Abstract
Type 2 diabetes (T2D) affects the health of millions of people worldwide. The identification of genetic determinants associated with changes in glycemia over time might illuminate biological features that precede the development of T2D. Here we conducted a genome-wide association study of longitudinal fasting glucose changes in up to 13,807 non-diabetic individuals of European descent from nine cohorts. Fasting glucose change over time was defined as the slope of the line defined by multiple fasting glucose measurements obtained over up to 14 years of observation. We tested for associations of genetic variants with inverse-normal transformed fasting glucose change over time adjusting for age at baseline, sex, and principal components of genetic variation. We found no genome-wide significant association (P < 5 × 10−8) with fasting glucose change over time. Seven loci previously associated with T2D, fasting glucose or HbA1c were nominally (P < 0.05) associated with fasting glucose change over time. Limited power influences unambiguous interpretation, but these data suggest that genetic effects on fasting glucose change over time are likely to be small. A public version of the data provides a genomic resource to combine with future studies to evaluate shared genetic links with T2D and other metabolic risk traits.
Subject terms: Medical genetics, Type 2 diabetes
Introduction
Type 2 diabetes mellitus (T2D), a disease characterized by persistent hyperglycemia, is a common and heritable complex disease affecting the health of millions of people worldwide1. Estimates from the World Health Organization indicate that 8.5% of the adult population had T2D in 2016, and this prevalence has been steadily increasing during the last three decades2.
Prospective epidemiological studies have demonstrated that the risk of T2D starts even in the normal fasting glucose range and exponentially increases in pre-diabetic ranges3–7. Relevant physiological perturbations producing a slow utilization of fasting glucose are likely to be present at stages of the disease as early as a decade before diagnosis8. The etiological causes of early glycemic perturbations are likely to be triggered by environmental and lifestyle factors9,10, but the precise biological mechanisms underpinning why people differently progress to hyperglycemia are unknown.
Recent large-scale genetic association meta-analyses have uncovered genetic variants cross-sectionally associated with T2D and related glycemic traits11–16. However, prospective data for genetic variant association discovery are scarce and findings have been inconsistent17–19. In this study, we conducted the largest genome-wide association study (GWAS) to date to identify genetic variants associated with fasting glucose changes over up to 14 years in 13,807 non-diabetic participants of European descent from nine cohorts.
Results
We included a total of 13,807 participants of European descent and free of diabetes at baseline and during the entire follow-up period with repeated fasting glucose levels measured at least at two time points over up to 14 years from nine cohorts. Characteristics of the study sample and follow-up, phenotype, and genotype information are presented in Table S1. The participants’ average age at baseline for each cohort ranged from 41 to 70 years old. The follow-up time varied by cohort, on average ranging from 5 to 25 years. The slope of fasting glucose was calculated based on available fasting glucose measurements during the follow-up and then the slopes were inverse normal transformed within each cohort, for harmonization. We refer to this transformed slope as fasting glucose change over time. At baseline, cohort-specific average fasting glucose ranged from 4.9 to 6.2 mmol/L in men and 4.8 to 5.9 mmol/L in women, respectively. Our primary analysis included all available participants. In an exploratory analysis, we investigated whether stratifying our sample by cohorts with long-term follow-up [≥10 years] or short-term follow-up [<10 years] identified pertinent signals.
In a genome-wide association meta-analysis we did not find evidence of genetic variants associated with fasting glucose change over time at genome-wide significance level (P < 5 × 10−8) (Supplemental Figs 1–3) nor evidence of inflated signals (Supplemental Figs 4–6) in the primary analysis including the entire sample or the sensitivity analysis. For the analysis with all samples, the most significant association with fasting glucose changes over time was an intronic variant at the ODZ4 locus (rs7114256; P = 8.78 × 10−7, Table 1, Fig. 1). There were five other suggestively associated (P < 5 × 10−6) variants for fasting glucose changes over time in three loci whose closest reference genes including ALLC (rs606243), NUDT12 (rs17496593, rs17496653, rs17562893) and ODZ4 (rs7103693) (Table 1, Supplemental Figs 7–8). In our exploratory analysis stratifying cohorts by follow-up time, there were a few suggestively associated variants with fasting glucose changes over time (Supplemental Tables 5–6). These included four loci whose closest reference genes were SNX16, BEGFA, GATA3 and CDKAL1 from short follow-up analysis with sample size up to 8,195 and ten loci whose closest reference genes were HCRTR2, WRN, SEPT9, SLC35B3, FAM84A, GRM8, MPP6, BAMBI, SSB, and C8orf31 from long follow-up analysis with sample size up to 3,669.
Table 1.
Genome-wide association results for genetic variants with an association p-value < 5 × 10−6.
| SNP | Chr | BPa | EAb | NEAb | EAFb | Beta | SE | P | Directionc | HetPVald | N | RefGene |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs7114256 | 11 | 78539553 | A | G | 0.92 | 0.129 | 0.03 | 8.78E-07 | +++++−?++ | 0.74 | 13,003 | ODZ4 |
| rs606243 | 2 | 4487817 | A | G | 0.74 | −0.078 | 0.02 | 1.42E-06 | ------??- | 0.36 | 11,862 | ALLC |
| rs17496593 | 5 | 104254353 | A | C | 0.91 | −0.111 | 0.02 | 2.12E-06 | -----+?-- | 0.21 | 13,005 | NUDT12 |
| rs17496653 | 5 | 104255187 | A | G | 0.09 | 0.110 | 0.02 | 2.58E-06 | +++++−?++ | 0.20 | 13,005 | NUDT12 |
| rs17562893 | 5 | 104266799 | T | G | 0.09 | 0.108 | 0.02 | 3.78E-06 | +++++−?++ | 0.19 | 12,994 | NUDT12 |
| rs7103693 | 11 | 78535307 | T | C | 0.08 | −0.120 | 0.03 | 4.19E-06 | -----+?-- | 0.79 | 13,003 | ODZ4 |
aPhysical Position (base pair) in build 36.
bEA: effect allele, NEA: non-effect allele, EAF: effect allele frequency.
cThe sign of EA effect and the order of Cohorts are BHS, COLAUS, DESIR, ERGO, FHS, HBCS, KORA, PREVEND, SARDIANA.
dHetPVal: P-value for testing for heterogeneity.
eRefGen: closest reference gene.
Figure 1.
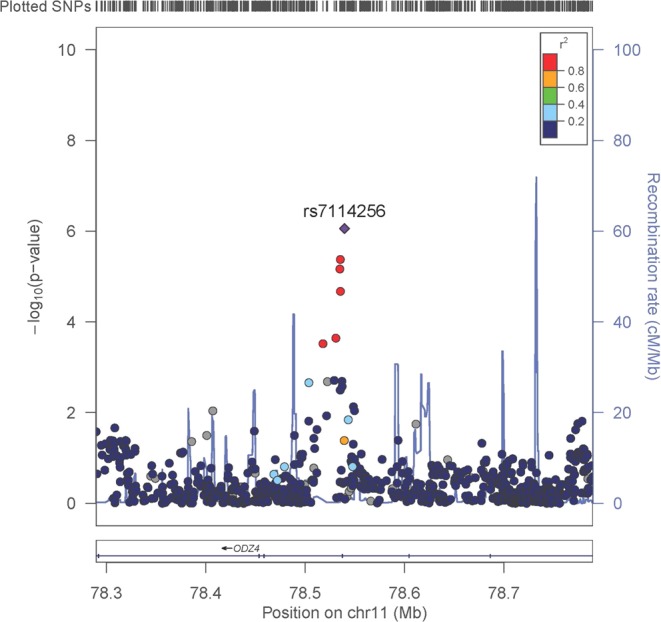
Regional association plot of rs7114256. Results from 500 kb regional associations for fasting glucose change over time, centered at rs7114256. The x axis denotes genomic position build 36 and the y axis denotes the −log(P-value) and recombination rate (blue line). The purple diamond symbol represents the most-associated SNP within the region, rs7114256. The color of each symbol indicates the LD value with rs7114256 based on the HapMap2 CEU sample.
Next, we investigated whether genetic variants available in our GWAS previously associated with T2D prevalence (82 SNPs)15 and cross-sectional glycemic traits based on our primary analysis with all available sample, including fasting glucose (32 SNPs)13, and HbA1c (58 SNPs)16, associated with fasting glucose change over time13,15,16. For T2D associated genetic variants15, we showed evidence of a nominal significant association between a variant at CDKN2A/B loci (rs10965248, P = 0.0192) and longitudinal fasting glucose change. In addition, two loci previously associated with cross-sectional FG in the latest GWAS13 for fasting glucose associated with longitudinal fasting glucose changes (GRB10; rs6943153, P = 0.0019 and PDX1; rs11619319, P = 0.0114), as well as five loci previously associated with HbA1c in the latest GWAS for HbA1c16 including TMC6 (rs2073285, P = 0.0019), CDH3 (rs4783565, P = 0.0057), ABO (rs579459, P = 0.0082), PDX1 (rs11619319, P = 0.0114), and HK1 (rs10823343, P = 0.0390) (Table 2, Supplemental Tables 2–4). After Bonferroni correction for conduct of 82, 32, and 58 tests for T2D risk, fasting glucose, and HbA1c, respectively none of these signals remained significant.
Table 2.
Association results of the genetic variants showing a nominal significant signal for fasting glucose change (p < 0.05) in known T2D or glycemic trait locia.
| SNP | Chr | BPb | Locus | EAb | NEAb | EAFb | Beta | SE | P | Directionc | HetPVald | N | HetISq |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fasting glucose loci | |||||||||||||
| rs6943153 | 7 | 50759073 | GRB10 | T | C | 0.30 | −0.044 | 0.01 | 0.002 | -------+- | 0.4257 | 13,800 | 1 |
| rs11619319 | 13 | 27385599 | PDX1 | A | G | 0.77 | 0.039 | 0.02 | 0.011 | +++++−+++ | 0.6785 | 13,807 | 0 |
| Type 2 diabetes loci | |||||||||||||
| rs10965248e | 9 | 22122878 | CDKN2A/B | A | G | 0.18 | −0.040 | 0.02 | 0.019 | ?--+--+-- | 0.06878 | 12,523 | 47 |
| HbA1c loci | |||||||||||||
| rs2073285 | 17 | 73628956 | TMC6 | T | C | 0.20 | 0.079 | 0.03 | 0.002 | +?++?+?+? | 0.05396 | 5,098 | 57 |
| rs4783565 | 16 | 67307691 | CDH3 | A | G | 0.32 | 0.043 | 0.02 | 0.006 | ?++++++?+ | 0.4491 | 11,356 | 0 |
| rs579459 | 9 | 135143989 | ABO | T | C | 0.78 | −0.041 | 0.02 | 0.008 | ------+-+ | 0.2827 | 13,779 | 18 |
| rs11619319 | 13 | 27385599 | PDX1 | A | G | 0.77 | 0.039 | 0.02 | 0.011 | +++++−+++ | 0.6785 | 13,807 | 0 |
| rs10823343 | 10 | 70761019 | HK1 | A | G | 0.75 | −0.037 | 0.02 | 0.039 | -+-----?? | 0.3766 | 8,809 | 6.7 |
aScott et al.13, Scott et al.15, Wheeler et al.16.
bBP:Physical Position (base pair) in build 36. EA: effect allele, NEA: non-effect allele, EAF: effect allele frequency.
cThe sign of EA effect and the order of Cohorts are BHS, COLAUS, DESIR, ERGO, FHS, HBCS, KORA, PREVEND, SARDIANA.
dHetPVal: P-value for testing for heterogeneity.
eUsing proxy SNP rs10965250 (r2 = 0.97 with rs10965248).
We conducted a power analysis with a sample size of 13,807 at the genome-wide significant threshold (5 × 10−8) to detect a genetic variant explaining at least 0.05% to 0.5% of the variation in the fasting glucose change over time. The results showed that our study has 80% power to detect genetic variants, which explain at least 0.28% of variation in change of fasting glucose over time (Fig. 2). This is equivalent to detect the genetic variants with minor allele frequency of 0.05 or 0.25 whose minimum effect corresponds to 0.17 or 0.09 standard deviation unit difference in the change of fasting glucose over time, respectively.
Figure 2.
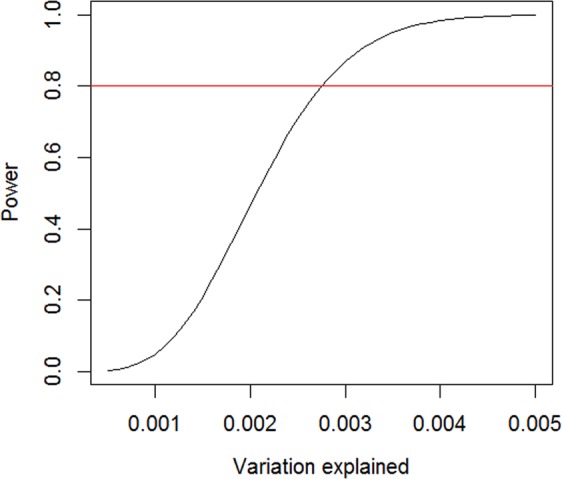
Power analysis. The relationship between power and variation explained in the trait of interest by a genetic variant with a sample size of 13,807 at a significance level of 5 × 10−8. The y-axis represents the power and the x-axis the variance explained by a genetic variant. The horizontal red line represents the power of 80%. This Figure shows that we had 80% power to detect a genetic variant that explained at least 0.28% of variation in fasting glucose change over time.
Discussion
We tested whether common genetic variants were associated with fasting glucose change over time in a GWAS including 13,807 initially non-diabetic participants from nine cohorts of European descent with repeated fasting glucose measures over up to 14 years, We found three suggestive associated variants at sub genome significance level (near ODZ4, ALLC, and NUDT12), and eight nominally associated previously known T2D-glycemia GWAS loci (CDKN2A/B, GRB10, PDX1, TMC6, CDH3, ABO, PDX1, and HK1) but none reached genome-wide significance for association or survived adjustment for multiple testing. We have placed the GWAS results data sets from this analysis on line at the Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) website (https://www.magicinvestigators.org) and T2D knowledge portal (http://www.type2diabetesgenetics.org) to provide a genomic resource to further combine with futures studies and evaluate shared genetic links with T2D and other metabolic risk traits.
To date, more than 120 genetic loci have been identified to be associated with cross-sectional glycemic outcomes in successive waves of large-scale genetic association studies12,14,15. These risk alleles are associated with glycemic phenotypes and predict incident T2D when aggregated into a genetic risk score12,20,21. However, findings from our study do not support that single common risk alleles have a substantive impact on longitudinal fasting glucose changes. If there are effects on glucose changes, they are likely to be small. This observation is in agreement with evidence from other intermediate phenotypes such as genetic variants associated with deterioration of lipid levels22–25, lung function26,27 or change in BMI28–31, where modest effects have been attributed to genetic factors on longitudinal trait changes, relative to single cross sectional trait measures. Regarding glycemic trajectories, no single genetic variants associated with glucose deterioration over time were detected in previous studies of European descent indivuals17 or in Han individuals19, although Han Chinese carrying a higher number of T2D increasing-risk variants showed a greater increase in FG over time compared with those carrying a lower number of T2D increasing-risk variants. One small single-cohort study identified five genomic regions associated at genome-wide significance with longitudinal change in fasting glucose (GCKR, G6PC2, GCK, SLC30A8, MTNR1B), but the study included diabetic individuals taking hypoglycemic medications, which almost certainly introduced confounding into genotype – glucose change associations18.
Our results may highlight the importance of environmental determinants of glycemic deterioration. Nevertheless, genetic determinants of changes in glycemia may remain relevant for people with rapid transition from pre-diabetes to diabetes, which the design of our study was not able to capture. A potential future strategy to identify these loci, if they exist, would be to focus on pre-diabetic individuals who progress to T2D and adjust for the effects of all known variants affecting cross-sectional blood glucose inter-individual variability32. A limitation to our study is that the meta-analysis only involved European participants, so the results may not be generalizable to other ancestry groups. The power of our study was relatively limited even though this is the largest existing meta-analysis of fasting glucose change. In addition, we identified challenges posed by phenotypic heterogeneity, e.g. different follow-up duration or different numbers of longitudinal data points. Larger sample sizes from new cohorts will be key to help confirm or refute the current findings. An alternative approach, if data are available in the future, would be to study longitudinal glycemia in large and homogeneous populations, with more homogeneous phenotypes especially with more consistent number of follow-up visits and similar follow-up duration to gain more statistical power.
In summary, a large GWAS did not identify common genetic variation genome-wide significantly associated with fasting glucose change over time. Such genetic effects, if present, are likely small. The data have been deposited as a public genetic epidemiological resource to aid the hunt for genetic determinants of T2D and its relevant physiology.
Methods and Materials
Study sample
We recruited in total 13,807 individuals of European descent free from T2D at baseline and during the entire follow-up period with repeated fasting glucose measurements at two or more time points from nine cohorts representing three continents (America, Europe and Australia). The participating cohorts include the Bogalusa Heart Study (BHS), the CoLaus study (COLAUS), the Data from the Epidemiological Study on the Insulin Resistance Syndrome study (DESIR), the Erasmus Rotterdam Gezondheid Onderzoek study (ERGO), Framingham Heart Study (FHS), the Helsinki Birth Cohort Study (HBCS), Cooperative Health Research in the Region of Augsburg (KORA), Prevention of Renal and Vascular End-stage Disease study (PREVEND), and the National Institute on Aging (NIA) SardiNIA Study (SARDINIA). The ethnicity information for each individual was based on questionnaires or assessed using genetic data (principal component analysis). Ethnic outliers detected by principal component analysis for European ethnicity were excluded from further analysis. Diabetes was defined as a fasting glucose level >7 mmol/l, or use of glucose lowering medication. The study conformed to the Declaration of Helsinki guidelines. Institutional Review Board and/or oversight committees approved the study in each participating cohort and all participants provided written informed consent (See Supplemental Text).
Genotyping, imputation and quality control
Genotyping was conducted as specified in the Table S1. Each study imputed their genotype to ~2.5 million Phase 2 HapMap CEU SNPs with imputation software, either IMPUTE or MACH33,34. We applied a quality control filter by removing SNPs with a minor allele frequency less than 1% and those with an imputation quality threshold proper_info < 0.4 for cohorts using IMPUTE and r2 > 0.3 for cohorts using MACH. We used imputed allelic dosage in our association analysis.
Phenotype
To calculate longitudinal fasting glucose slopes we used repeated FG measurements available in the longitudinal cohort studies. To harmonize FG measures, FG measures obtained from whole blood were converted to plasma levels by using a coefficient of 1.13 (FG in mmol/l). We then modelled the association for each individual between FG and duration of time between baseline measure and each follow-up measure. The resulting beta coefficients (slopes) were then pooled and inverse normal transformed. The transformed slopes were used as the trait ‘fasting glucose change’ for the genetic association analysis.
Association Analysis and Meta-analysis
For each participating cohort, we conducted genome-wide association analysis with transformed longitudinal fasting glucose changes adjusting for age at baseline, sex, and principal components of genetic variation to account for population stratification using linear regression with additive genetic effects for cohorts with unrelated samples. We performed mixed-effect model analysis with random effect to account for sample relatedness for cohorts with related samples. We then conducted inverse variance weighted meta-analysis of cohort-specific association results using METAL35. In an exploratory analysis, we stratified our analyses by study follow-up time and classified each cohort as having a long follow-up time (≥10 years) or a short follow-up time (<10 years). We applied genomic control correction to control type I error36. SNPs with a meta-analysis p-value ≤ 5 × 10−8 were considered to be genome-wide significant.
Interrogation of Published Loci for Type 2 Diabetes Related Traits
We tested the hypothesis that longitudinal fasting glucose slopes would be associated with previously-identified GWAS variants for T2D (128 SNPs)15, fasting glucose (32 SNPs)13, and HbA1c (60 SNPs)16. If the previously reported most-associated SNP was unavailable in the present analysis, we used a proxy SNP (LD r2 > 0.8) if available. After proxy searches we evaluated 172 loci, including 82 for T2D, 58 for HbA1c and 32 for FG. The Bonferroni corrected p-value threshold for these look-ups was set at 0.0003 (0.05/172).
Post-hoc power calculation
We conducted a post-hoc power analysis using Quanto software to investigate the power to detect 0.05% to 0.5% percent variation in phenotype explained by a genetic variant with a sample size of 13,807 at the genome-wide significant threshold (5 × 10−8).
Supplementary information
Acknowledgements
BHS. The Busselton Health Study acknowledges the generous support for the 1994/5 follow-up study from Healthway, Western Australia and the numerous Busselton community volunteers who assisted with data collection and the study participants from the Shire of Busselton. The Busselton Health Study is supported by The Great Wine Estates of the Margaret River region of Western Australia. The BHS gratefully acknowledges the assistance of the Western Australian DNA Bank (NHMRC Enabling Facility) with DNA samples and the support provided by the Ark (NHMRC Enabling Facility) for this study. CoLaus. The CoLaus study was and is supported by research grants from GlaxoSmithKline, the Faculty of Biology and Medicine of Lausanne, and the Swiss National Science Foundation (grants 33CSCO-122661, 33CS30-139468 and 33CS30-148401). The authors also express their gratitude to the participants in the Lausanne CoLaus study and to the investigators who have contributed to the recruitment, in particular research nurses Yolande Barreau, Anne-Lise Bastian, Binasa Ramic, Martine Moranville, Martine Baumer, Marcy Sagette, Jeanne Ecoffey and Sylvie Mermoud for data collection. We would also like to thank Gérard Waeber, Vincent Mooser and Dawn Waterworth, co-PIs of the study. Researchers must obtain approval from the Steering Committee of the CoLaus Study and from the Institutional Ethics Committee of the University in Lausanne, Switzerland. Researchers using the data are required to follow the terms of an Assistance Agreement containing a number of clauses designed to ensure protection of privacy and compliance with relevant laws. For further information go to www.colaus.ch or contact Peter Vollenweider (peter.vollenweider@chuv.ch). DESIR. The D.E.S.I.R. study has been funded by INSERM contracts with Caisse nationale de l’assurancemaladie des travailleurs salariés (CNAMTS), Lilly, Novartis Pharma, and sanofi-aventis; INSERM (Réseaux en Santé Publique, Interactions entre les déterminants de la santé, Cohortes Santé TGIR 2008); the Association Diabète Risque Vasculaire; the Fédération Française de Cardiologie; La Fondation de France; Association de Langue Française pour l’Etude du Diabète et des Maladies Métaboliques (ALFEDIAM)/Société Francophone de Diabétologie (SFD); l’Office national interprofessionnel des vins (ONIVINS); Ardix Medical; Bayer Diagnostics; Becton Dickinson; Cardionics; Merck Santé; Novo Nordisk; Pierre Fabre; Roche; Topcon. ERGO. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. The authors are grateful to the study participants, the staff from the Rotterdam Study and the participating general practitioners and pharmacists. Maryam Kavousi is supported by the VENI grant (91616079) from The Netherlands Organization for Health Research and Development (ZonMw). The generation and management of GWAS genotype data for the Rotterdam Study (RS I, RS II, RS III) was executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, Rotterdam, The Netherlands. The GWAS datasets are supported by the Netherlands Organisation of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012), the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) Netherlands Consortium for Healthy Aging (NCHA), project nr. 050-060-810. We thank Pascal Arp, Mila Jhamai, Marijn Verkerk, Lizbeth Herrera and Marjolein Peters, MSc, and Carolina Medina-Gomez, MSc, for their help in creating the GWAS database, and Karol Estrada, PhD, Yurii Aulchenko, PhD, and Carolina Medina-Gomez, MSc, for the creation and analysis of imputed data. FHS. This work was in part supported by the grant R01 DK078616, U01 DK078616 and K24 DK080140. Jordi Merino was supported by a postdoctoral fellowship funded by the European Commission Horizon 2020 program and Marie Skłodowska-Curie actions (H2020-MSCA-IF- 2015-703787). Also, supported using data and resources from the FHS of the National Heart Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine. The analyses reflect intellectual input and resource development from the FHS investigators participating in the SNP Health Association Resource (SHARe) project. This work was also partially supported by the National Heart, Lung and Blood Institute’s Framingham Heart Study (Contract No. N01‐HC‐25195) and its contract with Affymetrix, Inc for genotyping services (Contract No. N02‐HL‐6‐4278). HBCS. Helsinki Birth Cohort Study has been supported by grants from the Academy of Finland, the Finnish Diabetes Research Society, Folkhälsan Research Foundation, Novo Nordisk Foundation, Finska Läkaresällskapet, Juho Vainio Foundation, Signe and Ane Gyllenberg Foundation, University of Helsinki, Ministry of Education, Ahokas Foundation, Emil Aaltonen Foundation. HBCS Acknowledgements: We thank all study participants as well as everybody involved in the Helsinki Birth Cohort Study. Helsinki Birth Cohort Study has been supported by grants from the Academy of Finland, the Finnish Diabetes Research Society, Folkhälsan Research Foundation, Novo Nordisk Foundation, Finska Läkaresällskapet, Juho Vainio Foundation, Signe and Ane Gyllenberg Foundation, University of Helsinki, Ministry of Education, Ahokas Foundation, Emil Aaltonen Foundation. KORA. The KORA research platform (KORA, Cooperative Research in the Region of Augsburg) was initiated and financed by the Helmholtz Zentrum München – German Research Center for Environmental Health, Neuherberg, Germany and supported by grants from the German Federal Ministry of Education and Research (BMBF), the Federal Ministry of Health (Berlin, Germany), the Ministry of Innovation, Science, Research and Technology of the state North Rhine-Westphalia (Düsseldorf, Germany), and the Munich Center of Health Sciences (MC Health) as part of LMUinnovativ. This research was supported by the European Union’s Seventh Framework Programme (FP7-Health-F5-2012) under grant agreement no. 305280 (MIMOmics), by the Helmholtz-Russia Joint Research Group (HRJRG) 310, and by the German Center for Diabetes Research (DZD). We thank all members of field staffs who were involved in the planning and conduct of the MONICA/KORA Augsburg studies. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. PREVEND. PREVEND genetics is supported by the Dutch Kidney Foundation (Grant E033), the EU project grant GENECURE (FP-6 LSHM CT 2006 037697), the National Institutes of Health (grant 2R01LM010098), The Netherlands organisation for health research and development (NWO-Groot grant 175.010.2007.006, NWO VENI grant 916.761.70, ZonMw grant 90.700.441), the Netherlands Heart Foundation (grant NHS2010B280) and the Dutch Inter University Cardiology Institute Netherlands (ICIN). Verweij is supported by NWO VENI (016.186.125) and by a Marie Sklodowska-Curie GF (call: H2020-MSCA-IF-2014, Project ID: 661395). We would like to thank the Center for Information Technology of the University of Groningen for their support and for providing access to the Peregrine high performance computing cluster. SARDINIA. None.
Author Contributions
Cohort PI B.B., S.B., J.G.E., P.F., T.H., J.H.2, M.K., A.L.J., J.B.M., A.W.M., L.J.P., A.P., R.R.2, P.V.D.H., C.M.V.D., P.V. Genotyping P.F., H.G., J.H.1, M.M.N., P.V.D.H., N.V.1, P.V. Phenotyping S.J.L.B., J.B., T.C., H.G., J.H.2, A.L.J., E.K., J.F., J.L., A.W.M., A.P., W.R., R.R.1, P.V.D.H. Data Analysis K.S.B., J.L.B.G., M.C., T.C., D.D., J.D., H.G., A.I., Z.K., J.L., C.T.L., I.M., L.M., C.M., I.P., L.J.R.T., G.R., R.R.1, D.V.R., C.S., N.V.1, N.V.2, S.M.W., L.Y. Interpretation J.D., J.G.E., J.F., H.G., A.L.J., E.K., J.L., C.T.L., C.M., I.P., J.B.M., J.M., A.P., N.B.N. Writing I.B., D.D., J.D., C.T.L., J.B.M., J.M., N.B.N. All coauthors reviewed and approved the final version. Note: J.H.1: Jennie Hui, J.H.2: Joseph Hung, N.V.1: Niek Verweij, N.V.2: Nicole Vogelzangs, R.R.1: Rico Rueedi, R.R.2: Ronan Roussel.
Data Availability
We have publicly deposited the summary results statistics on line at the Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) website (https://www.magicinvestigators.org) and T2D knowledge portal (http://www.type2diabetesgenetics.org).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ching-Ti Liu and Jordi Merino contributed equally.
Contributor Information
Ching-Ti Liu, Email: ctliu@bu.edu.
Nabila Bouatia-Naji, Email: nabila.bouatia-naji@inserm.fr.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-45823-7.
References
- 1.NCD Risk Factor Collaboration (NCD-RisC) Worldwide trends in diabetes since 1980: a pooled analysis of 751 population-based studies with 4.4 million participants. Lancet (London, England) 2016;387:1513–30. doi: 10.1016/S0140-6736(16)00618-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.GLOBAL REPORT ON DIABETES WHO Library Cataloguing-in-Publication Data. ISBN978, 92–4 (2016).
- 3.de Vegt F, et al. Relation of impaired fasting and postload glucose with incident type 2 diabetes in a Dutch population: The Hoorn Study. JAMA. 2001;285:2109–13. doi: 10.1001/jama.285.16.2109. [DOI] [PubMed] [Google Scholar]
- 4.Tirosh A, et al. Normal fasting plasma glucose levels and type 2 diabetes in young men. N. Engl. J. Med. 2005;353:1454–62. doi: 10.1056/NEJMoa050080. [DOI] [PubMed] [Google Scholar]
- 5.Mason CC, Hanson RL, Knowler WC. Progression to Type 2 Diabetes Characterized by Moderate Then Rapid Glucose Increases. Diabetes. 2007;56:2054–2061. doi: 10.2337/db07-0053. [DOI] [PubMed] [Google Scholar]
- 6.Choi SH, et al. Hemoglobin A1c as a Diagnostic Tool for Diabetes Screening and New-Onset Diabetes Prediction: A 6-year community-based prospective study. Diabetes Care. 2011;34:944–949. doi: 10.2337/dc10-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tabák AG, Herder C, Rathmann W, Brunner EJ, Kivimäki M. Prediabetes: a high-risk state for diabetes development. Lancet (London, England) 2012;379:2279–90. doi: 10.1016/S0140-6736(12)60283-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tabák AG, et al. Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: an analysis from the Whitehall II study. Lancet. 2009;373:2215–2221. doi: 10.1016/S0140-6736(09)60619-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuomilehto J, et al. Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N. Engl. J. Med. 2001;344:1343–50. doi: 10.1056/NEJM200105033441801. [DOI] [PubMed] [Google Scholar]
- 10.Knowler WC, et al. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N. Engl. J. Med. 2002;346:393–403. doi: 10.1056/NEJMoa012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupuis J, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010;42:105–16. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris AP, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012;44:981–90. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott RA, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat. Genet. 2012;44:991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuchsberger C, et al. The genetic architecture of type 2 diabetes. Nature. 2016;536:41–47. doi: 10.1038/nature18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scott RA, et al. An Expanded Genome-Wide Association Study of Type 2 Diabetes in Europeans. Diabetes. 2017;66:2888–2902. doi: 10.2337/db16-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wheeler E, et al. Impact of common genetic determinants of Hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: A transethnic genome-wide meta-analysis. PLOS Med. 2017;14:e1002383. doi: 10.1371/journal.pmed.1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Webster RJ, Warrington NM, Beilby JP, Frayling TM, Palmer LJ. The longitudinal association of common susceptibility variants for type 2 diabetes and obesity with fasting glucose level and BMI. BMC Med. Genet. 2010;11:140. doi: 10.1186/1471-2350-11-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasmussen-Torvik LJ, et al. Impact of repeated measures and sample selection on genome-wide association studies of fasting glucose. Genet. Epidemiol. 2010;34:665–73. doi: 10.1002/gepi.20525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Q, et al. Cross-sectional and longitudinal replication analyses of genome-wide association loci of type 2 diabetes in Han Chinese. PLoS One. 2014;9:e91790. doi: 10.1371/journal.pone.0091790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meigs JB, et al. Genotype Score in Addition to Common Risk Factors for Prediction of Type 2 Diabetes. N. Engl. J. Med. 2008;359:2208–2219. doi: 10.1056/NEJMoa0804742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vassy JL, et al. Polygenic type 2 diabetes prediction at the limit of common variant detection. Diabetes. 2014;63:2172–82. doi: 10.2337/db13-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu Y, et al. Exploring genetic determinants of plasma total cholesterol levels and their predictive value in a longitudinal study. Atherosclerosis. 2010;213:200–5. doi: 10.1016/j.atherosclerosis.2010.08.053. [DOI] [PubMed] [Google Scholar]
- 23.Costanza MC, Beer-Borst S, James RW, Gaspoz J-M, Morabia A. Consistency between cross-sectional and longitudinal SNP: blood lipid associations. Eur. J. Epidemiol. 2012;27:131–8. doi: 10.1007/s10654-012-9670-1. [DOI] [PubMed] [Google Scholar]
- 24.Varga TV, et al. Genetic determinants of long-term changes in blood lipid concentrations: 10-year follow-up of the GLACIER study. PLoS Genet. 2014;10:e1004388. doi: 10.1371/journal.pgen.1004388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varga TV, et al. Novel genetic loci associated with long-term deterioration in blood lipid concentrations and coronary artery disease in European adults. Int. J. Epidemiol. 2017;46:1211–1222. doi: 10.1093/ije/dyw245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang W, et al. Large-scale genome-wide association studies and meta-analyses of longitudinal change in adult lung function. PLoS One. 2014;9:e100776. doi: 10.1371/journal.pone.0100776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.John C, et al. Genetic variants affecting cross-sectional lung function in adults show little or no effect on longitudinal lung function decline. Thorax. 2017;72:400–408. doi: 10.1136/thoraxjnl-2016-208448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delahanty LM, et al. Genetic Predictors of Weight Loss and Weight Regain After Intensive Lifestyle Modification, Metformin Treatment, or Standard Care in the Diabetes Prevention Program. Diabetes Care. 2012;35:363–366. doi: 10.2337/dc11-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandholt CH, et al. The effect of GWAS identified BMI loci on changes in body weight among middle-aged Danes during a five-year period. Obesity (Silver Spring). 2014;22:901–8. doi: 10.1002/oby.20540. [DOI] [PubMed] [Google Scholar]
- 30.Papandonatos GD, et al. Genetic Predisposition to Weight Loss and Regain With Lifestyle Intervention: Analyses From the Diabetes Prevention Program and the Look AHEAD Randomized Controlled Trials. Diabetes. 2015;64:4312–4321. doi: 10.2337/db15-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahmad S, et al. Established BMI-associated genetic variants and their prospective associations with BMI and other cardiometabolic traits: the GLACIER Study. Int. J. Obes. 2016;40:1346–1352. doi: 10.1038/ijo.2016.72. [DOI] [PubMed] [Google Scholar]
- 32.Ivarsdottir EV, et al. Effect of sequence variants on variance in glucose levels predicts type 2 diabetes risk and accounts for heritability. Nat. Genet. 2017;49:1398–1402. doi: 10.1038/ng.3928. [DOI] [PubMed] [Google Scholar]
- 33.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 35.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341X.1999.00997.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We have publicly deposited the summary results statistics on line at the Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) website (https://www.magicinvestigators.org) and T2D knowledge portal (http://www.type2diabetesgenetics.org).


