Abstract
Purpose
Congenital hypogonadotropic hypogonadism (CHH) is a rare genetic disorder mostly characterized by gonadotropins release and/or action deficiencies. Both isolated (idiopathic hypogonadotropic hypogonadism) and syndromic (Kallmann) forms are identified depending on the olfactory ability. Clinical and genetic heterogeneities of CHH have been widely explored, thus improving our understanding of the disease’s pathophysiology. This work aims to (1) provide a detailed clinical and hormonal description of normosmic CHH patients and (2) identify the mutation linked to the studied phenotype.
Participants and methods
We investigated three affected patients with normosmic CHH, belonging to a consanguineous Tunisian family. Patients underwent an insulin-induced hypoglycemia test. We performed whole exome sequencing to identify the causal mutation.
Results
At first diagnosis, a total gonadotropic deficiency was identified in all patients. The insulin-induced hypoglycemia test has also revealed a reduced cortisol secretion and complete growth hormone deficiency. At 20.8 years, one female exhibited a spontaneous recovery of the hypothalamic–pituitary–adrenal axis function, unlike her affected siblings who still depend on corticosteroid replacement therapy. Herein, we identified a novel homozygous nonstop mutation (c.1195T>C) in KISS1R gene in all affected subjects. This mutation led to the substitution of the physiologic stop codon by an arginine (p.X399R).
Conclusions
Our study highlights the importance of the KISS1R signaling, in gonadotropin-releasing hormone neurons, in the control of reproductive function. Additionally, our data suggests a complex central and peripheral metabolic control of puberty, through the hypothalamic KISS1R signaling. We suggest a mutual link between the hypothalamic–pituitary–gonadal, –adrenal, and –somatotropic axes.
Keywords: KISS1R, Normosmic congenital hypogonadotropic hypogonadism, Nonstop mutation, Kisspeptin, GnRH, Whole exome sequencing
Introduction
Congenital hypogonadotropic hypogonadism [CHH (MIM # 615267)] refers to a set of rare clinically and genetically heterogeneous reproductive disorders. CHH is characterized by absent, partial, or delayed puberty caused by deficient production, secretion, or action of gonadotropin-releasing hormone (GnRH) [1]. Clinically, it is typically examined, in both genders, during the second or third decades of life with a lack of secondary sexual characteristics, pubertal maturation, and fertility. The hormonal diagnosis relies on low circulating levels of gonadotropins (luteinizing hormone (LH) and follicle-stimulating hormone (FSH)), in association with sex steroids (testosterone and estradiol) deficiencies [1]. Moreover, CHH includes two forms, namely Kallmann syndrome (KS), with altered odor perception [2], and normosmic idiopathic hypogonadotropic hypogonadism (IHH), which are each characterized by a different genetic architecture. Autosomal dominant, autosomal recessive, and X-linked classic Mendelian transmission have been reported in CHH families from various ethnical backgrounds [3, 4]. Notably, incomplete penetrance, phenocopy, and variable clinical features have been observed even within the same pedigree with normosmic IHH or KS [4]. These complex CHH phenotypes could be explained by an oligogenic architecture [4]. In fact, it is thought that mutations recognized so far in about 30 genes [4] account for about 30% of all CHH cases [3]. On the other hand, oligogenic inheritance is responsible for 10–20% [5]. Several mutations in genes encoding receptors depict an autosomal recessive familial inheritance [GnRHR (more than 60 reported families), PROKR2 (approximately 20% of pedigrees), KISS1R (27 families)…] [4].
In the present study, we provide a detailed clinical and hormonal description of all affected members with normosmic CHH, belonging to a consanguineous Tunisian family. Using whole exome sequencing (WES), we identified a novel homozygous nonstop mutation in the KISS1R gene.
Materials and methods
Subjects
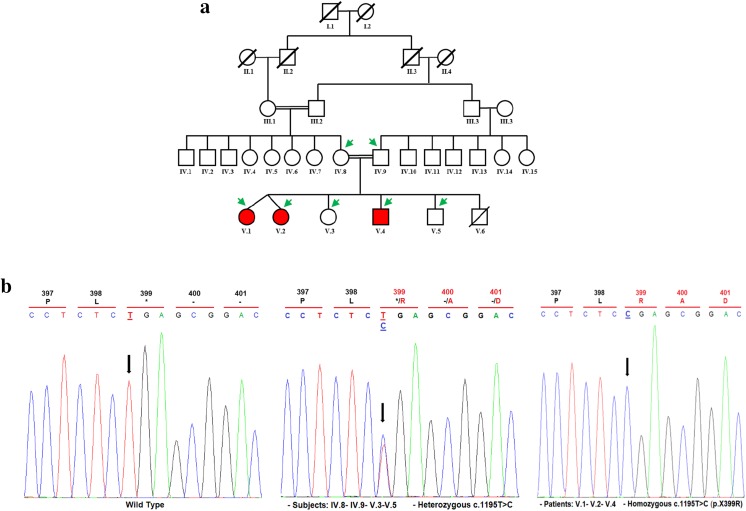
This study was approved by the ethics committee of Hedi Chaker University Hospital of Sfax, in Tunisia. Informed consent was obtained from members belonging to a consanguineous Tunisian family. This family consists of three affected siblings and four unaffected members (Fig. 1a).
Fig. 1.
KISS1R mutation underlying normosmic congenital hypogonadotropic hypogonadism with reduced cortisol and absence of GH responses to an insulin-induced hypoglycemia test. a Pedigree of the studied family. The generations within the family are indicated by Roman numerals. Squares and circles represent male and female respectively. Normal individuals are shown as clear symbol, whereas the affected individuals are shown as filled (red) symbol. Green arrows indicate available DNA members. b Validation of the c.1195T>C mutation by Sanger sequencing. Results of automatic DNA KISS1R sequencing covering the c.1195T>C homozygous mutation in the propositus (V.1, V.2, and V.4), compared to unaffected (IV.8, IV.9, V.3, V.5) family members and control subject are presented. Mutated position on the chromatographs is depicted with an arrow. Amino acid substitution is indicated above each sequence. A genetic counseling for carrier members (V.3 and V.5) is mandatory
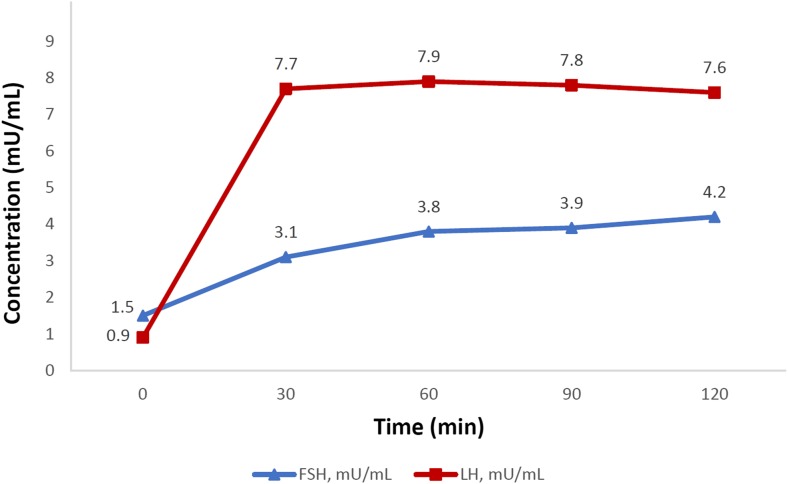
Patients were consulted for complete absence of spontaneous pubertal development and abnormal progression of growth. They were referred to undergo clinical and genetic investigations (Table 1, Fig. 1, Fig. 2). As a first diagnosis, the clinical investigation included the following parameters: chronologic age, body mass index (BMI), family history, karyotype, olfactory test, and magnetic resonance imaging (MRI) of the hypothalamic–pituitary region. The bone age was assessed using the Greulich-Pyle method. Serum levels of thyroid-stimulating hormone (TSH), free thyroxine 4 (FT4), adrenocorticotropic hormone (ACTH), cortisol, prolactin, growth hormone (GH), insulin-like growth factor-1 (IGF-1), luteinizing hormone (LH), follicle-stimulating hormone (FSH), and sex steroid hormones (testosterone and estradiol) were measured at baseline in all patients. Simultaneously, the insulin-induced hypoglycemia test (0.1 U/kg body weight, intravenously) was used to evaluate the hypothalamic–pituitary–adrenal (HPA) and hypothalamic–pituitary–somatotropic (HPS) axes. Stimulated serum levels of cortisol and GH were determined at sequential time points (15, 30, 60, 90, and 120 min post-stimulation). Stimulatory gonadotrophin-releasing hormone (GnRH) test (100 μg, intravenously) was used to evaluate the hypothalamic–pituitary–gonadal axis (HPG) only in patient V.4. Serum FSH and LH levels were measured every 30 min over a period of 120 min after GnRH administration (Fig. 2). The Synacthen test (1 μg, intravenously) was used to re-evaluate the HPA axis only in patient V.1 (Table 1).
Table 1.
Clinical and hormonal follow-up of affected patients
| Parameters | Patient V.1 | Patient V.2 | Patient V.4 |
|---|---|---|---|
| Gender | Female | Female | Male |
| Chronologic age (years) | 16.6 | 16.6 | 13.1 |
| Bone age (years) | 13 | 13 | 12 |
| BMI (kg/m2) | 19.5 | 19.5 | 19.8 |
| Karyotype | 46, XX | 46, XX | 46, XY |
| Brain and pituitary MRI | Normal | Normal | Normal |
| TSH (μU/mL) | 1.26 | 2.12 | 2.74 |
| FT4 (pmol/mL) | 14.6 | 14.8 | 13.3 |
| Prolactin (ng/mL) | 13.54 | 10.58 | 12.47 |
| ACTH (pg/mL) | 16 | 27.45 | 41.7 |
| IGF-1 (ng/mL) | NE | NE | 153 |
| Basal LH (mU/mL) | 0.16 | 0.06 | 0.34 |
| Basal FSH (mU/mL) | 1.5 | 0.6 | 1.6 |
| Basal testosterone (ng/mL) | 0.31 | < 0.45 | 0.13 |
| Basal estradiol (pg/mL) | 16 | 15 | < 10 |
| Response to insulin-induced hypoglycemia test (0.1 U/Kg, 30 min) | |||
| Plasma glucose (mmol/L) | |||
| Basal | 4.59 | 4.29 | 4.96 |
| Nadir | 2.44 | 3.1 | 3.05 |
| Cortisol (ng/mL) | |||
| Basal | 75 | 89 | 89 |
| Peak | 115 | 113 | 112 |
| GH (ng/mL) | |||
| Basal | 0.13 | 0.06 | 0.33 |
| Peak | 3.16 | 1.7 | 0.28 |
| Response to Synacthen test (1 μg) | |||
| Cortisol (ng/mL) | |||
| Basal | 142.6 | NE | NE |
| Peak 30–60 min | 207.2–325.05 | NE | NE |
Normal reference ranges are mentioned in “Laboratory testing” section. A positive response of cortisol to an insulin-induced hypoglycemia test (0.1 U/Kg body weight, intravenously, at 30 min post-stimulation), or stimulatory Synacthen test (1 μg, at 30 min and 1 h post-stimulation), when the cortisol peak is more than 180 ng/mL. A complete somatotropin deficiency was diagnosed when the GH peak is less than 5 ng/mL in response to hypoglycemia stimulatory test.
Abbreviations: BMI body mass index, TSH thyroid-stimulating hormone, FT4 free thyroxine, ACTH adrenocorticotropic hormone, IGF-1 insulin-like growth factor-1, LH luteinizing hormone, FSH follicle-stimulating hormone, GH growth hormone, NE not evaluated
Fig. 2.
GnRH test performed in V.4 patient at 14.5 years. GnRH (100 μg) was administrated intravenously before initiation of any hormonal treatment
Laboratory testing
Blood samples for biochemical determinations were collected between 7 and 9 AM after an overnight fast. Plasma TSH (normal reference range 0.27–4.2 μU/mL), FT4 (12–22 pmol/L), prolactin (males 3.46–19.4 ng/mL; women (non-pregnant) 4.79–23.3 ng/mL), ACTH (7.2–63.3 pg/mL), cortisol ((6 h–10 h) 60–184 ng/mL), FSH (males 1.5–12.4 mU/mL; women (follicular phase) 3.5–12.5 mU/mL), LH (males 1.7–8.6 mU/mL; women (follicular phase): 2.4–12.6 mU/mL), testosterone (males 2.49–8.36 ng/mL; women 0.084–0.481 ng/mL), estradiol (males 25.8–60.7 pg/mL; women (follicular phase) 12.4–233 pg/mL), and GH (males 0.076–10.8 ng/mL; women 0.12–8.06 ng/mL) concentrations were measured using an electrochemiluminescence immunoassay (ECLIA) method. The plasma glucose (3.89–5.83 mmol/L) was measured using an enzymatic hexokinase method. IGF-1 (13 years, 104–596 ng/mL) was determined by chemiluminescent quantitative measurement. Samples were analyzed on a Cobas 6000, Roche® analyzer following the manufacturer’s instructions for calibration and sample processing.
DNA extraction
Genomic DNA was extracted from the peripheral blood of seven family members and 100 unrelated unaffected Tunisian individuals using the standard phenol–chloroform procedure [6].
Next generation sequencing (NGS), bioinformatics filtering, and in silico prediction
We applied whole exome sequencing (WES) approach to identify the disease-associated gene in three members (IV.8, V.1, and V.2). WES (HiSeq2500, Illumina, San Diego, CA, USA) was performed at 100× sequencing mean with paired-end 2 × 100 reads generating more than 6 Gb data per sample. More than 80% of generated bases were of Q30 value. For exome enrichment, we used oligonucleotide probes of SureSelectXT Human All Exon 5th version (Agilent Technologies, Santa Clara, CA, USA) designed for human exons, and sheared genomic DNA was enriched for the regions of interest by hybridization with the capture probes. Generated sequences were then analyzed after quality control for variant calling and annotation. Sequencing read alignment (GRCh37/hg19), variant calling, and annotation were performed using the Genome Analysis Toolkit (GATK) variant calling and Variation and Mutation Annotation Toolkit (VariMat) programs. A filtering pipeline was established to remove frequent and benign polymorphisms. A wide number of candidate pathogenic variants, likely related to the clinical features of the disease, were retained. According to an autosomal recessive inheritance pattern, variants retained in affected (V.1 and V.2) and in unaffected (IV.8) members were filtered by the function class (missense, nonsense, splice sites, in-frame, or frameshift insertion/deletion) and classified by computational (in silico) predictive programs (Polymorphism Phenotyping v2 (PolyPhen-2), Sorting Intolerant From Tolerant (SIFT), Human Splice Finder (HSF)). Pathogenicity analysis of variants and disease annotation were carried out using OncoMed, Catalogue Of Somatic Mutations In Cancer (Cosmic), and according to ClinVar, Online Mendelian Inheritance in Man (OMIM), Human Gene Mutation Database (HGMD), genome-wide association studies (GWAS), and SwissVar databases.
Mutation screening
Transmission of candidate variants was analyzed by Sanger sequencing using suitable primers designed by primer3 webserver (http://bioinfo.ut.ee/primer3/). Genomic sequence of the KISS1R gene including the c.1195T>C mutation was amplified using the following primers: KISS1R-E5F: CTTCCTGGGCTCGCACTT; KISS1R-E5R: GCACCGAACGTCACAAGAG. PCRs were performed in a final volume of 50 μl, containing 100 ng of genomic DNA, 0.2 mM of each deoxynucleotide triphosphate (dNTP), 1.5 mM MgCl2, 10 μM of each primer, 1% DMSO, 1 × PCR buffer, and 1 unit of Taq DNA polymerase (Invitrogen). The PCR conditions were 95 °C for 10 min; 35 cycles of 95 °C for 45 s, 60 °C for 40 s, and 72 °C for 20 s; and a final extension at 72 °C for 5 min. Amplicons were visualized on 2% agarose gels and purified using the PureLink Quick Gel Extraction Kit (Thermo Fisher Scientific). Sequencing was performed using BigDye Terminator V3.1 Cycle Sequencing Kits and ABI 3100-4 Genetic Analyzer (Applied Biosystems, Foster City, CA), according to the manufacturer’s recommendations. Bioedit sequence alignment editor (V 7.1.3) was used for electropherogram analysis.
The DdeI restriction pattern was used to screen unaffected individuals. The c.1195T>C mutation abolishes a DdeI restriction site (Thermo Fisher Scientific). Digestion of PCR products was performed according to the manufacturer’s instructions. Then, the digested fragments were separated and visualized on a 3% agarose gels.
GenBank accession numbers
Human KISS1-R Protein: NP_115940; Human KISS1-R mRNA: NM_032551.
Results
Clinical characteristics
Patients (V.1, V.2, and V.4) were born at term, with normal anthropometric parameters. The clinical and hormonal investigations, at diagnosis, of all patients are outlined in Table 1 and Fig. 2.
The V.4 proband (46, XY) was brought to medical attention during the neonatal period due to repeated episodes of hypoglycemia, microphallus, and small intra-scrotal testes. No history of surgery or medical interventions was reported for neonatal and childhood period. At 13.1 years, his height was 1.42 m (−1.8 standard deviation (SD)) and his weight was 40 kg (50th Percentile) with a BMI of 19.8 kg/m2. His bone age was 12 years and he exhibited an absence of spontaneous pubertal progression (Tanner Stage 1). He was admitted for further investigation of the anterior pituitary function (Table 1, Fig. 2). Serum TSH, FT4, prolactin, IGF-1, and ACTH levels were all within the normal ranges. However, he had a reduced cortisol [89 (basal) to 112 ng/mL (peak)] and absence of GH (0.33 to 0.28 ng/mL) responses to an insulin-induced hypoglycemia test (3.05 mmol/L at 30 min). He had low basal testosterone (0.13 ng/mL) with low LH (0.34 mU/mL) and normal FSH (1.6 mU/mL) levels. The pattern of the LH response to GnRH stimulation showed a quick (at 30 min) and slightly high increase (7.7 mU/mL), persistent for 120 min, whereas the response of FSH was normal (4.2 mU/mL, at 120 min) (Fig. 2). Thus, the diagnosis of the hypothalamic CHH was confirmed. At age of 15.2 years, he underwent testosterone enanthate replacement therapy (50–250 mg, monthly, intramuscular injections), which induced significant changes in testicular volumes and pubertal progression to Tanner Stage 4. Hydrocortisone treatment was administered orally at a dose of 15 mg per day.
The twin sisters (V.1 and V.2; 46, XX) of the propositus were consulted for lack of breast development and primary amenorrhea (Tanner Stage 1). Physical examination, at 16.6 years, revealed a height of 1.52 m (− 2 SD), a weight of 45 kg (10th percentile), and a BMI of 19.5 kg/m2, for both. Their bone ages were 13 years. Pelvic ultrasonography for both patients revealed small uterus and ovaries (1.2 cm for both left and right) with the absence of follicles. Hormonal evaluation of HPG axis revealed low basal levels of LH (0.16 and 0.06 mU/mL, respectively for V.1 and V.2), FSH (1.5 and 0.6 mU/mL), and estradiol (16 and 15 pg/mL). Furthermore, they had a reduced cortisol response (from 75 to 115 ng/mL, and from 89 to 113 ng/ml, respectively) to the hypoglycemia stimulatory test (2.44 and 3.1 mmol/L, at 30 min), with normal ACTH levels. We noted an absence of GH response (from 0.13 to 3.16 ng/mL, and from 0.06 to 1.7 ng/mL, respectively) to the same stimulatory test. Serum TSH, FT4, and prolactin levels were all within normal ranges. At 17.1 years, both of them started an exogenous cyclic estrogen-progestin hormone therapy inducing pubertal progression to Tanner Stage 4 and regular cyclic menstruation. They were also treated with hydrocortisone at a dose of 15 mg per day.
The V.1 patient was re-evaluated, upon a follow-up visit at 20.8 years, and a Synacthen test was performed (Table 1). The hormonal evolution of V.1 patient was marked by a spontaneous recovery of the corticotropic function [basal cortisol level 142.6 ng/mL; peaks 207.2 (30 min), and 325.05 ng/ml (60 min)], after about 5 years of regular treatment with corticosteroid. The other siblings still depend on corticosteroid replacement therapy.
It is vital to note that there was no known familial history of delayed puberty, sterility, or anosmia. The proband’s parents as well as V.3 and V.5 members have normally reached puberty.
Identification of a novel KISS1R mutation
Using WES for two affected and one unaffected individuals’ DNA, as previously described in “Materials and methods” section, we identified a novel homozygous mutation (c.1195T>C) within KISS1R gene (Fig. 1b). Validation by Sanger sequencing showed that all affected patients were homozygous whereas all unaffected individuals were heterozygous. The novel allele (c.1195T>C), identified in the studied family, was absent in 100 unrelated healthy Tunisian individuals, the dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/) database, Human Gene Mutation Database (HGMD; http://www.hgmd.cf.ac.uk/ac/index.php), Exome Variant Server database (EVS, 6448 exomes; http://evs.gs.washington.edu/EVS/), Exome Aggregation Consortium (ExAC, 60,706 exomes; http://exac.broadinstitute.org), and the Genome Aggregation Database (GnomAD, 123,136 exomes and 15,496 whole genomes; http://gnomad.broadinstitute.org). This nonstop mutation, leading to the substitution of the expected physiologic stop codon by an arginine (p.X399R), results in the continuation of the open reading frame to the poly(A) signal.
Discussion
Clinical manifestations of CHH are complex and highly heterogeneous. For this reason, the WES approach is elected as a powerful tool to identify the causal pathogenic gene(s) and variants. Although more than 30 CHH genes have been reported, a higher genetic heterogeneity is expected [4]. In addition, the molecular genetic causes of CHH often remain unresolved, implying the existence of a growing list of genes to be accused.
A detailed clinical and hormonal description of three affected members, belonging to a consanguineous Tunisian family, suffering from normosmic CHH, reduced cortisol, and absence of GH responses to an insulin-induced hypoglycemia test at first diagnosis is the gist of the current work. Herein, we report a novel homozygous nonstop mutation (c.1195T>C), found in KISS1R gene, using WES. All unaffected members were heterozygous. The c.1195T>C mutation was proved to lead to the substitution of the expected stop codon of KISS1R by an arginine (p.X399R).
The Human KISS1R gene maps to chromosome 19p13.3. It consists of five exons and encodes seven transmembrane domains with a total of 398 amino acids. The natural ligand for KISS1R, Kisspeptin-1, has been identified by three independent groups [7–9]. The full stimulation of KISS1R signaling has been attributed to C-terminal decapeptide Kisspeptin-1 (112–121) [9]. Indeed, it was not until 2003 that the reproductive dimension of this gene could be recognized [10, 11]. A huge concern has been stimulated among clinicians and several scientists around the world ever since. Interestingly, mutations in the KISS1R gene constitute a rare cause of CHH. All hitherto identified mutations extend the length of the receptor without any particular hotspot regions. In addition, deleterious mutations (missense, insertion/deletion, premature stop codon, nonstop mutations) have been reported in patients from different ethnic backgrounds, with failure in GnRH secretion, pubertal maturation, and sterility [10–19]. Variable clinical phenotypes and hormonal profiles have been observed in patients with inactivating KISS1R mutations. Moreover, an activating mutation was identified in a girl with idiopathic central precocious puberty, under the autosomal dominant inheritance pattern [20]. Furthermore, oligogenic models of CHH transmission were reported in combination with heterozygous mutations of the KISS1R gene [21–23].
KISS1R gene c.1195T position has been previously reported substituted by adenine (c.1195T>A), replacing then the stop codon by an arginine residue (p.X399R). Seminara et al. ( [10] and Brioude et al. [17] explained the normosmic isolated CHH appearance among sporadic patients from different ethnic backgrounds (respectively Afro American and French Caucasian) with the presence of p.X399R in compound heterozygote respectively with p.R331X and p.L102P [10, 17]. The p.X399R mutation has been characterized by Seminara’s team as deleterious, in terms of inositol phosphate accumulation [10].
At the time of puberty onset, our subjects presented clinical features and hormonal profiles of complete normosmic Hypogonadotropic Hypogonadism. They all failed to undergo spontaneous pubertal onset and sexual maturation. In literature, the phenotypic analysis of patients reported with KISS1R-inactivating mutations was expressed in different degrees with either complete or partial gonadotropic deficiency [10–12, 14, 15, 19].
The hormonal evaluation of the activity of the HPG axis in our patients showed low gonadotropins and sex steroids concentrations (Table 1). The patient V.4 was sensitive to the first GnRH stimulation (Fig. 2). A positive gonadotropins response was detected at 30 min and persisted for 120 min after stimulation. This finding was compared to the response of reported patients carrying the p.X399R mutation in the compound heterozygous state. The Afro-American man [10] was sensitive to exogenous GnRH intake and his dose–response curve shifted to the left. However, in the French Caucasian man [17], the pulsatile LH release was not detected up to 16 days of pulsatile GnRH administration. Previous studies have reported variable responses of gonadotropins to GnRH testing (absent, very weak, relatively normal, high or quick), in unrelated patients harboring the same KISS1R mutation or even at the intra-familial level [10, 13–17].
The heterogeneity in the clinical presentation of patients with KISS1R mutations yielded interesting results. In our male patient, the presence of microphallus and testicular atrophy was noticed in the neonatal period, which is explained by severe gonadotropins and testosterone deficiencies during the prenatal period. Indeed, although the incomplete development of external genitalia (microphallus, testicular atrophy, or cryptorchidism) have been reported in male patients with KISS1R mutations in previous studies [14, 19], others displayed normal external genitalia [16]. For instance, two male patients were previously reported with abnormal differentiation of external genitalia [13, 18], the reported phenotype still may be either due to an unknown effect of KISS1R or a mutation in other gene(s) that govern the sexual differentiation. Interestingly, small testes have been reported among male KISS1R-deficient mice with a profound hypogonadotropic hypogonadism phenotype and an inability to reproduce [10, 24]. These findings suggest that the KISS1R gene mutations effects could be early detected among male patients. Recently, Shahab et al. [25] reported a bi-allelic KISS1R mutation (p.R297L) in a boy with IHH presented with microphallus and bilateral cryptorchidism and normal timing of adolescent puberty. The authors suggested that the “mini-puberty” of infancy is more dependent upon Kisspeptin-induced and GnRH-induced LH secretion rather than adolescent puberty [25]. On the other hand, the first clinical clues in females diagnosed with inactivating KISS1R mutations have been manifested at the time of puberty onset by the absence or incompletion of breast development, primary amenorrhea, and underdevelopment of ovaries [14–18]. Consequently, the early clinical diagnosis in newborn girls is challenging. However, early uterine and vaginal morphologies follow-up could provide new elements for the diagnosis of female children in KISS1R gene mutated families. Indeed, female KISS1R-deficient mice showed delayed vaginal opening [10, 24]. Therefore, full understanding of the kisspeptin signaling mechanisms in both genders, during the evolution of the reproductive axis in early phases, could unveil new insight of CHH diagnosis and therapy even in association with related reproductive disorders.
Inactivating KISS1R mutations have been reported in patients with isolated CHH [10–19]. In general, patients of both genders suffering from inactivating KISS1R mutations respond to sex steroids replacement therapy by triggering and maintaining the pubertal process [12–18], as similar to our patients, and to the continuous exogenous administration of pulsatile GnRH, by inducing fertility [13, 17].
Our patients, with homozygote nonstop mutation (p.X399R) in the KISS1R gene, display a reduced cortisol and absence of GH responses to an insulin-induced hypoglycemia test. Surprisingly, after about 5 years of regular treatment with corticosteroid, patient V.1 exhibited a spontaneous recovery of the function of her HPA axis, unlike the remaining siblings who still depended on corticosteroid replacement therapy.
According to the short-term induced hypoglycemia hyperinsulinemia condition, we suggest that a metabolic regulation of puberty in our patients through the KISS1R signaling may involve both intermediate and specific pathways depending on gender. These findings suggest the existence of a complex neuroanatomical and functional neuronal network between the HPG, HPA, and HPS axes, through the hypothalamic KISS1R signaling, during the pubertal transition period, that may integrate central, as well as peripheral, metabolic hormonal clues. These pathways are suggested to be implicated in the metabolic control of the onset of puberty.
In conclusion, KISS1R mutations are associated with a broad phenotypic spectrum. Therefore, there are no striking genotype-phenotype correlations of gene mutations. We identified a novel mutation in KISS1R gene (c.1195T>C) among patients with corticotroph and somatotroph deficiencies, and then a spontaneous recovery of the function of the HPA axis in one patient.
Acknowledgments
The authors are indebted to all members of the patients’ family for their invaluable cooperation in this genetic study. The authors are grateful to Dr. Lilia Affes, Riadh Ben Marzoug, and Dr. Safwen Moalla for providing necessary help. Sincere thanks to Dr. Kamel MaalouL, translator and English professor, for constructive criticism of the manuscript.
Funding
This work was supported by the Ministry of Higher Education and Scientific Research of Tunisia (LR15CBS07 budget) and the scientific research grant from the University of Sharjah (UAE) (Seed Research Project/R-286-2016).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Boehm U, Bouloux P-M, Dattani MT, de Roux N, Dodé C, Dunkel L, Dwyer AA, Giacobini P, Hardelin JP, Juul A, Maghnie M, Pitteloud N, Prevot V, Raivio T, Tena-Sempere M, Quinton R, Young J. European Consensus Statement on congenital hypogonadotropic hypogonadism—pathogenesis, diagnosis and treatment: expert consensus document. Nat Rev Endocrinol. 2015;11(9):547–564. doi: 10.1038/nrendo.2015.112. [DOI] [PubMed] [Google Scholar]
- 2.Franco B, Guioli S, Pragliola A, Incerti B, Bardoni B, Tonlorenzi R, Carrozzo R, Maestrini E, Pieretti M, Taillon-Miller P, Brown CJ, Willard HF, Lawrence C, Persico MG, Camerino G, Ballabio A. A gene deleted in Kallmann’s syndrome shares homology with neural cell adhesion and axonal path-finding molecules. Nature. 1991;353(6344):529–536. doi: 10.1038/353529a0. [DOI] [PubMed] [Google Scholar]
- 3.Topaloglu AK, Kotan LD. Genetics of hypogonadotropic hypogonadism. Endocr Dev. 2016;29:36–49. doi: 10.1159/000438841. [DOI] [PubMed] [Google Scholar]
- 4.Maione L, Dwyer AA, Francou B, Guiochon-Mantel A, Binart N, Bouligand J, Young J. GENETICS IN ENDOCRINOLOGY: genetic counseling for congenital hypogonadotropic hypogonadism and Kallmann syndrome: new challenges in the era of oligogenism and next-generation sequencing. Eur J Endocrinol. 2018;178(3):R55–R80. doi: 10.1530/EJE-17-0749. [DOI] [PubMed] [Google Scholar]
- 5.Topaloğlu AK. Update on the Genetics of idiopathic hypogonadotropic hypogonadism. J Clin Res Pediatr Endocrinol. 2017;9(Suppl 2):113–122. doi: 10.4274/jcrpe.2017.S010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawasaki ES. Sample preparation from blood, cells, and other fluids. NY: Academic Press; 1990. pp. 146–152. [Google Scholar]
- 7.Kotani M, Detheux M, Vandenbogaerde A, Communi D, Vanderwinden JM, le Poul E, Brézillon S, Tyldesley R, Suarez-Huerta N, Vandeput F, Blanpain C, Schiffmann SN, Vassart G, Parmentier M. The metastasis suppressor gene KiSS-1 encodes kisspeptins, the natural ligands of the orphan G protein-coupled receptor GPR54. J Biol Chem. 2001;276(37):34631–34636. doi: 10.1074/jbc.M104847200. [DOI] [PubMed] [Google Scholar]
- 8.Ohtaki T, Shintani Y, Honda S, Matsumoto H, Hori A, Kanehashi K, Terao Y, Kumano S, Takatsu Y, Masuda Y, Ishibashi Y, Watanabe T, Asada M, Yamada T, Suenaga M, Kitada C, Usuki S, Kurokawa T, Onda H, Nishimura O, Fujino M. Metastasis suppressor gene KiSS-1 encodes peptide ligand of a G-protein-coupled receptor. Nature. 2001;411(6837):613–617. doi: 10.1038/35079135. [DOI] [PubMed] [Google Scholar]
- 9.Muir AI, Chamberlain L, Elshourbagy NA, Michalovich D, Moore DJ, Calamari A, Szekeres PG, Sarau HM, Chambers JK, Murdock P, Steplewski K, Shabon U, Miller JE, Middleton SE, Darker JG, Larminie CGC, Wilson S, Bergsma DJ, Emson P, Faull R, Philpott KL, Harrison DC. AXOR12, a novel human G protein-coupled receptor, activated by the peptide KiSS-1. J Biol Chem. 2001;276(31):28969–28975. doi: 10.1074/jbc.M102743200. [DOI] [PubMed] [Google Scholar]
- 10.Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS, Jr, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, Zahn D, Dixon J, Kaiser UB, Slaugenhaupt SA, Gusella JF, O'Rahilly S, Carlton MBL, Crowley WF, Jr, Aparicio SAJR, Colledge WH. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349(17):1614–1627. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- 11.de Roux N, Genin E, Carel J-C, Matsuda F, Chaussain J-L, Milgrom E. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci U S A. 2003;100(19):10972–10976. doi: 10.1073/pnas.1834399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Semple RK, Achermann JC, Ellery J, Farooqi IS, Karet FE, Stanhope RG, O’Rahilly S, Aparicio SA. Two novel missense mutations in g protein-coupled receptor 54 in a patient with hypogonadotropic hypogonadism. J Clin Endocrinol Metab. 2005;90(3):1849–1855. doi: 10.1210/jc.2004-1418. [DOI] [PubMed] [Google Scholar]
- 13.Lanfranco F, Gromoll J, von Eckardstein S, Herding EM, Nieschlag E, Simoni M. Role of sequence variations of the GnRH receptor and G protein-coupled receptor 54 gene in male idiopathic hypogonadotropic hypogonadism. Eur J Endocrinol. 2005;153(6):845–852. doi: 10.1530/eje.1.02031. [DOI] [PubMed] [Google Scholar]
- 14.Tenenbaum-Rakover Y, Commenges-Ducos M, Iovane A, Aumas C, Admoni O, de Roux N. Neuroendocrine phenotype analysis in five patients with isolated hypogonadotropic hypogonadism due to a L102P inactivating mutation of GPR54. J Clin Endocrinol Metab. 2007;92(3):1137–1144. doi: 10.1210/jc.2006-2147. [DOI] [PubMed] [Google Scholar]
- 15.Nimri R, Lebenthal Y, Lazar L, Chevrier L, Phillip M, Bar M, Hernandez-Mora E, de Roux N, Gat-Yablonski G. A novel loss-of-function mutation in GPR54/KISS1R leads to hypogonadotropic hypogonadism in a highly consanguineous family. J Clin Endocrinol Metab. 2011;96(3):E536–E545. doi: 10.1210/jc.2010-1676. [DOI] [PubMed] [Google Scholar]
- 16.Breuer O, Abdulhadi-Atwan M, Zeligson S, Fridman H, Renbaum P, Levy-Lahad E, Zangen DH. A novel severe N-terminal splice site KISS1R gene mutation causes hypogonadotropic hypogonadism but enables a normal development of neonatal external genitalia. Eur J Endocrinol. 2012;167(2):209–216. doi: 10.1530/EJE-12-0127. [DOI] [PubMed] [Google Scholar]
- 17.Brioude F, Bouligand J, Francou B, Fagart J, Roussel R, Viengchareun S, Combettes L, Brailly-Tabard S, Lombès M, Young J, Guiochon-Mantel A. Two families with normosmic congenital hypogonadotropic hypogonadism and biallelic mutations in KISS1R (KISS1 receptor): clinical evaluation and molecular characterization of a novel mutation. PLoS One. 2013;8(1):e53896. doi: 10.1371/journal.pone.0053896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demirbilek H, Ozbek MN, Demir K, Kotan LD, Cesur Y, Dogan M, Temiz F, Mengen E, Gurbuz F, Yuksel B, Topaloglu AK. Normosmic idiopathic hypogonadotropic hypogonadism due to a novel homozygous nonsense c.C969A (p.Y323X) mutation in the KISS1R gene in three unrelated families. Clin Endocrinol. 2015;82(3):429–438. doi: 10.1111/cen.12618. [DOI] [PubMed] [Google Scholar]
- 19.Francou B, Paul C, Amazit L, Cartes A, Bouvattier C, Albarel F, Maiter D, Chanson P, Trabado S, Brailly-Tabard S, Brue T, Guiochon-Mantel A, Young J, Bouligand J. Prevalence of KISS1 receptor mutations in a series of 603 patients with normosmic congenital hypogonadotrophic hypogonadism and characterization of novel mutations: a single-centre study. Hum Reprod. 2016;31(6):1363–1374. doi: 10.1093/humrep/dew073. [DOI] [PubMed] [Google Scholar]
- 20.Teles MG, Bianco SDC, Brito VN, Trarbach EB, Kuohung W, Xu S, Seminara SB, Mendonca BB, Kaiser UB, Latronico AC. A GPR54-activating mutation in a patient with central precocious puberty. N Engl J Med. 2008;358(7):709–715. doi: 10.1056/NEJMoa073443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miraoui H, Dwyer AA, Sykiotis GP, Plummer L, Chung W, Feng B, Beenken A, Clarke J, Pers TH, Dworzynski P, Keefe K, Niedziela M, Raivio T, Crowley WF, Jr, Seminara SB, Quinton R, Hughes VA, Kumanov P, Young J, Yialamas MA, Hall JE, van Vliet G, Chanoine JP, Rubenstein J, Mohammadi M, Tsai PS, Sidis Y, Lage K, Pitteloud N. Mutations in FGF17, IL17RD, DUSP6, SPRY4, and FLRT3 are identified in individuals with congenital hypogonadotropic hypogonadism. Am J Hum Genet. 2013;92(5):725–743. doi: 10.1016/j.ajhg.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Känsäkoski J, Fagerholm R, Laitinen E-M, Vaaralahti K, Hackman P, Pitteloud N, Raivio T, Tommiska J. Mutation screening of SEMA3A and SEMA7A in patients with congenital hypogonadotropic hypogonadism. Pediatr Res. 2014;75(5):641–644. doi: 10.1038/pr.2014.23. [DOI] [PubMed] [Google Scholar]
- 23.Nair S, Jadhav S, Lila A, Jagtap V, Bukan A, Pandit R, Ekbote A, Dharmalingam M, Kumar P, Kalra P, Gandhi P, Walia R, Sankhe S, Raghavan V, Shivane V, Menon P, Bandgar T, Shah N. Spectrum of phenotype and genotype of congenital isolated hypogonadotropic hypogonadism in Asian Indians. Clin Endocrinol. 2016;85(1):100–109. doi: 10.1111/cen.13009. [DOI] [PubMed] [Google Scholar]
- 24.Colledge WH. Transgenic mouse models to study Gpr54/kisspeptin physiology. Peptides. 2009;30(1):34–41. doi: 10.1016/j.peptides.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 25.Shahab M, Lippincott M, Chan Y-M, Davies A, Merino PM, Plummer L, Mericq V, Seminara S. Discordance in the dependence on Kisspeptin signaling in mini puberty vs adolescent puberty: human genetic evidence. J Clin Endocrinol Metab. 2018;103(4):1273–1276. doi: 10.1210/jc.2017-02636. [DOI] [PMC free article] [PubMed] [Google Scholar]