, Turnbull LW, Lowry M, et al. In vivo quantification of citrate concentration and water T2 relaxation time of the pathologic prostate gland using 1H MRS and MRI. Magn. Reson. Imaging. 1997;15(10):1177–1186. [DOI] [PubMed] [Google Scholar]
- 41.Verma S, Rajesh A, Morales H, et al. Assessment of aggressiveness of prostate cancer: correlation of apparent diffusion coefficient with histologic grade after radical prostatectomy. AJR Am. J. Roentgenol 2011;196(2):374–381. [DOI] [PubMed] [Google Scholar]
- 42.Langer DL, van der Kwast TH, Evans AJ, et al. Prostate cancer detection with multi-parametric MRI: logistic regression analysis of quantitative T2, diffusion-weighted imaging, and dynamic contrast-enhanced MRI. J. Magn. Reson. Imaging JMRI. 2009;30(2):327–334. [DOI] [PubMed] [Google Scholar]
- 43.Yamauchi FI, Penzkofer T, Fedorov A, et al. Prostate cancer discrimination in the peripheral zone with a reduced field-of-view T2-mapping MRI sequence. Magn. Reson. Imaging. 2015;33(5):525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Houdt PJ, Agarwal HK, van Buuren LD, et al. Performance of a fast and high-resolution multi-echo spin-echo sequence for prostate T2 mapping across multiple systems. Magn. Reson. Med 2018;79(3):1586–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giganti F, Gambarota G, Moore CM, et al. Prostate cancer detection using quantitative T2 and T2-weighted imaging: The effects of 5-alpha-reductase inhibitors in men on active surveillance. J. Magn. Reson. Imaging. 2018;47(6):1646–1653. [DOI] [PubMed] [Google Scholar]
- 46.Woo S, Kim SY, Cho JY, et al. Preoperative Evaluation of Prostate Cancer Aggressiveness: Using ADC and ADC Ratio in Determining Gleason Score. AJR Am. J. Roentgenol 2016;207(1):114–120. [DOI] [PubMed] [Google Scholar]
- 47.Vargas HA, Akin O, Franiel T, et al. Diffusion-weighted endorectal MR imaging at 3 T for prostate cancer: tumor detection and assessment of aggressiveness. Radiology. 2011;259(3):775–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thörmer G, Otto J, Horn L-C, et al. Non-invasive estimation of prostate cancer aggressiveness using diffusion-weighted MRI and 3D proton MR spectroscopy at 3.0 T. Acta Radiol 2015;56(1):121–128. [DOI] [PubMed] [Google Scholar]
- 49.Donati OF, Mazaheri Y, Afaq A, et al. Prostate Cancer Aggressiveness: Assessment with Whole-Lesion Histogram Analysis of the Apparent Diffusion Coefficient. Radiology. 2013;271(1):143–152. [DOI] [PubMed] [Google Scholar]
- 50.Chatterjee A, Watson G, Myint E, et al. Changes in Epithelium, Stroma, and Lumen Space Correlate More Strongly with Gleason Pattern and Are Stronger Predictors of Prostate ADC Changes than Cellularity Metrics. Radiology. 2015;277(3):751–762. [DOI] [PubMed] [Google Scholar]
- 51.Langer DL, van der Kwast TH, Evans AJ, et al. Intermixed Normal Tissue within Prostate Cancer: Effect on MR Imaging Measurements of Apparent Diffusion Coefficient and T2—Sparse versus Dense Cancers. Radiology. 2008;249(3):900–908. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.