Abstract
We previously reported that ionizing radiation (IR) mediates cell death through the induction of CUGBP elav-like family member 2 (CELF2), a tumor suppressor. CELF2 is an RNA binding protein that modulates mRNA stability and translation. Since IR induces autophagy, we hypothesized that CELF2 regulates autophagy-mediated colorectal cancer (CRC) cell death. For clinical relevance, we determined CELF2 levels in The Cancer Genome Atlas (TCGA). Role of CELF2 in radiation response were carried out in CRC cell lines by immunoblotting, immunofluorescence, autophagic vacuole analyses, RNA stability assay, quantitative PCR and electron microscopy. In vivo studies were performed in a xenograft tumor model. TCGA analyses demonstrated that compared to normal tissue, CELF2 is expressed at significantly lower levels in CRC, and is associated with better overall five-year survival in patients receiving radiation. Mechanistically, CELF2 increased levels of critical components of the autophagy cascade including Beclin-1, ATG5 and ATG12 by modulating mRNA stability. CELF2 also increased autophagic flux in CRC. IR significantly induced autophagy in CRC which correlates with increased levels of CELF2 and autophagy associated proteins. Silencing CELF2 with siRNA, mitigated IR induced autophagy. Moreover, knockdown of CELF2 in vivo conferred tumor resistance to IR. These studies elucidate an unrecognized role for CELF2 in inducing autophagy and potentiating the effects of radiotherapy in CRC.
Keywords: RNA binding protein, Colorectal Cancer, Autophagy, Radiotherapy
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer. The incidence of those diagnosed under the age of 50 has been steadily rising by 1.8% each year1. Surgical resection, followed by adjuvant chemotherapy and radiotherapy is the primary mode of treatment for advanced stages of CRC. A recent report showed that neoadjuvant radiation had a survival benefit in patients over 502; yet, there was no increase in survival rates in younger patients. Understanding the biology of radiotherapy will improve survivorship in patients with CRC metastases.
Ionizing radiation (IR) induces cancer cell death through several mechanisms including apoptosis and autophagy. We previously reported that prostaglandin E2 facilitates CRC resistance to IR by suppressing the expression of RNA binding protein CELF2 (CUG binding protein, elav-like family member 2)3. RNA binding proteins play a major role in the post-transcriptional regulation of the DNA damage response4. We previously reported that CELF2 expression is significantly reduced in colorectal cancers5. Ectopic expression of CELF2 in CRC cells induces cell death and potentiates the effects of chemotherapy6,7. IR induces CELF2 expression, resulting in cell death by suppressing translation of cyclooxygenase-2 (COX-2) mRNA8.
We hypothesized that CELF2 plays a role in IR-induced autophagic cell death. First, we determined the clinical relevance of CELF2 expression across multiple cancer types using the TCGA datasets. We demonstrate that an increase in CELF2 levels, through ectopic expression or IR exposure, is associated with increased stability and expression of various autophagy-associated proteins. These studies demonstrate a unique role for RNA binding protein CELF2 in facilitating cell death through the regulation of cellular autophagy.
Materials and Methods
Cells and Reagents
CRC lines HCT116, and SW480, obtained from ATCC (Manassas, VA), and were authenticated using a STR-based method. Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Corning, Corning, NY) with 10% heat-inactivated FBS (Sigma-Aldrich, St. Louis, MO) without antibiotics at 37 °C in the presence of 5% CO2. Chloroquine diphosphate salt was obtained from Sigma Aldrich (St. Louis, MO). Primary antibodies targeting CELF2 (AV40324), and Atg12 (#4180), Beclin-1 (#4122), Atg5 (#12994) were obtained from Sigma-Aldrich (St. Louis, MO) and Cell Signaling (Danvers, MA), respectively. Secondary Anti-rabbit IgG Dylight 488 (#35553), used for immunofluorescence, and Hoescht 33342 nuclear counter stain were obtained from ThermoFisher (Waltham, MA).
In vitro knockdown of CELF2 was conducted using three pooled siRNA transcripts obtained from Santa Cruz Biotechnology, (CELF2: sc-44554; Control: sc-44236, Santa Cruz Biotechnology, Dallas, TX). The full-length coding region of human CELF2 was amplified by RT-PCR from HCT-116 cells and expressed as amino-terminal FLAG epitope-tagged proteins in plasmid pCMV-Tag2B (a cytomegalovirus immediate early promoter driven expression vector) after cloning at the HindIII and XhoI restriction sites.
2.2. Analysis of TCGA Datasets
Data from the CRC patients was mined from the publicly available TCGA cohort colorectal adenocarcinoma (COADREAD) samples report. Clinical and CELF2 mRNA expression data (polyA + IlluminaHiSeq) for TCGA solid tumors from patients with known radiotherapy status were downloaded from the UCSC Xena Browser website (https://xenabrowser.net/heatmap/). All genomic data analyzed herein were generated by the TCGA Research Network (http://cancergenome.nih.gov/). We analyzed data for CELF2 expression data from 361 CRC specimens and 51 site matched normal tissue.
In vitro assays
For IR experiments, cells were plated in a 10 cm2 dish (1×106 cells) and were irradiated using a J.L. Shepherd and Associates Mark I Model 68A cesium-137 source irradiator (dose rate of 2.9 Gy/min). Colony formation studies, immunofluoresence and immunoblotting were carried out as previously described25.
RNA stability assays were performed for autophagy related proteins Beclin-1, ATG5 and ATG12 in CELF2 overexpressing or parental cells. Stability of mRNA was assessed following addition of actinomycin D (10 μg/ml), a potent inhibitor of mRNA synthesis. After 0–8 h of actinomycin D treatment, total mRNA was extracted and subjected to quantitative PCR. Data are presented relative to control cells at the time of addition of actinomycin D.
Autophagic vacuole analyses with monodansylcadaverine (MDC)
Following induction of autophagy by amino acid starvation, the cells were incubated with 0.05 mM MDC in PBS at 37°C for 10 minutes9. After incubation, cells were washed four times with PBS and collected in 10 mM Tris-HCl, pH 8 containing 0.1% Triton X-100. Intracellular MDC was measured (excitation wavelength 380 nm, emission filter 525 nm) in a Packard Fluorocount microplate reader. To normalize the measurements to the number of cells present in each well, a solution of ethidium bromide was added to a final concentration of 0.2 mM and the DNA fluorescence was measured (excitation wavelength 530 nm, emission filter 590 nm). The MDC incorporated was expressed as specific activity (arbitrary units).
RNA Immunoprecipitation
For immunoprecipitation, cell lysates were harvested in a 1% formaldehyde solution, and prepared by sonication. CELF2 antibody (Sigma) was used to immunoprecipitate CELF2 protein. Subsequently, the pellet and supernatant were incubated at 70°C for 1 hour to reverse the cross-links. Total RNA was isolated from the precipitate and supernatant, and subjected to RT-PCR analyses10.
RNA analyses using RT-PCR and quantitative PCR
Cells were plated in 10 cm2 dish (1 × 106 cells) and subjected to IR or treatment conditions. RNA was extracted from cells using TRIzol reagent (Fisher Scientific), per the manufacturer’s instructions. For quantitative PCR, reverse transcription using SuperScript II with random hexonucleotide primers (Life Technologies, Carlsbad, CA) was used. cDNA was amplified by using Jumpstart Taq polymerase (Sigma-Aldrich) and SYBR Green nucleic acid stain (Life Technologies). Threshold crossing values for each gene were normalized to β-actin gene expression.
Electron Microscopy
Cells were fixed in 2.5% glutaraldehyde in 0.1M sodium cacodylate buffer (pH 7.4) for 1 h at room temperature, then washed 3 times in cacodylate buffer. Cells were then fixed in 1% osmium tetroxide in 0.1M cacodylate buffer for 1 h at room temperature. Following this, 70 nm sections were cut on a ‘Reichert Ultracut S’ ultramicrotome. The sections were subsequently stained with 4% uranyl acetate for 10 min and Reynald’s lead citrate for 1.5 min. Sections were imaged at 80 kV on a JEOL 1200 EX transmission electron microscope.
Tumor Xenograft
Animal studies were performed in accordance with institutional guidelines by the University of Kansas Medical Center under an Institutional Animal Care and Use Committee approved protocol. The right flank of athymic male mice were injected with HCT116, 1 × 106 cells. Tumor bearing mice were irradiated at a dose of 2 Gy on days 13, 14, 15, 16, and 17. Both scrambled and siCELF2 were incorporated into 1,2-dioleoyl-sn-Glycero-3-Phosphocholine (DOPC) lipid capsules (Avanti Polar Lipids). Packaged siRNA was delivered by intratumoral injection (5 μM, 50 μL injection) on days 13, 14, 15, 16, and 17 (n=5/group). Tumor volume was measured with a vernier caliper, and using the formula, length (longer dimension) x width2 × 0.5 mm3. After the experiment, tumors were excised, weighed and processed for immunoblot.
Statistical Analyses
For TCGA datasets, analyses were carried out using R version 3.2.3 (9) and the R packages car, Rmisc, survival, and survminer, and graphs were made using ggplot2. Welch’s two-sample t-test was used to compare CRC data to normal, and one-way analysis of variance (ANOVA) and Tukey’s post hoc test was used to compare CELF2 expression in CRC stages to normal tissues. Cox Proportional Hazards was used in analysis of overall and recurrence free survival.
Statistics calculations were performed using Graphpad Prism Software (v. 6.03). Student t-tests are used to calculate statistical significance between treatment groups, and data are reported as mean ± standard error of the mean (SEM), unless otherwise noted. Non-parametric, Mann-Whitney test was used to assess the significance in tumor volumes and weights, and biomarker determinations. Statistical significance between test groups determined by p<0.05. All experiments were validated by two or more biological repeats.
Results
Increased CELF2 expression associates with better prognosis in patients undergoing radiotherapy.
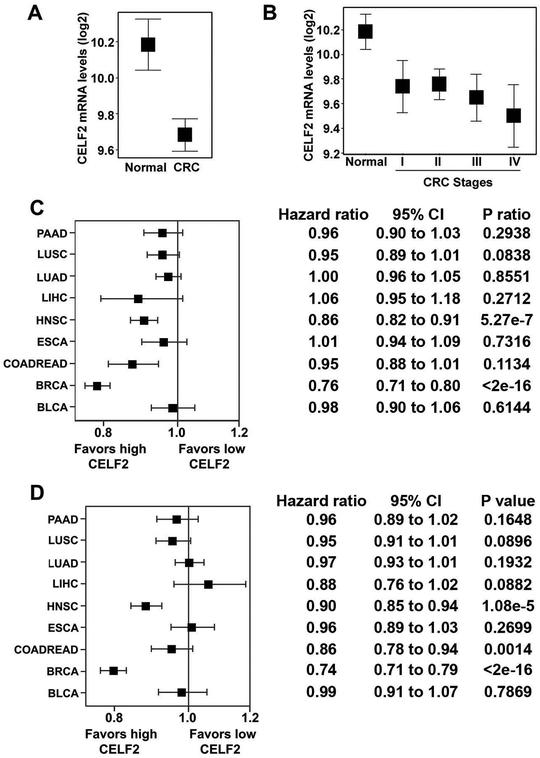
To assess alterations in CELF2 mRNA expression during CRC progression, we probed The Cancer Genome Atlas (TCGA) gene expression data from 410 CRC patients and 51 site-matched normal tissue specimens. Compared to normal tissue, CRC tumors expressed significantly less CELF2 (1.5 fold-decrease, p=2.036×10−9, Fig 1A); however, no significant differences were observed in CELF2 levels between different pathological stages (p>0.05 by Games-Howell Post-hoc Test; Fig 1B). We next wanted to assess whether CELF2 correlated with better outcome in patients undergoing radiotherapy. Since the CRC cohort contained only 31 radiation-treated patients with CELF2 mRNA expression data, we included additional TCGA solid tumor cohorts with gene expression and radiation exposure data, which increased total patient number to 4498. We determined the hazard ratios of five-year overall survival and disease recurrence in radiation patients according to CELF2 mRNA expression in many solid cancers (Fig 1C–D). Increased CELF2 expression was associated with better overall survival over five-years in TCGA cohorts of colorectal cancer (CRC), breast cancer (BRCA), and head and neck cancer (HNSC) patients receiving radiation. Interestingly, while increased expression did not correlate with better recurrence free survival for radiation treated CRC patients, BRCA and HNSC recurred less frequently following radiation when CELF2 expression increased.
Fig. 1. CELF2 is down-regulated in CRC and correlates with better prognosis.
(A) TCGA analyses of CELF2 levels determined by RNAseq of tissue. n=51 cancer-free normal and n=361 CRC patients. CELF2 expression is down-regulated in CRC as compared to normal colorectal tissue (p = 4.263e-8).
(B) CELF2 expression decreases by stage in CRC. At later stages of CRC, there is a greater difference in CELF2 expression compared to normal tissue than at earlier stages (Stage IV to normal, p=0.0006144, n=61; Stage III to normal, p=0.0033, n=123; Stage II to normal, p=0.0252, n=161; Stage I to normal, p=0.0587, n=65; n=51 cancer-free normal).
(C) Increased CELF2 expression associated with better 5-year overall survival in bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), CRC (COADREAD), esophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), and pancreatic adenocarcinoma (PAAD).
(D) Disease recurrence across multiple TCGA cohorts of patients having undergone radiotherapy. While increased expression did not associate with better recurrence free survival for radiation treated CRC patients, breast and head and neck cancer recurred less frequently following radiation when CELF2 mRNA expression was increased. All error bars shown are 95% confidence intervals.
CELF2 Induces Autophagy in CRC
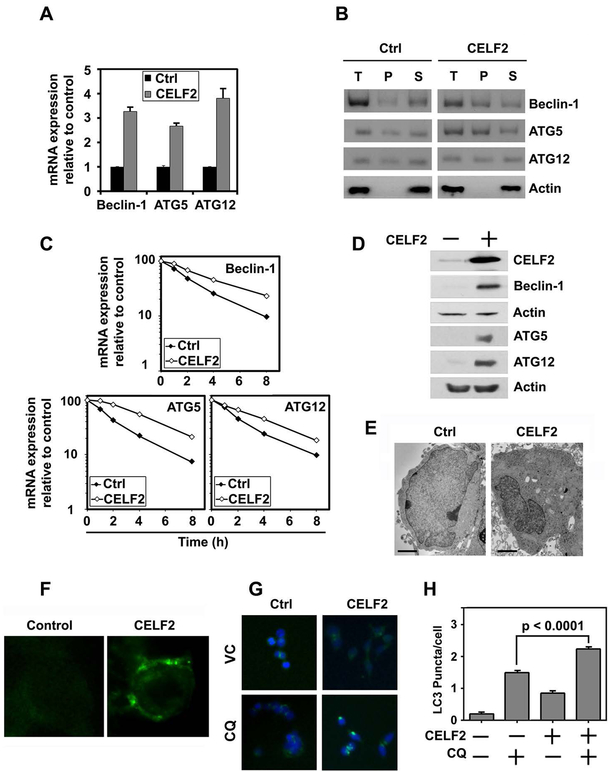
Given the role of CELF2 in reducing cancer mortality, we sought to understand whether CELF2 affects autophagy. Indeed, when we overexpressed CELF2 in HCT116 cells, which has low basal expression of CELF211, we observed a robust increase in autophagy related proteins Beclin-1 (BECN1), ATG5 and ATG12 mRNAs (Fig 2A). Previously, we have demonstrated that CELF2 is a RNA binding protein that binds to AU-rich sequences in 3’untranslated regions of mRNAs for COX-2 and Mcl-1, and upon binding increases the stability of these mRNAs8,12. Therefore, we determined whether CELF2 binds to the three mRNAs. For this, we performed a RNA-immunoprecipitation-coupled RT-PCR analyses using CELF2 as the bait. There were increased levels of Beclin-1, ATG5 and ATG12 mRNAs in the precipitate of CELF2 overexpressing cells compared to controls suggesting that CELF2 interacts with the mRNA (Fig 2B). Given the increased binding, we next determined the stability of the three mRNAs in response to CELF2 overexpression. Real-time RT-PCR analyses of total mRNA from cells demonstrated that overexpression of CELF2 induced the stability of all three mRNAs (Fig 2C). Half-life of the mRNAs following CELF2 expression was increased by at least two-fold. Previously, we had shown that CELF2 binding to COX-2 and Mcl-1 mRNAs results in suppression of translation. Hence, we performed western blot analyses to determine Beclin-1, ATG5 and ATG12 protein levels. CELF2 overexpression lead to increased expression of all three proteins (Fig 2D). Thus, CELF2 regulates autophagy through the stabilization of mRNA of autophagy-related proteins in CRC cells.
Fig. 2. CELF2 overexpression induces autophagy.
(A) CELF2 overexpression increases expression of BECN1, ATG5, and ATG12. Cumulative data of two biological repeats plated in triplicate.
(B) RNA immunoprecipitation of CELF2 overexpressing cells or vector Control (Ctrl) demonstrates direct interaction of CELF2 with BECN1, ATG5 and ATG12. T=total RNA Lysate, P=CELF2 bound precipitate, S=Supernatant.
(C) CELF2 overexpression increases stability of Beclin-1, ATG5 and ATG12 mRNA transcripts.
(D) CELF2 overexpression induces autophagy associated proteins, Beclin-1, Atg5, and Atg12, in CRC cells. Representative immunoblot demonstrating CELF2 overexpression in HCT116 cells.
(E) CELF2 overexpression induces autophagosomes. Transmission electron microscope image of CELF2 overexpressing (CELF2) HCT116 cells demonstrates increased autophagic puncta in O/E cells. Scale bars represent 2 μm.
(F) Immunofluorescence imaging of monodansyl cadeverin-labeled autophagosome vacuoles in CELF2 overexpressing HCT116 cells.
(G) LC3 puncta are induced in CELF2 overexpressing cells. Immunofluorescence (LC3 puncta are green, and nuclei stained blue with hoescht stain) in wild-type (WT) or CELF2 OE HCT116 treated with or without CQ (80 μM for 2 hours) to inhibit autophagic flux. Images taken at 200x magnification.
(H) LC3 puncta were quantified by blinded observer in at least 30 cells per group, and graphs represent cumulative results from duplicate experiments. Error bars represent ± SEM.
Autophagy is morphologically marked by the appearance of autophagosomes13. By transmission electron microscopy, we observed a robust increase in autophagosomes within the cell (Fig 2E). We further validated these observations by assessing the incorporation of monodansyl cadaverine (MDC), a lysomotropic dye that serves as an autophagy marker9, in the CELF2 overexpressing cells (Fig 2F). Additionally, immunofluorescent visualization of LC3 puncta demonstrates a significant increase in LC3 puncta per cell with CELF2 overexpression (Fig 2G,H).
IR induces autophagy in CRC cells
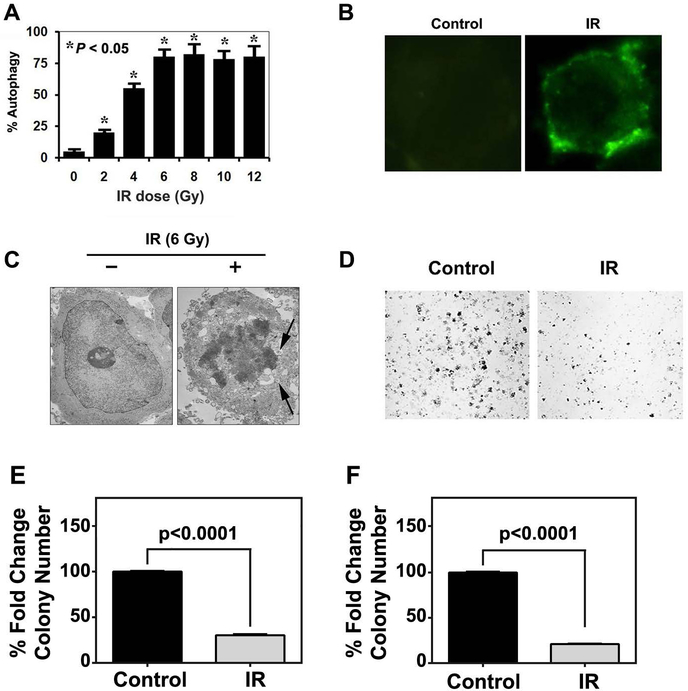
Autophagy mediates IR toxicity by acting as a mechanism of cell death14. We first confirmed that IR induces autophagic cell death in CRC cell lines. We previously determined that 6 Gy induces significant cell death7. We next performed a dose escalation and observed dose-dependent increase in MDC incorporation (Fig 3A). Maximum incorporation was observed at 6 GY IR. A representative image of MDC incorporation is presented in Fig 3B. We further confirmed that this by the presence of autophagosomes by transmission electron microscopy (Fig 3C). To determine the long-term effect of IR-induced autophagy, we performed clonogenicity assays and observed significantly lower number and size of colonies following 6 Gy IR (Fig 3D–F). Together, these data suggest that colon cancer cells undergo autophagic cell death following IR exposure.
Fig. 3. IR induces autophagy.
(A) Quantification of monodansyl cadeverin (MDC)-incorporation following increasing doses of IR treatment (0–12 Gy). There is a dose-dependent increase in MDC incorporation until 6 Gy IR.
(B) Representative Immunofluorescence imaging of monodansyl cadeverin-labeled autophagosome vacuoles in HCT116 following either 0 or 6 Gy IR treatment.
(C) Transmission electron micrograph demonstrating autophagosomes in 6 Gy IR treated HCT116 cells.
(D) Bright field images of HCT116 colonies following either 0 or 6 Gy IR treatment.
(E) Quantification of colony number from HCT116 cells following 0 or 6 IR treatment.
(F) Quantification of colony size from HCT116 cells following 0 or 6 IR treatment.
IR induces CELF2 in CRC cells
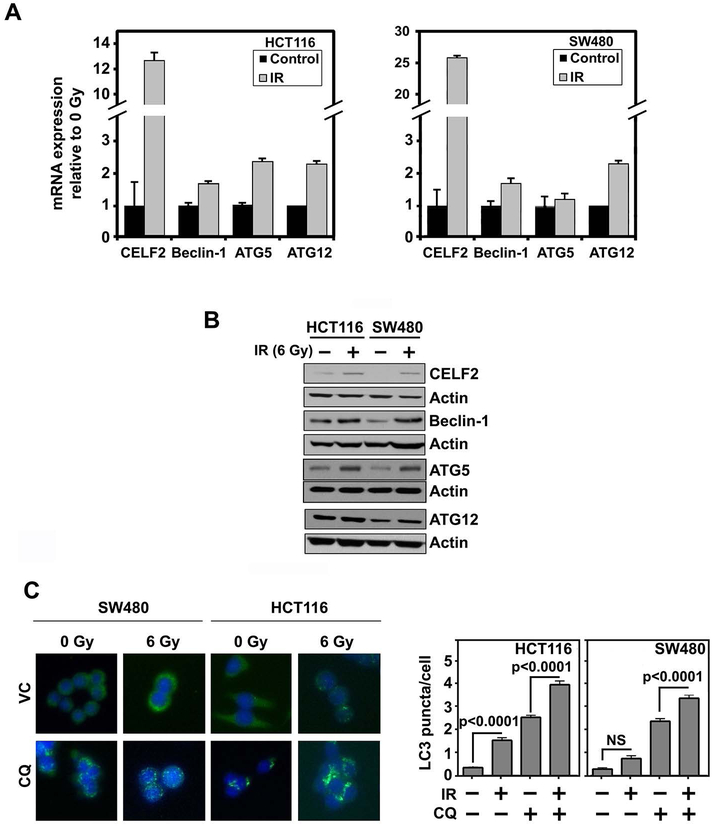
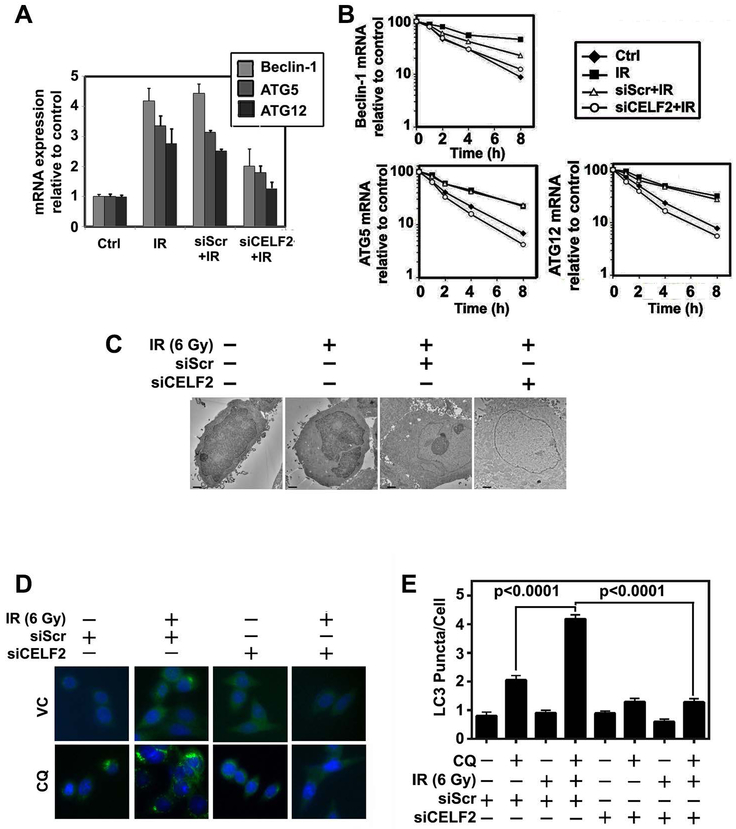
We determined the effect of 6 Gy IR on expression of CELF2 and the three autophagy related genes Beclin-1, ATG5 and ATG12 in CRC cell lines. In addition to robust induction of CELF2, mRNA and protein levels of autophagy related genes increased following 6 Gy IR exposure (Fig 4A,B). We also confirmed increased autophagosome formation by LC3 puncta (Fig 4C). Further, on inhibition of the autophagic flux with chloroquine (CQ), we observed an increase in LC3 puncta in irradiated cells compared to the control cells (Fig 3D). These data further demonstrate enhanced autophagy in response to IR, which corresponds with increased CELF2.
Fig. 4. IR induces CELF2 and autophagy.
(A) Expression of CELF2, Beclin-1, Atg5, and Atg12 mRNA in CRC cell lines (HCT116 and SW480) with 0 or 6 Gy IR. Graph depicts cumulative results of two biological repeats plated in triplicate.
(B) Representative immunoblot of HCT116 or SW480 with 0 or 6 Gy IR demonstrates increased expression of CELF2, Beclin-1, Atg5, and Atg12 in irradiated cells.
(C) Immunofluorescence of LC3 puncta (green) in CRC cells (nuclei stained blue with hoescht stain) treated with CQ (80 μM for 2 hours). Graphs represent cumulative results from duplicate experiments of at least 30 cells counted per group. Error bars represent ± SEM.
CELF2 Mediates IR-Induced Autophagy
We sought to understand if CELF2 was necessary for IR induced autophagy. Knockdown of CELF2 with siRNA mitigated IR-induced expression of BECN1, ATG5, and ATG12 mRNA as compared to scrambled siRNA (Fig 5A). Additionally, the mRNA transcripts had increased stability following exposure of cells to IR, and knockdown of CELF2 attenuated the increase in mRNA stability (Fig 5B). These data demonstrate that IR induced increased stability of BECN1, ATG5, and ATG12 mRNA is due to the binding of CELF2 to these transcripts. We also demonstrate that CELF2 knockdown attenuated IR-induced LC3 puncta and autophagosome formation compared to the control siRNA treated cells (Fig 5C,D). These data demonstrate the direct connection between CELF2 and IR-induced autophagic cell death.
Fig. 5. CELF2 is required for IR-induced autophagy.
(A) mRNA expression of Beclin-1, Atg5, and Atg12 in 0 Gy or IR (6 Gy) treated HCT116 with either CELF2 knockdown by siRNA (siCELF2) or scrambled control siRNA (siScr). Graph depicts cumulative results of two biological repeats.
(B) RNA stability in the context of 0 Gy or IR (6 Gy) treated HCT116 with either siCELF2 or siScr demonstrates decreased stability of autophagy related transcripts, Beclin-1, Atg5, and Atg12, with CELF2 knockdown.
(C) Representative electron microscopy image of 0 Gy or IR (6 Gy) HCT116 cells with either scrambled control siRNA (siScr) or CELF2 targeted siRNA (siCELF2). Scale bar represents 1 μm.
(D) Immunofluorescence of LC3 puncta (green) in HCT116 cells treated with either 0 Gy or 6 Gy IR and siScr or siCELF2 (nuclei stained blue with hoescht stain) treated with CQ (80 μM for 2 hours) was quantified.
(E) The graph represents cumulative results from duplicate experiments of at least 30 cells counted per group. Error bars represent ± SEM.
CELF2 mediates IR-dependent tumor reduction in vivo
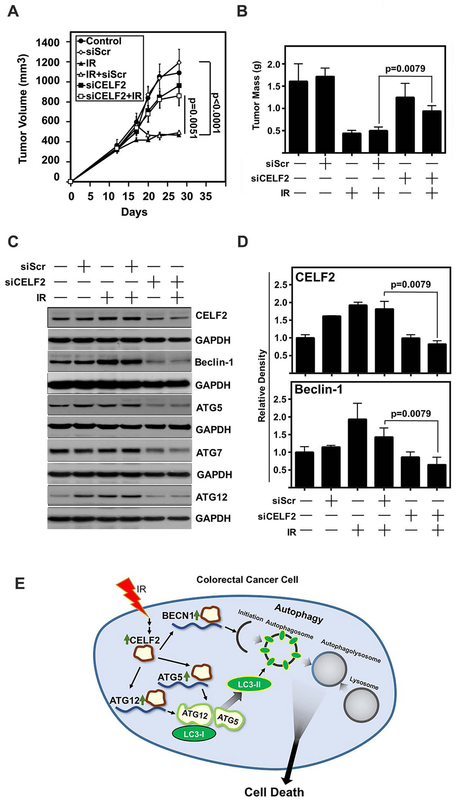
To determine whether CELF2 is essential for IR-mediated inhibition of tumor growth, siRNA specific to CELF2 was delivered intratumorally into HCT116 xenografts receiving 2 Gy IR. As expected, IR significantly reduced the tumor volume in control tissues or in those where a scrambled siRNA was delivered (Fig 6A). CELF2 knockdown mitigated the antitumor effects of radiation (Fig 4A). Endpoint tumor weights demonstrate significantly decreased mass in animals treated with scrambled siRNA and IR, while CELF2 knockdown before IR treatment resulted in greater tumor weights (Fig 6B). Excised tumors demonstrated an increase in autophagy associated proteins upon IR treatment (Fig 6C,D). These data lead to the conclusion that IR induced CELF2 mediates increased autophagic flux in the tumor and corresponds with an autophagy dependent cell death mechanism.
Fig. 6. CELF2 is required for IR-mediated tumor growth suppression in vivo.
(A) Tumor volume of HCT116 xenograft tumors in nude mice. Mice were treated with IR (2 Gy) on days 13, 14, 15, 16, and 17. Scrambled (Scr) or CELF2 siRNA was delivered in DOPC lipid capsules on days 13, 14, 15, 16, and 17. n=10 tumors in control, siScr, siScr + IR, and siCELF2 groups, n=12 tumors in IR alone and siCELF2 + IR. Tumor volume determined by vernier calipers and error bars represent ± SEM.
(B) Tumor weight of excised tumors at the conclusion of the experiment (n=5 per group). Graph depicts mean weight and error bars represent ± SEM.
(C) Immunoblot of excised representative tumors demonstrates increase of CELF2, Beclin-1, Atg5 and Atg12 in IR treated tumors, with a reduction in siCELF2 treated tumors.
(D) Cumulative results of densitometric analysis of CELF2 and Beclin-1 immunoblots of excised HCT116 xenografts relative to GAPDH as loading control. N=5 per group.
(E) Schematic representation of IR induction of CELF2 and subsequent initiation of autophagic cell death.
Discussion
In this study, we observe the critical role of IR induced CELF2 in stabilizing autophagy related mRNA transcripts BECN1, ATG5, and ATG12 and promoting an autophagy associated cell death mechanism (Fig 6E). Autophagy mediates cell death in both normal and pathologic conditions15–17. We identify in this study the role of IR in reducing tumor volume in vivo, which coincides with increased autophagic flux, suggesting an autophagy associated cell death mechanism.
We found ATG5 to be stabilized by CELF2, and this may provide a bridge between autophagy-mediated and apoptosis-mediated cell death. ATG5 plays a dual role in autophagy and apoptosis. ATG5 is cleaved by calpain, and translocates from the cytosol to the mitochondria where it triggers cytochrome C release, leading to apoptotic cell death18. Additionally, ATG5 is induced by radiation or chemotherapy mediated DNA damage, and translocates to the nucleus where it participates in mitotic catastrophe19. Thus, our observation of CELF2 stabilizing ATG5 in this paper explains a more complete mechanism by which we have previously observed CELF2 to mediate apoptosis and mitotic catastrophe. In line with this, our lab has observed the role of IR induced CELF2 in mediating other cell death mechanisms, notably mitotic catastrophe and apoptosis3. Autophagy may precede apoptosis20, may coincide with apoptosis21, or may be a cell death mechanism independent of apoptosis22. Although closely aligned with each other, autophagy and apoptosis are two distinct cellular mechanisms. This is exemplified by the ability of cells to undergo apoptosis without the autophagic machinery, and by the fact that cells can undergo autophagic flux without apoptotic machinery22. Therefore, in light of previous studies, our results establish a death mechanism that coincides with autophagy induction through CELF2 stabilization of autophagy transcripts13,23. This mixed phenotype of cell death mechanisms is in agreement with other reports of autophagy associated cell death24.
Previous reports have demonstrated a number of roles for CELF211. Although expressed ubiquitously in normal tissues, CELF2 is markedly reduced in tumors11. It serves to stabilize COX-2 mRNA, while preventing its translation8. Since COX-2 expression promotes CRC progression, the ability to inhibit translation is a major tumor suppressing mechanism of CELF225,26. However, in the current studies, CELF2 positively regulates translation of the three autophagy related transcripts BECN1, ATG5, and ATG12. Differential regulation of translation by CELF2 is intriguing and will need further investigation. Additionally, in other tissues, CELF2 is responsible for maintaining MAP kinase signaling during T-cell activation27, and is differentially expressed throughout embryonic development12,28. This study builds upon such findings and sheds light of an important physiological role for CELF2 in stabilizing autophagy related transcripts in irradiated CRC and promoting autophagic machinery translation.
In conclusion, IR induces CELF2 expression in CRC. Overexpression of CELF2 or IR treatment results in an increase in autophagy. CELF2 increases the stability of autophagy associated Beclin-1, ATG5 and ATG12 proteins. Inhibition of CELF2, confers resistance to IR in CRC, highlighting a critical role of CELF2 in radiation response.
Grant Support
This work was supported by the NIH grants U01AI138323 to SS, CA182872 to SA, and CA227838 to SMT, the Kansas Bioscience Authority, Thomas P. O’Sullivan IV and Marina O’Sullivan Family Fund and Braden’s Hope Foundation Grant to SA. The National Cancer Institute Cancer Center Support Grant to the University of Kansas Cancer Center, P30CA168524.
Abbreviations
- CRC
Colorectal Cancer
- BRCA
breast cancer
- HNSC
head and neck cancer
- CELF2
CUGBP elav-like family member 2
- COX2
cyclooxygenase 2
- BECN1
Beclin 1
- MDC
monodansylcadaverine
- IR
ionizing radiation
- CQ
chloroquine
Footnotes
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- 1.Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA: A Cancer Journal for Clinicians. 2017:n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 2.Wu L, Pang S, Yao Q, et al. Population-based study of effectiveness of neoadjuvant radiotherapy on survival in US rectal cancer patients according to age. Scientific reports. 2017;7(1):3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Natarajan G, Ramalingam S, Ramachandran I, et al. CUGBP2 downregulation by prostaglandin E2 protects colon cancer cells from radiation-induced mitotic catastrophe. American Journal of Physiology - Gastrointestinal and Liver Physiology. 2008;294(5):G1235–G1244. [DOI] [PubMed] [Google Scholar]
- 4.Kai M Roles of RNA-Binding Proteins in DNA Damage Response. International Journal of Molecular Sciences. 2016;17(3):310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramalingam S, Ramamoorthy P, Subramaniam D, Anant S. Reduced Expression of RNA Binding Protein CELF2, a Putative Tumor Suppressor Gene in Colon Cancer. Immuno-gastroenterology. 2012;1(1):27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stojcheva N, Schechtmann G, Sass S, et al. MicroRNA-138 promotes acquired alkylator resistance in glioblastoma by targeting the Bcl-2-interacting mediator BIM. Oncotarget. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jakstaite A, Maziukiene A, Silkuniene G, et al. Upregulation of cugbp2 increases response of pancreatic cancer cells to chemotherapy. Langenbeck’s Archives of Surgery. 2016;401(1):99–111. [DOI] [PubMed] [Google Scholar]
- 8.Mukhopadhyay D, Houchen CW, Kennedy S, Dieckgraefe BK, Anant S. Coupled mRNA Stabilization and Translational Silencing of Cyclooxygenase-2 by a Novel RNA Binding Protein, CUGBP2. Molecular Cell. 2003;11(1):113–126. [DOI] [PubMed] [Google Scholar]
- 9.Biederbick A, Kern HF, Elsasser HP. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. European journal of cell biology. 1995;66(1):3–14. [PubMed] [Google Scholar]
- 10.Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002;26(2):182–190. [DOI] [PubMed] [Google Scholar]
- 11.Ramalingam S, Ramamoorthy P, Subramaniam D, Anant S. Reduced Expression of RNA Binding Protein CELF2, a Putative Tumor Suppressor Gene in Colon Cancer. Immuno-gastroenterology. 2012;1(1):27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramaniam D, Ramalingam S, Linehan DC, et al. RNA Binding Protein CUGBP2/CELF2 Mediates Curcumin-Induced Mitotic Catastrophe of Pancreatic Cancer Cells. PloS one. 2011;6(2):e16958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galluzzi L, Bravo-San Pedro JM, Vitale I, et al. Essential versus accessory aspects of cell death: recommendations of the NCCD 2015. Cell Death Differ. 2015;22(1):58–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palumbo S, Comincini S. Autophagy and ionizing radiation in tumors: The “survive or not survive” dilemma. Journal of Cellular Physiology. 2013;228(1):1–8. [DOI] [PubMed] [Google Scholar]
- 15.Zhu J-h, Horbinski C, Guo F, Watkins S, Uchiyama Y, Chu CT. Regulation of Autophagy by Extracellular Signal-Regulated Protein Kinases During 1-Methyl-4-Phenylpyridinium-Induced Cell Death. The American Journal of Pathology. 2007;170(1):75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wen Y, Zand B, Ozpolat B, et al. Antagonism of tumoral prolactin receptor promotes autophagy-related cell death. Cell reports. 2014;7(2):488–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131(6):1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yousefi S, Perozzo R, Schmid I, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nature cell biology. 2006;8:1124. [DOI] [PubMed] [Google Scholar]
- 19.Maskey D, Yousefi S, Schmid I, et al. ATG5 is induced by DNA-damaging agents and promotes mitotic catastrophe independent of autophagy. Nat Commun. 2013;4:2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laane E, Tamm KP, Buentke E, et al. Cell death induced by dexamethasone in lymphoid leukemia is mediated through initiation of autophagy. Cell Death Differ. 2009;16(7):1018–1029. [DOI] [PubMed] [Google Scholar]
- 21.Mohseni N, McMillan SC, Chaudhary R, Mok J, Reed BH. Autophagy promotes caspase-dependent cell death during Drosophila development. Autophagy. 2009;5(3):329–338. [DOI] [PubMed] [Google Scholar]
- 22.Shimizu S, Kanaseki T, Mizushima N, et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6(12):1221–1228. [DOI] [PubMed] [Google Scholar]
- 23.Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12(1):1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galluzzi L, Aaronson SA, Abrams J, et al. Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell Death Differ. 2009;16(8):1093–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chulada PC, Thompson MB, Mahler JF, et al. Genetic disruption of Ptgs-1, as well as Ptgs-2, reduces intestinal tumorigenesis in Min mice. Cancer Res. 2000;60(17):4705–4708. [PubMed] [Google Scholar]
- 26.Wang D, DuBois RN. The Role of COX-2 in Intestinal Inflammation and Colorectal Cancer. Oncogene. 2010;29(6):781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez NM, Agosto L, Qiu J, et al. Widespread JNK-dependent alternative splicing induces a positive feedback loop through CELF2-mediated regulation of MKK7 during T-cell activation. Genes Dev. 2015;29(19):2054–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blech-Hermoni Y, Stillwagon SJ, Ladd AN. Diversity and conservation of CELF1 and CELF2 RNA and protein expression patterns during embryonic development. Dev Dyn. 2013;242(6):767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]