Abstract
Purpose
Follicle-stimulating hormone receptor (FSHR) expression in granulosa cells is critical in enabling follicles to achieve accelerated growth. Although FSHR expression has been reported to be epigenetically regulated, the mechanism is unclear. Cooperation between oocytes and granulosa cells is also essential for normal follicular growth. Among oocyte-derived factors, bone morphogenetic protein 15 (BMP15) promotes follicular growth and is suggested to have epigenetic effects. We examined the role of BMP15 in the acquirement of FSHR in human granulosa cells.
Methods
Immortalized non-luteinized human granulosa (HGrC1) cells were stimulated with trichostatin A (TSA) or BMP15 to analyze FSHR expression, histone modifications, and USF1/2 binding at the FSHR promoter region. Histone acetyl transferase (HAT) activity and phosphorylation of Smad 1/5/8 and p38 MAPK were examined with or without BMP15, SB203580, and LDN193189. CYP19A1 expression and estradiol production were also studied.
Results
TSA and BMP15 induced FSHR mRNA expression in a dose-dependent manner and histone modifications were observed with increased binding of USF1/2. BMP15 increased FSHR protein expression, which was suppressed by LDN193189. BMP15 increased phosphorylation of Smad 1/5/8 and significantly increased HAT activity, which was inhibited by LDN193189, but not by SB203580. BMP15 increased phosphorylation of p38 MAPK and USF1. LDN193189 suppressed BMP15-induced phosphorylation of both p38 MAPK and USF1, whereas SB203580 suppressed the phosphorylation of USF1. BMP15 increased CYP19A1 mRNA expression and estradiol production.
Conclusion
BMP15 induced FSHR expression in human granulosa cells through Smad and non-Smad pathways. This mechanism of FSHR induction by BMP15 may be utilized for controlling follicular growth.
Electronic supplementary material
The online version of this article (10.1007/s10815-019-01469-y) contains supplementary material, which is available to authorized users.
Keywords: Granulosa, Bone morphogenetic protein 15, Follicle stimulating hormone, Small mothers against decapentaplegic
Introduction
Follicle-stimulating hormone receptor (FSHR) is essential for follicle growth and ovulation in the ovary. Expression of FSHR in granulosa cells is increased as healthy follicles grow, whereas it is decreased in atretic follicles [1]. FSHR in granulosa cells is expressed in a stage-dependent manner of follicle growth. FSHR is undetectable in primordial or primary follicles initially but is observed in small antral follicles at a later stage. Granulosa cells increase the production of FSHR until follicles reach the pre-ovulatory stage. Only a few selected follicles acquire FSHR expression enabling gonadotropin-dependent growth, while most follicles remain quiescent. Expression of FSHR in granulosa cells is precisely regulated to induce normal folliculogenesis. However, the exact regulatory mechanism of FSHR expression remains unclear.
Luteinizing hormone receptor (LHR), another gonadotropin receptor, is known to be regulated in an epigenetic manner [2, 3]. LHR and FSHR possess a similar DNA structure suggesting a similar epigenetic regulatory mechanism for FSHR [4]. Epigenetic regulation, such as histone modification, plays a critical role in gene expression. Acetylation of histones mainly reduces the affinity between DNA and histones to access the chromatin structure [5]. This allows RNA polymerase and transcription factors to bind to DNA easily, thereby promoting transcription. Trichostatin A (TSA), a histone deacetylase inhibitor, has been shown to induce changes of histone modifications contributing to increasing LHR expression [3].
At the same time, regulation from the coresident oocyte is another possible regulatory mechanism of FSHR expression in granulosa cells. The interaction between granulosa cells and oocytes, and their subsequent coordinated growth is vital for normal follicle growth. This interaction is known to be mediated in part by paracrine factors produced by both granulosa cells and oocytes [6]. In the follicle, cytokines belonging to the transforming growth factor-β (TGF-β) family participate in the regulation of cell proliferation and apoptosis and differentiation in a variety of tissues, acting as key regulators of folliculogenesis. Among these regulators, factors derived from the oocytes are bone morphogenetic protein 15 (BMP15) and growth differentiation factor 9 (GDF9) [7, 8]. BMP15 and GDF9 are known to regulate follicle development, ovulation rate, and oocyte quality [9–12].
More than 20 BMPs have been identified. Some are reported to exert epigenetic effects. BMP2, BMP6, and BMP7 phosphorylate small mothers against decapentaplegic (Smad) 1/5/8 and induce histone acetyltransferase activity (HAT) to epigenetically regulate expressions of heart development-related genes [13]. Some BMPs, such as BMP4, BMP6, and BMP7, are known to induce FSHR gene expression [14, 15]. However, these factors are not specifically derived from the oocyte. Considering the close interaction between granulosa cells and oocytes during follicle growth, it is reasonable to suspect that oocyte-derived factors may be involved in the regulation of FSHR expression in granulosa cells. BMP15 has been shown to activate the Smad 1/5/8 signaling pathway [16, 17], suggesting a possible role of BMP15 in the epigenetic regulation of FSHR expression. BMPs are also known to act through the p38 MAPK signaling pathway, a non-Smad pathway, as well as through the Smad pathway [18]. In non-granulosa cells, previous reports have shown that p38 mitogen-activated protein kinase (MAPK) induces upstream stimulatory factor 1 (USF1) phosphorylation [19, 20].
BMP15 has been reported to produce conflicting effects on FSHR mRNA expression in human and rat [21, 22]. However, human granulosa cells used for in vitro studies are mostly obtained during the oocyte retrieval step in in vitro fertilization (IVF) cycles. These granulosa cells have typically been luteinized, which drastically alters the characteristics of granulosa cells, making it difficult to draw conclusions on expression profiles of gonadotropin receptors. Presently, there are no reports on the effects of BMP15 on non-luteinized human granulosa cells, which reflect granulosa cells of growing follicles. With the use of immortalized non-luteinized human granulosa cells (HGrC1), we have been able to examine the characteristics of non-luteinized granulosa cells [17].
The purpose of this study was to elucidate the regulation of FSHR expression of growing follicles in human granulosa cells. Referring to the epigenetic regulatory mechanism of LHR, which shares a similar structure to FSHR, we first examined if FSHR can also be epigenetically regulated with TSA. We then focused on the molecular mechanism of BMP15-induced FSHR expression, including its epigenetic effects.
Materials and methods
Reagents
Human recombinant BMP15 was obtained from R&D Systems, Inc. (Minneapolis, MN, USA). The histone deacetylase inhibitor, trichostatin A (TSA) was purchased from Sigma (St. Louis, MO, USA). SB203580, a p38 MAPK inhibitor, was obtained from Calbiochem (Merck KGaA, Darmstadt, Germany). LDN193189, an Alk2, 3 antagonist, was obtained from Sigma.
Cell culture, RNA isolation, and real-time RT-PCR
HGrC1 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Sigma) containing 10% fetal bovine serum (FBS; Biological Industries Ltd., Beit HaEmek, Israel), 100 IU/mL of penicillin, 100 μg/mL of streptomycin, and 25 mg/L of amphotericin B at 37 °C in 5% CO2 and 5% O2 air. HGrC1 cells were plated onto 6-well multi-dishes (Nunclon DELTA Surface; Nunclon, Roskilde, Denmark), 100-mm tissue culture dishes, or 150-mm tissue culture dishes (Iwaki, Tokyo, Japan). Into each well of 6-well plates, 60 × 104 were plated. The cells reached 100% confluence in approximately 2 days, reaching 132 × 104 cells per well. The confluent HGrC1 cells obtained from the 6-well multi-dishes were cultured in DMEM without FBS overnight. The cells were then treated with 0–1000 ng/ml TSA, 0–500 ng/ml BMP15, or 5 IU/mL FSH for 48 h. To observe CYP19A1 expression and estradiol production, 10 μM androstenedione (4-androstene-3,17-dione; Sigma) were added to the medium. Total RNA was isolated from cells using the RNeasy Mini kit (QIAGEN Inc., Valencia, CA, USA) according to the manufacturer’s protocol. Reverse transcription reactions with 1 μg of total RNA were performed with a first-strand cDNA synthesis kit (ReverTra Ace α; Toyobo Co. Ltd., Osaka, Japan). Real-time RT-PCR was performed using the Thermal Cycler Dice (Takara Bio Inc., Tokyo, Japan) and KOD SYBR qPCR Mix (Toyobo Co. Ltd) using 96-well 0.2-mL thin-wall PCR tubes. The real-time RT-PCR mixture contained 2× KOD SYBR qPCR Mix, 0.2 μM PCR primers and 2 μL of complementary DNA in a total volume of 25 μL. PCR primers were 5′ TTT CAA GAA CAA GGA TCC ATT CC 3′ (forward) and 5′ CCT GGC CCT CAG CTT CTT AA 3′ (reverse) for FSHR, 5′ GGT GAG AGA GAC ATA AAG ATT G 3′ (forward) and 5′ TTC AGG ATA ATG TTT GTC CC 3′ (reverse) for CYP19A1, and 5′ GCA CCG TCA AGG CTG AGA AC 3′ (forward) and 5′ TGG TGA AGA CGC CAG TGG A 3′ (reverse) for GAPDH. The PCR profile included an initial incubation at 95 °C for 30 s followed by 45 cycles of denaturation at 95 °C for 5 s, annealing at 55 °C for 10 s and extension at 72 °C for 20 s. All real-time RT-PCR was performed in duplicate. To eliminate the possibility of non-specific amplification or primer-dimer formation, the dissociation curves were checked for a single peak. The ratio of FSHR mRNA to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA was calculated by applying the 2−ΔΔCt technique.
Western blot analysis
The confluent HGrC1 cells obtained from the 6-well multi-dishes were cultured in DMEM without FBS overnight. The cells were then treated with or without 500 ng/ml BMP15, 10 μM LDN193189, or 10 μM SB203580 for 48 h for performing FSHR analysis. For Smad 1/5/8, p38, and USF1 phosphorylation analysis, cells were pretreated with 10 μM LDN193189 or 10 μM SB203580 for 2 h before stimulation with 500 ng/ml BMP15 for 0–90 min. The cells were lysed in a radioimmunoprecipitation buffer (10 mmol/L Tris-HCl, pH 7.4, 150 mmol/L NaCl, 1% Nonidet P-40, 5 mmol/L EDTA, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1.2% aprotinin, 5 μmol/L leupeptine, 4 μmol/L antipain, 1 mmol/L phenylmethylsulfonylfluoride, and 0.1 mmol/L Na3VO4). The cell lysates were clarified by centrifugation at 13,000g at 4 °C for 15 min, for protein extraction. Equal amounts of proteins were mixed with 2× sample buffer (4% sodium dodecyl sulfate (SDS), 10% beta-mercaptoethanol, and 20% glycerol in 0.125 M Tris, pH 6.8) containing bromophenol blue and boiled for 5 min. The samples were loaded and separated via 10% SDS-polyacrylamide gel electrophoresis (PAGE). Proteins separated by SDS-PAGE were transferred to polyvinylidene difluoride membranes (Immobilon-P transfer membrane) and blocked for 1 h with blocking buffer (5% non-fat milk in Tris-buffered saline containing 0.5% Tween-20 (TBST) for 1 h. Next, the membranes were incubated overnight at 4 °C with primary antibodies for anti-FSHR Ab (no. 22665-1-AP, 1:1000; Protein Tech, Chicago, IL), anti-Smad 1/5/8 Ab (N-18-R, 1:5000; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), antiphospho-Smad 1/5/8 Ab (no. 9511, 1:500; Cell Signaling Technology, Inc., Danvers, MA), anti-p38 MAPK Ab (sc-7149, 1:500; Santa Cruz Biotechnology, Inc.), antiphospho-p38MAPK Ab (no. 4511, 1:1000; Cell Signaling Technology, Inc.), anti-USF1 Ab (no. 32414, 1:1000; Signalway Antibody LLC, College Park, MD, USA), and antiphospho-USF1 Ab (no. 12651, 1:1000; Signalway Antibody LLC), and anti-actin (1:10000; no. 017–24573, Wako Pure Chemical Industries Ltd., Tokyo, Japan). Membranes were washed three times with Tween/PBS for 10 min, and then incubated with anti-rabbit immunoglobulin G (IgG) (1:1000; Cell Signaling Technology) for 1 h. Finally, membranes were treated with ECL-Western blot detecting reagent (Amersham Biosciences Corp., Piscataway, NJ) for visualization and analyzed using the public domain ImageJ program version v1.50i (https://imagej.nih.gov/ij/). The bands were quantified by densitometry and normalized to the appropriate loading controls.
ChIP-qPCR
The levels of acetylation of histone and transcriptional factor binding to the promoters of FSHR genes were evaluated by the chromatin immunoprecipitation (ChIP) assay. ChIP experiments were performed using the ChIP-IT Express Enzymatic kit (Active motif, Carlsbad, CA, USA). Briefly, 1.5 × 107 HGrC1 cells were cultured in DMEM without FBS for overnight. Then, the cells were treated with or without 200 ng/ml TSA for 24 h. The cells were also stimulated with or without 250 ng/ml BMP15 for 24 h. The cells were then fixed by 1% formaldehyde at room temperature for 10 min and were lysed using a Dounce homogenizer. The fixed chromatin was further subjected to enzymatic digestion. Fifty microliters of the soluble chromatin were removed to check the DNA concentration and for confirmation that the chromatin had been sheared correctly using gel electrophoresis. Equal amounts of digested chromatin were used for each immunoprecipitation with the respective antibodies against transcription factors or histone modifications. Antibodies for histone 3 lysine 9 acetylation (H3K9ac), histone 3 lysine 14 acetylation (H3K14ac), histone 4 lysine 8 acetylation (H4K8ac), histone 4 lysine 12 acetylation (H4K12ac), histone 4 lysine 16 acetylation (H4K16ac), upstream stimulatory factor 1 and 2 (USF1 and USF2), and normal rabbit IgG (negative control) were used. After washing the precipitated complexes, the cross-linking of DNA and proteins was reversed by heating the samples at 95 °C for 15 min. Samples were then digested with proteinase K at 37 °C for 1 h. Real-time PCR analyses were performed on the ChIP-precipitated DNA and input DNA. The expression levels of the FSHR gene promoter region was calculated using 2−ΔΔCt technique. The values were normalized according to the percent input method, where each values are divided by the values obtained from an input sample, thereby producing data as % input (formula for % input: 100 × 2^(Ct(adjusted input) − Ct(IP)). This data shows the relative binding of the specific histone/transcription factor to the FSHR gene promoter region.
The primers for the FSHR gene promoter region were 5′ AGC TTA TCT TGC CTG GAA 3′ (forward) and 5′ TCT GAC TTG AGA ACT GGT AG 3′ (reverse). Antibodies for H3K9ac, H3K14ac, H4K8ac, H4K12ac, and H4K16ac were obtained from Active Motif. Antibodies for USF1 and USF2 were obtained from Santa Cruz Biotechnology Inc. Antibodies for normal rabbit IgG was obtained from Cell Signaling Technology, Inc.
HAT activity assays
The confluent HGrC1 cells obtained from 100 mm tissue culture dishes were cultured in DMEM overnight without FBS. Later, the cells were stimulated with or without 250 ng/ml BMP15, 10 μM SB203580, or 10 μM LDN193189 for 24 h. Nuclear extract of the stimulated cells was prepared using the nuclear extraction kit (BioVision, Mountain View, CA) following the manufacturer’s protocol. Activity of histone acetyltransferases (HAT) in the nuclear fraction was quantified by an ELISA kit (BioVision) according to the manufacturer’s instructions and was read by an enzyme micro-plate reader (Viento808IU; BioTek Instruments, Inc., Winooski, VT, USA) at 440 nm.
ECLIA
The confluent HGrC1 cells on 6-well multi-dishes were cultured in DMEM without FBS overnight. Subsequently, 10 μM androstenedione was added in the culture medium, and the cells were treated with or without 500 ng/ml BMP15 or 5 IU/ml human recombinant FSH for 48 h, to detect the presence of estradiol in the culture medium. Estradiol concentrations in the medium were analyzed by using electrochemiluminescent immunoassay (ECLIA) by SRL, INC. (Tokyo, Japan).
Statistical analysis
All experiments were repeated thrice, and data represent mean ± SD. All data were analyzed by SPSS software, (SPSS version 24, Chicago, IL, USA). Statistical tests used in this study were t test, Mann–Whitney U test, Welch’s t test, and one-way ANOVA. p < 0.05 was considered statistically significant.
Results
Effects of TSA on FSHR expression in HGrC1 cells
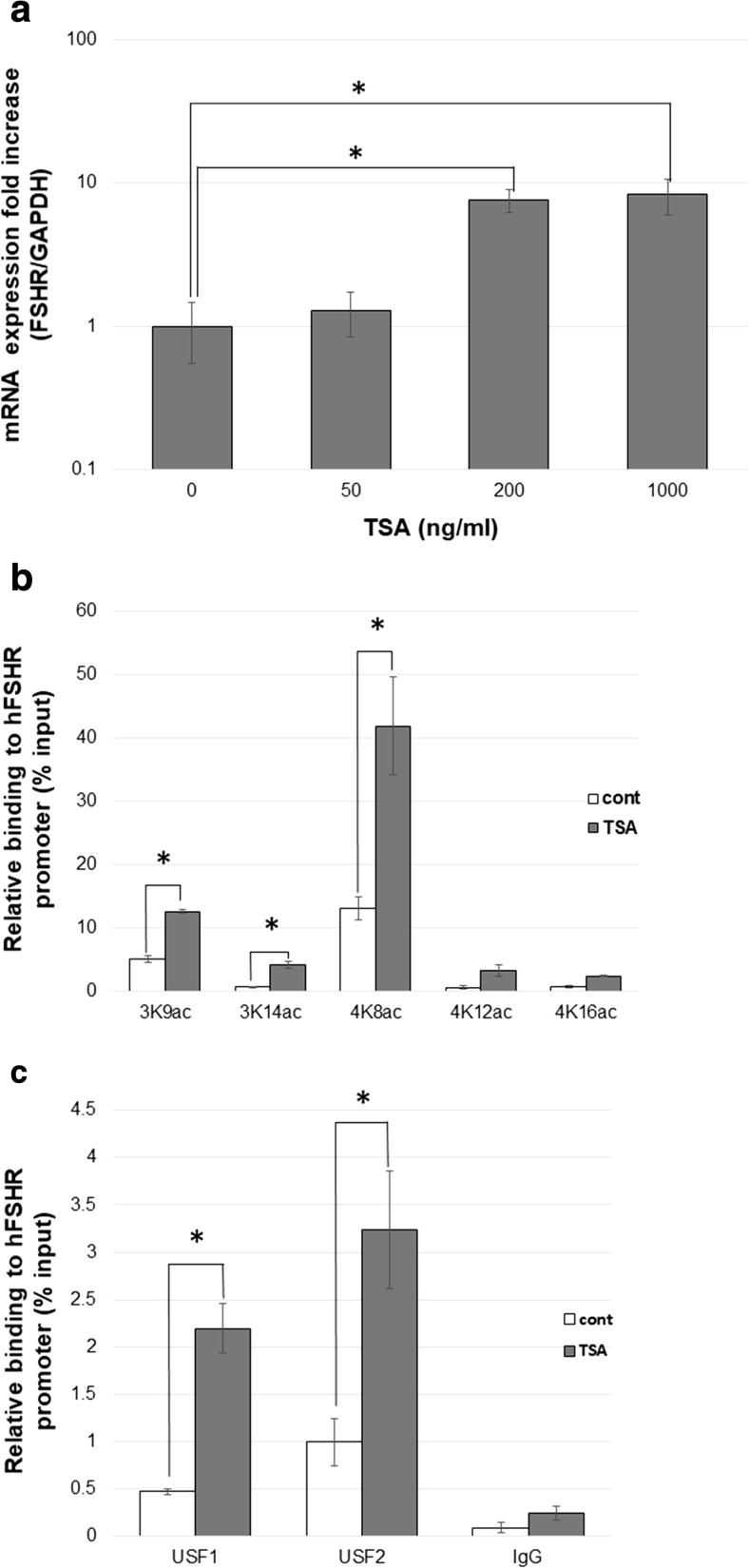
We examined the effects of histone modification on FSHR expression by stimulating HGrC1 cells with TSA. TSA significantly induced FSHR mRNA expression in a dose-dependent manner (Fig. 1a). Five histone modifications, namely H3K9ac, H3K14ac, H4K8ac, H4K12ac, and H4K16ac were studied by ChIP analysis to examine the effects of TSA. These acetylation sites have been reported to be differently involved in the control of the LHR gene expression in a cell type–specific manner [3]. TSA significantly elevated the acetylation levels of H3K9, H3K14, and H4K8 (Fig. 1b). USF1 and USF2 are known to bind to the FSHR promoter region and act as the key transcription factors for the FSHR gene [23]. Our results of ChIP assays indicated that TSA significantly increased both USF1 and USF2 binding to the FSHR promoter (Fig. 1c). These results suggest that human FSHR mRNA expression is upregulated by histone acetylation at the FSHR promoter region, which allows increased binding of transcription factors USF1/2.
Fig. 1.
TSA induces FSHR expression in HGrC1. a HGrC1 cells were treated with 0–1000 ng/ml for 48 h. TSA induced FSHR mRNA expression in a concentration-dependent manner. b ChIP-qPCR: Five histone (H3K9, H3K14, H4K8, H4K12, and H4K16) modifications were compared in HGrC1 cells with or without 200 ng/ml TSA for 24 h. Acetylation at H3K9, H3K14, and H4K8 were significantly elevated with TSA stimulation. c ChIP-qPCR: transcriptional factor (USF1 and USF2) binding to the FSHR promoter region were analyzed in HGrC1 cells with or without 200 ng/ml TSA for 24 h. Both transcriptional factor binding were significantly elevated with TSA stimulation. All data are shown as mean ± SD of three experiments. * p < 0.05 vs. control. TSA trichostatin A
Effects of BMP15 on FSHR expression in HGrC1 cells
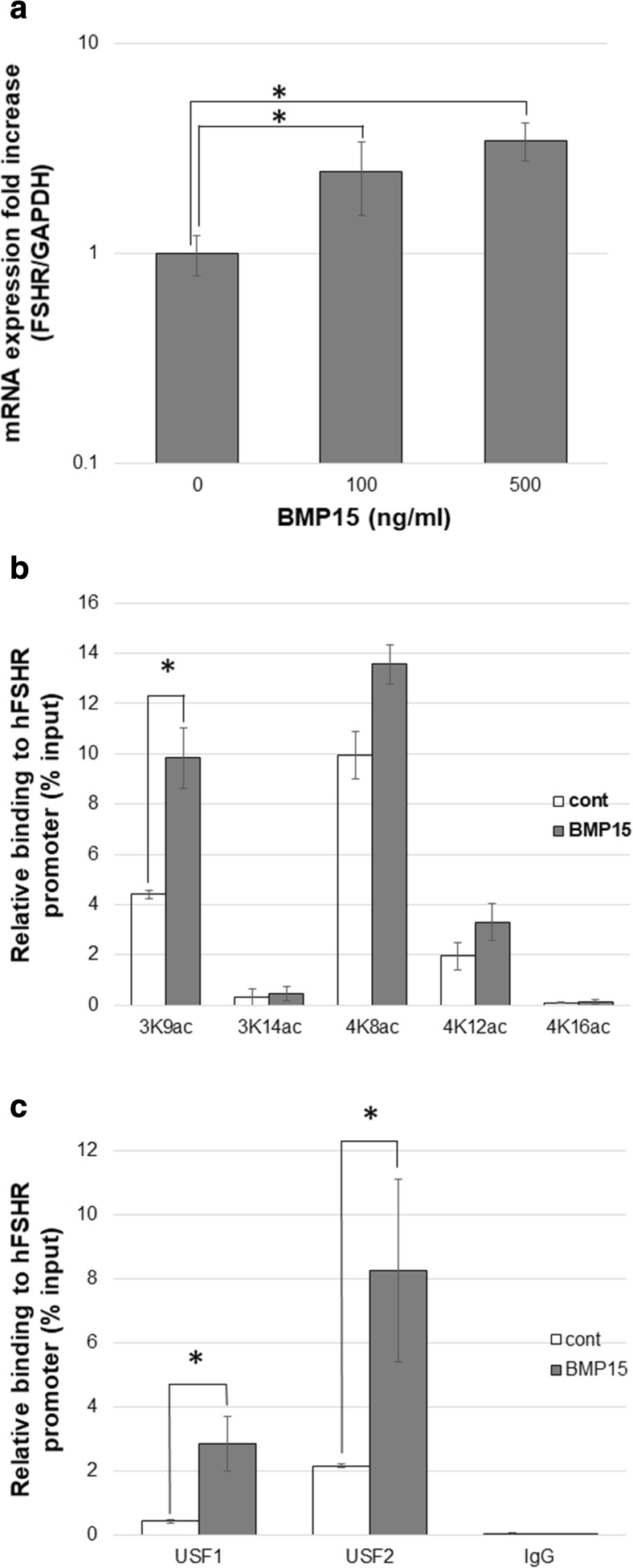
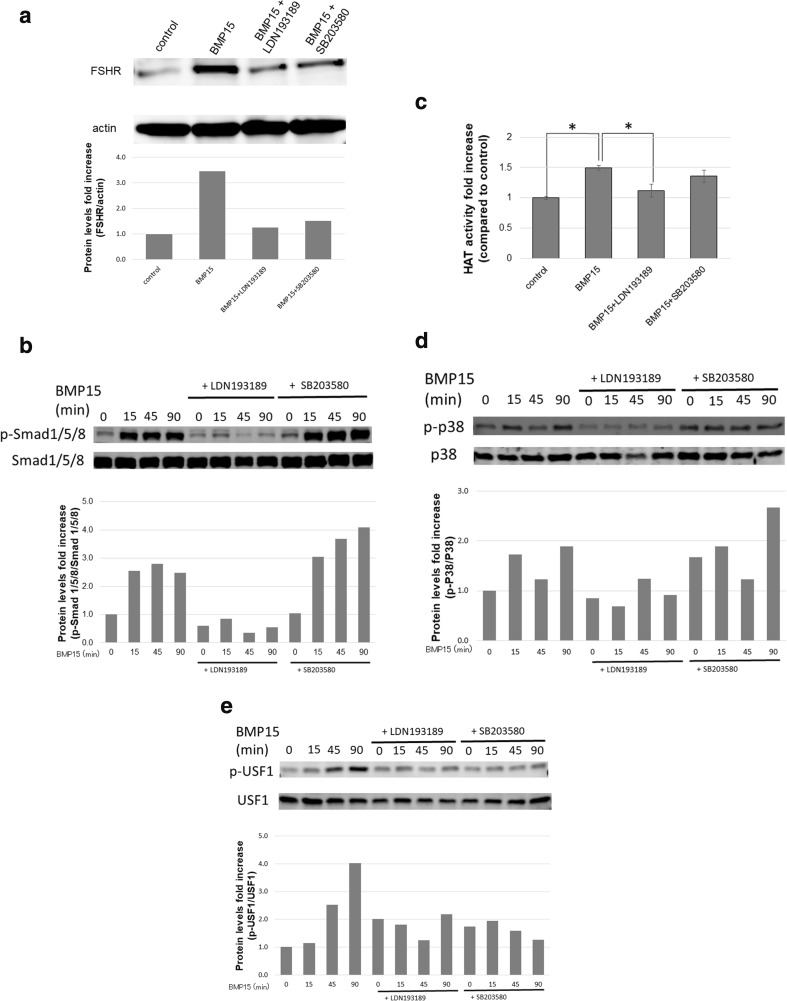
BMP15, produced by the oocytes, has been reported to play a key role in follicle development [11]. We therefore examined the effects of BMP15 on FSHR expression in HGrC1 cells. FSHR mRNA expression was significantly increased with BMP15 treatment in a dose-dependent manner (Fig. 2a). ChIP analysis showed that acetylation at H3K9 was elevated with BMP15 in HGrC1 cells, similar to the effects of TSA (Fig. 2b). USF1 and USF2 binding to the FSHR promoter region were both elevated with BMP15 (Fig. 2c). FSHR protein expression was also increased with BMP15, whereas costimulation with SB203580 and LDN193189 suppressed the induction by BMP15 (Fig. 3a). LDN193189, which acts as a BMP15 receptor blocker, suppressed BMP15-induced FSHR expression to control level. However, SB203580, a p38 inhibitor, suppressed BMP15-induced FSHR expression to a lesser extent.
Fig. 2.
BMP15 induces FSHR expression in HGrC1. a HGrC1 cells were treated 0–500 ng/ml BMP15 for 48 h. BMP15 induced FSHR mRNA expression in a concentration-dependent manner. b ChIP-qPCR: five histone (H3K9, H3K14, H4K8, H4K12, and H4K16) modifications were compared in HGrC1 cells with or without 250 ng/ml BMP15 for 24 h. Acetylation at H3K9 was significantly elevated with BMP15. c ChIP-qPCR: USF1 and USF2 binding to the FSHR promoter region were analyzed in HGrC1 cells with or without 250 ng/ml BMP15 for 24 h. Both transcriptional factor binding were significantly elevated with BMP15 stimulation. Data are shown as mean ± SD of three experiments. * p < 0.05
Fig. 3.
BMP15 induces the Smad and non-Smad pathway. a HGrC1 cells were treated with or without 500 ng/ml BMP15, 10 μM LDN193189, or 10 μM SB203580 for 48 h. BMP15 increased FSHR protein expression. However, LDN193189 suppressed the effect, similar to that of the control. SB203580 suppressed the effect but to a lesser extent as compared to LDN193189. b HGrC1 cells were treated with or without 500 ng/ml BMP15 for 0–90 min. The cells were pretreated with LDN193189 and SB203580 for 2 h. BMP15 increased phosphorylation of Smad 1/5/8. The effect was inhibited by LDN193189, but not by SB203580. c HGrC1 cells were stimulated with or without 250 ng/ml BMP15, 10 μM SB203580, or 10 μM LDN193189 for 24 h. BMP15 significantly increased HAT activity compared to the control. The effect was inhibited by LDN193189, but not by SB203580. Data are shown as mean ± SD of three experiments. * p < 0.05. d BMP15 increased phosphorylation of p38 MAPK, whereas the addition of LDN193189 suppressed this effect. SB203580 addition exerted no effect. e BMP15 increased phosphorylation of USF1, whereas the addition of LDN193189 suppressed this effect. SB addition suppressed BMP15-induced phosphorylation of USF1. In Western blotting, we analyzed the ratio of FSHR to actin, p-Smad 1/5/8 to Smad 1/5/8, p-p38 to p38, and p-USF1 to USF1 by using ImageJ to show densitometry
Effects of BMP15 on the Smad pathway
To examine how BMP15 induces gene transcription through the Smad pathway, Smad 1/5/8 phosphorylation, HAT activity, and histone modifications were studied. BMP15 increased phosphorylation of Smad 1/5/8; this effect was subsequently inhibited by LDN193189 (Fig. 3b). However, SB203580 did not alter the effects of BMP15 on Smad 1/5/8. Furthermore, HAT activity was significantly increased with BMP15, which was inhibited by LDN193189 but remained unchanged by SB203580 (Fig. 3c). These results suggest that through the phosphorylation of Smad 1/5/8, BMP15 induces HAT activity, resulting in histone modifications that are favorable for FSHR expression.
Effects of BMP15 on the non-Smad pathway and downstream effectors of FSHR
To examine how BMP15 induces gene transcription through the non-Smad pathway, the effects of BMP15 on p38 MAPK and USF1 were analyzed. Phosphorylation of p38 MAPK was suppressed by LDN193189, whereas SB203580 did not show this effect (Fig. 3d). Phosphorylation of USF1 was increased by BMP15, whereas the addition of LDN193189 and SB203580 both suppressed this effect (Fig. 3e). SB203580 is reported to suppress the downstream targets of phosphorylated p38, without affecting the phosphorylation of p38 (Fig. 3d) [24].
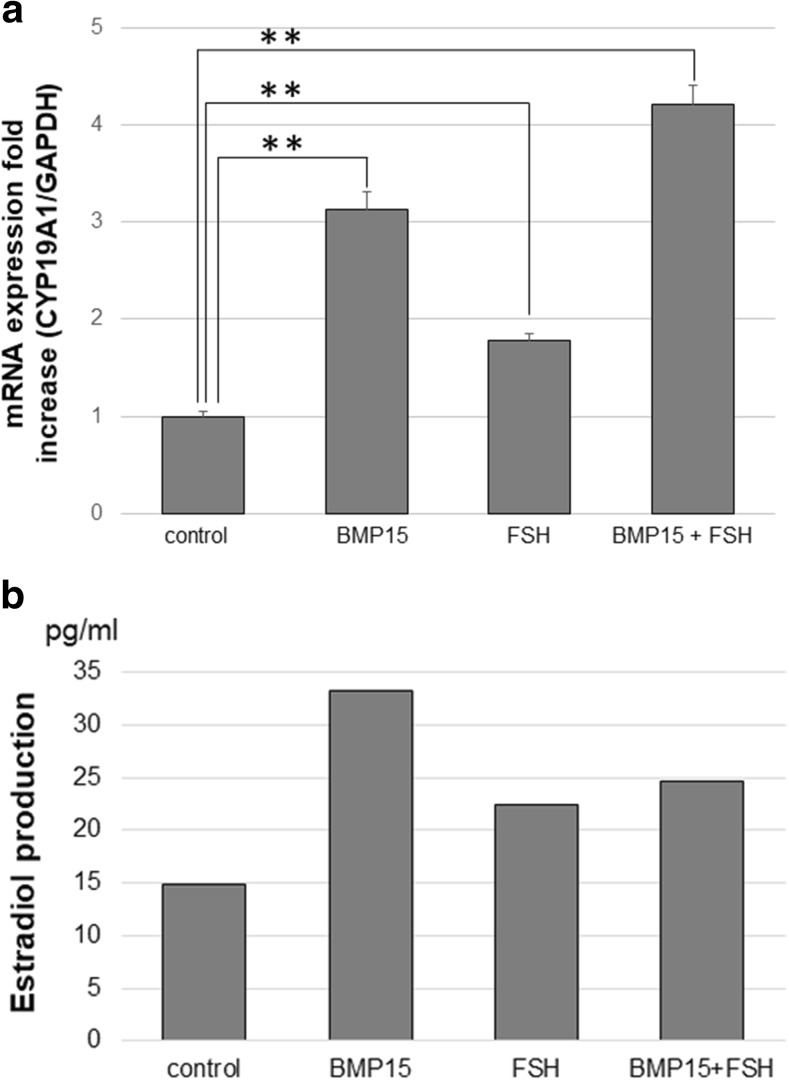
FSH acts exclusively through the FSHR [25]. FSH is known to induce CYP19A1, which is a key enzyme in converting androstenedione to estradiol [26]. BMP15 significantly induced CYP19A1 mRNA levels. Additive effects were seen with BMP15 and FSH (Fig. 4a). Estradiol production was increased with BMP15 stimulation (Fig. 4b).
Fig. 4.
BMP15 induces the downstream effectors of FSHR. a The HGrC1 cells were treated with or without 500 ng/ml BMP15 or 5 IU/ml human recombinant FSH for 48 h, after 10 μM androstenedione was added in the culture medium. The HGrC1 cells were stimulated, after 10 μM androstenedione was added in the culture medium. BMP15 increased CYP19A1 mRNA expression, while increase with FSH was minimal. Additive effects of BMP15 and FSH were observed. b BMP15 increased estradiol production. All data are shown as mean ± SD of three experiments. **p < 0.001 vs. control
Discussion
The purpose of the present study was to examine the molecular mechanism and its link to the epigenetic machinery necessary for BMP 15-induced FSHR expression in human granulosa cells. We demonstrated that BMP15 increases FSHR mRNA expression through both the Smad pathway and non-Smad p38 MAPK pathways. For the transcription factor of FSHR (USF1) to bind to the FSHR promoter region, the promoter region DNA must be structurally accessible and USF1 must be phosphorylated. Through the Smad pathway, BMP15 phosphorylates Smad 1/5/8 to epigenetically alter the DNA structure such that it favors USF1 binding. Through the non-Smad p38 MAPK pathway, BMP15 induces the phosphorylation of USF1, which is necessary for USF1 binding.
BMP15 is a follicle growth factor derived from oocytes that is reported to be involved in the regulation of FSHR expression [21, 22]. Dimeric BMP ligands bind to type II receptors (BMPRII, ActRIIa, and ActRIIb) to phosphorylate type I receptors (ALK2, ALK3, and ALK6). These activated receptors transfer signals by phosphorylating the transcription factors Smad 1/5/8. The phosphorylated Smads form a heterodimeric complex with Smad 4 in the nucleus, and this complex modulates gene expression. Besides Smads, growing evidence imply that MAPKs are involved in transmitting intracellular signaling of BMPs [27]. It has been reported that LDN193189 (an ALK 2, 3 antagonist) inhibits both Smads and p38 MAPK pathways [28].
Recently, epigenetic regulations, such as histone modification and DNA methylation, have been studied for regulating gene expression. Acetylation of histones mainly makes the chromatin structure accessible and promotes transcription [5]. Studies using human cancer cell lines demonstrated that an epigenetic regulatory step is a crucial mechanism in LHR expression [2, 3], which possesses a pronounced structural similarity to the FSHR. In our study, TSA (a histone deacetylase inhibitor) induced FSHR mRNA expression in HGrC1 cells with increasing histone acetylation and transcription factor USF1/2 binding at the FSHR promoter region (Fig. 1). The Smad pathway is linked with histone acetylation BMP signaling [13]. In our study, BMP15 increased Smad 1/5/8 phosphorylation (Fig. 3b) and HAT activity (Fig. 3c). BMP15 increased acetylation of histone H3 and H4 at the FSHR promoter region (Fig. 2b) and increased the binding of USF1/2 to the FSHR promoter region (Fig. 2c). These results suggest that BMP15 may have increased acetylation of histones at the FSHR promoter region, via Smad 1/5/8 phosphorylation, modifying the chromatin structure of the FSHR promoter region to be favorable for USF1/2 binding, resulting in increased FSHR expression. BMP15 may be a physiological key mediator for the epigenetic regulation of FSHR expression.
BMP15 increased phosphorylation of p38 MAPK and USF1 (Fig. 3d). p38 MAPK has been demonstrated to be an important downstream effector of BMPs in cells other than granulosa cells [18]. USF1 has been also demonstrated to be a downstream target of activated p38 MAPK, leading to the appearance of a phosphorylated USF1 form [19, 20]. Our results showed that BMP15 elevated the phosphorylation levels of USF1/2 by activating the non-Smad p38 MAPK pathway. Through the increased binding of USF1/2, in addition to its elevated phosphorylation, BMP15 may activate USF1/2-mediated transcription, resulting in enhanced FSHR expression.
Furthermore, we assessed the downstream effectors of the FSHR. FSH signaling is known to act exclusively through the FSHR. FSH increases estradiol production from granulosa cells of growing follicles, primarily by inducing CYP19A1. CYP19A1 is the key aromatase which converts androstenedione to estradiol. The addition of FSH to the culture medium slightly elevated CYP19A1 mRNA expression levels in HGrC1 cells, whereas BMP15 showed a significant elevation (Fig. 4a). Since basal activity of the FSH signaling may have already been present in the control group, the addition of FSH slightly increased FSH-induced CYP19A1 expression. BMP15 may have been able to significantly increase CYP19A1 expression through its ability to increase FSHR, allowing accelerated FSH signaling. Consequently, the stimulation with both BMP15 and FSH resulted in inducing CYP19A1 expression levels further, showing an additive effect. As a result, estradiol production was also shown to be elevated (Fig. 4b). However, the elevation and additive effect may have been muted because estradiol production occurs considerably downstream from BMP15 signaling, making its detection using the cell culture technique difficult.
Our study is limited, in that the in vivo effects were not examined. However, our in vitro results provide valuable information for transition to clinical treatment. Although TSA, a non-specific histone deacetylase inhibitor, significantly elevated FSHR expression, it may not yet be practicable for clinical use due to its possible adverse effects. Our study pointed out that BMP15, a biological signaling factor with a major role in the ovaries, produces epigenetic effects related to the expression of FSHR, and may be efficacious as a biological substitute of TSA. However, we could only demonstrate that BMP15 induces increased binding of USF1/2, rather than the phosphorylated USF1/2, because there are no commercial phosphorylated USF antibodies, which could be used for the ChIP test. Subsequent in vivo studies are being planned.
The induction of FSHR expression in granulosa cells by the BMP15-induced pathways may be valuable for ovarian tissue culture to induce follicular growth ex-vivo. It may also be effective for IVF treatments in “poor responders,” who are typically unresponsive to FSH, resulting in retrieval of few, if not any, oocytes. By inducing FSHR in the residual ovarian follicles, more mature follicles may be induced for oocyte retrieval. It is well known that most chemotherapeutic agents are toxic to growing follicles, leading to ovarian follicle depletion. Therefore, early and increased follicle induction may be achieved via BMP-induced pathways for oncofertility patients who require immediate chemotherapy after oocyte retrieval. The BMP-induced pathways may on the other hand also be inhibited in order to suppress follicular growth during chemotherapy.
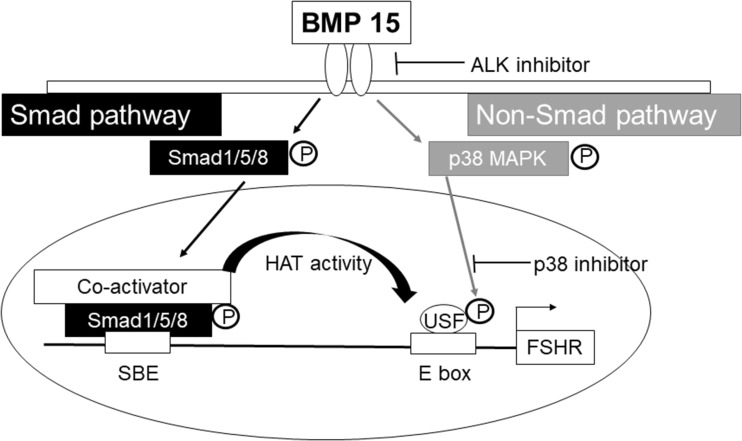
In conclusion, BMP15 induces FSHR expression in human granulosa cells through Smad and non-Smad pathways (Fig. 5), leading to increased follicular growth and oocyte ovulation. This mechanism of FSHR induction by BMP15 may be utilized for controlling follicular growth, both in induction and in suppression.
Fig. 5.
The mechanism of BMP15-induced FSHR expression through the Smad and non-Smad pathway. For the transcription factor of FSHR (USF1) to bind to the FSHR promoter region, the promoter region DNA must be structurally accessible and USF1 must be phosphorylated. Through the Smad pathway, BMP15 phosphorylates Smad 1/5/8 to epigenetically alter the DNA structure favorably for binding of USF1 to FSHR. Through the non-Smad p38 MAPK pathway, BMP15 induces the phosphorylation of USF1 necessary for binding to FSHR
Electronic supplementary material
(PNG 395 kb)
(PNG 475 kb)
(PNG 332 kb)
(PNG 403 kb)
(PNG 378 kb)
(PNG 433 kb)
Funding
This study was funded by Grants-in-Aid for Scientific Research (18K16767) to Tomoko Nakamura and (15H04984) to Akira Iwase from the Japan Society for the Promotion of Science.
Compliance with ethical standards
Disclosure statement
The authors have nothing to disclose.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Saint-Dizier M, Malandain E, Thoumire S, Remy B, Chastant-Maillard S. Expression of follicle stimulating hormone and luteinizing hormone receptors during follicular growth in the domestic cat ovary. Mol Reprod Dev. 2007;74(8):989–996. doi: 10.1002/mrd.20676. [DOI] [PubMed] [Google Scholar]
- 2.Liao M, Zhang Y, Dufau ML. Protein kinase Calpha-induced derepression of the human luteinizing hormone receptor gene transcription through ERK-mediated release of HDAC1/Sin3A repressor complex from Sp1 sites. Mol Endocrinol. 2008;22(6):1449–1463. doi: 10.1210/me.2008-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y, Fatima N, Dufau ML. Coordinated changes in DNA methylation and histone modifications regulate silencing/derepression of luteinizing hormone receptor gene transcription. Mol Cell Biol. 2005;25(18):7929–7939. doi: 10.1128/MCB.25.18.7929-7939.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heckert LL, Daley IJ, Griswold MD. Structural organization of the follicle-stimulating hormone receptor gene. Mol Endocrinol. 1992;6(1):70–80. doi: 10.1210/mend.6.1.1738373. [DOI] [PubMed] [Google Scholar]
- 5.Fu B, Wang H, Wang J, Barouhas I, Liu W, Shuboy A, Bushinsky DA, Zhou D, Favus MJ. Epigenetic regulation of BMP2 by 1,25-dihydroxyvitamin D3 through DNA methylation and histone modification. PLoS One. 2013;8(4):e61423. doi: 10.1371/journal.pone.0061423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGee EA, Hsueh AJ. Initial and cyclic recruitment of ovarian follicles. Endocr Rev. 2000;21(2):200–214. doi: 10.1210/edrv.21.2.0394. [DOI] [PubMed] [Google Scholar]
- 7.McGrath SA, Esquela AF, Lee SJ. Oocyte-specific expression of growth/differentiation factor-9. Mol Endocrinol. 1995;9(1):131–136. doi: 10.1210/mend.9.1.7760846. [DOI] [PubMed] [Google Scholar]
- 8.Dube JL, Wang P, Elvin J, Lyons KM, Celeste AJ, Matzuk MM. The bone morphogenetic protein 15 gene is X-linked and expressed in oocytes. Mol Endocrinol. 1998;12(12):1809–1817. doi: 10.1210/mend.12.12.0206. [DOI] [PubMed] [Google Scholar]
- 9.Otsuka F, McTavish KJ, Shimasaki S. Integral role of GDF-9 and BMP-15 in ovarian function. Mol Reprod Dev. 2011;78(1):9–21. doi: 10.1002/mrd.21265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juengel JL, McNatty KP. The role of proteins of the transforming growth factor-beta superfamily in the intraovarian regulation of follicular development. Hum Reprod Update. 2005;11(2):143–160. doi: 10.1093/humupd/dmh061. [DOI] [PubMed] [Google Scholar]
- 11.Persani L, Rossetti R, Di Pasquale E, Cacciatore C, Fabre S. The fundamental role of bone morphogenetic protein 15 in ovarian function and its involvement in female fertility disorders. Hum Reprod Update. 2014;20(6):869–883. doi: 10.1093/humupd/dmu036. [DOI] [PubMed] [Google Scholar]
- 12.Gilchrist RB, Lane M, Thompson JG. Oocyte-secreted factors: regulators of cumulus cell function and oocyte quality. Hum Reprod Update. 2008;14(2):159–177. doi: 10.1093/humupd/dmm040. [DOI] [PubMed] [Google Scholar]
- 13.Shi J, Zhao W, Pan B, Zheng M, Si L, Zhu J, Liu L, Tian J. Alcohol exposure causes overexpression of heart development-related genes by affecting the histone H3 acetylation via BMP signaling pathway in cardiomyoblast cells. Alcohol Clin Exp Res. 2017;41(1):87–95. doi: 10.1111/acer.13273. [DOI] [PubMed] [Google Scholar]
- 14.Shi J, Yoshino O, Osuga Y, Nishii O, Yano T, Taketani Y. Bone morphogenetic protein 7 (BMP-7) increases the expression of follicle-stimulating hormone (FSH) receptor in human granulosa cells. Fertil Steril. 2010;93(4):1273–1279. doi: 10.1016/j.fertnstert.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 15.Rajesh G, Mishra SR, Paul A, Punetha M, Vidyalakshmi GM, Narayanan K, Bag S, Bhure SK, Singh Chouhan V, Maurya VP, Singh G, Sarkar M. Transcriptional and translational abundance of bone morphogenetic protein (BMP) 2, 4, 6, 7 and their receptors BMPR1A, 1B and BMPR2 in buffalo ovarian follicle and the role of BMP4 and BMP7 on estrogen production and survival of cultured granulosa cells. Res Vet Sci. 2018;118:371–388. doi: 10.1016/j.rvsc.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 16.Moore RK, Otsuka F, Shimasaki S. Molecular basis of bone morphogenetic protein-15 signaling in granulosa cells. J Biol Chem. 2003;278(1):304–310. doi: 10.1074/jbc.M207362200. [DOI] [PubMed] [Google Scholar]
- 17.Bayasula IA, Kiyono T, Takikawa S, Goto M, Nakamura T, et al. Establishment of a human nonluteinized granulosa cell line that transitions from the gonadotropin-independent to the gonadotropin-dependent status. Endocrinology. 2012;153(6):2851–2860. doi: 10.1210/en.2011-1810. [DOI] [PubMed] [Google Scholar]
- 18.Noth U, Tuli R, Seghatoleslami R, Howard M, Shah A, Hall DJ, et al. Activation of p38 and Smads mediates BMP-2 effects on human trabecular bone-derived osteoblasts. Exp Cell Res. 2003;291(1):201–211. doi: 10.1016/s0014-4827(03)00386-0. [DOI] [PubMed] [Google Scholar]
- 19.Corre S, Primot A, Baron Y, Le Seyec J, Goding C, Galibert MD. Target gene specificity of USF-1 is directed via p38-mediated phosphorylation-dependent acetylation. J Biol Chem. 2009;284(28):18851–18862. doi: 10.1074/jbc.M808605200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galibert MD, Carreira S, Goding CR. The Usf-1 transcription factor is a novel target for the stress-responsive p38 kinase and mediates UV-induced tyrosinase expression. EMBO J. 2001;20(17):5022–5031. doi: 10.1093/emboj/20.17.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogura-Nose S, Yoshino O, Osuga Y, Shi J, Hiroi H, Yano T, Taketani Y. Anti-mullerian hormone (AMH) is induced by bone morphogenetic protein (BMP) cytokines in human granulosa cells. Eur J Obstet Gynecol Reprod Biol. 2012;164(1):44–47. doi: 10.1016/j.ejogrb.2012.05.017. [DOI] [PubMed] [Google Scholar]
- 22.Otsuka F, Yamamoto S, Erickson GF, Shimasaki S. Bone morphogenetic protein-15 inhibits follicle-stimulating hormone (FSH) action by suppressing FSH receptor expression. J Biol Chem. 2001;276(14):11387–11392. doi: 10.1074/jbc.M010043200. [DOI] [PubMed] [Google Scholar]
- 23.Hermann BP, Hornbaker K, Rice DA, Sawadogo M, Heckert LL. In vivo regulation of follicle-stimulating hormone receptor by the transcription factors upstream stimulatory factor 1 and upstream stimulatory factor 2 is cell specific. Endocrinology. 2008;149(10):5297–5306. doi: 10.1210/en.2007-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar S, Jiang MS, Adams JL, Lee JC. Pyridinylimidazole compound SB 203580 inhibits the activity but not the activation of p38 mitogen-activated protein kinase. Biochem Biophys Res Commun. 1999;263(3):825–831. doi: 10.1006/bbrc.1999.1454. [DOI] [PubMed] [Google Scholar]
- 25.George JW, Dille EA, Heckert LL. Current concepts of follicle-stimulating hormone receptor gene regulation. Biol Reprod. 2011;84(1):7–17. doi: 10.1095/biolreprod.110.085043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gore-Langton RE, Dorrington JH. FSH induction of aromatase in cultured rat granulosa cells measured by a radiometric assay. Mol Cell Endocrinol. 1981;22(2):135–151. doi: 10.1016/0303-7207(81)90087-3. [DOI] [PubMed] [Google Scholar]
- 27.Gallea S, Lallemand F, Atfi A, Rawadi G, Ramez V, Spinella-Jaegle S, Kawai S, Faucheu C, Huet L, Baron R, Roman-Roman S. Activation of mitogen-activated protein kinase cascades is involved in regulation of bone morphogenetic protein-2-induced osteoblast differentiation in pluripotent C2C12 cells. Bone. 2001;28(5):491–498. doi: 10.1016/s8756-3282(01)00415-x. [DOI] [PubMed] [Google Scholar]
- 28.Boergermann JH, Kopf J, Yu PB, Knaus P. Dorsomorphin and LDN-193189 inhibit BMP-mediated Smad, p38 and Akt signalling in C2C12 cells. Int J Biochem Cell Biol. 2010;42(11):1802–1807. doi: 10.1016/j.biocel.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PNG 395 kb)
(PNG 475 kb)
(PNG 332 kb)
(PNG 403 kb)
(PNG 378 kb)
(PNG 433 kb)