Abstract
Background:
One mechanism underlying the development of alcoholic liver disease is over activation of the innate immune response. Recent investigations indicate that the lysosomal pathway of autophagy down regulates the inflammatory state of hepatic macrophages, suggesting that macrophage autophagy may regulate innate immunity in alcoholic liver disease. The function of macrophage autophagy in the development of alcoholic liver disease was examined in studies employing mice with a myeloid-specific decrease in autophagy.
Methods:
Littermate control and Atg5Δmye mice lacking Atg5-dependent myeloid autophagy were administered a Lieber-DeCarli control (CD) or ethanol (ED) diet alone or together with lipopolysaccharide (LPS) and examined for the degree of liver injury and inflammation.
Results:
Knockout mice with decreased macrophage autophagy had equivalent steatosis but increased mortality and liver injury from ED alone. Increased liver injury and hepatocyte death also occurred in Atg5Δmye mice administered ED and LPS in association with systemic inflammation as indicated by elevated serum levels of proinflammatory cytokines. Hepatic macrophage and neutrophil infiltration was unaffected by decreased autophagy, but levels of proinflammatory cytokine gene induction were significantly increased in the livers but not adipose tissue of knockout mice treated with ED and LPS. Inflammasome activation was increased in ED/LPS-treated knockout mice resulting in elevated interleukin (IL)-1β production. Increased IL-1β promoted alcoholic liver disease as liver injury was decreased by administration of an IL-1 receptor antagonist.
Conclusions:
Macrophage autophagy functions to prevent liver injury from alcohol. This protection is mediated in part by down regulation of inflammasome dependent and independent hepatic inflammation. Therapies to increase autophagy may be effective in this disease through anti-inflammatory effects on macrophages.
Keywords: Alcoholic liver disease, Inflammasome, Interleukin-1β, Lipopolysaccharide, Tumor necrosis factor
INTRODUCTION
Investigations into the mechanisms of hepatotoxic liver injury have demonstrated that hepatocyte injury and death are mediated in large part by the secondary effects of injurious factors produced by the associated hepatic and systemic innate immune response (Krenkel and Tacke, 2017). The liver presents a unique immune environment for two reasons – an anatomical location that leads to continuous exposure to absorbed intestinal materials including immune activators such as bacterial lipopolysaccharide (LPS), and the presence of a large number of resident tissue macrophages. Gut-produced pathogen-associated molecular patterns such as LPS activate hepatic resident Kupffer cells and infiltrating macrophages recruited after injury. The importance of LPS in the etiology of alcoholic liver disease has been demonstrated by studies in which mice lacking the LPS receptor toll-like receptor 4 are protected from alcohol-induced injury (Uesugi et al., 2001).
The normal function of macrophage activation is to promote the repair of tissue damage. In many types of liver injury, however, over activation of the innate immune response occurs by unknown mechanisms and acts to potentiate liver injury including that in alcoholic liver disease (Maher et al., 2008; Adams et al., 2010; Szabo et al., 2012; Wang et al., 2012). Activated proinflammatory macrophages produce cytotoxic factors such as the cytokines tumor necrosis factor (TNF) and interleukin (IL)-1β that promote liver injury and inflammation in alcoholic liver disease (Iimuro et al., 1997; Yin et al., 1999; Petrasek et al., 2012). A delineation of the mechanisms that restrict proinflammatory macrophage activation is therefore important to understanding and potentially preventing liver injury from alcohol.
The lysosomal degradative pathway of macroautophagy, hereafter referred to as autophagy, regulates essential cellular processes in many cell types (Czaja, 2011). A number of liver diseases including that from alcohol have been shown to be associated with a decrease in hepatic autophagic function (Czaja et al., 2013). Studies have also indicated that autophagy plays an important regulatory function in inflammation. The initial link between autophagy and inflammation was the demonstrated ability of this pathway to down regulate inflammation secondary to microbial infections through sequestration and elimination of the pathogens (Levine et al., 2011; Deretic et al., 2013). Recent investigations in knockout mice with a macrophage-specific inhibition of autophagy have demonstrated in the liver that autophagy performs critical functions in down regulating sterile inflammation. We have shown that in the setting of obesity-induced hepatic steatosis macrophage autophagy is impaired and that this defect increases liver inflammation and injury from LPS by promoting proinflammatory M1 macrophage polarization (Liu et al., 2015). Alternatively in acute fulminant toxic liver injury from galactosamine/LPS decreased macrophage autophagy promoted hepatic inflammation and injury by increasing caspase 1-dependent inflammasome cleavage of pro-IL-1β to its active, secreted form (Ilyas et al., 2016). Similarly in chronic liver injury from carbon tetrachloride, impaired macrophage autophagy led to IL-1-dependent liver injury and fibrosis (Lodder et al., 2015).
In light of the emerging importance of autophagy in modulating the hepatic immune response, and the established role of dysregulated macrophage activation in alcoholic liver disease, we examined the function of macrophage autophagy in this disease by studying the effects of a selective inhibition of myeloid cell autophagy in mice fed an alcohol diet alone or together with an injection of LPS. Macrophage autophagy does not regulate the excessive lipid accumulation induced by alcohol, but with decreased macrophage autophagy there is increased liver injury and inflammation. Hyperactivation of both inflammasome independent and dependent macrophage activation occur with impaired autophagy. These findings identify macrophages as an important hepatic cell type in addition to hepatocytes in which levels of autophagy regulate the development of liver disease from alcohol.
MATERIALS AND METHODS
Animal Model
Mice were maintained under 12 h light/dark cycles with unlimited access to food and water prior to the period of Lieber-DeCarli diet feeding. All studies were performed in 10-14 week old female mice. Atg5F/F mice (Hara et al., 2006) containing floxed alleles for the autophagy gene Atg5 were crossed with LysM-Cre mice (Clausen et al., 1999) to generate Atg5Δmye mice with a myeloid cell-specific knockout of Atg5-dependent autophagy, as previously described (Liu et al., 2015). Both strains of transgenic mice are on a C57BL/6J background. Mouse genotypes were confirmed by PCR with established primers. For all studies Atg5Δmye mice littermates lacking the Cre transgene were used as controls.
Mice were fed a liquid Lieber-DeCarli diet. At start of the experiments all mice were placed on 5 days of control diet (Bio-Serv, Flemington, NJ; #F1259SP). Mice were then randomized to receive a 5% liquid ethanol diet (Bio-Serv; #F1258SP), or pair-fed an equal caloric amount of the control diet for 21 days. On the final day of feeding some mice received a single intraperitoneal injection of LPS (7.5 mg/kg; E. coli 0111:B4; Sigma, St. Louis, MO) as previously described (Lalazar et al., 2016). Mice were sacrificed at 6 h after LPS injection for analysis. Some mice were pretreated with an equal volume of normal saline (NS) vehicle or 25 mg/kg of the IL-1 receptor antagonist (IL-1Ra) anakinra (Amgen, Thousand Oaks, CA) 24 and 0.5 h before LPS administration. All mouse studies were approved by the Animal Care and Use Committees of the Albert Einstein College of Medicine or Emory School of Medicine and followed the National Institutes of Health guidelines for animal care.
Kupffer Cell Isolation and Culture
Mouse liver nonparenchymal cells were isolated by Liberase (Roche, Basal, Switzerland) perfusion and centrifuged at 50 g two times to remove all hepatocytes. Kupffer cells were isolated from the total nonparenchymal cell population by differential centrifugation through a 29% Nycodenz (Accurate Chemical & Scientific Corp., Westbury, NY) gradient at 1,380 g. Following washing in DMEM (Hyclone, Logan, UT), the cells were plated in DMEM, 10% fetal bovine serum (Gemini Bio-Products, West Sacramento, CA), 2% 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Fisher Scientific, fair Lawn, NJ), 1% penicillin/streptomycin (Gemini Bio-Products), 1% non-essential amino acids and 1% sodium pyruvate (GE Healthcare Life Sciences, Logan, UT). Non-adherent cells were removed after 1 h by changing the medium to RPMI-1640 (Fisher Scientific) with the same supplements. Cell purity was demonstrated by mRNA enrichment for macrophage genes and absence of expression for hepatocyte, cholangiocyte and hepatic stellate cell genes by quantitative real-time reverse transcription polymerase chain reaction. After overnight culture, the medium was changed in the cells, and some cells were treated with 200 nM bafilomycin A1 (Sigma) for 2 h.
Serum Assays
Serum alanine transaminase (ALT) and aspartate transaminase (AST) were measured by commercial kits (TECO Diagnostics, Anaheim, CA).
Histology
Portions of livers were fixed in 10% neutral formalin, stained with hematoxylin and eosin and graded in a blinded fashion by a single pathologist for the degree of steatosis or liver injury. The percentage of hepatic parenchyma with steatosis or apoptotic/necrotic injury was semi-quantitatively graded on a sliding scale of: 0, absent; 0.5, minimal; 1, mild; 1.5, mild to moderate; 2, moderate; 2.5, moderate to marked; and 3, marked.
Triglyceride Assay
Liver triglyceride content was determined using a kit according to the manufacturer’s instructions (Sigma) and normalized to liver weight.
Protein Isolation and Western Blotting
Total liver protein was isolated and western blotting performed as previously described (Wang et al., 2004; Wang et al., 2006). Membranes were probed with antibodies that recognized Atg5 (Cell Signaling, Beverly, MA; #12994), caspase 1 (Santa Cruz Biotechnology, Dallas, TX; #SC-514), caspase 3 (Cell Signaling; #9665), caspase 7 (Cell Signaling; #9492), IL-1β (R&D Systems, Minneapolis, MN; #AF-401-NA), microtubule associated protein 1 light chain 3 (LC3; Cell Signaling; #2775), sequestosome-1 (p62; Cell Signaling; #39749), and tubulin (Cell Signaling; #2148).
TUNEL Assay
Numbers of TUNEL positive cells were determined in liver sections with the commercial kit DeadEnd Colorimetric System (Promega, Madison, WI). Tissue sections were deparaffinized in xylene, gradually rehydrated in decreasing concentrations of ethanol, and the assay performed according to the manufacturer’s instructions. Under light microscopy, the numbers of TUNEL positive cells in 10 randomly selected high power fields (400X magnification) were counted per liver section.
Immunofluorescence Microscopy
Immunofluorescence staining for macrophages and neutrophils was performed on frozen liver sections as previously described (Ilyas et al., 2016). The antibodies employed were anti-CD68 (Abd Serotec, Raleigh, NC; #MCA1957GA), anti-Ly6G (Biolegend, San Diego, CA; #127602) and a Cy3-conjugated secondary antibody (Jackson ImmunoResearch, West Grove, PA; #111-165-152).
Cytokine Assay
Measurements of serum cytokines were by a multiplex kit MCYTOMAG-70K (Millipore, Burlington, MA) that employed Luminex® xMAP® Technology.
Quantitative Real-time Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
Total RNA was isolated using the commercial kits RNeasy Plus (QIAGEN, Valencia, CA) for liver and RNeasy Lipid Tissue (QIAGEN) for white adipose tissue. Reverse transcription with 1 μg of RNA was carried out in an Eppendorf Mastercycler (Hamburg, Germany) using a high capacity cDNA reverse transcription kit (ABI, Foster City, CA). Primer annealing was done at 25°C for 10 min, followed by elongation at 37°C for 2 h and inactivation of the enzyme at 85°C for 5 min. Negative controls (no added transcriptase) were performed in parallel. qRT-PCR for the genes Cd68, Ly6g, tumor necrosis factor (Tnf), interleukin-1β (Il1b), interleukin-1α (Il1a), interferonγ (Ifng), and glyceraldehyde 3-phosphate dehydrogenase (Gapdh) was performed in triplicate in a 7500 Fast Real-Time PCR System (ABI). The primers in Table 1 were employed and purchased from Integrated DNA Technologies (Coralville, IA). qRT-PCR was carried out using Power SYBR Green Master Mix (ABI). Taq polymerase was activated at 95°C for 10 min. The cycling parameters were denaturation at 95°C for 30 sec and extension at 60°C for 1 min (for 40 cycles). Data analysis was performed using the 2−ΔΔCT method for relative quantification. All samples were normalized to Gapdh.
Table 1.
qRT-PCR primer sequences.
| Cd68 | Forward | 5′-ATTGAGGAAGGAACTGGTGTAG-3′ |
| Reverse | 5′-CCTCTGTTCCTTGGGCTATAAG-3′ | |
| Gapdh | Forward | 5′-AGGTCGGTGTGAACGGATTTG-3′ |
| Reverse | 5′-TGTAGACCATGTAGTTGAGGTCA-3′ | |
| Il1a | Forward | 5′-CGCTTGAGTCGGCAAAGAAAT-3′ |
| Reverse | 5′-CTTCCCGTTGCTTGACGTTG-3′ | |
| Il1b | Forward | 5′-GCAACTGTTCCTGAACTCAACT-3′ |
| Reverse | 5′-ATCTTTTGGGGTCCGTCAACT-3′ | |
| Il4 | Forward | 5′-ATGGATGTGCCAAACGTCCT-3′ |
| Reverse | 5′-AGCTTATCGATGAATCCAGGCA-3′ | |
| Il10 | Forward | 5′-CGGACTGCCTTCAGCCAGGTG-3′ |
| Reverse | 5′-CACAGGGGAGAAATCGATGACAGC-3′ | |
| Ifng | Forward | 5′-ATGAACGCTACACACTGCATC-3′ |
| Reverse | 5′-CCATCCTTTTGCCAGTTCCTC-3′ | |
| Ly6g | Forward | 5′-GCACATGAAAGAGGCAGTATTC-3′ |
| Reverse | 5′-GAGACATTGAACAGCACACATAG-3′ | |
| Tnf | Forward | 5′-CCCTCACACTCAGATCATCTTCT-3′ |
| Reverse | 5′-GCTACGACGTGGGCTACAG-3′ |
Statistical Analysis
Numerical results are reported as means ± S.E. All data are derived from at least three independent experiments which were conducted in separate litters of mice at different times. Sample numbers are specified for each experiment. Higher numbers of mice were used in the ethanol-treated groups than the control diet-fed groups in order to achieve statistically significant differences. Statistical significance among control and treated groups was determined by one way analysis of variance (ANOVA). A log rank test was used to compare differences in survival among the four groups. Statistical significance was defined as P<0.05.
RESULTS
Inhibition of Macrophage Autophagy Increases Liver Injury
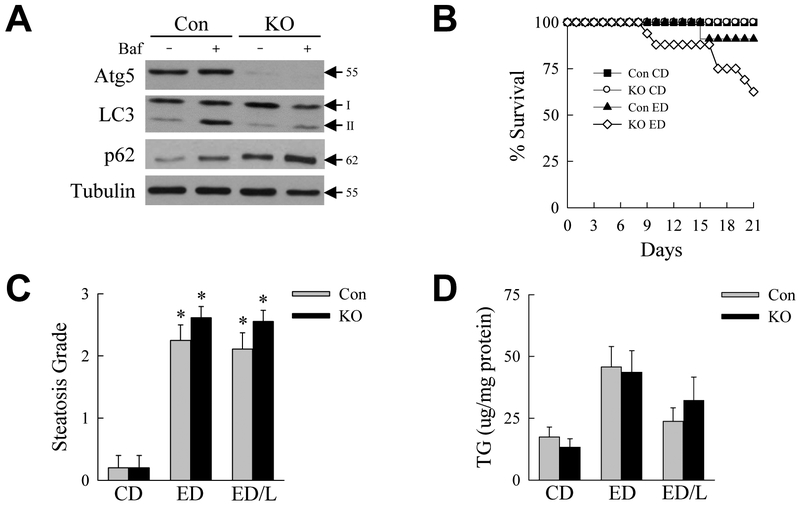
To examine the effects of macrophage autophagy Atg5Δmye mice were generated with a myeloid-specific knockout of the critical autophagy gene Atg5 and fed an ethanol diet. The hepatic macrophage knockout was confirmed by western blotting of Kupffer cells isolated from untreated littermate control and Atg5Δmye knockout mice. Kupffer cells from knockout mice have markedly decreased Atg5 levels (Fig. 1A). As a result cells from Atg5Δmye mice have reduced autophagic flux as demonstrated by decreased levels of LC3-II after treatment with the lysosomal inhibitor bafilomycin A1 as compared to cells from control mice (Fig. 1A). Untreated knockout cells also have elevated p62 levels that increased to a lesser extent with bafilomycin A1 treatment as compared to control cells, consistent with a decrease in autophagy (Fig. 1A). Littermate control and Atg5Δmye mice were fed a Lieber-DeCarli control diet (CD) or ethanol diet (ED) for 21 days. To better examine the effects of the alcoholic liver disease cofactor LPS in mice with decreased macrophage autophagy, some ED-fed mice were also administered a single dose of LPS at the end of the 21-day ED feeding period and analyzed 6 h after LPS injection (ED/L).
Fig. 1.
Mice with decreased macrophage autophagy fed an ED have increased mortality but equivalent steatosis. (A) Immunoblots of total protein from hepatic macrophages from control mice (Con) and Atg5Δmye knockout (KO) mice probed for Atg5, LC3, p62 and tubulin as a loading control. Cells were untreated or treated with bafilomycin A1 (Baf) for 2 h. The Atg5 band represents the Atg5-Atg12 conjugate form. Molecular weights and LC3-I and LC3-II are indicated by arrows. (B) Survival over 21 days of feeding with control diet (CD) or ethanol diet (ED) in control and knockout mice (KO ED mice P<0.01 as compared to Con CD mice, and P<0.03 as compared to Con ED mice; n=10-24). (C) Semi-quantitative histological grades of steatosis and (D) hepatic triglyceride content in the livers of control and knockout mice fed CD, ED or ED with LPS injection (ED/L) (*P<0.00004 as compared to control mice on CD; n=5-19).
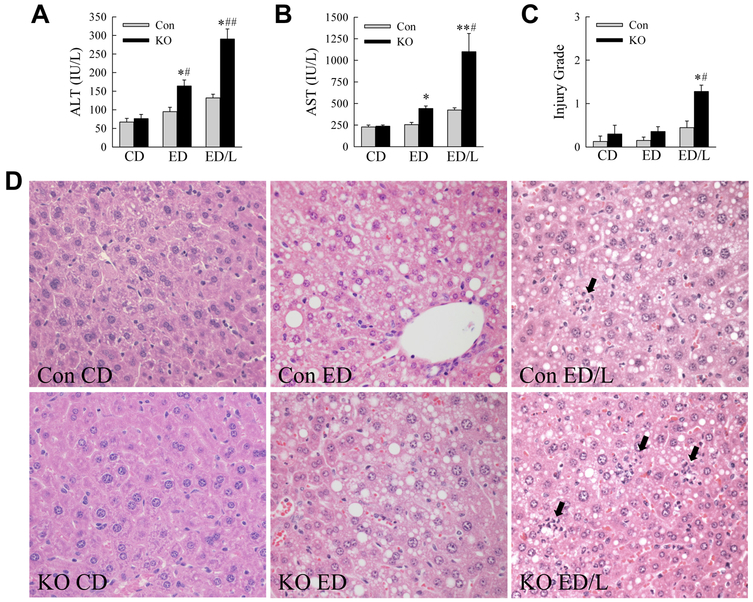
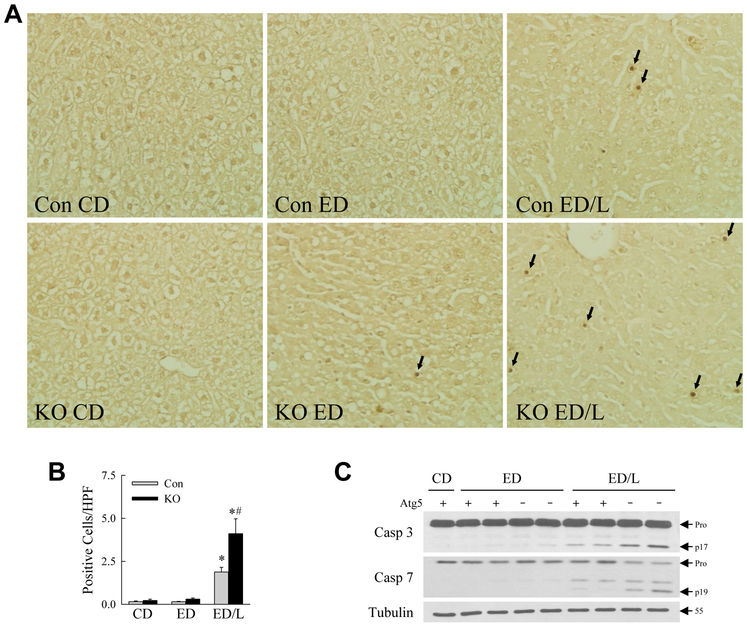
Over the 21-day feeding period all CD-fed mice survived whereas ED feeding caused mortality that was significantly increased in the knockouts (Fig. 1B). No deaths occurred in any of the groups over the time period after LPS administration. In surviving mice excessive lipid accumulation occurred with ED feeding as indicated by increases in the degree of steatosis by semi-quantitative, blinded histological grading (Fig. 1C) and hepatic triglyceride content (Fig. 1D), although only the increase in steatosis was statistically significant. Decreased macrophage autophagy had no effect on the amount of hepatic steatosis or triglyceride content (Fig. 1C,D). In control mice the serum markers of liver injury ALT and AST were unaffected by ED alone, but significantly increased with ED/L treatment (Fig. 2A,B). Levels of both ALT and AST were significantly elevated in Atg5Δmye mice as compared to littermate controls with both ED alone and ED/L (Fig. 2A,B). Liver injury was modest by histology, but histological grading confirmed that Atg5Δmye mice had greater liver injury than control mice with ED feeding, but the effect was only statistically significant with ED/L treatment (Fig. 2C). Increased cell death was present in ED/L-treated Atg5Δmye mice as demonstrated by the numbers of TUNEL-positive cells in these livers (Fig. 3A,B). TUNEL-positive cells were evenly distributed throughout the hepatic lobules. Apoptosis occurred in ED/L-treated knockout mice as revealed by increased levels of active, cleaved effector caspase 3 and 7 in knockout mice on immunoblotting (Fig. 3C). These findings demonstrate that macrophage autophagy functions to prevent liver injury from alcohol feeding particularly in the setting of LPS stimulation.
Fig. 2.
Inhibition of macrophage autophagy increases alcohol-induced liver injury. (A) Serum ALT levels in littermate control (Con) and Atg5Δmye knockout (KO) mice fed a control diet (CD) or ethanol diet (ED) alone or with LPS (ED/L) (*P<0.002, as compared to Con CD mice; #P<0.01, ##P<0.00001 as compared to Con mice with the same treatment; n=7-18). (B) Serum AST levels in the same mice (*P<0.01, **P<0.0000001 as compared to Con CD mice; #P<0.000001 as compared to Con ED/L mice; n=7-18). (C) Histological grade of liver injury in the mice (*P<0.01 as compared to Con CD mice; #P<0.03 as compared to Con mice with the same treatment; n=5-9). (D) Representative hematoxylin and eosin stained liver sections from control and knockout mice with the indicated treatments (400X). Arrows indicate areas of injury and inflammation.
Fig. 3.
Knockout mice have increased hepatic apoptosis from ED/L treatment. (A) Representative images of TUNEL-stained livers from control (Con) and knockout (KO) mice that received the indicated treatments. Arrows indicate TUNEL-positive cells (400X). (B) Quantification of the numbers of TUNEL positive cells per high power field (HPF) (*P<0.01 as compared to Con CD; #P<0.0003 as compared to Con ED/L; n=5-9). (C) Immunoblots of total hepatic protein from control (Atg5 +) and knockout (Atg5 -) mice probed for caspase 3 (Casp 3), caspase 7 (Casp 7) and tubulin. The procaspase (Pro) and cleaved caspase 3 (p17) and caspase 7 (p19) forms and the molecular weight of tubulin are indicated by arrows. Immunoblots are representative of findings from 3 independent experiments.
Knockout Mice have a Greater Systemic Proinflammatory Response to LPS
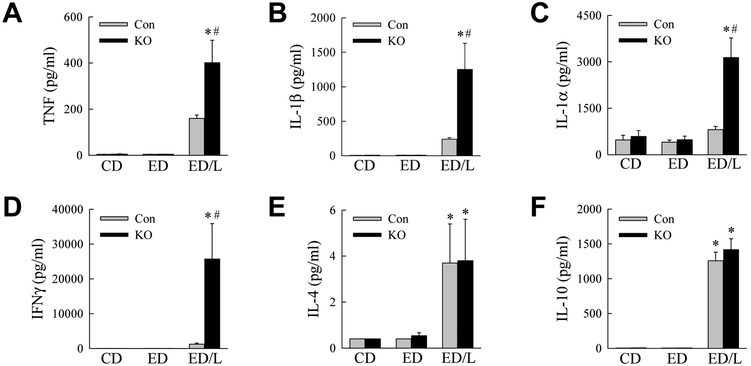
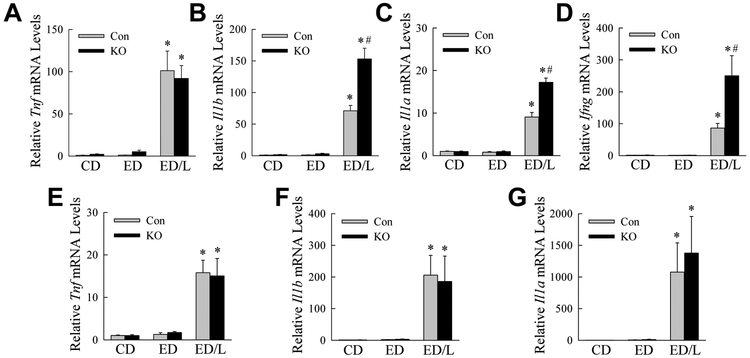
Our previous studies demonstrated that with decreased autophagy, hepatic macrophages assume a more proinflammatory phenotype (Liu et al., 2015; Ilyas et al., 2016). To assess whether systemic inflammation occurred with ED feeding in knockout mice, serum cytokine levels were examined. The proinflammatory cytokines TNF, IL-1β, IL-1α and IFNγ were all increased by ED/L but not ED alone in both control and knockout mice (Fig. 4A-D). Serum levels of all four cytokines were significantly greater in the knockout mice (Fig. 4A-D). In contrast, the anti-inflammatory cytokines IL-4 and IL-10 were significantly increased with ED/LPS treatment, but induction were equivalent in control and knockout mice (Fig. 4E,F). ED/L treatment therefore led to an increased proinflammatory macrophage response in knockout mice.
Fig. 4.
Knockout mice have increased serum levels of proinflammatory but not anti-inflammatory cytokines. Serum from control diet (CD), ethanol diet (ED) and ethanol diet/LPS (ED/L) treated control (Con) and knockout (KO) mice was assayed for protein levels of (A) TNF, (B) IL-1β, (C) IL-1α, (D) IFNγ, (E) IL-4 and (F) IL-10 (*P<0.001, as compared to Con CD; #P<0.0003 as compared to Con ED/L; n=6-17).
Loss of Atg5-dependent Macrophage Autophagy did not Affect Hepatic Inflammatory Cell Number
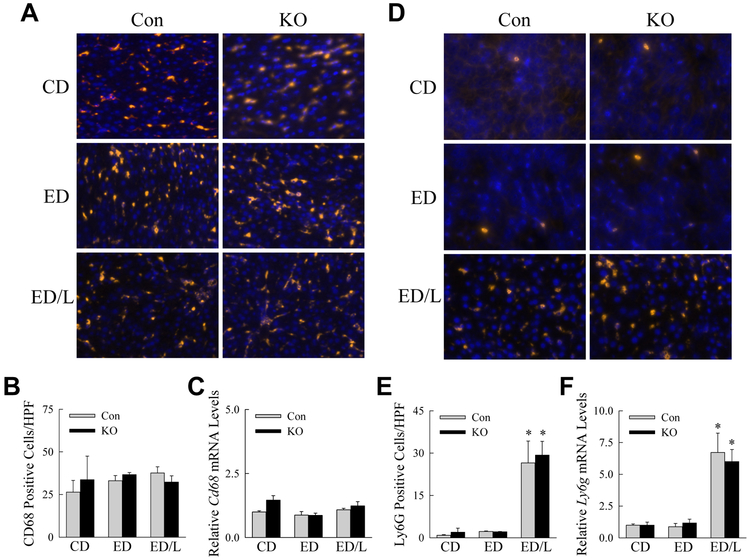
To determine whether systemic inflammation in knockout mice was due to increased numbers of hepatic inflammatory cells, livers were examined for the effects of decreased autophagy on the relative numbers of hepatic macrophages and neutrophils. Immunofluorescence staining for the macrophage marker CD68 demonstrated that macrophage number was unaffected by ED feeding or LPS administration and equivalent in control and knockout mice (Fig. 5A,B). Cd68 mRNA levels in the livers were also unchanged among the groups (Fig. 5C). In contrast, low numbers of hepatic neutrophils were present in control and ED-fed mice, but a marked neutrophil infiltration occurred with LPS administration (Fig. 5D,E). The decrease in macrophage autophagy did not alter the extent of neutrophil recruitment by immunofluorescence (Fig. 5D,E), or the induction of hepatic Ly6g mRNA by LPS (Fig. 5F). The liver injury and systemic inflammation in ED-fed knockout mice was not secondary to an increase in the numbers of hepatic inflammatory cells.
Fig. 5.
Numbers of hepatic macrophages and neutrophils are unaffected by a decrease in autophagy. (A) Immunofluorescence images of liver sections stained for CD68 from control (Con) and knockout (KO) mice that received the indicated treatments (400X). (B) Numbers of macrophages by counts of CD68-positive cells per high power field (HPF; n=3-5). (C) Hepatic mRNA levels for Cd68 (n=5-10). (D) Immunofluorescence images of the same livers for Ly6G. (E) Quantification of the numbers of LyG6-positive neutrophils (*P<0.04 as compared to Con CD; n=3-5). (F) Liver Ly6g mRNA levels (*P<0.005 as compared to Con CD; n=5-10).
ED-fed Atg5Δmye Mice have an Increased Hepatic Proinflammatory Response to LPS
To determine the effects of the loss of Atg5-dependent macrophage autophagy specifically on liver inflammation, hepatic cytokine gene induction was examined by qRT-PCR. Cytokine mRNA expression was significantly increased in both control and in Atg5Δmye mice only with ED/L treatment (Fig. 6A-D). Tnf mRNA induction was equivalent in control and knockout mice (Fig. 6A), however, the other proinflammatory cytokines Il1b, Il1a and Ifng were significantly increased in knockout mice as compared to control mice (Fig. 6B-D).
Fig. 6.
Atg5Δmye mice have increased hepatic proinflammatory gene expression with ED/L treatment. Hepatic mRNA levels for the proinflammatory genes (A) Tnf, (B) Il1b, (C) Il1a, and (D) Ifng (*P<0.00001, as compared to Con CD; #P<0.002 as compared to Con ED/L; n=5-12). White adipose tissue mRNA levels for (E) Tnf, (F) Il1b, and (G) Il1a (*P< 0.01 as compared to Con CD; n=4-8).
Adipose tissue has been implicated as a source of proinflammatory cytokines in alcoholic liver disease in mice (Lin et al., 1998; Sebastian et al., 2011) and humans (Naveau et al., 2010; Voican et al., 2015). Epididymal white adipose tissue was therefore also examined for the effects of decreased macrophage autophagy on cytokine production. ED/L induced adipose proinflammatory cytokine genes but the effects were equivalent in control and knockout mice (Fig. 6E-G). Macrophage sensitization to a more proinflammatory phenotype from ED/L is therefore specific to hepatic macrophages.
Increased Inflammasome Activation Leads to Hepatic Injury in Knockout Mice
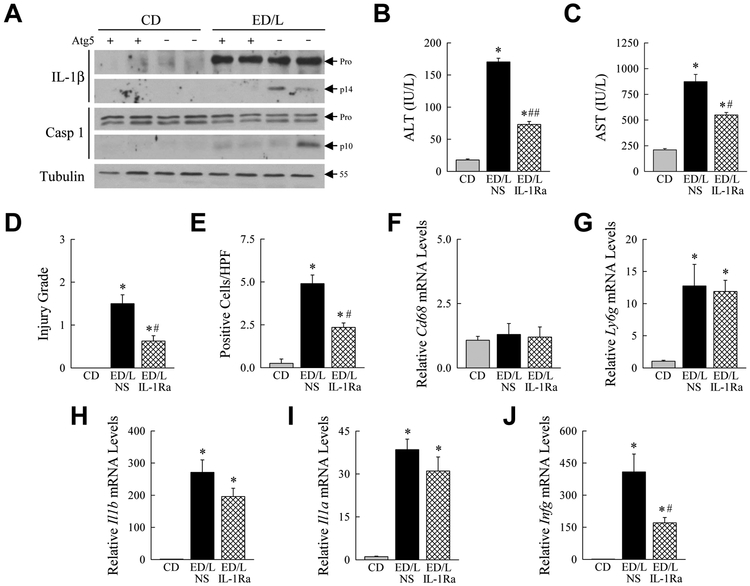
In response to inflammatory stimuli, macrophage IL-1β is generated by increased gene transcription and translation followed by cleavage of pro-IL-1β to its active secreted form by caspase 1 in the intracellular structure the inflammasome (Kubes and Mehal, 2012). Knockout mice had increased Il1b gene induction but the increase in serum IL-1β protein suggested that autophagy-deficient hepatic macrophages had increased inflammasome activation as well. Immunoblots of total liver protein confirmed that control and knockout mice had similar increases in pro-IL-1β protein in response to ED/L (Fig. 7A). However, knockout mouse livers had a marked increase in the amount of active, cleaved IL-1β (Fig. 7A), confirming that the liver is a source of increased serum IL-1β due to increased inflammasome activation. Although levels were variable, some ED/L-treated knockout livers had detectable increased cleaved caspase 1 (Fig. 7A), consistent with elevated inflammasome activation.
Fig. 7.
Liver injury in knockout mice is caused by inflammasome generated IL-1β. (A) Western blots of total liver protein from control (Atg5 +) and knockout (Atg5 -) mice administered CD or ED/L probed for IL-1β, caspase 1 and tubulin. The two panels for IL-1β and caspase 1 represent shorter (top panels) and longer (bottom panels) exposures of the same immunoblot. Arrows indicate pro-IL-1β and pro-caspase 1 (Pro), cleaved IL-1β (p14) and caspase 1 (p10), and the molecular weight of tubulin. Serum (B) ALT and (C) AST levels in Atg5Δmye mice administered CD or ED/L and pretreated with normal saline vehicle (NS) or IL-1Ra (*P<0.001 as compared to CD mice; #P<0.001, ##P<0.000001 as compared to NS-injected mice; n=4). (D) Histological grade of liver injury in the mice (*P<0.02 as compared to CD mice; #P<0.004 as compared to NS-injected mice; n=4). (E) Numbers of TUNEL-positive cells in the same livers (*P<0.006 as compared to CD mice; #P<0.002 as compared to NS-injected mice; n=4). mRNA levels in total liver for Cd68 (F) and Ly6g (G) (*P<0.001 as compared to CD mice; n=4). Liver mRNA levels for Il1b (H), Il1a (I), and Ifng (J) (*P<0.001 as compared to CD mice; #P<0.02 as compared to NS-injected mice; n=4).
Inflammasome-generated IL-1β has been shown to promote liver injury in the setting of a fatty liver (Miura et al., 2010; Petrasek et al., 2012). We therefore examined whether IL-1β generated by deficient macrophage autophagy promoted increased liver injury in knockout mice. Mice were administered an IL-1Ra that competes with IL-1β and IL-1α for cellular receptor binding to block IL-1 signaling (Dinarello, 2009). IL-1Ra administration reduced ED/L-induced liver injury in knockout mice as reflected in significantly decreased serum ALT and AST levels (Fig. 7B,C), histological injury score (Fig. 7D), and numbers of TUNEL-positive cells (Fig. 7E). Blocking IL-1 had no effect on macrophage or neutrophil numbers as determined by mRNA levels for Cd68 (Fig. 7F), and Ly6g (Fig. 7G), respectively. Examination of hepatic cytokine gene expression revealed a trend to decreased inflammation with a 20-60% decrease in proinflammatory gene mRNA levels, although only the decrease in Ifng was statistically significant (Fig. 7H,I,J). Impaired macrophage autophagy therefore promoted liver injury in the setting of ED feeding by amplifying inflammasome production of IL-1β that injured the liver.
DISCUSSION
Due in part to the difficulties in measuring autophagic function in vivo, and the differences among cell culture and animal models that have been studied, a number of different effects of alcohol on autophagy have been reported (Li et al., 2014). In models of acute binge alcohol administration, autophagy is increased in the liver (Ding et al., 2010; Thomes et al., 2015). However, with chronic ethanol administration as occurs in alcoholic liver disease autophagic function is decreased (Thomes et al., 2015). Delineating the consequences of decreased autophagy in alcoholic liver disease is therefore important to understanding the pathophysiology of this disease. Previous studies have focused on the effects of deficient autophagy in hepatocytes on alcoholic liver disease (Ding et al., 2010). Activation of the innate immune response is critical in the development of this disease, but the role of macrophage autophagy in alcoholic liver disease has not been investigated. The findings of this study demonstrate that macrophage autophagy regulates liver injury in this disease in part though the down regulation of proinflammatory macrophage activation.
Macrophage autophagy was protective against liver injury from ethanol alone as knockout mice had increased mortality and liver transaminases when fed an ED. These effects cannot be attributed to an increased innate immune response, as elevated inflammation was not detected in the surviving knockout mice fed an ED alone. It is possible that the deaths occurred in individual mice who mounted an increased innate immune response, as the inflammatory state in these mice could not be examined. Alternatively decreased autophagy could have promoted death from other causes such as infection, although this etiology seems unlikely in a sterile animal facility.
Studies of ED and LPS co-administration were conducted because of the known importance of LPS-induced inflammation in alcoholic liver disease, and the failure of the Lieber-DeCarli diet to induce significant inflammation as seen in human alcoholic liver disease. Deficient macrophage autophagy markedly increased liver injury and cell death in association with an increased hepatic and systemic proinflammatory innate immune response. The mechanism of this effect was not an increase in hepatic inflammatory cell number. Rather there was an increased proinflammatory cytokine response with no change in anti-inflammatory cytokine induction. Both inflammasome dependent and independent cytokines were increased in knockout mice indicating multiple mechanisms by which decreased autophagy increased inflammation. These findings are consistent with our previous findings that autophagy can down regulate proinflammatory M1 macrophage polarization or inflammasome activation in hepatic macrophages (Liu et al., 2015; Ilyas et al., 2016). Unlike other models of liver injury, with ED feeding both proinflammatory mechanisms drive an over active immune response. The proinflammatory effect of impaired autophagy promoted alcoholic liver disease as inhibition of the biological effects of the proinflammatory cytokine IL-1 that was elevated in knockout mice decreased liver injury.
With decreased macrophage autophagy there was increased systemic as well as hepatic inflammation with ED/L administration. Recent clinical studies in human alcoholic liver disease have demonstrated the important contribution of systemic inflammation to decompensation and poor survival in chronic liver disease including that from alcohol (Claria et al., 2016). Patients with alcoholic liver disease have increased morbidity and mortality from sepsis (Gustot et al., 2017). Decreased macrophage autophagy may be a mechanism of both increased systemic inflammation and sensitivity to sepsis in alcoholics. Interestingly the effects of the decrease in macrophage autophagy were specific for hepatic macrophages and did not occur in adipose macrophages. This finding may reflect the collective effects of hepatic ethanol metabolism and the decrease in macrophage autophagy that occurs in liver but not fat tissue. However, in murine studies of the physiological decrease in autophagy induced by aging an increase in inflammation also occurred specifically in the liver and not white adipose tissue (Fontana et al., 2013). There may therefore exist undefined differences in the function of autophagy in hepatic versus adipose macrophages.
Factors other than alcohol itself may decrease hepatic macrophage autophagy and promote the development of alcoholic liver disease. Aging is associated with reduced levels of autophagy in the liver (Rubinsztein et al., 2011), and specifically in macrophages (Stranks et al., 2015). Diet-induced and genetic obesity decreases autophagic function in the liver (Singh et al., 2009; Yang et al., 2010). High fat diet feeding decreases autophagy in macrophages (Liu et al., 2015), and promotes proinflammatory macrophage activation in alcohol-fed mice (Xu et al., 2011). Interestingly, aging (Raynard et al., 2002; Wang et al., 2014) and obesity (Naveau et al., 1997; Ruhl and Everhart, 2005; Hart et al., 2010) are recognized cofactors that promote the development of alcoholic liver disease. Our findings suggest that the mechanism by which aging and obesity promote alcoholic liver disease may in part be due to the mechanism of decreased macrophage autophagy.
Human alcoholic liver disease currently lacks any proven therapy. Studies of the function of autophagy in hepatocytes have indicated that with chronic alcohol ingestion hepatocyte autophagy is decreased and that increasing autophagy ameliorates disease (Ding et al., 2010). Our findings suggest that an alternative mechanism by which therapies to augment autophagy may be effective in this disease is through increasing macrophage autophagy to down regulate injurious inflammation. Inducers of autophagy that have successfully treated murine alcoholic liver disease also decreased inflammation (Lin et al., 2013), and our findings suggest that this effect may have resulted in part from the beneficial induction of autophagy in macrophages. That a single agent might have the dual benefit of both decreasing cellular injury and down regulating inflammation in this disease suggests that the strategy of inducing autophagy could be a very effective approach to the treatment of alcoholic liver disease.
ACKNOWLEDGEMENT
This work was supported in part by NIH grant R01AA022601 (MJC). We thank Rebecca A. Czaja for the statistical analysis.
Footnotes
CONFLICT OF INTEREST
The authors have nothing to disclose.
Contributor Information
Ghulam Ilyas, Division of Digestive Diseases, Department of Medicine, Emory University School of Medicine, Atlanta, GA.
Francesca Cingolani, Division of Digestive Diseases, Department of Medicine, Emory University School of Medicine, Atlanta, GA.
Enpeng Zhao, Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, NY.
Kathryn Tanaka, Department of Pathology, Albert Einstein College of Medicine, Bronx, NY.
Mark J. Czaja, Division of Digestive Diseases, Department of Medicine, Emory University School of Medicine, Atlanta, GA.
REFERENCES
- Adams DH, Ju C, Ramaiah SK, Uetrecht J, Jaeschke H (2010) Mechanisms of immune-mediated liver injury. Toxicol Sci 115:307–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claria J, Stauber RE, Coenraad MJ, Moreau R, Jalan R, Pavesi M, Amoros A, Titos E, Alcaraz-Quiles J, Oettl K, Morales-Ruiz M, Angeli P, Domenicali M, Alessandria C, Gerbes A, Wendon J, Nevens F, Trebicka J, Laleman W, Saliba F, Welzel TM, Albillos A, Gustot T, Benten D, Durand F, Gines P, Bernardi M, Arroyo V, Consortium CSIOTE-C, The European Foundation for the Study of Chronic Liver F (2016) Systemic inflammation in decompensated cirrhosis: characterization and role in acute-on-chronic liver failure. Hepatology 64:1249–1264. [DOI] [PubMed] [Google Scholar]
- Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I (1999) Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res 8:265–277. [DOI] [PubMed] [Google Scholar]
- Czaja MJ (2011) Functions of autophagy in hepatic and pancreatic physiology and disease. Gastroenterology 140:1895–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaja MJ, Ding WX, Donohue TM, Friedman SL, Kim JS, Komatsu M, Lemasters JJ, Lemoine A, Lin JD, Ou JH, Perlmutter DH, Randall G, Ray RB, Tsung A, Yin XM (2013) Functions of autophagy in normal and diseased liver. Autophagy 9:1131–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Saitoh T, Akira S (2013) Autophagy in infection, inflammation and immunity. Nat Rev Immunol 13:722–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello CA (2009) Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol 27:519–550. [DOI] [PubMed] [Google Scholar]
- Ding WX, Li M, Chen X, Ni HM, Lin CW, Gao W, Lu B, Stolz DB, Clemens DL, Yin XM (2010) Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology 139:1740–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Zhao E, Amir M, Dong H, Tanaka K, Czaja MJ (2013) Aging promotes the development of diet-induced murine steatohepatitis but not steatosis. Hepatology 57:995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustot T, Fernandez J, Szabo G, Albillos A, Louvet A, Jalan R, Moreau R, Moreno C (2017) Sepsis in alcohol-related liver disease. J Hepatol 67:1031–1050. [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441:885–889. [DOI] [PubMed] [Google Scholar]
- Hart CL, Morrison DS, Batty GD, Mitchell RJ, Davey Smith G (2010) Effect of body mass index and alcohol consumption on liver disease: analysis of data from two prospective cohort studies. BMJ 340:c1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iimuro Y, Gallucci RM, Luster MI, Kono H, Thurman RG (1997) Antibodies to tumor necrosis factor alfa attenuate hepatic necrosis and inflammation caused by chronic exposure to ethanol in the rat. Hepatology 26:1530–1537. [DOI] [PubMed] [Google Scholar]
- Ilyas G, Zhao E, Liu K, Lin Y, Tesfa L, Tanaka KE, Czaja MJ (2016) Macrophage autophagy limits acute toxic liver injury in mice through down regulation of interleukin-1β. J Hepatol 64:118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenkel O, Tacke F (2017) Liver macrophages in tissue homeostasis and disease. Nat Rev Immunol 17:306–321. [DOI] [PubMed] [Google Scholar]
- Kubes P, Mehal WZ (2012) Sterile inflammation in the liver. Gastroenterology 143:1158–1172. [DOI] [PubMed] [Google Scholar]
- Lalazar G, Ilyas G, Malik SA, Liu K, Zhao E, Amir M, Lin Y, Tanaka KE, Czaja MJ (2016) Autophagy confers resistance to lipopolysaccharide-induced mouse hepatocyte injury. Am J Physiol Gastrointest Liver Physiol 311:G377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW (2011) Autophagy in immunity and inflammation. Nature 469:323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang S, Ni HM, Huang H, Ding WX (2014) Autophagy in alcohol-induced multiorgan injury: mechanisms and potential therapeutic targets. Biomed Res Int 2014:498491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CW, Zhang H, Li M, Xiong X, Chen X, Chen X, Dong XC, Yin XM (2013) Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. J Hepatol 58:993–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HZ, Yang SQ, Zeldin G, Diehl AM (1998) Chronic ethanol consumption induces the production of tumor necrosis factor-alpha and related cytokines in liver and adipose tissue. Alcohol Clin Exp Res 22:231S–237S. [DOI] [PubMed] [Google Scholar]
- Liu K, Zhao E, Ilyas G, Lalazar G, Lin Y, Haseeb M, Tanaka KE, Czaja MJ (2015) Impaired macrophage autophagy increases the immune response in obese mice by promoting proinflammatory macrophage polarization. Autophagy 11:271–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder J, Denaes T, Chobert MN, Wan J, El-Benna J, Pawlotsky JM, Lotersztajn S, Teixeira-Clerc F (2015) Macrophage autophagy protects against liver fibrosis in mice. Autophagy 11:1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher JJ, Leon P, Ryan JC (2008) Beyond insulin resistance: innate immunity in nonalcoholic steatohepatitis. Hepatology 48:670–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Kodama Y, Inokuchi S, Schnabl B, Aoyama T, Ohnishi H, Olefsky JM, Brenner DA, Seki E (2010) Toll-like receptor 9 promotes steatohepatitis by induction of interleukin-1β in mice. Gastroenterology 139:323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveau S, Cassard-Doulcier AM, Njike-Nakseu M, Bouchet-Delbos L, Barri-Ova N, Boujedidi H, Dauvois B, Balian A, Maitre S, Prevot S, Dagher I, Agostini H, Grangeot-Keros L, Emilie D, Perlemuter G (2010) Harmful effect of adipose tissue on liver lesions in patients with alcoholic liver disease. J Hepatol 52:895–902. [DOI] [PubMed] [Google Scholar]
- Naveau S, Giraud V, Borotto E, Aubert A, Capron F, Chaput JC (1997) Excess weight risk factor for alcoholic liver disease. Hepatology 25:108–111. [DOI] [PubMed] [Google Scholar]
- Petrasek J, Bala S, Csak T, Lippai D, Kodys K, Menashy V, Barrieau M, Min SY, Kurt-Jones EA, Szabo G (2012) IL-1 receptor antagonist ameliorates inflammasome-dependent alcoholic steatohepatitis in mice. J Clin Invest 122:3476–3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynard B, Balian A, Fallik D, Capron F, Bedossa P, Chaput JC, Naveau S (2002) Risk factors of fibrosis in alcohol-induced liver disease. Hepatology 35:635–638. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Marino G, Kroemer G (2011) Autophagy and aging. Cell 146:682–695. [DOI] [PubMed] [Google Scholar]
- Ruhl CE, Everhart JE (2005) Joint effects of body weight and alcohol on elevated serum alanine aminotransferase in the United States population. Clin Gastroenterol Hepatol 3:1260–1268. [DOI] [PubMed] [Google Scholar]
- Sebastian BM, Roychowdhury S, Tang H, Hillian AD, Feldstein AE, Stahl GL, Takahashi K, Nagy LE (2011) Identification of a cytochrome P4502E1/Bid/C1q-dependent axis mediating inflammation in adipose tissue after chronic ethanol feeding to mice. J Biol Chem 286:35989–35997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ (2009) Autophagy regulates lipid metabolism. Nature 458:1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranks AJ, Hansen AL, Panse I, Mortensen M, Ferguson DJ, Puleston DJ, Shenderov K, Watson AS, Veldhoen M, Phadwal K, Cerundolo V, Simon AK (2015) Autophagy controls acquisition of aging features in macrophages. J Innate Immun 7:375–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo G, Petrasek J, Bala S (2012) Innate immunity and alcoholic liver disease. Dig Dis 30 Suppl 1:55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomes PG, Trambly CS, Fox HS, Tuma DJ, Donohue TM Jr. (2015) Acute and chronic ethanol administration differentially modulate hepatic autophagy and transcription factor EB. Alcohol Clin Exp Res 39:2354–2363. [DOI] [PubMed] [Google Scholar]
- Uesugi T, Froh M, Arteel GE, Bradford BU, Thurman RG (2001) Toll-like receptor 4 is involved in the mechanism of early alcohol-induced liver injury in mice. Hepatology 34:101–108. [DOI] [PubMed] [Google Scholar]
- Voican CS, Njike-Nakseu M, Boujedidi H, Barri-Ova N, Bouchet-Delbos L, Agostini H, Maitre S, Prevot S, Cassard-Doulcier AM, Naveau S, Perlemuter G (2015) Alcohol withdrawal alleviates adipose tissue inflammation in patients with alcoholic liver disease. Liver Int 35:967–978. [DOI] [PubMed] [Google Scholar]
- Wang H, Ma L, Yin Q, Zhang X, Zhang C (2014) Prevalence of alcoholic liver disease and its association with socioeconomic status in north-eastern China. Alcohol Clin Exp Res 38:1035–1041. [DOI] [PubMed] [Google Scholar]
- Wang HJ, Gao B, Zakhari S, Nagy LE (2012) Inflammation in alcoholic liver disease. Annu Rev Nutr 32:343–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Schattenberg JM, Rigoli RM, Storz P, Czaja MJ (2004) Hepatocyte resistance to oxidative stress is dependent on protein kinase C-mediated down-regulation of c-Jun/AP-1. J Biol Chem 279:31089–31097. [DOI] [PubMed] [Google Scholar]
- Wang Y, Singh R, Lefkowitch JH, Rigoli RM, Czaja MJ (2006) Tumor necrosis factor-induced toxic liver injury results from JNK2-dependent activation of caspase-8 and the mitochondrial death pathway. J Biol Chem 281:15258–15267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Lai KKY, Verlinsky A, Lugea A, French SW, Cooper MP, Ji C, Tsukamoto H (2011) Synergistic steatohepatitis by moderate obesity and alcohol in mice despite increased adiponectin and p-AMPK. J Hepatol 55:673–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Li P, Fu S, Calay ES, Hotamisligil GS (2010) Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab 11:467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin M, Wheeler MD, Kono H, Bradford BU, Gallucci RM, Luster MI, Thurman RG (1999) Essential role of tumor necrosis factor alpha in alcohol-induced liver injury in mice. Gastroenterology 117:942–952. [DOI] [PubMed] [Google Scholar]