Abstract
Background:
Neural stem cells (NSCs) generate most of the neurons of the adult brain in humans, during the mid-first through second trimester period. This critical neurogenic window is particularly vulnerable to prenatal alcohol exposure, which can result in diminished brain growth. Previous studies showed that ethanol exposure does not kill NSCs, but, rather, results in their depletion by influencing cell cycle kinetics and promoting aberrant maturation, in part, by altering NSC expression of key neurogenic miRNAs. NSCs reside in a complex microenvironment rich in extracellular vesicles, shown to traffic miRNA cargo between cells.
Methods:
We profiled the miRNA content of extracellular vesicles from control and ethanol-exposed ex vivo neurosphere cultures of fetal NSCs. We subsequently examined the effects of one ethanol-sensitive miRNA, miR-140–3p, on NSC growth, survival, and maturation.
Results:
Ethanol exposure significantly elevates levels of a subset of miRNAs in secreted extracellular vesicles. Overexpression of one of these elevated miRNAs, miR-140–3p and its passenger strand relative, miR-140–5p, significantly increased the proportion of S-phase cells while decreasing the proportion of G0/G1 cells compared to controls In contrast, while miR-140–3p knockdown had minimal effects on the proportion of cells in each phase of the cell cycle, knockdown of miR-140–5p significantly decreased the proportion of cells in G2/M phase. Furthermore, miR-140–3p overexpression, during mitogen-withdrawal-induced NSC differentiation, favors astroglial maturation at the expense of neural and oligodendrocyte differentiation.
Conclusion:
Collectively, the dysregulated miRNA content of extracellular vesicles following ethanol exposure may result in aberrant neural progenitor cell growth and maturation, explaining brain growth deficits associated with prenatal alcohol exposure.
Keywords: microRNA, extracellular vesicles, Exosomes, Fetal Alcohol, Neural Stem Cells
Introduction:
Prenatal alcohol exposure (PAE) can result in a cluster of craniofacial, neuro-cognitive, and growth deficits that are collectively termed Fetal Alcohol Spectrum Disorders (FASDs) (Streissguth and O’Malley, 2000). Despite published prevention guidelines (ACOG, 2011), PAE is common (Popova et al., 2017) and FASDs are thought to affect between 1.1 and 5% of school-aged children in the US (May et al., 2018), and may account for between 2 and 3% of all births, worldwide (Roozen et al., 2016). Self-reported PAE is particularly high during the 1st and 2nd trimesters of pregnancy (SAMHSA, 2013), due in part to the high prevalence of unplanned pregnancies in the US (Finer and Zolna, 2016). The period encompassing the mid-1st trimester through the end of the 2nd trimester, when inadvertent alcohol exposure is most likely to occur, is a particularly important period of fetal brain vulnerability, because, neural stem cells (NSCs) generate most of the neurons of the adult brain during this period (Bystron et al., 2008). Consequently, interfering with neurogenesis during this critical developmental window is likely to produce persistent deficits in brain function.
Previous research shows that PAE in preclinical models, during 1st and 2nd trimester-equivalent periods of brain development, decreases brain weight (Maier et al., 1999). However, while ethanol results in significant cell death in differentiating neurons (Cheema et al., 2000, McAlhany et al., 2000, Mooney and Miller, 2003), NSCs that give rise to these neurons are themselves resistant to cell death following ethanol exposure (Vangipuram and Lyman, 2010, Prock and Miranda, 2007, Santillano et al., 2005). Instead, we and others previously found that ethanol exposure results in increased cell proliferation and NSC growth, resulting in the depletion of NSCs in favor of more mature neural progenitor populations (Santillano et al., 2005, Vangipuram and Lyman, 2010, Miller, 1989, Miller and Nowakowski, 1991, Tingling et al., 2013, Camarillo and Miranda, 2008).
The mechanisms that mediate PAE’s effects on NSC growth and maturation are likely to be complex and remain to be elucidated. MicroRNAs (miRNAs), a class of small non-protein-coding RNAs that inhibit protein translation through their ability to guide the binding of RNA-induced silencing complexes (RISC) to target mRNAs (Miranda, 2014), have emerged as one promising candidate (as reviewed in (Mahnke et al., 2018)). Previous studies from our group and others have shown that certain miRNAs are responsive to ethanol exposure, and that these miRNAs control several ethanol-sensitive developmental processes, such as survival and proliferation in fetal NSCs, and may mediate the effects of PAE on early brain development (Sathyan et al., 2007, Tsai et al., 2014, Pappalardo-Carter et al., 2013, Tal et al., 2012).
Self-renewal and maturation within the fetal neuro-epithelial NSC niche may also be synchronized by paracrine signals that are vulnerable to ethanol exposure. Recent studies suggest that miRNAs are not only localized within the cell, but are also present in circulation and extracellular milieu (Pirola Carlos et al., 2012). Interestingly, cellular miRNAs may be packaged within small membrane-bound extracellular vesicles (EVs) for release (Sohel, 2016, Zhang et al., 2015). EVs are also known to contain protein, lipid, as well as RNAs, and may participate in both paracrine and endocrine intercellular communication, whereby EVs released from one cell fuse with, and release their content into, neighboring or distant cells without any direct cell-to-cell contact (Mathivanan et al., 2010, Osteikoetxea et al., 2015, Llorente et al., 2013, Subra et al., 2007, Taylor and Gercel-Taylor, 2013, Baglio et al., 2015, Lu and Risch, 2016). Currently, two of the heavily studied categories of extracellular vesicles are exosomes and microvesicles. Exosomes are EVs, in the size of 50–200 nanometers, generated from early endosomes that mature in the multi-vesicular bodies and are released to the extracellular space through exocytosis. In contrast, microvesicles are 50–2000 nanometer EVs, generated by direct budding of the cellular plasma membrane, and shed into the extracellular environment (Lawson et al., 2016, Théry et al., 2002, El Andaloussi et al., 2013, Thery et al., 2009, Iraci et al., 2016).
Though NSCs are known to be rich producers of EVs, the role of EVs in NSC development and differentiation, and in the NSC response to pathological stress, is unknown (Morton et al., 2018, Janas et al., 2016). For this reason, we performed EV isolation from NSCs. We then compared the EV miRNA profiles of control and ethanol-exposed NSCs, identifying a subset of miRNAs whose expression levels were significantly induced by ethanol treatment. One of them, miR-140–3p, is a microRNA known to control mesenchymal stem cell fate commitment (Li et al., 2014, Salone et al., 2014, Karlsen et al., 2014), that we have previously demonstrated to be alcohol- and nicotine-sensitive (Balaraman et al., 2012). Using gain- and loss-of-function strategies, we show that over-expression of miR-140–3p increased NSC growth. We further showed that miR-140–3p influences expression of key differentiation-associated mRNA transcripts. Thus, the elevated expression of miR-140–3p, as observed in EVs derived from ethanol-treated NSCs, favors the accumulation of GFAP and reduction of GLAST glial progenitors, and is consistent with our previous findings that ethanol inhibits neurogenesis (Santillano et al., 2005, Camarillo and Miranda, 2008, Camarillo et al., 2007). Therefore, the dysregulated miRNA content of NSC-EVs, following ethanol exposure, may underlie some of the aberrant neural maturation associated with FASDs.
Materials and Methods
Ex Vivo Fetal Mouse Neurosphere Culture Model
Neural stem cells (NSCs) from the dorsal telencephalic vesicles of mixed male and female gestational day (GD) 12.5 C57BL/6N mouse fetuses (Envigo; Catalog #4440F) were propagated as non-adherent spheres in serum-free mitogenic media, as previously published (Santillano et al., 2005, Prock and Miranda, 2007, Sathyan et al., 2007, Camarillo et al., 2007, Tsai et al., 2014). All animal procedures were approved by the Texas A&M University Laboratory Animal Care Committee.
Ethanol Treatment
NSCs seeded at a density of 2 × 106 cells per T25 flask, with each flask being defined as a single sample, were randomly assigned to a control (0 mg/dL, 0mM) or high (320mg/dl, 70 mM) ethanol exposure group. Before use, 190 proof ethanol (Sigma; Catalog # 493538) was diluted into fresh culture medium, and ethanol concentrations in culture-conditioned medium were measured by gas chromatography for each experiment. Both control and ethanol-treated flasks were tightly capped, with phenolic caps and parafilm sealed, to prevent ethanol loss in the culture medium throughout the three-day exposure period. Gas chromatographic analyses of our ethanol exposure group indicated that alcohol concentrations in the media (270–350mg/dL; 58mM-76mM) approximated the blood alcohol concentration attained in heavy drinkers (Adachi et al., 1991, Perper et al., 1986). Mitogen-withdrawal-driven differentiation was achieved by seeding neurospheres onto laminin-coated (ThermoFisher; Catalog # 23017015) culture dishes in the absence of EGF and LIF, but with FGF, as described previously (Camarillo et al., 2007, Camarillo and Miranda, 2008, Miranda et al., 2008). This paradigm results in the preferential formation of neuronal-lineage-committed migratory cells.
Extracellular Vesicle Isolation
Extracellular vesicle fractions were isolated from ethanol-treated and control neurospheres and cultures using the ExoQuick-TC™ Exosome Precipitation Solution according to manufacturer instructions (System Biosciences; Catalog # ExoTC10A-1). Samples were then passed through a 0.2um sterile filter with polyethersulfone membrane (VWR; Catalog # 28145–501) to exclude particles with diameters larger than 200 nanometers.
Nanoparticle Tracking Analysis
The size and concentration of extracellular vesicles were measured by nanoparticle tracking analysis (Nanosight LM10; Malvern). Briefly, the NanoSight instrument can be used to track, detect, and measure the size and concentration of EVs through detection of light scatter generated by nanoparticles in suspension (Dragovic et al., 2011, Gardiner et al., 2013). This light scatter is video-captured, with frame-by-frame analysis used to determine the particle Brownian motion and travel distance, allowing for calculation of particle diffusion and hydrodynamic diameter. Gibco® 1 x PBS buffer (Thermo Fisher; Catalog # 14190144) was used for dilution of isolated EV samples. 1ml of diluted EV sample was slowly syringe-injected loaded into the Nanosight LM10 chamber. For each sample, a minimum of three 60s videos, with a minimum of 1000 tracks per video, were recorded and analyzed. All settings were kept constant between samples for the sake of consistency and accuracy.
Western Blot Analysis
Protein from neurospheres and isolated EVs was extracted using 1xRIPA lysis buffer (EMD Millipore; Reference # 20–188), with addition of Halt protease & phosphatase inhibitor cocktail (Thermo Fisher Scientific; Product # 78442). Extracted protein concentration was determined using Pierce BCA protein assay kit (Thermo Fisher Scientific; Catalog # 23225). 25ug of protein was size-fractionated on a 4–12% Bis-Tris Gel, ran at 150V for 60 minutes, and blotted to a PVDF membrane using iBlot transfer system (Thermo Fisher Scientific; Catalog # IB301001). Subsequently, the membrane was blocked for one hour with 2.5% goat serum and 5% nonfat dry milk in Tris-buffered saline that contains 0.1% Tween-20 (TTBS), then incubated overnight with polyclonal rabbit anti-CD63 antibodies diluted 1:1000 (System Biosciences; Catalog # EXOAB-CD63A-1). CD63 is an established surface marker for EVs. The blot was washed and incubated with an HRP-conjugated goat anti-rabbit IgG (Invitrogen) at dilution 1:1000 for one hour, then developed using PerkinElmer Western Lightning Plus-ECL (PerkinElmer; Catalog # NEL103001EA) and visualized using a CCD camera (Fluorchem Q, Alpha Innotech).
Transmission Electron Microscopy (TEM) and Immuno-gold Labeling of EVs
For the validation of our EV isolation, TEM and immunogold labeling were used. TEM sample preparation and imaging were performed at the Texas A&M Microscopy and Imaging Center, following an established protocol (Théry et al., 2006). For immuno-gold labeling, EV surface marker CD63 (polyclonal rabbit anti-CD63 antibodies diluted 1:100 System Biosciences; Catalog # EXOAB-CD63A-1) was used.
Briefly, 5ul of resuspended EV-containing pellet was deposited on Formvar-carbon coated electron-microscopy grids for 20 minutes and fixed in 1% glutaraldehyde for 5 minutes. Grids were washed by floating on 100ul of PBS/50 mM glycine. The grids were then transferred to a drop of blocking buffer (PBS/5% (w/v) BSA) for 10 minutes and floated on a 5-μl drop with the first antibody diluted at 1:1000 in the PBS/1% (w/v) BSA and incubated for 30 minutes. Grids were washed by floating on washing buffer (PBS/0.1% (w/v) BSA). Grids were washed with PBS and incubate on 5-μl drops of 12 nm goat anti-Rabbit colloidal gold (Jackson ImmunoResearch Lab; Catalog # 111–205-144) diluted 1:5000 in the PBS/1% (w/v) BSA for 20 minutes. EV samples were contrasted with a mixture of 2% methyl cellulose and 4% uranyl-acetate by floating on a 50-μl drop for 10 minutes on ice. Residual staining solution was wicked off by blotting with Whatman no. 1 filter paper. Grids were air-dried before being examined in a JEOL 1200 EX transmission electron microscope (JEOL USA Inc.) at an acceleration voltage of 100 kV. Images of EVs were recorded at calibrated magnifications using 3k slow-scan CCD camera (model 15C, SIA).
RNA Isolation, miRNA qPCR Array, and mRNA qPCR
RNA from neurospheres and EVs was isolated using the miRNeasy mini kit (Qiagen; Catalog # 217004). cDNA synthesis was performed using the qScript™ cDNA SuperMix (Qiagen; Catalog # 95048) or the Universal cDNA Synthesis Kit II (Exiqon; Catalog # 203301). EV miRNA was reverse transcribed using a 10ul mix of cDNA and ExiLENT SYBR® Green master mix (Exiqon: Catalog # 203421), which was loaded into each well of a microRNA Ready-to-Use PCR Mouse & Rat panel I+II, V4.M qPCR array (Exiqon; Catalog # 203615) to assess and measure 752 unique miRNAs as described previously (Balaraman et al., 2013, Balaraman et al., 2014, Balaraman et al., 2016). qPCR analysis was done on an Applied Biosystem ViiA 7 Real-time PCR system (ABI/Life Technologies, Grand Island, NY) (Balaraman et al., 2014, Selvamani et al., 2014). miRNA expression values were normalized to the 5 most invariant miRNAs across samples for qPCR arrays (mmu-miR-99b-5p, mmu-miR-384–5p, mmu-miR-151–3p, mmu-miR-25–3p, mmu-miR-676–3p), or U6 for the ex vivo differentiation or overexpression and antagomir studies.
For mRNA transcript quantification, the presented data correspond to the mean 2-ΔΔCt after being normalized to β-actin. Primers were designed to span exon-exon junctions. For each primer pair, thermal stability curves were assessed for evidence of a single amplicon. The length of each amplicon was verified using agarose gel electrophoresis, and amplicon identity was verified by Sanger sequencing. A list of primers and their sequences is presented in Table 1.
Table 1:
List of Primers Used
| Targets (Mus musculus) | Forward Sequence | Reverse Sequence | Product Length | NCBI Accession Number |
|---|---|---|---|---|
| ACTB | CTCTGGCTCCTAGCACCATGAAGA | GTAAAACGCAGCTCAGTAACAGTCCG | 200bp | NM_007393 |
| Nestin | CTCAGATCCTGGAAGGTGGG | GCAGAGTCCTGTATGTAGCCA | 81bp | NM_016967 |
| Olig2 | GAACCCCGAAAGGTGTGGAT | TTCCGAATGTGAATTAGATTTGAGG | 105bp | NM_016967 |
| NeuN | AACCAGCAACTCCACCCTTC | CGAATTGCCCGAACATTTGC | 118bp | NM_001285437 |
| DCX | ACCTGACCCGATCCTTGTCT | ACATAGCTTTCCCCTTCTTCCA | 121bp | NM_001110222 |
| PDGFRα | CGTGCTTGGTCGGATTTTGG | CAGGTTGGGACCGGCTTAAT | 83bp | NM_001083316 |
| MAP2 | CTGGAGGTGGTAATGTGAAGATTG | TCTCAGCCCCGTGATCTACC | 84bp | NM_001039934 |
| GFAP | CTAACGACTATCGCCGCCAA | CAGGAATGGTGATGCGGTTT | 297bp | NM_001131020 |
| GLAST | CAACGAAACACTTCTGGGCG | CCAGAGGCGCATACCACATT | 260bp | NM_148938 |
miRNA Transfection
Mimics for the differentially expressed EV miRNAs, at a concentration of 25nM (Dharmacon miRNA mimics), were transfected into NSCs, using Lipofectamine RNAiMAX (ThermoFisher; Catalog # 13778) according to the manufacturer’s instructions. Similarly, hairpin inhibitors for mmu-miR-140 were transfected into NSCs at a concentration of 25nM (Dharmacon). miRIDIAN microRNA Mimic Negative Control #1 (Dharmacon; Catalog # CN-001000–01) and miRIDIAN microRNA Hairpin Inhibitor Negative Control #1 (Catalog # IN-001005–01-05) were used as controls.
MTT Cell Viability Assay
The MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) colorimetric assay (ThermoFisher; Catalog # M6494) for cell metabolic activity, measured by formation of a colored formazan reaction product, was used to assess the number of viable cells. 72 hours following transfection with miRNAs, NSCs were incubated for 3 hours with 6mM MTT. The reaction product was subsequently solubilized with 10% SDS in 0.01N HCl for 3 hours. Absorbance intensities were measured at 570nM, using Tecan Infinite 200 microplate reader (Tecan; SKU# 8344–50-0005).
Cell Cycle Analysis
At 48 hours post-transfection, DNA synthesis was assessed by pulse-labeling cells with 10μM EdU (5-ethynyl-2’-deoxyuridine) for 1 hour. Cells were immediately harvested, and cell cycle analysis was performed with the Click-iT® EdU Alexa Fluor® 488 Flow Cytometry Assay Kit (Thermo Fisher, Cat # C10420), in conjunction with 7-Amino-Actinomycin D (Thermo Fisher, Cat # 00–6993-50), according to manufacturer instructions, using the Beckman Coulter® Gallios 2/5/3 Flow Cytometer. Data was analyzed using Kaluza software (Beckman Coulter).
Cell Death Analysis
At 48 hours post-transfection, the Promega Caspase-Glo® 3/7 Assay Systems (Promega, Cat # G8091) was used to quantify apoptotic cell death.
Ingenuity Pathway Analysis
Ingenuity pathway analysis was conducted as previously described (Balaraman et al., 2016). Briefly, IPA miRNA Target Filter® was used to identify experimentally validated miRNA-target gene interactions or potential miRNA gene targets with a high predicted confidence of interaction (context score < −0.4 [11, 12]). Subsequently, IPA’s Core Analysis workflow was used to conduct functional network analysis, to identify gene regulatory networks overrepresented amongst predicted miRNA targets.
Single Cell Gene Expression
Confluent neurospheres were trypsinized using 0.25% EDTA-free trypsin (Thermofisher #15090046) for 5 minutes at room temperature with occasional pipetting. Trypsin was subsequently inactivated with 10% FBS and cells were filtered through a 40-micron filter to ensure single-cell suspensions. The cells were then spun down followed by an ice-cold PBS wash, and 10,000 cells were reserved for ice-cold methanol fixation. Cells were rehydrated according to manufacturer instructions immediately preceding the 10X Genomics Single cell Protocol.
We used the chromium single cell 3’ reagent kit (10X Genomics; CG000183) to prepare a single cell master mix using an input of 7000 rehydrated cells. The single cell master mixes were subsequently loaded into Chromium Chip B (10X Genomics; PN-1000074) and gel beads in emulsion were prepared according to the manufacturer protocol followed by RT, cDNA amplification and library preparation step. The libraries were sequenced on NextSeq 500 (Illumina) (400 million single-end reads sequencing depth).
The Cell Ranger analysis pipeline was used to demultiplex raw base call (BCL) files generated by the sequencer into FASTQ files, perform genome alignment, filtering, and unique molecular index (UMI) quantification utilizing the Chromium cellular barcodes to generate gene-barcode UMI matrices. The gene-barcode UMI matrices were used as input for analysis using R’s “Seurat” package, during which UMIs were log normalized.
For the correlation analyses, we calculated the spearman’s correlation between Wwp2 (the miR140 host gene, miR140HG) and a set of markers for stem cell identity and differentiation in Wwp2/miR140HG-positive cells (a total of 972 Wwp2/miR140HG-positive cells out of 13,770 assessed cells, with a range of Wwp2/miR140HG expression from 0.11 to 2.99 log10 normalized UMI counts, representing a 753-fold range in expression). The correlation plot was constructed using the “corrplot” package in ‘R’.
Statistical Analysis
Statistical analyses, including linear regression, student’s t-test, or one-way ANOVA with either Sidak’s or Dunnet’s post-hoc analysis, were conducted using the GraphPad Prism software, version 6.00 for Windows.
Results
Mouse Neural Stem Cells are Abundant Producers of Extracellular Vesicles
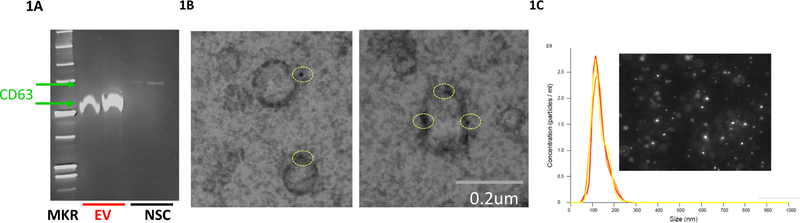
We isolated EVs from the culture supernatant of murine NSCs in neurosphere cultures. An immunoblot showed that, compared to parent cells, the presumptive EV fraction from our NSCs supernatant was enriched for the EV marker CD63 (Figure 1A). Electron micrographs indicated that the NSC-derived EVs were mainly round-shaped vesicles, with immuno-gold staining demonstrating positive staining for CD63 on the surface of our isolated EVs (Figure 1B). NSCs secreted an abundant number of extracellular vesicles, ranging from ~ 1010 to 1011 vesicles per initial cell count of ~2 × 106 cells, after 72 hours of culture (Figure 1C and Supplementary Figure 2B). There was no significant difference in vesicle size between EVs that were positively stained for CD63 and those that were not (Supplementary Figures 1A and 1B). Nanoparticle tracking analysis (NTA) indicated the NSC EV diameters ranged in size from 50–150nm, with the diameter median and mode being ~130nm, consistent with the known size range (50–200nm) of exosomes (Figure 1C) (Mathivanan et al., 2010, Théry et al., 2002, Janas et al., 2016).
Figure 1. NSCs are Abundant Producers of EVs.
1A) Immunoblot of CD63 expression in EVs and NSCs. Differences in CD63 molecular weight between EV and NSC fraction are due to polyethylene glycol in the Exoquick-TC™ preparation.
1B) TEM image of EVs immunolabelled with CD63. Yellow circles indicate puncta with CD63 labelling.
1C) Nanosight image of EVs derived from NSCs Mkr=molecular weight marker
Ethanol Exposure Alters the microRNA Content of NSC-Derived EVs
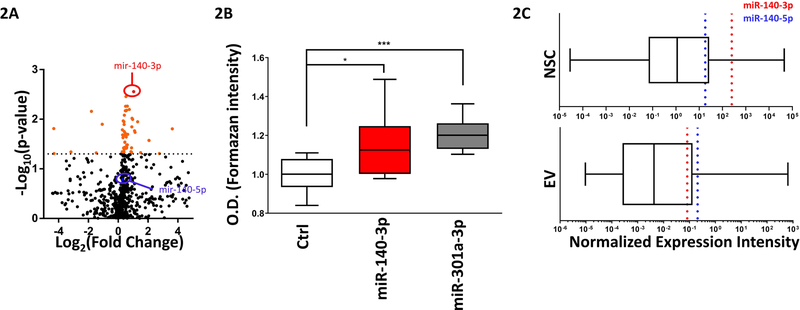
Ethanol exposure did not result in a significant change in the size and concentration of EVs derived from neurosphere cultures, or the expression of CD63 on EVs (Supplementary Figures 2A-2C). However, ethanol exposure of NSCs at 320mg/dL altered the profile of miRNAs packaged within EVs. Out of the 652 expressed miRNAs, 53 miRNAs were differentially expressed (Unpaired t-test, p<0.05) in the EV fraction of ethanol-treated NSCs compared to controls, not accounting for FDR correction (Figure2A, Table 2). We observed that the majority of differentially expressed miRNAs in EVs, 47 out of 53 miRNAs, were significantly upregulated by ethanol treatment relative to controls, whereas only 6 miRNAs were significantly downregulated. Interestingly, of the top 30% most abundantly expressed EV miRNAs exhibiting a greater than 1.5-fold change between treatment conditions, only 4 were significantly increased in the EV fraction of ethanol-treated NSCs compared to controls when controlling for the false discovery rate (Unpaired t-test, q<0.05; Table 3). Collectively, our data suggests that a small cluster of miRNAs in NSC-derived EVs are significantly altered with ethanol exposure.
Figure 2. Ethanol Alters the miRNA Profile in NSC-derived EVs.
2A) Volcano plot of miRNA expression in EVs from control and ethanol treated NSCs. X-axis depicts Log2 transformed fold change of Ethanol/Control miRNA expression. Orange data points above the dotted line represent miRNAs significantly affected by ethanol exposure. The red dot signifies miR-140–3p and the blue dot signifies miR-140–5p. n=5 samples per group
2B) MTT absorbance values following control, miR-140–3p, and miR-310a-3p overexpression in NSCs. n=10 samples per group.
2C) Relative abundance of miR-140–3p (red-dotted line) and miR-140–5p (blue-dotted line) compared to other miRNAs within NSCs (top) and NSC-derived EVs (bottom).
*p<0.05, ***p<0.001 by Student’s t-test
Table 2:
Table of Significantly Altered miRNAs in EVs from Ethanol-Treated NSCs
| miRNA | Log2(Etoh/Ctrl) | p-value |
|---|---|---|
| mmu-miR-140–3p | 1.038047 | 0.002772 |
| rno-miR-338–3p | 0.5185305 | 0.003492 |
| mmu-miR-340–5p | 0.6101994 | 0.005452 |
| mmu-miR-709 | 0.4990811 | 0.005491 |
| mmu-miR-15b-3p | 0.7437596 | 0.006361 |
| mmu-miR-378a-5p | 0.5595576 | 0.006606 |
| mmu-miR-181a-5p | 0.4760198 | 0.006757 |
| mmu-miR-187–3p | 0.5162219 | 0.006777 |
| rno-miR-667–3p | −1.796813 | 0.006934 |
| mmu-miR-378a-3p | 0.4153968 | 0.008311 |
| mmu-miR-486–3p | 0.9300453 | 0.009593 |
| mmu-miR-674–5p | 0.9715856 | 0.010089 |
| mmu-miR-185–5p | 0.3336599 | 0.0108 |
| mmu-miR-301a-3p | 0.3874377 | 0.011227 |
| mmu-miR-467d-3p | 1.302656 | 0.011264 |
| mmu-miR-669l-5p | −1.072414 | 0.012656 |
| rno-miR-344a-3p | 0.3923947 | 0.012793 |
| mmu-miR-29a-5p | 0.9157254 | 0.014249 |
| mmu-miR-668–3p | −4.322045 | 0.015463 |
| mmu-miR-127–5p | 3.628419 | 0.01561 |
| mmu-miR-15a-5p | 0.2828906 | 0.016326 |
| mmu-miR-137–3p | 1.097612 | 0.017092 |
| mmu-miR-331–3p | 0.4010352 | 0.017681 |
| mmu-miR-30c-2–3p | 1.131432 | 0.018304 |
| mmu-miR-339–5p | 0.4689127 | 0.01912 |
| mmu-miR-322–3p | 0.6379156 | 0.020104 |
| mmu-miR-503–3p | 0.2956948 | 0.020702 |
| rno-miR-501–5p | 0.4597853 | 0.021145 |
| mmu-miR-30a-5p | 0.5354863 | 0.02288 |
| mmu-miR-16–5p | 0.3169613 | 0.023537 |
| mmu-miR-467c-5p | 2.060594 | 0.026732 |
| rno-miR-125b-2–3p | 2.074389 | 0.026822 |
| mmu-miR-872–5p | 0.4429964 | 0.028883 |
| mmu-miR-23a-3p | 0.4438826 | 0.029259 |
| rno-miR-17–1-3p | 5.706707 | 0.030089 |
| mmu-miR-338–5p | 0.6141916 | 0.032902 |
| mmu-miR-30a-3p | 0.5727178 | 0.033067 |
| mmu-miR-195a-5p | 0.4338778 | 0.036908 |
| mmu-miR-139–5p | 0.5740866 | 0.037107 |
| rno-miR-340–3p | 0.4686742 | 0.037293 |
| rno-miR-465–5p | 0.8976653 | 0.037783 |
| mmu-miR-132–3p | 0.3586375 | 0.03788 |
| mmu-miR-30e-3p | 0.5565696 | 0.038382 |
| rno-miR-345–5p | 0.4033093 | 0.040369 |
| mmu-miR-146b-5p | 0.8567061 | 0.045176 |
| mmu-miR-135a-5p | 0.2700475 | 0.045312 |
| mmu-miR-684 | −3.171892 | 0.045519 |
| rno-miR-324–3p | 0.3913724 | 0.045842 |
| mmu-miR-671–5p | 0.8974715 | 0.046466 |
| mmu-miR-1968–5p | 1.504539 | 0.047055 |
| mmu-miR-150–5p | −1.468246 | 0.047752 |
| mmu-miR-1936 | −4.304381 | 0.049028 |
| mmu-miR-540–3p | 2.762398 | 0.049127 |
Table 3:
Table of Most Abundant miRNAs in EVs from Ethanol-Treated NSCs
| miRNA | Log2(Etoh/Ctrl) | p-value | q-value |
|---|---|---|---|
| mmu-miR-140–3p | 1.08176 | 0.002772 | 0.03785 |
| mmu-miR-15b-3p | 0.754359 | 0.006361 | 0.03785 |
| mmu-miR-340–5p | 0.60876 | 0.005452 | 0.03785 |
| mmu-miR-674–5p | 0.942559 | 0.010089 | 0.045024 |
| mmu-miR-29a-5p | 0.88916 | 0.014249 | 0.05087 |
| mmu-miR-322–3p | 0.62676 | 0.020104 | 0.059809 |
| mmu-miR-30a-3p | 0.60816 | 0.033066 | 0.084319 |
| mmu-miR-30e-3p | 0.59616 | 0.038382 | 0.08564 |
| mmu-miR-29a-3p | 0.78976 | 0.051292 | 0.096661 |
| mmu-miR-1983 | 0.76416 | 0.054152 | 0.096661 |
| mmu-miR-129–5p | 0.90616 | 0.063557 | 0.103135 |
| mmu-miR-326–3p | 0.77416 | 0.108994 | 0.140517 |
| mmu-miR-17–3p | 0.676559 | 0.10497 | 0.140517 |
| mmu-miR-34b-5p | 0.636559 | 0.117668 | 0.140517 |
| mmu-miR-129–1-3p | 0.614159 | 0.118081 | 0.140517 |
| rno-miR-99a-3p | 0.63276 | 0.181754 | 0.190842 |
| mmu-miR-29c-3p | 0.60136 | 0.180988 | 0.190842 |
| mmu-miR-500–3p | 0.61616 | 0.215239 | 0.213446 |
| mmu-miR-488–3p | 3.29636 | 0.296455 | 0.278512 |
| mmu-miR-98–5p | 3.04196 | 0.350318 | 0.312659 |
miR-140–3p Promotes NSC Proliferation
Given that ethanol exposure has profound effects on NSC and neuroepithelial growth (Santillano et al., 2005), we next investigated whether the ethanol-sensitive EV miRNAs themselves had effects on NSC growth. To this end, we overexpressed 20 of the most significantly altered EV miRNAs in NSCs and found that only two of these miRNAs, mir-140–3p and miR-301a-3p, significantly increased NSC growth (Unpaired t-test, p=0.012 and p=0.0006, n=8 samples per group; Figure 2B and Supplementary Figure 3). Interestingly, miR-140–3p is also the most significantly increased miRNA in the EVs of ethanol treated NSCs (Unpaired t-test, p=0.0028), with both miR-140–3p and its passenger strand, miR-140–5p, also expressed at high levels within NSCs and NSC-derived EVs (Figure 2C). Consistent with our array data, follow-up qRT-PCR analysis in neurosphere culture samples, confirmed significant elevation of miR-140–3p within NSCs and the NSC-derived EV fraction following 3-days of ethanol exposure (ANOVA F(1,16)=26.04, p=0.0001, Supplementary Figure 4). However, at 5 days of ethanol exposure, we did not observe any change in miR-140–3p (2 independent biological replicates, all p’s >0.5) in contrast to our previous observations (Balaraman et al., 2012), that prolonged ethanol suppressed intracellular miR-140–3p expression, a difference that may perhaps be attributable to differences in sourcing of animals for each study.
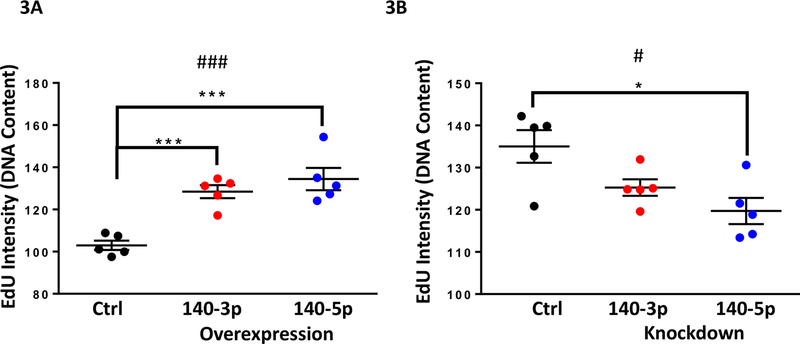
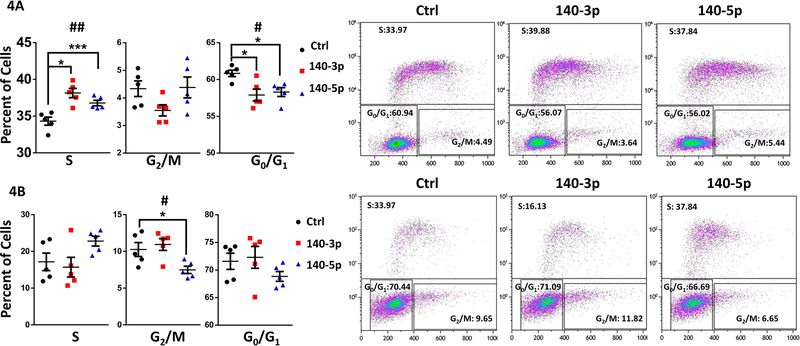
We further investigated the effects of miR-140 overexpression or inhibition on NSC growth through its effects on the cell cycle (for data on efficacy of overexpression and knockdown, see Supplementary Figures 5A-5D). After pulse-labeling cells with the nucleotide analog EdU for 1-hour, we found that both miR-140–3p and miR-140–5p overexpression significantly increased EdU incorporation, suggesting an increased rate of DNA-synthesis (ANOVA F(2,12)=19.58, p=0.0002; Figure 3A). Likewise, transfection with miR-140–3p and miR-140–5p antagomirs significantly decreased EdU incorporation (ANOVA F(2,12)=6.309, p=0.013), with post-hoc analysis indicating that miR-140–5p antagomir significantly reduced EdU incorporation (Dunnett’s post-hoc, p=0.009; Figure 3B). We also observed that both miRNA 140–3p and miR-140–5p overexpression significantly increased the proportion of actively dividing cells in S-phase (ANOVA F(2,12)=13.42, p=0.0009), while decreasing the proportion of cells in G0/G1-phase (ANOVA F(2,12)=6.421, p=0.013; Figure 4A). Contrastingly, the antagomir to miR-140–3p had minimal effects on cell cycle, whereas only the antagomir to miR-140–5p had a significant effect of decreasing the proportion of cells in G2/GM-phase (ANOVA F(2,12)=5.616, p=0.019; Dunnett’s post-hoc, p=0.047; Figure 4B). Furthermore, neither overexpression nor inhibition of miR-140–3p or miR-140–5p resulted in increased NSC apoptosis (Figures 5A and 5B) as measure by a Caspase 3/7 activation assay. Collectively, our data suggests that miR-140–3p, mirroring the effects of ethanol exposure (Santillano et al., 2005), promotes NSC proliferation through its actions on the cell cycle, without inducing cell death.
Figure 3. miR-140–3p and miR-140–5p Regulate the Rate of DNA Synthesis.
3A) EdU Intensity of NSCs following miR-140–3p or miR-140–5p overexpression
3B) EdU Intensity of NSCs following miR-140–3p or miR-140–5p inhibition
n=5 samples per group, #p<0.05, ###p<0.001 by ANOVA. *p<0.05, ***p<0.001 by Dunnett’s post-hoc.
Figure 4. miR-140 Influences Cell Cycle Dynamics.
Proportion of cells in S, G0/G1, or G2/M phase of the cell cycle following miR-140–3p and miR-140–5p overexpression (4A) or inhibition (4B). Representative flow cytometry images are displayed on the right panel.
n=5 samples per group, #p<0.05, ##p<0.01 by ANOVA. *p<0.05, ***p<0.001 by Dunnett’s post-hoc.
Figure 5. miR-140 Does Not Influence Apoptotic Cell Death.
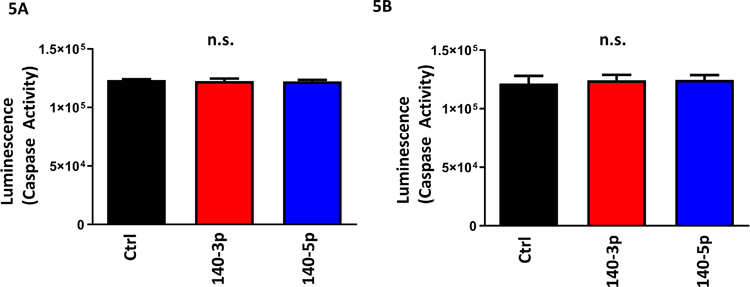
Quantification of caspase 3/7 activity following miR-140–3p and miR-140–5p overexpression (4A) or inhibition (4B).
n=5 samples per group, n.s. indicates no significance.
miR-140–3p Overexpression During Neural Differentiation Promotes an Aberrant Astrogliogenic Fate
To determine whether miR-140 might also influence NSC maturation, we first examined miR-140 levels during a 3-day mitogen (EGF/LIF)-withdrawal-induced NSC differentiation paradigm. Following the withdrawal of FGF, and in the presence of the extracellular matrix protein laminin, neurospheres become adherent. Subsequently, progenitors with a bi-polar morphology, resembling migratory neurons, proceed to migrate away from the neurosphere core and express neuronal markers like neurofilament (Figure 6A). In previous studies, we showed that these migratory cells express neuronal lineage markers and represent early neuronal lineage commitment (Camarillo et al., 2007, Camarillo and Miranda, 2008). We found that, in control, non-ethanol-exposed cultures, intracellular miR-140–3p expression significantly decreased during differentiation (ANOVA F(2,42)=6.76, p=0.003), whereas miR-140–5p expression was not affected by differentiation (Figures 6B and 6C).
Figure 6. miR-140–3p Expression Decreases During NSC differentiation.
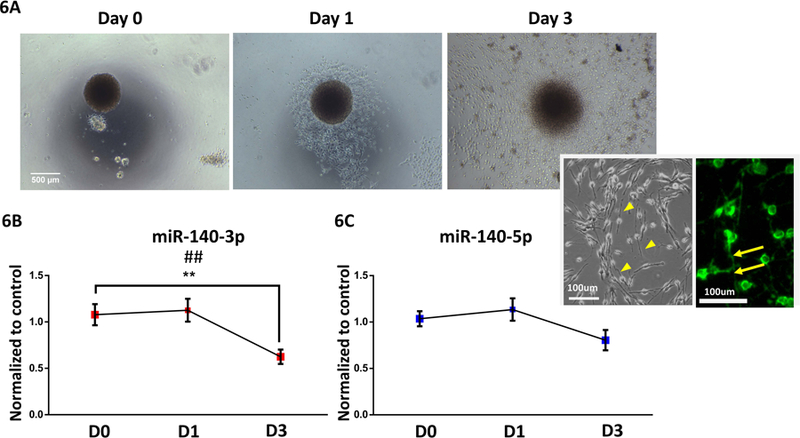
6A) Photomicrographs of a neurosphere over a 3-day mitogen-withdrawal-induced differentiation paradigm. Inset images depict the bipolar morphological appearance and immunolabeling with anti-neurofilament antibody of maturing, migratory neurons. Arrow heads demarcate axonal processes of bipolar neurons and arrows indicate a neurofilament labeled axon.
6B) miR-140–3p and 6C) mir-140–5p expression in NSCs over a three-day mitogen-withdrawal-induced differentiation paradigm
n=15 samples per group, ##p<0.01 by ANOVA, **p<0.01 by Dunnett’s post-hoc. D0, D1, and D3 indicate days 0, 1, and 3 of differentiation, respectively.
Interestingly, when miR-140–3p was overexpressed during NSC differentiation (Supplementary Figures 6A and 6B), we noted that on day 3 of differentiation, the mRNA expression of GFAP, an astrocytic marker, was increased (day x treatment interaction, ANOVA F(2,23)=6.468, p=0.0059; Sidak’s post-hoc, p=0.0006), while the expression of GLAST mRNA was decreased (day x treatment interaction, ANOVA F(2,23)=16.79, p<0.0001; Sidak’s post-hoc, p<0.0001, Figures 7A and 7B). Mir-140–3p overexpression also significantly decreased mRNA expression for the early neuronal differentiation marker, DCX (day x treatment interaction, ANOVA F(2,23)=7.88, p=0.003; Sidak’s post-hoc, p=0.0005), as well as for the later neuronal lineage commitment marker, NeuN (day x treatment interaction, ANOVA F(2,23)=9.49, p=0.001; Sidak’s post-hoc, p=0.002, Figures 7C and 7D). There was also a significant decrease in mRNA expression for the oligodendrocytic marker, PDGFRα (day x treatment interaction, ANOVA F(2,23)=5.98, Sidak’s post-hoc, p=0.0081; Figure 7E). While there was only a marginally significant interaction effect (ANOVA F(2,23)=2.85, p=0.078) between differentiation and miR-140–3p overexpression on expression of a second oligodendrocyte marker, Olig2, a planned comparison indicated that miR-140–3p overexpression also significantly decreased Olig2 mRNA expression on day 3 (Sidak’s post-hoc, p=0.027; Figure 7F).
Figure 7. miR-140–3p Influences NSC Differentiation.
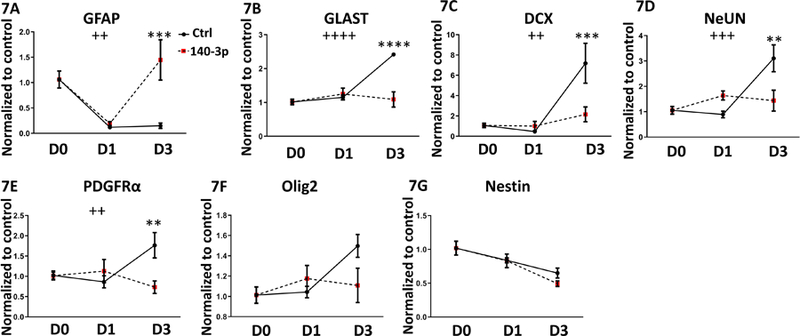
Expression of 7A) GFAP, 7B) Glast, 7C) DCX, 7D) NeUN, 7E) PDGFRα, 7F) Olig2, and 7G) Nestin in NSCs following miR-140–3p overexpression in a three-day mitogen-withdrawal-induced differentiation paradigm.
n=5 samples per group, ++p<0.01, +++p<0.001, ++++p<0.0001 interaction effect between differentiation and miR-140–3p overexpression, ANOVA. **p<0.01, ***p<0.001, ****p<0.0001 by Sidak’s post-hoc. D0, D1, and D3 indicate days 0, 1, and 3 of differentiation, respectively.
Compared to miR-140–3p, miR-140–5p overexpression had a more restricted effect on differentiation. MiR-140–5p decreased GLAST expression on day 3 of differentiation (day x treatment interaction, ANOVA F(2,22)=12.87, p=0.0002, Sidak’s post-hoc, p<0.0001), but did not affect GFAP expression (Figures 8A and 8B). While there were minimal effects on neural differentiation (Figures 8C and 8D), Olig2 (day x treatment interaction, ANOVA F(2,22)=4.80, p=0.019; Sidak’s post-hoc, p=0.005) and PDGFRα (day x treatment interaction, ANOVA F(2,22)=4.12, p=0.03,; Sidak’s post-hoc, p=0.04) were also significantly decreased on day 3 of differentiation compared to controls (Figures 8E and 8F). Interestingly, we observed a main effect of miR-140–5p overexpression, resulting in decreased Nestin mRNA expression throughout the differentiation period (ANOVA F(1,22)=5.807, p=0.025; Figure 8G). Collectively, our data show that miRNA-140–3p overexpression results in aberrant astroglial-directed differentiation, while suppressing neuronal and oligodendrocytic differentiation.
Figure 8. miR-140–5p Influences NSC Differentiation.
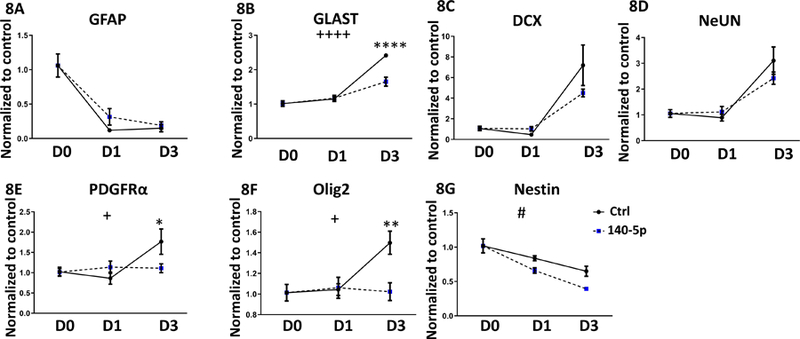
Expression of 8A) GFAP, 8B) Glast, 8C) DCX, 8D) NeUN, 8E) PDGFRα, 8F) Olig2, and 8G) Nestin in NSCs following miR-140–5p overexpression in a three-day mitogen-withdrawal-induced differentiation paradigm.
n=5 samples per group, ++p<0.01, +++p<0.001, ++++p<0.0001 interaction effect between differentiation and miR-140–5p overexpression, ANOVA. #p<0.05 main effect of miR-140–5p overexpression, ANOVA. **p<0.01, ***p<0.001, ****p<0.0001 by Sidak’s post-hoc. D0, D1, and D3 indicate days 0, 1, and 3 of differentiation, respectively.
miR-140–3/5p are encoded as intronic miRNAs within the host gene, Wwp2 (miR-140HG). We therefore used Wwp2/miR-140HG expression to identify presumptive miR140-transcribing NSCs and assess, in this subpopulation, using a single-cell RNAseq approach, the relationship between miR-140HG expression and markers of maturation. We found that Wwp2/miR-140HG was expressed in 972 out of a total of 13,770 assessed cells, suggesting that ~7% of all neural progenitor cells contribute to the high levels of miR-140–3p in EVs. Moreover, we found that even in Wwp2/miR-140HG-positive cells, there was a large, 753-fold range (0.11–2.99 log10 RPKM) in Wwp2/miR140HG expression, suggesting the possibility of a similarly large dynamic range in miR140–3p expression as well. Consistent with our overexpression studies, the expression of miR-140 host transcript was significantly negatively correlated with Nestin expression and positively correlated with GFAP expression indicating that cells that highly express miR-140HG also highly express GFAP (Figure 9A).
Figure 9. miR-140 host gene expression is correlated with glial markers.
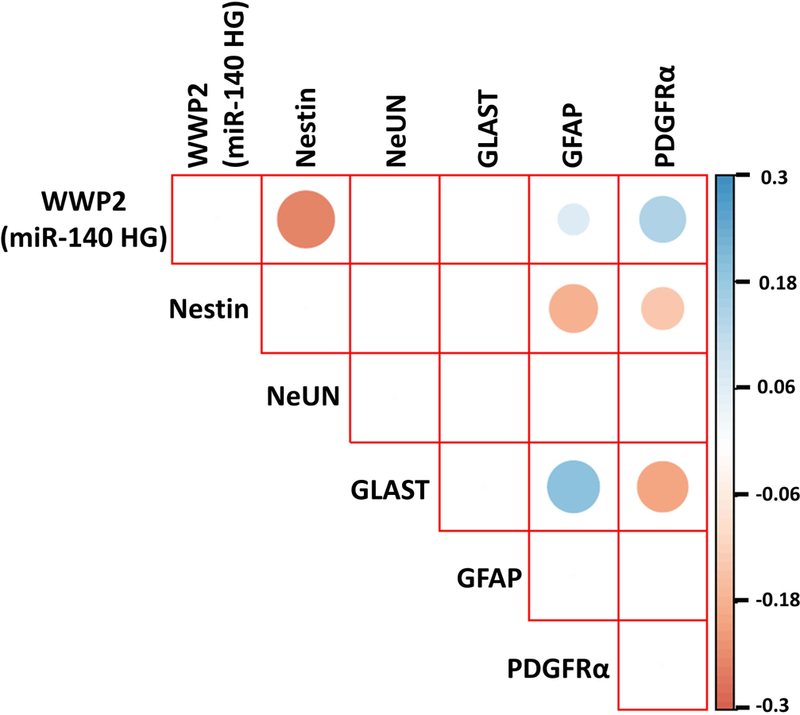
Correlation plot of WWP2, the mir-140 host gene (miR-140 HG), with different markers of neural and glial maturation.
Discussion
Extracellular vesicles represent an important mode of paracrine communication due to their ability to interact with various cell types and tissues. Specifically, tumor invasion and metastases can be promoted by metalloproteases released from tumor cell-derived EVs (Muralidharan-Chari et al., 2010). Cancer cells have also been observed to propagate oncogenes and their cancer-associated transforming phenotype through EVs (Al-Nedawi et al., 2008). Recent studies have also shown that EVs within the central nervous system serve as a mode of intercellular communication for both neurons and glia by trafficking growth factors and other molecules (Morel et al., 2013, Ratajczak et al., 2006, Morton et al., 2018, Muralidharan-Chari et al., 2010).
Stem cells also produce an abundance of EVs (Drago et al., 2013, Katsuda et al., 2013, Tetta et al., 2013). Studies have shown that EVs released from stem cells, including NSCs, traffic cargo (including proteins, mRNAs, and miRNAs) to target cells, influencing their behavior. A recent study demonstrated that CD63-GFP-expressing NSC-derived EVs were readily up-taken by embryonic NSCs and astrocytes in vitro (Yoshimura et al., 2018). Functionally, NSC-derived EVs have been observed to transfer IFN-γ to activate Stat1 signaling in target cells (Cossetti et al., 2014). EVs may also play an important role in maintaining the stem cell phenotype. Indeed, EVs released by stem cells have been credited for their pro-regenerative ability by enhancing cell proliferation, inhibiting apoptosis, and promoting immune tolerance of recipient cells (Grange et al., 2017, Gai et al., 2016, Zhan et al., 2015, Bruno et al., 2016). Our previous studies have documented that ethanol exposure significantly reduced the numbers of cells expressing stem cell markers CD117, CD133, Sca-1, and ABCG2, and suggested that ethanol depletes neuroepithelial cells by promoting premature maturation (Santillano et al., 2005). Interestingly, EVs are known to be released from cells as a response to physiologic stress and environmental stimuli (H Rashed et al., 2017). Thus, it is feasible that ethanol’s effects on the developing neuroepithelium may be mediated through its actions on NSC-derived EVs.
In this study, we characterized the effects of ethanol exposure on our NSC-derived EV cargo and the implication of these effects on NSC growth and maturation. We found that while ethanol exposure did not alter numbers or sizes of NSC-derived EVs, it significantly altered their miRNA content, with miR-140–3p being the most significantly increased miRNA. We, and others, previously reported that intracellular expression of miR-140–3p is both ethanol and nicotine sensitive (Balaraman et al., 2012, Huang and Li, 2009). In this study, we found that a 72-hour period of ethanol exposure increased miR140–3p levels in both NSCs, and in NEC-derived EVs.
Furthermore, we observed that miR-140–3p overexpression significantly increased NSC proliferation through its effects on the cell cycle, mirroring observed effects of ethanol exposure (Santillano et al., 2005) on NSC growth. In the context of a stereotypic mitogen-withdrawal-stimulated NSC maturation paradigm, miR-140–3p promoted aberrant GFAP-mRNAhi/GLAST-mRNAlo astroglial differentiation, while suppressing neuronal and oligodendroglial lineage markers. Single-cell RNAseq analysis demonstrated a similar positive association between expression of mRNA transcripts from the Wwp2/miR-140HG locus which encodes miR-140–3p, and the expression of GFAP, but not GLAST, suggesting that alcohol amplifies a normally existing relationship between miR-140–3p and gliogenesis. The presence of neurons is required for GLAST expression in astrocytes (Swanson et al., 1997, Perego et al., 2000). Consequently, the loss of neuronal linage following miR-140–3p overexpression may indirectly result in aberrant astrocytic maturation and contribute to aberrant astrocyte function that has been associated with prenatal alcohol exposure (Wilhelm et al., 2018). The loss of oligodendroglial markers following miR-140–3p overexpression is also consistent with the appearance of white matter abnormalities in FASD (Norman et al., 2009) and loss of oligodendrocytes (Newville et al., 2017) in models of PAE. Thus, dysregulated miRNA content of neural progenitor EVs following ethanol exposure may underlie some of the aberrant brain maturation associated with FASD. While we did not investigate the direct targets of miR-140–3p and miR-140–5p that mediated their effects on NSC differentiation, it is likely that these miRNAs influence multiple pathways involved in neural maturation. In fact, Ingenuity Pathway Analysis™ indicates that the predicted targets of miR-140–3p are overrepresented in several developmental pathways including for example, the PTEN and 14–3-3 protein pathways (Supplementary Figure 7), which is extensively involved in NSC maturation, migration, and cell-division (Duan et al., 2015, Wen et al., 2013, Lee et al., 2019, Gregorian et al., 2009, Toyo-oka et al., 2014). Furthermore, this study examined the effects of miRNAs on neural development in isolation. Emerging research has shown that miRNAs are likely to work in concert to mediate disease pathology, including in FASD growth parameters (Tseng et al., 2019).
MicroRNAs in EVs have recently been implicated in ethanol activation of inflammatory neurotoxicity (Coleman et al., 2017) and hepatotoxicity (Massey et al., 2018) in the adult, and extracellular miRNAs have been identified as biomarkers for the severity of prenatal alcohol effects in infants (Balaraman et al., 2016). This initial study to characterize the effects of ethanol exposure on the miRNA content of EVs derived from fetal NSCs likely feeds into an emerging hypothesis that packaged extracellular RNAs are significant contributors to ethanol effects in both development and the adult. Given that EVs are an important mode of paracrine communication for NSCs and may play an important role in NSC renewal and differentiation, alterations in the miRNA content of EVs may underlie aberrant neural maturation in response to environmental factors including PAE. Future studies should focus on determining how ethanol influences packaging of NSC-derived EVs as well as their trafficking to target cells. Ultimately, exploiting the paracrine role of EVs may be a means to mitigate the effects of PAE, as has been shown with other pathologies (Wang et al., 2017, Bian et al., 2014, Kim et al., 2016).
Supplementary Material
Acknowledgments:
We thank KunHee Han and Dong-Ki Kim and Darwin Prockop at the Texas A&M University (TAMU) Institute for Regenerative Medicine for Nanoparticle Tracking Analysis, as well as Min Woo Sung and Stanislav Vitha in the TAMU Microscopy and Imaging Center for Transmission Electron Microscopy imaging. We would also like to thank Andrew Hillhouse and Kranti Koganti at the TAMU Institute for Genome Sciences and Society. This work was supported by a grant from the NIH, R01 AA024659 (RCM) and F31AA026505 (AMT).
Footnotes
Conflict of Interest Statement:
The authors declare no conflict of interest.
References:
- ACOG 2011. Committee opinion no. 496: At-risk drinking and alcohol dependence: obstetric and gynecologic implications. Obstet Gynecol, 118, 383–8. [DOI] [PubMed] [Google Scholar]
- ADACHI J, MIZOI Y, FUKUNAGA T, OGAWA Y, UENO Y & IMAMICHI H 1991. Degrees of alcohol intoxication in 117 hospitalized cases. J Stud Alcohol, 52, 448–53. [DOI] [PubMed] [Google Scholar]
- AL-NEDAWI K, MEEHAN B, MICALLEF J, LHOTAK V, MAY L, GUHA A & RAK J 2008. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol, 10, 619–24. [DOI] [PubMed] [Google Scholar]
- BAGLIO SR, ROOIJERS K, KOPPERS-LALIC D, VERWEIJ FJ, PÉREZ LANZÓN M, ZINI N, NAAIJKENS B, PERUT F, NIESSEN HWM, BALDINI N & PEGTEL DM 2015. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Research & Therapy, 6, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BALARAMAN S, LUNDE ER, SAWANT O, CUDD TA, WASHBURN SE & MIRANDA RC 2014. Maternal And Neonatal Plasma MicroRNA Biomarkers For Fetal Alcohol Exposure In An Ovine Model. Alcoholism, clinical and experimental research, 38, 1390–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BALARAMAN S, SCHAFER JJ, TSENG AM, WERTELECKI W, YEVTUSHOK L, ZYMAK-ZAKUTNYA N, CHAMBERS CD & MIRANDA RC 2016. Plasma miRNA Profiles in Pregnant Women Predict Infant Outcomes following Prenatal Alcohol Exposure. PLOS ONE, 11, e0165081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BALARAMAN S, TINGLING JD, TSAI P-C & MIRANDA RC 2013. Dysregulation of microRNA Expression and Function Contributes to the Etiology of Fetal Alcohol Spectrum Disorders. Alcohol Research : Current Reviews, 35, 18–24. [PMC free article] [PubMed] [Google Scholar]
- BALARAMAN S, WINZER-SERHAN UH & MIRANDA RC 2012. Opposing actions of ethanol and nicotine on microRNAs are mediated by nicotinic acetylcholine receptors in fetal cerebral cortical-derived neural progenitor cells. Alcoholism, clinical and experimental research, 36, 1669–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BIAN S, ZHANG L, DUAN L, WANG X, MIN Y & YU H 2014. Extracellular vesicles derived from human bone marrow mesenchymal stem cells promote angiogenesis in a rat myocardial infarction model. J Mol Med (Berl), 92, 387–97. [DOI] [PubMed] [Google Scholar]
- BRUNO S, PORTA S & BUSSOLATI B 2016. Extracellular vesicles in renal tissue damage and regeneration. Eur J Pharmacol, 790, 83–91. [DOI] [PubMed] [Google Scholar]
- BYSTRON I, BLAKEMORE C & RAKIC P 2008. Development of the human cerebral cortex: Boulder Committee revisited. Nature Reviews Neuroscience, 9, 110. [DOI] [PubMed] [Google Scholar]
- CAMARILLO C, KUMAR LS, BAKE S, SOHRABJI F & MIRANDA RC 2007. Ethanol regulates angiogenic cytokines during neural development: Evidence from an in vitro model of mitogen-withdrawal induced cerebral cortical neuroepithelial differentiation. Alcoholism, clinical and experimental research, 31, 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAMARILLO C & MIRANDA RC 2008. Ethanol exposure during neurogenesis induces persistent effects on neural maturation: evidence from an ex vivo model of fetal cerebral cortical neuroepithelial progenitor maturation. Gene expression, 14, 159–171. [PMC free article] [PubMed] [Google Scholar]
- CHEEMA ZF, WEST JR & MIRANDA RC 2000. Ethanol induces Fas/Apo [apoptosis]-1 mRNA and cell suicide in the developing cerebral cortex. Alcohol Clin Exp Res, 24, 535–543. [PubMed] [Google Scholar]
- COLEMAN LG JR., ZOU J & CREWS FT 2017. Microglial-derived miRNA let-7 and HMGB1 contribute to ethanol-induced neurotoxicity via TLR7. J Neuroinflammation, 14, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COSSETTI C, IRACI N, MERCER TR, LEONARDI T, ALPI E, DRAGO D, ALFARO-CERVELLO C, SAINI HK, DAVIS MP, SCHAEFFER J, VEGA B, STEFANINI M, ZHAO C, MULLER W, GARCIA-VERDUGO JM, MATHIVANAN S, BACHI A, ENRIGHT AJ, MATTICK JS & PLUCHINO S 2014. Extracellular vesicles from neural stem cells transfer IFN-gamma via Ifngr1 to activate Stat1 signaling in target cells. Mol Cell, 56, 193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DRAGO D, COSSETTI C, IRACI N, GAUDE E, MUSCO G, BACHI A & PLUCHINO S 2013. The stem cell secretome and its role in brain repair. Biochimie, 95, 2271–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DRAGOVIC RA, GARDINER C, BROOKS AS, TANNETTA DS, FERGUSON DJ, HOLE P, CARR B, REDMAN CW, HARRIS AL, DOBSON PJ, HARRISON P & SARGENT IL 2011. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine, 7, 780–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DUAN S, YUAN G, LIU X, REN R, LI J, ZHANG W, WU J, XU X, FU L, LI Y, YANG J, ZHANG W, BAI R, YI F, SUZUKI K, GAO H, ESTEBAN CR, ZHANG C, IZPISUA BELMONTE JC, CHEN Z, WANG X, JIANG T, QU J, TANG F & LIU G-H 2015. PTEN deficiency reprogrammes human neural stem cells towards a glioblastoma stem cell-like phenotype. Nature communications, 6, 10068–10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EL ANDALOUSSI S, MÄGER I, BREAKEFIELD XO & WOOD MJA 2013. Extracellular vesicles: biology and emerging therapeutic opportunities. Nature Reviews Drug Discovery, 12, 347. [DOI] [PubMed] [Google Scholar]
- FINER LB & ZOLNA MR 2016. Declines in Unintended Pregnancy in the United States, 2008–2011. N Engl J Med, 374, 843–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAI C, CARPANETTO A, DEREGIBUS MC & CAMUSSI G 2016. Extracellular vesicle-mediated modulation of angiogenesis. Histol Histopathol, 31, 379–91. [DOI] [PubMed] [Google Scholar]
- GARDINER C, FERREIRA YJ, DRAGOVIC RA, REDMAN CWG & SARGENT IL 2013. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. Journal of Extracellular Vesicles, 2, 19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRANGE C, IAMPIETRO C & BUSSOLATI B 2017. Stem cell extracellular vesicles and kidney injury. Stem Cell Investig, 4, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GREGORIAN C, NAKASHIMA J, LE BELLE J, OHAB J, KIM R, LIU A, SMITH KB, GROSZER M, GARCIA AD, SOFRONIEW MV, CARMICHAEL ST, KORNBLUM HI, LIU X & WU H 2009. Pten deletion in adult neural stem/progenitor cells enhances constitutive neurogenesis. The Journal of neuroscience : the official journal of the Society for Neuroscience, 29, 1874–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- H RASHED M, BAYRAKTAR E, G KH, ABD-ELLAH MF, AMERO P, CHAVEZ-REYES A & RODRIGUEZ-AGUAYO C 2017. Exosomes: From Garbage Bins to Promising Therapeutic Targets. Int J Mol Sci, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUANG W & LI MD 2009. Nicotine modulates expression of miR-140*, which targets the 3’-untranslated region of dynamin 1 gene (Dnm1). Int J Neuropsychopharmacol, 12, 537–46. [DOI] [PubMed] [Google Scholar]
- IRACI N, LEONARDI T, GESSLER F, VEGA B & PLUCHINO S 2016. Focus on Extracellular Vesicles: Physiological Role and Signalling Properties of Extracellular Membrane Vesicles. International Journal of Molecular Sciences, 17, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JANAS AM, SAPOŃ K, JANAS T, STOWELL MHB & JANAS T 2016. Exosomes and other extracellular vesicles in neural cells and neurodegenerative diseases. Biochimica et Biophysica Acta (BBA) - Biomembranes, 1858, 1139–1151. [DOI] [PubMed] [Google Scholar]
- KARLSEN TA, JAKOBSEN RB, MIKKELSEN TS & BRINCHMANN JE 2014. microRNA-140 targets RALA and regulates chondrogenic differentiation of human mesenchymal stem cells by translational enhancement of SOX9 and ACAN. Stem Cells Dev, 23, 290–304. [DOI] [PubMed] [Google Scholar]
- KATSUDA T, KOSAKA N, TAKESHITA F & OCHIYA T 2013. The therapeutic potential of mesenchymal stem cell-derived extracellular vesicles. Proteomics, 13, 1637–53. [DOI] [PubMed] [Google Scholar]
- KIM DK, NISHIDA H, AN SY, SHETTY AK, BARTOSH TJ & PROCKOP DJ 2016. Chromatographically isolated CD63+CD81+ extracellular vesicles from mesenchymal stromal cells rescue cognitive impairments after TBI. Proc Natl Acad Sci U S A, 113, 170–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAWSON C, VICENCIO JM, YELLON DM & DAVIDSON SM 2016. Microvesicles and exosomes: new players in metabolic and cardiovascular disease. J Endocrinol, 228, R57–71. [DOI] [PubMed] [Google Scholar]
- LEE H, THACKER S, SARN N, DUTTA R & ENG C 2019. Constitutional mislocalization of Pten drives precocious maturation in oligodendrocytes and aberrant myelination in model of autism spectrum disorder. Translational psychiatry, 9, 13–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI T, LI H, LI T, FAN J, ZHAO RC & WENG X 2014. MicroRNA expression profile of dexamethasone-induced human bone marrow-derived mesenchymal stem cells during osteogenic differentiation. J Cell Biochem, 115, 1683–91. [DOI] [PubMed] [Google Scholar]
- LLORENTE A, SKOTLAND T, SYLVANNE T, KAUHANEN D, ROG T, ORLOWSKI A, VATTULAINEN I, EKROOS K & SANDVIG K 2013. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim Biophys Acta, 1831, 1302–9. [DOI] [PubMed] [Google Scholar]
- LU L & RISCH HA 2016. Exosomes: potential for early detection in pancreatic cancer. Future Oncol, 12, 1081–90. [DOI] [PubMed] [Google Scholar]
- MAHNKE AH, SALEM NA, TSENG AM, CHUNG DD & MIRANDA RC 2018. Nonprotein-coding RNAs in Fetal Alcohol Spectrum Disorders. Prog Mol Biol Transl Sci, 157, 299–342. [DOI] [PubMed] [Google Scholar]
- MAIER SE, MILLER JA & WEST JR 1999. Prenatal binge-like alcohol exposure in the rat results in region-specific deficits in brain growth. Neurotoxicol Teratol, 21, 285–91. [DOI] [PubMed] [Google Scholar]
- MASSEY VL, QIN L, CABEZAS J, CABALLERIA J, SANCHO-BRU P, BATALLER R & CREWS FT 2018. TLR7-let-7 Signaling Contributes to Ethanol-Induced Hepatic Inflammatory Response in Mice and in Alcoholic Hepatitis. Alcohol Clin Exp Res, 42, 2107–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATHIVANAN S, JI H & SIMPSON RJ 2010. Exosomes: Extracellular organelles important in intercellular communication. Journal of Proteomics, 73, 1907–1920. [DOI] [PubMed] [Google Scholar]
- MAY PA, CHAMBERS CD, KALBERG WO, ZELLNER J, FELDMAN H, BUCKLEY D, KOPALD D, HASKEN JM, XU R, HONERKAMP-SMITH G, TARAS H, MANNING MA, ROBINSON LK, ADAM MP, ABDUL-RAHMAN O, VAUX K, JEWETT T, ELLIOTT AJ, KABLE JA, AKSHOOMOFF N, FALK D, ARROYO JA, HERELD D, RILEY EP, CHARNESS ME, COLES CD, WARREN KR, JONES KL & HOYME HE 2018. Prevalence of Fetal Alcohol Spectrum Disorders in 4 US Communities. JAMA, 319, 474–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCALHANY REJ, WEST JR & MIRANDA RC 2000. Glial-derived neurotrophic factor (GDNF) prevents ethanol-induced apoptosis and JUN kinase phosphorylation. Developmental Brain Research, 119, 209–216. [DOI] [PubMed] [Google Scholar]
- MILLER M 1989. Effects of prenatal exposure to ethanol on neocortical development: II. Cell proliferation in the ventricular and subventricular zones of the rat. The Journal of Comparative Neurology, 287 326–338. [DOI] [PubMed] [Google Scholar]
- MILLER MW & NOWAKOWSKI RS 1991. Effect of prenatal exposure to ethanol on the cell cycle kinetics and growth fraction in the proliferative zones of fetal rat cerebral cortex. Alcohol Clin Exp Res, 15, 229–32. [DOI] [PubMed] [Google Scholar]
- MIRANDA RC 2014. MicroRNAs and ethanol toxicity. Int Rev Neurobiol, 115, 245–84. [DOI] [PubMed] [Google Scholar]
- MIRANDA RC, SANTILLANO DR, CAMARILLO C & DOHRMAN D 2008. Modeling the impact of alcohol on cortical development in a dish: strategies from mapping neural stem cell fate. Methods Mol Biol, 447, 151–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOONEY SM & MILLER MW 2003. Ethanol-induced neuronal death in organotypic cultures of rat cerebral cortex. Brain Res Dev Brain Res, 147, 135–41. [DOI] [PubMed] [Google Scholar]
- MOREL L, REGAN M, HIGASHIMORI H, NG SK, ESAU C, VIDENSKY S, ROTHSTEIN J & YANG Y 2013. Neuronal exosomal miRNA-dependent translational regulation of astroglial glutamate transporter GLT1. J Biol Chem, 288, 7105–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORTON MC, NECKLES VN, SELUZICKI CM, HOLMBERG JC & FELICIANO DM 2018. Neonatal Subventricular Zone Neural Stem Cells Release Extracellular Vesicles that Act as a Microglial Morphogen. Cell Rep, 23, 78–89. [DOI] [PubMed] [Google Scholar]
- MURALIDHARAN-CHARI V, CLANCY JW, SEDGWICK A & D’SOUZA-SCHOREY C 2010. Microvesicles: mediators of extracellular communication during cancer progression. J Cell Sci, 123, 1603–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEWVILLE J, VALENZUELA CF, LI L, JANTZIE LL & CUNNINGHAM LA 2017. Acute oligodendrocyte loss with persistent white matter injury in a third trimester equivalent mouse model of fetal alcohol spectrum disorder. Glia, 65, 1317–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NORMAN AL, CROCKER N, MATTSON SN & RILEY EP 2009. Neuroimaging and fetal alcohol spectrum disorders. Dev Disabil Res Rev, 15, 209–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OSTEIKOETXEA X, BALOGH A, SZABÓ-TAYLOR K, NÉMETH A, SZABÓ TG, PÁLÓCZI K, SÓDAR B, KITTEL Á, GYÖRGY B, PÁLLINGER É, MATKÓ J & BUZÁS EI 2015. Improved Characterization of EV Preparations Based on Protein to Lipid Ratio and Lipid Properties. PLoS ONE, 10, e0121184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAPPALARDO-CARTER DL, BALARAMAN S, SATHYAN P, CARTER ES, CHEN WJ & MIRANDA RC 2013. Suppression and epigenetic regulation of MiR-9 contributes to ethanol teratology: evidence from zebrafish and murine fetal neural stem cell models. Alcohol Clin Exp Res, 37, 1657–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEREGO C, VANONI C, BOSSI M, MASSARI S, BASUDEV H, LONGHI R & PIETRINI G 2000. The GLT-1 and GLAST glutamate transporters are expressed on morphologically distinct astrocytes and regulated by neuronal activity in primary hippocampal cocultures. J Neurochem, 75, 1076–84. [DOI] [PubMed] [Google Scholar]
- PERPER JA, TWERSKI A & WIENAND JW 1986. Tolerance at high blood alcohol concentrations: a study of 110 cases and review of the literature. J Forensic Sci, 31, 212–21. [PubMed] [Google Scholar]
- PIROLA CARLOS J, GIANOTTI TOMAS F, CASTAÑO GUSTAVO O & SOOKOIAN S 2012. Circulating MicroRNA‐122 signature in nonalcoholic fatty liver disease and cardiovascular disease: A new endocrine system in metabolic syndrome. Hepatology, 57, 2545–2547. [DOI] [PubMed] [Google Scholar]
- POPOVA S, LANGE S, PROBST C, GMEL G & REHM J 2017. Estimation of national, regional, and global prevalence of alcohol use during pregnancy and fetal alcohol syndrome: a systematic review and meta-analysis. Lancet Glob Health, 5, e290–e299. [DOI] [PubMed] [Google Scholar]
- PROCK TL & MIRANDA RC 2007. Embryonic Cerebral Cortical Progenitors Are Resistant to Apoptosis, but Increase Expression of Suicide Receptor DISC-complex Genes and Suppress Autophagy Following Ethanol Exposure. Alcoholism, clinical and experimental research, 31, 694–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RATAJCZAK J, WYSOCZYNSKI M, HAYEK F, JANOWSKA-WIECZOREK A & RATAJCZAK MZ 2006. Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia, 20, 1487–95. [DOI] [PubMed] [Google Scholar]
- ROOZEN S, PETERS GJ, KOK G, TOWNEND D, NIJHUIS J & CURFS L 2016. Worldwide Prevalence of Fetal Alcohol Spectrum Disorders: A Systematic Literature Review Including Meta-Analysis. Alcohol Clin Exp Res, 40, 18–32. [DOI] [PubMed] [Google Scholar]
- SALONE V, HENRIONNET C, BRANLANT C, GILLET P & PINZANO A 2014. Both miRNA-29b downregulation and miRNA-140 overexpression drive respectively MSC proliferation and chondrogenic differentiation in collagen scaffold. Osteoarthritis and Cartilage, 22, S319–S320. [Google Scholar]
- SAMHSA. 2013. The NSDUH Report: 18 percent of pregnant women drink alcohol during early pregnancy. NSDUH Report [Online]. Available: http://www.samhsa.gov/data/sites/default/files/spot123-pregnancy-alcohol-2013/spot123-pregnancy-alcohol-2013.pdf [Accessed 2013].
- SANTILLANO DR, KUMAR LS, PROCK TL, CAMARILLO C, TINGLING JD & MIRANDA RC 2005. Ethanol induces cell-cycle activity and reduces stem cell diversity to alter both regenerative capacity and differentiation potential of cerebral cortical neuroepithelial precursors. BMC Neuroscience, 6, 59–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SATHYAN P, GOLDEN HB & MIRANDA RC 2007. Competing Interactions between Micro-RNAs Determine Neural Progenitor Survival and Proliferation after Ethanol Exposure: Evidence from an <em>Ex Vivo</em> Model of the Fetal Cerebral Cortical Neuroepithelium. The Journal of Neuroscience, 27, 8546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SELVAMANI A, WILLIAMS MH, MIRANDA RC & SOHRABJI F 2014. Circulating miRNA profiles provide a biomarker for severity of stroke outcomes associated with age and sex in a rat model. Clin Sci (Lond), 127, 77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOHEL MH 2016. Extracellular/Circulating MicroRNAs: Release Mechanisms, Functions and Challenges. Achievements in the Life Sciences, 10, 175–186. [Google Scholar]
- STREISSGUTH AP & O’MALLEY K 2000. Neuropsychiatric implications and long-term consequences of fetal alcohol spectrum disorders. Semin Clin Neuropsychiatry, 5, 177–90. [DOI] [PubMed] [Google Scholar]
- SUBRA C, LAULAGNIER K, PERRET B & RECORD M 2007. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie, 89, 205–12. [DOI] [PubMed] [Google Scholar]
- SWANSON RA, LIU J, MILLER JW, ROTHSTEIN JD, FARRELL K, STEIN BA & LONGUEMARE MC 1997. Neuronal regulation of glutamate transporter subtype expression in astrocytes. J Neurosci, 17, 932–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAL TL, FRANZOSA JA, TILTON SC, PHILBRICK KA, IWANIEC UT, TURNER RT, WATERS KM & TANGUAY RL 2012. MicroRNAs control neurobehavioral development and function in zebrafish. FASEB J, 26, 1452–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAYLOR D & GERCEL-TAYLOR C 2013. The origin, function, and diagnostic potential of RNA within extracellular vesicles present in human biological fluids. Frontiers in Genetics, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TETTA C, GHIGO E, SILENGO L, DEREGIBUS MC & CAMUSSI G 2013. Extracellular vesicles as an emerging mechanism of cell-to-cell communication. Endocrine, 44, 11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THERY C, OSTROWSKI M & SEGURA E 2009. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol, 9, 581–593. [DOI] [PubMed] [Google Scholar]
- THÉRY C, ZITVOGEL L & AMIGORENA S 2002. Exosomes: composition, biogenesis and function. Nature Reviews Immunology, 2, 569. [DOI] [PubMed] [Google Scholar]
- TINGLING JD, BAKE S, HOLGATE R, RAWLINGS J, NAGSUK PP, CHANDRASEKHARAN J, SCHNEIDER SL & MIRANDA RC 2013. CD24 expression identifies teratogen-sensitive fetal neural stem cell subpopulations: evidence from developmental ethanol exposure and orthotopic cell transfer models. PLoS One, 8, e69560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TOYO-OKA K, WACHI T, HUNT RF, BARABAN SC, TAYA S, RAMSHAW H, KAIBUCHI K, SCHWARZ QP, LOPEZ AF & WYNSHAW-BORIS A 2014. 14–3-3ε and ζ Regulate Neurogenesis and Differentiation of Neuronal Progenitor Cells in the Developing Brain. The Journal of Neuroscience, 34, 12168–12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TSAI P-C, BAKE S, BALARAMAN S, RAWLINGS J, HOLGATE RR, DUBOIS D & MIRANDA RC 2014. MiR-153 targets the nuclear factor-1 family and protects against teratogenic effects of ethanol exposure in fetal neural stem cells. Biology Open, 3, 741–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TSENG AM, MAHNKE AH, WELLS AB, SALEM NA, ALLAN AM, ROBERTS VH, NEWMAN N, WALTER NA, KROENKE CD, GRANT KA, AKISON LK, MORITZ KM, CHAMBERS CD, MIRANDA RC & COLLABORATIVE INITIATIVE ON FETAL ALCOHOL SPECTRUM, D. 2019. Maternal circulating miRNAs that predict infant FASD outcomes influence placental maturation. Life science alliance, 2, e201800252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VANGIPURAM SD & LYMAN WD 2010. Ethanol alters cell fate of fetal human brain-derived stem and progenitor cells. Alcohol Clin Exp Res, 34, 1574–83. [DOI] [PubMed] [Google Scholar]
- WANG N, CHEN C, YANG D, LIAO Q, LUO H, WANG X, ZHOU F, YANG X, YANG J, ZENG C & WANG WE 2017. Mesenchymal stem cells-derived extracellular vesicles, via miR-210, improve infarcted cardiac function by promotion of angiogenesis. Biochim Biophys Acta Mol Basis Dis, 1863, 2085–2092. [DOI] [PubMed] [Google Scholar]
- WEN Y, LI W, CHOUDHURY GR, HE R, YANG T, LIU R, JIN K & YANG SH 2013. Astroglial PTEN Loss Disrupts Neuronal Lamination by Dysregulating Radial Glia-guided Neuronal Migration. Aging Dis, 4, 113–26. [PMC free article] [PubMed] [Google Scholar]
- WILHELM CJ, HASHIMOTO JG, ROBERTS ML, ZHANG X, GOEKE CM, BLOOM SH & GUIZZETTI M 2018. Plasminogen activator system homeostasis and its dysregulation by ethanol in astrocyte cultures and the developing brain. Neuropharmacology, 138, 193–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOSHIMURA A, ADACHI N, MATSUNO H, KAWAMATA M, YOSHIOKA Y, KIKUCHI H, ODAKA H, NUMAKAWA T, KUNUGI H, OCHIYA T & TAMAI Y 2018. The Sox2 promoter-driven CD63-GFP transgenic rat model allows tracking of neural stem cell-derived extracellular vesicles. Disease Models & Mechanisms, 11, dmm028779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHAN C, MA CB, YUAN HM, CAO BY & ZHU JJ 2015. Macrophage-derived microvesicles promote proliferation and migration of Schwann cell on peripheral nerve repair. Biochem Biophys Res Commun, 468, 343–8. [DOI] [PubMed] [Google Scholar]
- ZHANG J, LI S, LI L, LI M, GUO C, YAO J & MI S 2015. Exosome and Exosomal MicroRNA: Trafficking, Sorting, and Function. Genomics, Proteomics & Bioinformatics, 13, 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.